Abstract
Cigarette smoking, a powerful mixture of chemical oxidants, is the strongest environmental risk factor for developing age-related macular degeneration (AMD), the most common cause of blindness among the elderly in western societies. Despite intensive study, the full impact of smoking on the retinal pigment epithelium (RPE), a central cell type involved in AMD pathobiology, remains unknown. The relative contribution of the known dysfunctional pathways to AMD, at what stage they are most pathogenic, or whether other processes are relevant, is poorly understood, and furthermore, whether smoking activates them, is unknown. We performed global RNA-sequencing of the RPE from C57BL/6J mice exposed to chronic cigarette smoke for 6 months to identify potential pathogenic and cytoprotective pathways. The RPE transcriptome induced by chronic cigarette smoking exhibited a mixed response of marked suppression of the innate immune response including type I and II interferons and upregulation of cell differentiation and morphogenic gene clusters, suggesting an attempt by the RPE to maintain its differentiated state despite smoke-induced injury. Given that mice exposed to chronic smoke develop early features of AMD, these novel findings are potentially relevant to the transition from aging to AMD.
Keywords: aging, age-related macular degeneration, differentiation, innate immunity, RNA sequencing, smoking
Graphical Abstract
Introduction
It is perplexing that people choose to smoke cigarettes, given that it contains nearly 5000 chemical oxidants and toxins[1], and more than 16 million Americans are afflicted with a smoking-induced disease such as cancer, heart disease, stroke, chronic obstructive pulmonary disease, diabetes, and/or age-related macular degeneration (AMD)[2]. The total economic cost of smoking is over $300 billion per year to cover direct medical care and lost work productivity[2]. AMD is the world’s leading cause of blindness among the elderly, with 196 million people currently afflicted worldwide, and with the aging population, this number is predicted to expand to 288 million in 2040[3]. In the US alone, 11 million people have AMD, a number similar to those with all invasive cancers combined, and more than double those with Alzheimer’s disease[4].
The National Eye Institute’s AMD Pathobiology group recently concluded that identifying all of the pathogenic signals, prioritizing their contribution relative to one another, and establishing the disease stage when the predominant signals initiate the disease process, will enable effective treatment design for each stage of AMD[5]. The AREDS2 formulation slows intermediate AMD progression, and anti-VEGF therapies have transformed treatment of exudative AMD[6-8]. Since preventing or curing early/intermediate AMD would eliminate the burdens of advanced AMD and reduce the accompanying financial cost, understanding the pathogenic signals that induce the transition from aging to early AMD will enable targeted treatment for this disease stage. In addition to advanced age and genetic susceptibility[9-11], smoking is a major risk factor for age-related macular degeneration (AMD)[12]. Retinal pigment epithelial (RPE) cell atrophy is a hallmark feature of early AMD, and the RPE, in particular, appears to be a target of cigarette smoke in AMD[13, 14]. Most studies have characterized the acute response to cigarette smoke by the RPE, and these studies have focused on a specific pathway. While valuable, a broad perspective of the most relevant pathogenic and cytoprotective responses by the RPE to chronic cigarette smoke exposure is still poorly understood.
Rod photoreceptor dysfunction and death in association with RPE morphologic derangement and the appearance of binucleated nuclei in the perifoveal macula are among the earliest changes in AMD[15-18]. With a similar RPE and rod/cone density as the human perifovea[19], mice are a reasonable model for studying early AMD changes. Our lab and others have previously reported that when mice are exposed to cigarette smoke for 6 months, the RPE developed marked ultrastructural derangement with increased apoptosis that is reminiscent of early AMD[20-22]. How the RPE atrophies, what pathogenic pathways are activated, and what cytoprotective responses fail as a consequence of cigarette smoking are not well characterized. Each cell’s function is dictated in large part, by its transcriptional program. To identify the key pathogenic pathways and cytoprotective responses by the RPE to chronic cigarette smoke, we exposed C57BL/6J mice to cigarette smoke for a period of 6 months, and evaluated their transcriptomic response by RNA-sequencing.
