Summary
Background
Distinguishing a urinary tract infection (UTI) from asymptomatic bacteriuria (ASB) in children with neuropathic bladders is difficult. Currently used markers of infection, such as the routine urinalysis, lack specificity for UTI in this population. The urinary microbiome may help differentiate these states.
Objective
The objective of this work was to describe the baseline microbiome in children with neuropathic bladders, and to determine if differences exist among the urine microbiomes of children with neuropathic bladders who have negative urine cultures, ASB, or UTI.
Study design
This is a cross-sectional study of children with neuropathic bladders who use clean intermittent catheterization for bladder management who had a urine culture sent as part of clinical management. Residual urine, initially collected via catheter for urine culture, was obtained for use in this work. Microbial DNA was isolated, and the V4 region of the 16SrRNA gene sequenced. The relative abundance of each bacteria was measured in each group. Alpha diversity, measured by Chao1 and the Shannon Diversity Index, was also measured in each group. PERMANOVA was used to compare the microbiota between groups.
Results
36 children with neuropathic bladders were included in this study (UTI = 11, ASB = 19, negative cultures = 4). The most abundant bacteria were unspecified Enterobacteriaceae, Klebsiella, Staphylococcus, Streptococcus, and Enterococcus. Children who catheterize their urethra have a higher proportion of Staphylococcus, while the urine microbiome of those who catheterize through a Mitrofanoff consists predominantly of members of the family Enterobacteriaceae. Given the low numbers of patients with Mitrofanoffs and augmented bladders, we did not statistically compare the urine microbiomes between these patients. There was no difference in either alpha diversity or the overall microbiota between children with neuropathic bladders with UTI, ASB, and negative cultures.
Discussion
In this pilot cohort of children with neuropathic bladders, bacteria that are members of the family Enterobacteriaceae are the most predominant bacteria in the urine microbiomes. There was no difference in the urine microbiome between those with UTI, ASB, and negative cultures. Route of catheterization may affect the composition of the urine microbiome, although due to limited sample size, this was not confirmed statistically.
Conclusion
There was no difference in the urine microbiome between patients with negative urine cultures, ASB, and UTI. Further work is needed to determine if the urine microbiome varies based on either the route of catheterization or the presence of augmented bladder.
Keywords: Urinary tract infection, Neuropathic bladder, Pediatrics, Urine microbiome
Summary Figure Graphical Abstract
Principal coordinate analysis chart demonstrating a lack of clustering between patients with no growth, asymptomatic bacteriuria (ASB), and urinary tract infection (UTI).

Introduction
Children with neuropathic bladders are at high risk for urinary tract infections (UTIs). However, accurate diagnosis is difficult as there are no widely-accepted Definitions of UTI in these children [1,2]. The UTI definition proposed by the American Academy of Pediatrics, which uses the combination of a positive urine culture and pyuria as indicative of a UTI in the general pediatrics population [3], does not apply to the neuropathic bladder patient population. Indeed, both asymptomatic bacteriuria (ASB) and pyuria are common in children with neuropathic bladder [4,5]. In the absence of a standardized definition of UTI, clinicians are left without guidance on how to diagnose UTIs in this population, leading to variability in the management of suspected UTIs in these children [6] and likely overtreatment of ASB.
Examination of the urine microbiome, the community of bacteria within the bladder that is identifiable using sequencing or expanded culturing techniques, may help improve the diagnosis of UTI [7,8]. Multiple authors have demonstrated associations between changes in the urine microbiome with various clinical conditions, including lower urinary tract symptoms [9], urge incontinence [10], and bladder cancer [11]. Furthermore, additional work has found that urine microbial diversity is decreased in a cohort of patients who developed UTI[12], while another case series described an increase in microbial diversity that preceded the onset of UTI in a patient with neuropathic bladder [13]. Therefore, it is possible that a deeper understanding of the urine microbiome may help differentiate UTI from ASB. However, while the urine microbiome of adult patients with neuropathic bladders has been described[14,15], to our knowledge, there are no published reports of the urine microbiome in children with neuropathic bladders, and there have been no cross-sectional comparisons of the urine microbiome in people with neuropathic bladder with and without UTI. Therefore, there are limited data available to guide future work on the urine microbiome in this population. The primary objective of this study was to describe the baseline microbiome in children with neuropathic bladders and to identify factors that may account for variation in the urine microbiome. The secondary objective was to determine if differences exist among the urine microbiomes of children with neuropathic bladders who have negative urine cultures, ASB, or UTI.