Materials and Methods
Animals and treatments
All experimental protocols used in this study were in accordance with National Institute Health (NIH) guidelines and were approved by the Johns Hopkins University Animal Care and Use Committee. Briefly, an equal number of 2-month female and male C57BL/6J mice (RD8 negative) were placed in a smoking chamber for 2.5 hours per day, 5 days per week for 6 months, as described previously[20], or raised in a filtered air environment for 6 months.
Tissue preparation
After mice were sacrificed and eyes were enucleated, one eye was dissected to remove the RPE/choroid, which was prepared for RNA or protein extraction. The other eye was fixed in 2.5% glutaraldehyde and 1% paraformaldehyde in 0.08 M cacodylate buffer for transmission electron microscopy (TEM). The central 2x2 mm tissue temporal to the optic nerve was postfixed with 1% osmium tetroxide, dehydrated, and embedded in Poly/Bed 812 resin (Polysciences, Inc., Warrington, PA).
Ultrastructural analysis
Ultrathin sections were stained with uranyl acetate and lead citrate and examined with a JEM-100 CX electron microscope (JEOL, Tokyo, Japan).
RNA extraction and Library Preparation
Total RNA from mouse RPE/choroid using triplicate biological replicates for air or smoking were isolated using RNeasy Mini Kit (Qiagen, Valencia, CA), with on-column DNA digestion by RNase-free DNase, following the manufacturer's instructions. RNA integrity was verified using Agilent 4200 TapeStation (Agilent, Santa Clara, CA). Stranded RNA-seq library construction was carried out using 100 ng of RNA with the TruSeq Stranded mRNA Sample Preparation Kit (Illumina, San Diego, CA). Libraries were paired-end sequenced to 126 bases using a HiSeq 2500 Sequencing System (Illumina, San Diego, CA).
RNA-Sequencing Analysis
The analysis pipeline has been described previously[23, 24] and was performed using mouse genome GRCm38.p6 with Ensembl v98 annotation. The gene-level differential expression analysis was performed between control (Air) and cigarette smoking (CS) treatment using the exact test from the edgeR v3.26.6[25] package in R (https://r-project.org). Genes were kept for analysis if all replicates of either group expressed at 1.0 count per million (CPM) or higher. Genes were defined as significantly differentially expressed (DEG) between Air and CS, if the absolute fold change exceeded 1.5 and had a Benjamini-Hochberg false discovery rate (FDR) of less than 5%. Functional gene enrichment was performed using gProfileR v0.6.7[26] with Gene Ontology (GO) Biological Process[27] gene sets. Reduced redundancy representation was performed using the most child term of any significant (less than 1% FDR) group of terms. Gene Set Enrichment Analysis (GSEA) was performed using the Hallmark Pathways from the GSEA database [28] using the fgsea v1.10.0 [https://doi.org/10.1101/060012] package in R. Potential protein–protein interactions of differentially expressed genes was analyzed using the STRING database (version 11.0) [29, 30]. The resulting dataframe object was modified to be plotted as enrichment plot with R package DOSE[31].
Cell culture
The established human ARPE-19 cell line[32] was maintained in Dulbecco's Modified Eagle Medium:F12 50/50 mix, supplemented with 10% inactivated fetal bovine serum and 2 mM L-glutamine, at 37 °C in a humidified atmosphere containing 5% CO2. Cells were seeded at 50,000 cells/cm2 in 12-well plates for 2 days followed by 1 day of serum starvation. Cells were treated with cigarette smoke extract (CSE) for up to 24 hr, and RNA was isolated for RT-qPCR, or protein was extracted for immunoblotting.
RT-PCR
RT-qPCR was performed as previously described[33](Applied Biosystems, Foster City, CA) on a StepOne-Plus Real-Time PCR system (Applied Biosystems) using Primer sequences (Applied Biosystems). Data were analyzed by the comparative threshold cycle method, with Cyclophilin A as an internal control.
Immunoblot analysis
Western blot analysis was performed as previously described[33]. Briefly, RPE/choroid, whole cell lysates, or supernatant were prepared using RIPA buffer (Sigma, Inc., St. Louis, MO). Proteins were separated by 4–12% Bis-Tris sodium dodecyl sulphate polyacrylamide gel electrophoresis, transferred to nitrocellulose, and probed with primary antibodies and then secondary antibodies (Table S5).
Statistical analysis
Statistical analysis was carried out using the unpaired t-test, with GraphPad software (GraphPad Software, Inc., San Diego, CA). Significance is indicated by *p<0.05, **p<0.01, and ***p<0.001. Each experiment was repeated at least three times. Blots are selected as the representative one of specific group of experiments, and graphs represent the mean ± SEM of at least three independent experiments.
Results
The RPE undergoes morphologic and ultrastructural derangement with chronic cigarette smoke exposure
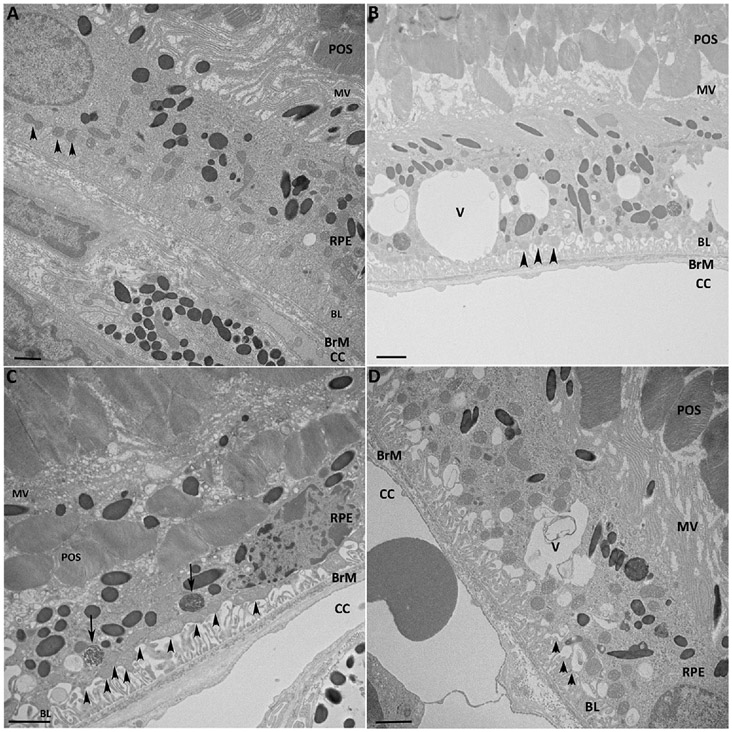
Mice exposed to chronic cigarette smoke develop significant oxidative injury and morphologic derangement to the RPE, as observed in early AMD[20-22]. To verify that our model reproduced these essential changes, 2-month old C57BL/6J mice were exposed to either cigarette smoke (CS) or air for 6 months, and the RPE/choroid was examined by TEM. Figure 1 shows the marked ultrastructural derangement to the RPE that includes mesenchymal cell shape, loss of apical microvilli and basal infoldings, and development of intracellular vacuoles, all of which are seen in human AMD[34-37].
Figure 1.
Ultrastructural changes to the RPE after chronic smoking. A. Normal morphology and ultrastructure of the RPE from a C57BL6J mouse raised in air. Apical microvilli (MV) surround photoreceptor outer segments (POS). The basolateral (BL) RPE has normal infoldings (arrowheads). A variety of ultrastructural changes are seen in the RPE from C57BL6J mice exposed to smoke for 6 months. B. The microvilli are shortened, multiple vacuoles (V) are seen within the cell body, and the basal infoldings are shortened and widened (arrowheads). C. More severe mesenchymal shape to the RPE cell with similar changes to the microvilli and basal infoldings as in (B). Undigested POS (arrows) are seen in the basal region of the cell. D. Vacuoles with membranous debris are seen. BrM, Bruch’s membrane, CC, choriocapillaris. Bar = 5 μm.