Materials and methods
Patients and samples:
Urine samples were initially collected to assess the predictive ability of a specific urine protein for the diagnosis of UTI in children with neuropathic bladders [16]. Patients were eligible for enrollment in the prior study if they were between the ages of 2 months and 21 years, had a diagnosis of neuropathic bladder that required management with clean intermittent catheterization and had a urine culture sent by the treating physician. A subset of these urine cultures were sent from patients undergoing routine urodynamics. All of the samples sent from asymptomatic patients were from children undergoing routine urodynamics and were not collected for evaluation for UTI. The remainder were sent for clinical concern for UTI. Residual urine was obtained from the clinical lab within 10 h of collection, centrifuged, and the supernatant aliquoted and frozen at −80° Celsius. Samples were considered for use in this work if more than 5 mL of urine supernatant that had not undergone any previous freeze–thaw cycles were available. All samples from patients with UTI that met these criteria were included in this work. ASB and samples that did not grow bacteria and that met the sample criteria (i.e. more than 5 ml, no previous freeze–thaw cycles) were matched to the UTI samples based on patient age and sex. This work was approved by the local Institutional Review Board, with a waiver of informed consent, as the samples were residual urine samples and deidentified at the time of microbiome analysis.
Definitions:
UTI was defined using an adapted version of the definition published by Madden-Fuentes et al. [2] Patients were considered to have a UTI if they had more than 10 urinary white blood cells, a positive urine culture, defined as greater than 50,000 colony-forming units/ml, and two or more of the following symptoms: fever (as defined as a temperature greater than 38° Celsius), abdominal pain, back pain, new or worsened incontinence, pain with catheterization, or malodorous or cloudy urine. ASB was defined as a patient with a positive urine culture that did not meet criteria for UTI.
DNA extraction, 16S rRNA Gene Amplification, and Sequencing:
DNA was isolated from urine samples using the QIAmp circulating nucleic acid kit with QIAamp columns (Qiagen, Germantown, Maryland) per the manufacturer’s instructions. After isolation, the DNA concentration was quantitated (NanoDrop 2000, Thermo Fisher Scientific) and quantified (Femto Human and Bacterial DNA quantification kits, Zymo Research). The V4 regions of the 16S rRNA genes were then amplified as previously described [17]. (using the KAPA Hifi Hot Start ready mix, KAPA Biosystems). UltraPure DEPC-treated water served as negative controls, which was processed in exactly the same way as the rest of the samples. After PCR, the products were purified (Agencourt AmPure XP Beads, Beckman Coulter genomics), and then underwent dual-indexing PCR (Nextera XT kit, Illumina), followed by further purification (Agencourt AmPure XP Beads, Beckman Coulter Genomics). The final V4 libraries with quantitated (Qubit 2.0 Fluorometer with the DNA Broad range assay kit, Thermo Fisher Scientific), and sized with a DNA 1000 kit on a 2100 Bioanalyzer (Agilent technologies). The dual-indexed Libraries were normalized, pooled to 4 nM, and sequenced on a Miseq sequencer using a Miseq v3 600 cycle kit for paired-end sequencing (Illumina). PhiX control (20%) (Illumina) was spiked in. Resultant Fastq files were checked for quality control with the FastQC software (Illumina) using a Phred quality score of 30 as a cut-off for analysis.
Bioinformatics:
Raw FASTQ files were trimmed and filtered for quality reads using Trimmomatic [18]. Clean sequences were aligned to the Greengenes [19] (13_8 97%, default QIIME1) representative operational taxonomic units (OTU) sequences using closed-reference OTU picking/Shotgun UniFrac workflow in QIIME1 [20]. Sequences were clustered into OTUs at the species level. Samples were subsampled (rarefaction analysis) to the smallest sample size (16,000 reads/sample) to remove the effect of sample size bias on community composition. Alpha-diversity was estimated with the Shannon index and Chao1 richness estimator using the rarefied OTU table [21]. Phylogenetic beta-diversity Unifrac metrics (unweighted and weighted unifrac) were calculated between pairs of samples. Differences between samples were explored using principal coordinates analysis (PCoA) and both Unifrac distances. Beta-diversity Unifrac indices were compared using permutational multivariate analysis of variance (Permanova) as implemented in the vegan R package [22] and the same predictors and covariables as above. All analyses were performed following the QIIME1 (version 1.9.1) pipeline. All relevant data are available from the corresponding author upon reasonable request.