Global transcriptional response by the RPE/choroid to chronic cigarette smoke
To gain an understanding of the prominent cellular pathways that are impacted by chronic smoke exposure, we performed RNA-seq of the RPE/choroid from mice exposed to either CS or air for 6 months. Samples were sequenced to a mean depth of 15.1 ± 1.1 million fragments per sample, of which 86.1 ± 1.2 million fragments aligned to known gene annotations that were used for quantitation (Figure S1A).
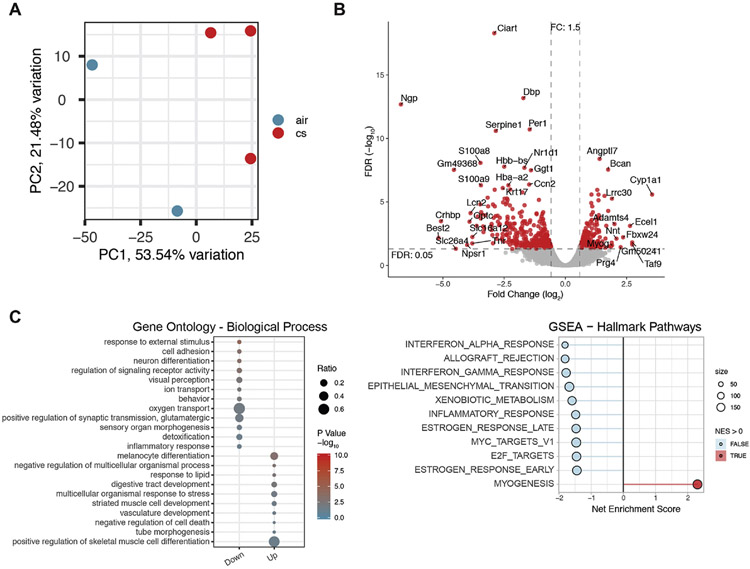
Gene expression values (CPM) were examined by principal component analysis (PCA) to evaluate replication of samples within the two groups (Figure S1B). Sample air.3 was determined to have a high level of retina contamination (Figure S1C) and was eliminated from the analysis (Figure 2A). As an initial characterization of the molecular alterations induced by chronic smoke exposure, we identified 558 differently expressed genes (DEGs) in the RPE/choroid between the CS and air treated groups, as represented by the Volcano plot (Figure 2B). The complete DEG results are listed in Table S1.
Figure 2.
Overview of differentially expressed genes. A. Principle Component Analysis for all expressed genes, totaling 15,559 genes. The samples clustered into two distinct groups of mice exposed to chronic cigarette smoking (CS) and air. B. Volcano plot of DEGs shows separation between air and CS treated mice. C. Gene Ontology enrichment of biological processes (BP) with chronic smoking exposure. Down-regulated genes from smoking are pooled in cluster1, and up-regulated genes in cluster2. The ratio of genes changed in each BP is reflected by the dot size. p-value is reflected by dot color, with red as the most confident change. D. Gene set enrichment analysis using the Hallmark Pathways. Pathways enriched in the down-regulated genes are indicated with a negative net enrichment score (light blue) and those enriched in the up-regulated genes have a positive enrichment score (red). Size indicates the number of genes in the pathway.
A number of pathologic pathways are impaired with smoking; these include oxidative stress response, mitochondrial function, innate immune response, and extracellular matrix regulation[5]. Many of the significant DEGs are associated with these pathways. For example, Cyp1a1 is induced by chronic smoking and likely represents a protective response to smoking since it is involved in xenobiotic metabolism, specifically degrades nicotine[38], and is linked to Aryl Hydrocarbon Receptor Signaling, which is also associated with xenobiotic metabolism and detoxification[39]. Nnt couples the hydride transfer between NAD(H) and NADP(+) to proton translocation across the inner mitochondrial membrane, using energy from the mitochondrial proton gradient to produce high concentrations of NADPH that is used for free radical neutralization[40]. Adamts4, which was increased 4.0-fold by smoking relative to air, is a metalloproteinase that specifically degrades extracellular matrix proteins Aggrecan and Bcan, which were also increased 3.4 fold [41]. Interestingly, mice with an Adamts4 mutation exhibit RPE dedifferentiation with reduced pigmentation and RPE-specific gene expression[42].