Statistical Analysis:
Continuous variables were compared between groups using either ANOVA and post hoc Tukey or Kruskal–Wallis and post hoc Dunn for variables that were not normally distributed. Chi-square was used to compare categorical variables between groups. To examine the abundance of bacteria in patients with either Mitrofanoff alone, Mitrofanoff and enterically augmented bladder, or neither, a subgroup of patients without UTI was examined to describe the microbiomes in these patient groups with the potential confounding effect of UTI. As the number of patients were too small for any meaningful statistical testing, descriptive statistics were used to describe the composition of these groups.
Results
Thirty-four children with neuropathic bladders were included in this study (UTI = 11, ASB = 19, negative cultures = 4). There was no difference in age, sex, or etiology of neuropathic bladder between groups. There was also no difference in the number of patients with either a Mitrofanoff or augmented bladder between groups. There were more ASB patients with less than 10 urinary white blood cells compared to patients in the UTI group. There were no other differences in the proportion of patients with between 10 and 50, and more than 50 urinary white blood cells between groups. (Table 1)
Table 1.
Patients demographics and results of urine testing.
| No Growth (n = 4) | ASB (n = 19) | UTI (n = 11) | P-value | |
|---|---|---|---|---|
| Clinical Data | ||||
| Mean Age (years) | 15.0 (6) | 8.8 (5) | 11.0 (6) | 0.11 |
| Male | 3 (75) | 9 (47) | 7 (64) | 0.49 |
| Myelomeningocele | 0 (0) | 7 (37) | 2 (18) | 0.24 |
| Anorectal Malformation | 3 (75) | 7 (37) | 4 (36) | 0.34 |
| Tethered Cord | 1 (25) | 5 (26) | 2 (18) | 0.88 |
| Mitrofanoff | 0 (0) | 7 (37) | 4 (36) | 0.34 |
| Augmented Bladder | 0 (0) | 3 (16) | 1 (9) | 0.64 |
| Asymptomatic | 4 (100)* | 17 (90)* | 0 (0) | <0.01 |
| Results of Urine Testing | ||||
| <10 urine white blood cells | 2 (50) | 8 (47)* | 0 (0) | 0.02 |
| 10-50 urine white blood cells | 1 (25) | 5 (29) | 6 (55) | 0.35 |
| >50 urine white blood cells | 1 (25) | 4 (24) | 5 (46) | 0.45 |
| E. coli | – | 12 (63) | 6 (55) | – |
| Klebsiella pneumoniae | – | 3 (16) | 4 (36) | – |
| Enterobacter sp | – | 2 (11) | – | – |
| Enterococcus | – | 1 (5) | – | – |
| Staphylococcus | 1 (5) | – | – | |
| Citrobacter | – | – | 1 (9) | – |
Data presented as: n (%).
P-values not provided for urine culture results are this data is descriptive in nature.
p < 0.05 compared to UTI.
The relative abundance of bacteria in each patient is shown in Fig. 1. The relative abundance of bacteria in the urine microbiome based on gender is shown in Fig. 2. The five most abundant bacteria in the cohort of all 34 children were as follows: unspecified members of the family Enterobacteriaceae (mean abundance of 56%), Klebsiella (19%), Staphylococcus (7%), Streptococcus (3%), and non-specified members of the family Neisseriaceae (3%). The remaining bacteria all had a mean abundance of less than 2%. The relative abundance of bacteria in patients without UTI and with either Mitrofanoff alone, Mitrofanoff and augmented bladder, or neither is shown in Fig. 3. The predominant bacteria in 9 of the 16 patients without either Mitrofanoff or augmented bladder was unspecified members of the family Enterobacteriaceae, while Klebsiella was the most predominant bacteria in 3 patients, Staphylococcus most predominant in 3 patients, and non-specified members of the family Neisseriaceae the most predominant bacteria in 1 patient in the non-Mitrofanoff, non-augmented bladder group. The predominant bacteria in 3 of the 4 patients in the Mitrofanoff alone group were either unspecified members of the family Enterobacteriaceae (n = 1) or Klebsiella (n = 2). The most abundant bacteria in the remaining patient in the Mitrofanoff alone group was Enterococcus. Two of the three patients in the Mitrofanoff and augmented bladder group had unspecific members of Enterobacteriaceae, and one had Streptococcus as the most predominant bacteria in their urine (Fig. 3). We did not statistically compare the urine microbiomes based on the presence or absence of either a Mitrofanoff or augmented bladder as the number of patients in each group was too small to lend validity to this comparison.