Several of the most downregulated genes from chronic smoking are related to the immune response. For example, Ngp, a cystatin superfamily member, regulates inflammatory responses through TLR-4 and phagocytosis[43]. Npsr1 induces the production of pro-inflammatory cytokines TNF-α and interferon-γ[44]. Crhbp activates NF-kB activation to promote the inflammatory response[45]. Lcn2, an adipokine, and S100A8, a calcium binding protein of the S100 family, are both elevated during acute inflammation as a protective response, and recruit leukocytes and pro-inflammatory cytokines[46, 47]. The downregulation of these genes could contribute to an impaired protective immune response following chronic smoke exposure.
Gene enrichment analysis reveals impaired innate immune and induction of differentiation response after chronic smoking
To define how the RPE responds to chronic cigarette smoke exposure, an unbiased evaluation of the transcriptome was performed using gene ontology and gene set enrichment analysis to identify specific biological processes that were over-represented in DEGs (Figure 2C, D). The top suppressed processes are associated with defense response, including the anti-viral response from type I and II interferon (IFN). The specific genes in these categories are listed in Table S2. On the other hand, the top activated processes include morphogenesis, differentiation, and development genes. The over-representation of these genes is indicative of a transcriptional response by the RPE/choroid to recover essential functions that were impaired by chronic smoke exposure.
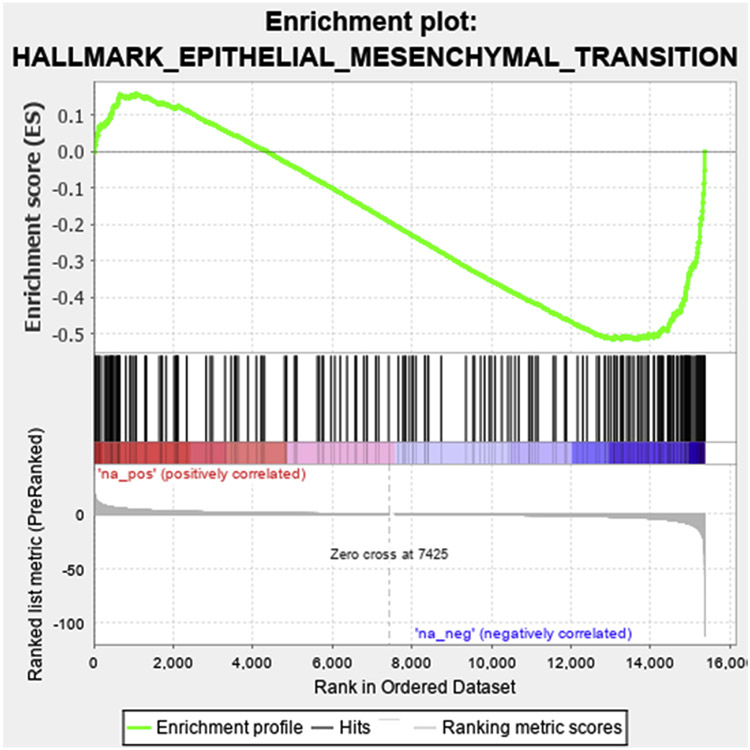
We recently reported that in early AMD, some RPE cells have entered epithelial mesenchymal transition (EMT), an adaptive transcriptional process that allows cells to survive a harsh microenvironment[48-51]. The transcriptional responses related to cell proliferation, cell migration, cell-cell adhesion in response to chronic smoking suggest prompted us to assess whether the RPE are entering EMT. The DEGs were ranked by their degree of differential expression in smoke relative to air control, and compared to the EMT gene set from GSEA database[28]. As illustrated in Figure 3, a set of DEGs was significantly related to the EMT gene signature, with a majority of these genes downregulated by chronic smoking (normalized enrichment score (NES) =−1.68, FDR = 0.0178). However, the expression pattern induced by smoking was for the most part, indicative of MET rather than EMT (Table S3). This expression pattern compliments the upregulation of genes related to differentiation and suggests that the RPE is attempting to maintain its epithelial state.
Figure 3.
Enrichment plot of differentially expressed genes (DEGs) induced by chronic smoking in the RPE that are related to Epithelial mesenchymal transition. DEGs were ranked from positive to negative fold change and compared to the Broad Institute’s GSEA EMT 184 gene set. The enrichment score is plotted, which shows that the majority of DEGs were downregulated.
String analysis reveals the influence of multiple signaling pathways after chronic smoking
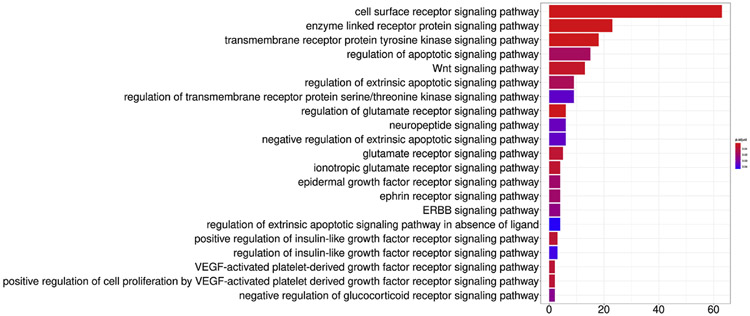
To identify signaling transduction pathways involved in the major processes identified by GO analysis, we performed String analysis of DEGs followed by enrichment analysis, selected terms with the keyword “pathway”, and identified the corresponding genes. Next, selecting the keyword “pathway” to find corresponding genes, which were intersected with the first neighborhood genes, performed enrichment analysis. Figure 4 shows the over-represented DEGs and their probability of being assigned to the specific signaling pathway. The cell surface receptor signaling, enzyme linked receptor protein signaling, transmembrane receptor protein tyrosine kinase signaling, regulation of apoptotic signaling, and the Wnt signaling pathway were the top five most likely signaling pathways involved after chronic smoke exposure.
Figure 4.
Signal transduction pathways involved in the major processes were identified by GO analysis and then String analysis, enrichment analysis by selecting the keyword “pathway” to identify the involved genes that were altered by chronic smoking. The signaling pathways with the strongest likelihood of involvement are indicated by the p values. The number of genes that were differentially expressed is also shown.
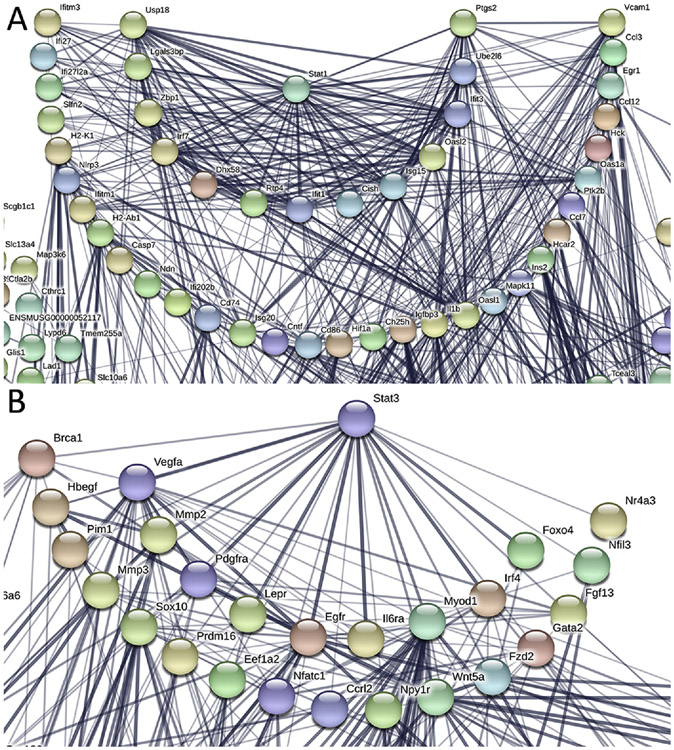
The top suppressed responses to chronic smoking, as noted above, included type I and II interferons (IFN), which are among the cell surface receptor signaling DEGs. Type I IFN responses can dampen type II IFN responses, and type II can reduce type I IFN immunity through STAT1 and STAT3 signaling[52, 53]. STAT1 and STAT3 have a reciprocal relationship in neurodegenerative diseases and tumor metastasis[54-56]. For example, STAT1 causes cell death signals, while STAT3 induces protective responses[54-56]. Because of the influence of STAT1 and STAT3 on IFN responses, we conducted String analysis to identify any potential functional interactions between these two pathways. Of the 1203 downregulated genes that were submitted for String analysis, 13 genes have close connectivity (combined probability score >0.7) and 31 genes have medium connectivity (combined probability score <0.7 but >0.4) with STAT1[57] (Figure 5A). The most connected genes, with their annotated gene function, are listed in Table S4.
Figure 5.