Figure 1.

Relative abundance of components of the urine microbiome for all patients in the cohort. The ten most predominant organisms are listed in the legend.
Figure 2.

Relative abundance of bacteria in the urine microbiome of males versus females. The ten most predominant organisms are listed in the legend.
Figure 3.

Relative abundance of bacteria in the urine microbiomes of patients with a Mitrofanoff and augmented bladder, Mitrofanoff alone, or neither Mitrofanoff or augmented bladder. The ten most predominant organisms are listed in the legend.
The relative abundance of bacteria in the urinary microbiomes in patients categorized as no growth, ASB, or UTI is shown in Fig. 4. The most abundant bacteria in the nogrowth group were Staphylococcus (38%), followed by nonspecified members of the family Neisseriaceae (21%), Enterobacteriaceae (17%) and then Gemella (5%). The most abundant bacteria in the ASB group were nonspecified members of the family Enterobacteriaceae (65%), Klebsiella (17%), Staphylococcus (5%), Streptococcus (4%), and Enterococcus (4%). The most abundant bacteria in the urine of UTI patients were Enterobacteriaceae (55%), Klebsiella (26%), Staphylococcus (11%), Streptococcus (6%) and Enterococcus (3%). Principal coordinates analysis shows a significant overlap of all three groups, without any notable clustering (Summary figure) PERMANOVA resulted in a p-value greater than 0.05, suggesting no significant difference in the community composition between these three groups.
Figure 4.
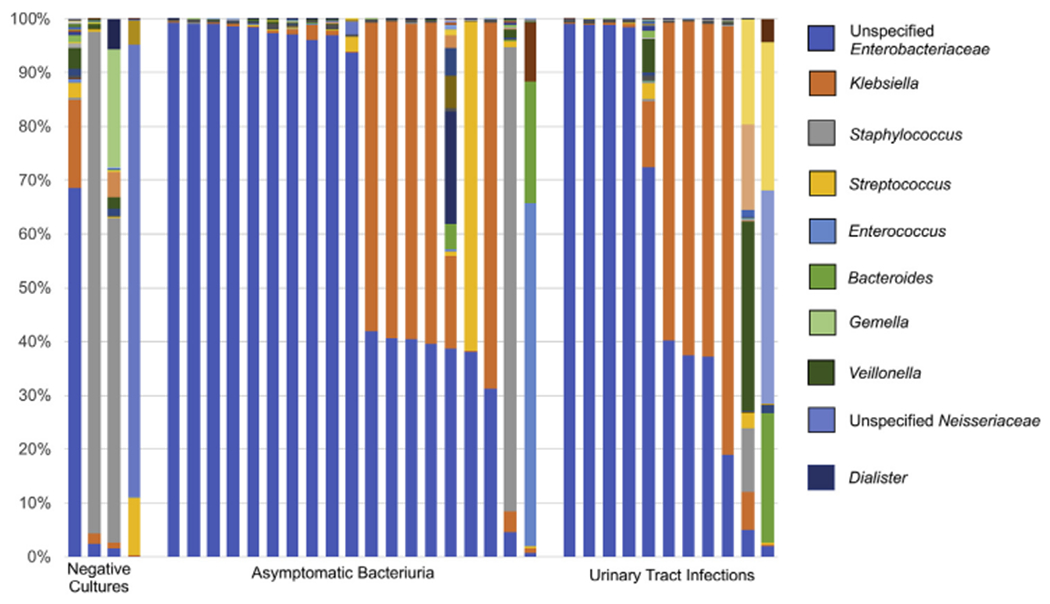
Relative abundance of bacteria in the urine microbiome of patients with negative urine cultures, asymptomatic bacteriuria (ASB), and urinary tract infections (UTI). The ten most predominant organisms are listed in the legend.
There was no significant difference in alpha diversity, as measured by either Chao1 or Shannon Diversity Index, between patients with no growth, ASB, and UTI (Supplemental Fig. 1). While median Chao1 was significantly higher in patients with negative urine cultures compared to those with positive urine cultures (351(interquartile range (IQR) 114) versus 140 (IQR:123), p = 0.03), there was no difference in the Shannon Diversity Index between patients with negative and positive urine cultures (1.6 (IQR: 1.54), 1.5 (IQR: 1.6), p = 0.28).
Discussion
Here, we report our findings regarding the composition of the urinary microbiome in children with neuropathic bladders. We show that the most abundant bacteria in our cohort were Enterobacteriaceae, Klebsiella, Staphylococcus, Streptococcus, and Enterococcus. Further, we demonstrate that there is no difference in alpha diversity, as measured by Chao1, or composition of the urine microbiome between the cohort of children with neuropathic bladder with UTI, ASB, and negative urine cultures.
There are limited data within the literature documenting the composition of the urine microbiome in children. A recently published study looked at the urine microbiome of pre-pubertal boys without neuropathic bladders [23]. The most abundant bacteria in that work, which include Staphylococcus, Varibaculum, Peptoniphilus, and Actinobaculum are different from those found in our patients, suggesting that the urine microbiome is distinct in children with neuropathic bladders compared to those with normally-functioning bladders. Other work focused on the safety of a single intravesical instillation of Lactobacillus reported urine microbiome findings from five children with neuropathic bladder. In this work, the preinstillation urine microbiomes are more similar to those we report here, with Escherichia and Streptococcus among the most abundant bacteria [24]. Similarly, a study of adults with neuropathic bladder found that the most abundant bacteria in the urine microbiome included both Escherichia and Klebsiella, which is similar to our findings[15].However, in these prior papers, there are much higher proportions of anaerobic bacteria reported than were found in our cohort. This discrepancy is most likely a result of the different types of urine used. In both of these publications, fresh whole urine was used. In the present study, we used previously-frozen, cell-free urine. It has been documented that cell-free urine can allow for the identification of many members of the microbiome, including many anaerobes, suggesting that the cell-free state of the urine was not responsible for this difference [25]. The relative lack of anaerobes in this work is more likely an effect of the single freeze/thaw cycle rather than indicative of a lack of urinary anaerobes in this population, although further work is needed to confirm this.
We did not show a difference in the microbial communities between patients with no growth, ASB, and UTI. The lack of difference between patients with negative cultures and those with either ASB or UTI could be the result of the small number of patients with negative cultures included in this work. This result needs to be confirmed in larger studies before more definitive conclusions can be made. However, our data does show a lack of difference between the ASB and UTI groups, as demonstrated by the complete overlap in these communities in the PCoA chart. There are several potential explanations for this finding. The first is that the urine microbiome only reflects the communities of bacteria within the urine, but not the host response to these bacteria. Studying the urine microbiome in isolation does not reflect the interaction between the host bladder tissue and the microbiome, which is likely a better indicator of the presence or absence of a UTI. A second possibility is that we have not found a difference in these groups due to clinical misclassification of our samples. The lack of a standardized and validated definition of UTI makes this a valid consideration. However, the most plausible explanation for the lack of a difference is that the urine microbiome in children with neuropathic bladders is unique to each patient, and longitudinal comparisons within patients are the most appropriate ways to determine whether a change occurs in the urine microbiome in the setting of UTI. This was demonstrated by Bossa et al. who documented a change in an individual’s urine microbiome in the setting of UTI, which subsequently normalized following treatment [13].
We also described the microbiome between patients with and without Mitrofanoff or bladder augmentation in patients without symptoms of a UTI. Although we did not statistically compare these groups due to the low number of patients, we were able to make a few observations about these groups. Three of the 16 patients with neither a Mitrofanoff nor augmented bladder had Staphylococcus as the predominant organism in their urine microbiomes, whereas Staphylococcus has a relative abundance of less than 2% in any of the patients with either Mitrofanoff or augmented bladder. Previous work has examined the augmented bladder tissue microbiome of patients before and after bladder augmentation with either ileum or colon. The authors did not find a difference in the local microbiome between the native and augmented bladders, and they did not find a difference between the bladder augmented with ileum or colon [26]. However, the authors did not examine urine, nor did they compare methods of catheterization, and therefore, these results are not directly comparable. Although our data suggest that the route of catheterization/altered bladder anatomy may affect the urine microbiome, confirmation of this observation is needed. Further, our data cannot differentiate the reason behind this potential difference, which may be due to the source of the tissue used for either the conduit or the bladder augmentation, the alternate route of catheterization, or simply the presence of the neuropathic bladder. While it is likely that future, more appropriately-powered studies may better elucidate this, our data is not sufficient to draw conclusions on this topic.
There are several limitations to this work, which include the small number of patients, the use of previously-frozen urine samples, and the use of cell-free urine. The small number of patients included in this cohort limits the ability to control for all possible confounders, including age, sex, duration of catheter use, or use of antibiotic prophylaxis or antibiotic irrigations. Further, we were unable to reliably obtain information regarding antibiotic exposure, and could not include this information in the analysis. We also did not include any urine samples from children with normally-functioning bladders as controls. There were no samples that would be an appropriate control collected and processed in the same manner as the other samples in this work. We did include control measures at the processing and sequencing stage, but we were unable to include control samples from the original cohort. An additional limitation, as discussed above, is the lack of a standardized definition of UTI in this population. This potential for misclassification bias may have altered the results of this work.
Conclusion
Here, we found that children with neuropathic bladders have urine microbiomes that are predominantly composed of members of the family Enterobacteriaceae. Additionally, patients who catheterize through their urethra may have a higher proportion of Staphylococcus, while the urine microbiome of those who catheterize through a Mitrofanoff predominantly consisted of members of the family Enterobacteriaceae. Finally, in this cross-sectional work, we did not find a difference in the urine microbiome among patients with negative urine cultures, ASB, and UTI. Further longitudinal work is needed to better characterize the change in the urine microbiome at the time of UTI.
Supplementary Material
Acknowledgments
Funding
This work was partially supported by K12-HD-001339 (NICHD). The funder did not have any role in either study design, data collection, interpretation, or analysis, the writing of this report, or the decision to submit this report for publication.
Abbreviations
- UTI
Urinary tract infection
- ASB
Asymptomatic bacteriuria
Footnotes
Ethical approval
This work is in compliance with the policy on ethical consent. IRB approval was obtained for this work, with waiver of informed consent as the urine samples used within this work were considered discarded specimens.
Conflicts of interest
The authors have no conflicts of interest relevant to this article to disclose.
Appendix A.: Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpurol.2020.02.005.
References
- [1].Elliott SP, Villar R, Duncan B. Bacteriuria management and urological evaluation of patients with spina bifida and neurogenic bladder: a multicenter survey. J Urol 2005;173:217–20. 10.1097/01.ju.0000146551.87110.f4. [DOI] [PubMed] [Google Scholar]
- [2].Madden-Fuentes R, McNamara E. Variation in Definitions of Urinary Tract Infections in Spina Bifida Patients: A Systematic Review. Pediatrics 2013;132:132–9. 10.1542/peds.2013-0557. [DOI] [PubMed] [Google Scholar]
- [3].Finnell SME, Carroll AE, Downs SM. Technical reporte–Diagnosis and management of an initial UTI in febrile infants and young children. Pediatrics 2011;128:e749–70. 10.1542/peds.2011-1332. [DOI] [PubMed] [Google Scholar]
- [4].Schlager T a, Dilks S, Trudell J, Whittam TS, Hendley JO.Bacteriuria in children with neurogenic bladder treated with intermittent catheterization: natural history. J Pediatr 1995;126:490–6. 10.1016/S0022-3476(95)70477-9. [DOI] [PubMed] [Google Scholar]
- [5].Forster CS, Haslam DB, Jackson E, Goldstein SL. Utility of a routine urinalysis in children who require clean intermittent catheterization. J Pediatr Urol 2017;13 10.1016/j.jpurol.2017.01.016. [DOI] [PubMed] [Google Scholar]
- [6].Forster CS, Jackson E, Goldstein SL. Variation among sub-specialists in the diagnosis of urinary tract infection in children with neurogenic bladders. J Pediatr Urol 2018. 10.1016/j.jpurol.2018.07.016. [DOI] [PubMed] [Google Scholar]
- [7].Price TK, Dune T, Hilt EE, Thomas-White KJ, Kliethermes S, Brincat C, et al. The Clinical Urine Culture: Enhanced Techniques Improve Detection of Clinically Relevant Microorganisms. J Clin Microbiol 2016;54:1216–22. 10.1128/JCM.00044-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wolfe AJ, Toh E, Shibata N, Rong R, Kenton K, FitzGerald M, et al. Evidence of uncultivated bacteria in the adult female bladder. J Clin Microbiol 2012;50:1376–83. 10.1128/JCM.05852-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Bajic P, Van Kuiken ME, Burge BK, Kirshenbaum EJ, Joyce CJ, Wolfe AJ, et al. Male Bladder Microbiome Relates to Lower Urinary Tract Symptoms. Eur Urol Focus 2018:1–7. 10.1016/j.euf.2018.08.001. [DOI] [PubMed] [Google Scholar]
- [10].Thomas-White KJ, Kliethermes S, Rickey L, Lukacz ES, Richter HE, Moalli P, et al. Evaluation of the urinary microbiota of women with uncomplicated stress urinary incontinence. Am J Obstet Gynecol 2017;216:55e1–55.e16. 10.1016/j.ajog.2016.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bucevic Popovic V, Šitum M, Chow C-ET, Chan LS, Roje B, Terzić J. The urinary microbiome associated with bladder cancer. Sci Rep 2018;8:12157 10.1038/s41598-018-29054-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Horwitz D, McCue T, Mapes AC, Ajami NJ, Petrosino JF, Ramig RF, et al. Decreased microbiota diversity associated with urinary tract infection in a trial of bacterial interference. J Infect 2015;71:358–67. 10.1016/S2215-0366(16)30284-X.Epidemiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bossa L, Kline K, McDougald D, Lee BB, Rice SA. Urinary catheter-associated microbiota change in accordance with treatment and infection status. PLoS One 2017;12:1–20. 10.1371/journal.pone.0177633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Fouts DE, Pieper R, Szpakowski S, Pohl H, Knoblach S, Suh M-J, et al. Integrated next-generation sequencing of 16S rDNA and metaproteomics differentiate the healthy urine microbiome from asymptomatic bacteriuria in neuropathic bladder associated with spinal cord injury. J Transl Med 2012;10:174. 10.1186/1479-5876-10-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Groah SL, Pérez-Losada M, Caldovic L, Ljungberg IH, Sprague BM, Castro-Nallar E, et al. Redefining Healthy Urine: A Cross-Sectional Exploratory Metagenomic Study of People With and Without Bladder Dysfunction. J Urol 2016;196:579–87. 10.1016/j.juro.2016.01.088. [DOI] [PubMed] [Google Scholar]
- [16].Forster CS, Jackson E, Ma Q, Bennett M, Shah SS, Goldstein SL.Predictive ability of NGAL in identifying urinary tract infection in children with neurogenic bladders. Pediatr Nephrol 2018; 33:1365–74. 10.1007/s00467-018-3936-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Forster CS, Hsieh MH, Pérez-Losada M, Caldovic L, Pohl H, Ljungberg I, et al. Lactobacillus rhamnosus GG is safe in children and adults with neuropathic bladder: A phase Ia clinical trial. J Spinal Cord Med 2019;18:1–8. 10.1080/10790268.2019.1616456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30: 2114–20. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 2012;6:610–8. 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7: 335–6. 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Faith D Conservation evaluation and phylogenetic diversity. Biol Conserv 1992;61:1–10. 10.1016/0006-3207(92)91201-3. [DOI] [Google Scholar]
- [22].VEGAN Dixon P., a package of R functions for community ecology. 2003. 10.1658/1100-9233 014[0927: VAPORF]20CO;2 2009. [DOI] [Google Scholar]
- [23].Kassiri B, Shrestha E, Kasprenski M, Antonescu C, Florea LD, Sfanos KS, et al. A Prospective Study of the Urinary and Gastrointestinal Microbiome in Prepubertal Males. Urology 2019;131: 204–10. 10.1016/j.urology.2019.05.031. [DOI] [PubMed] [Google Scholar]
- [24].Forster CS, Hsieh MH, Pérez-losada M, Pohl H, Ljungberg I, Sprague B, et al. Lactobacillus rhamnosus GG is safe in children and adults with neuropathic bladder: A phase Ia clinical trial. J Spinal Cord Med 2019;18:1–8. 10.1080/10790268.2019.1616456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Burnham P, Dadhania D, Heyang M, Chen F, Westblade LF, Suthanthiran M, et al. Urinary cell-free DNA is a versatile analyte for monitoring infections of the urinary tract. Nat Commun 2018;9:1–10. 10.1038/s41467-018-04745-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kispal ZFF, Vajda P, Kardos D, Klymiuk I, Castellani C, Singer G, et al. The local microbiome after pediatric bladder augmentation: intestinal segments and the native urinary bladder host similar mucosal microbiota. J Pediatr Urol 2019;15:30e1–7. 10.1016/j.jpurol.2018.07.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