String analysis of STAT1 and STAT3 signaling after chronic smoking in the RPE/choroid. The network was built based on high confidence evidence from both experimental protein-protein interaction and curated databases. The thickness of the lines that represent interactions is proportional to the STRING combined probability score. A. 44 genes are connected to Stat1 with chronic smoking. B. 25 genes are connected to Stat3 with chronic smoking.
Unlike STAT1, STAT3 mRNA levels in the RPE/choroid did not change after chronic smoke exposure. However, ontology enrichment analysis showed that a number of genes downstream of STAT3, such as extracellular matrix molecules, and known STAT3 inhibitors including SOCS1 and TMF1, were up regulated. We then performed String analysis of STAT3 with the 942 genes upregulated with smoke exposure. Of the 556 genes that were identified in the String database, 20 genes show strong (combined probability score >0.8) and 17 genes weak connection (combined probability score <0.5) with STAT3 (Figure 5B). The 20 most connected genes, with their annotated gene function, are listed in Table S4. Of these 20 genes, EGFR and PDGFRA are upstream of STAT3, suggesting that STAT3 signaling could be triggered by different cytokines (EGF/PDGF vs. IFNs) during smoking, and could activate downstream targets such as MMP2, MMP3, and MMP14[58, 59]. Furthermore, MITF, a transcription factor that cooperatively induces cellular transformation with STAT3[60, 61], was upregulated 1.85 fold in the RPE/choroid of mice exposed to chronic smoking. In addition, SOX10 and PAX3, which synergistically activate MITF, were also upregulated by smoking 1.5-fold and 2.0-fold, respectively. While STAT3 itself was not differentially expressed in the RPE/choroid with smoking, this transcriptional pattern suggests that a PDGFR/EGFR-STAT3 signaling pathway was activated, and this could be enhanced by MITF, SOX10, and PAX3. Taken together, the RNA-Seq data suggest that STAT1 signaling is suppressed while STAT3 signaling is increased.
STAT1 and STAT3 are reciprocally activated in the RPE/choroid after smoking
We next determined whether STAT1 and STAT3 signaling were indeed activated by measuring their phosphorylated forms. Following the trend of STAT1 mRNA, both STAT1 and pSTAT1 were reduced in the RPE/choroid of mice exposed to 6 months of smoking (Figure 6A). Likewise, STAT3 in the RPE was unchanged. However, pSTAT3 was increased in the RPE/choroid of mice with chronic smoking (Figure 6B), suggesting that STAT3 activity is increased and likely post-translationally regulated. To further assess activation of STAT3 signaling, we examined the downstream production of the ECM molecules MMP2 and MMP14 and detected their upregulation in the RPE/choroid of mice after 6 months of smoking (Figure 6C).
Figure 6.
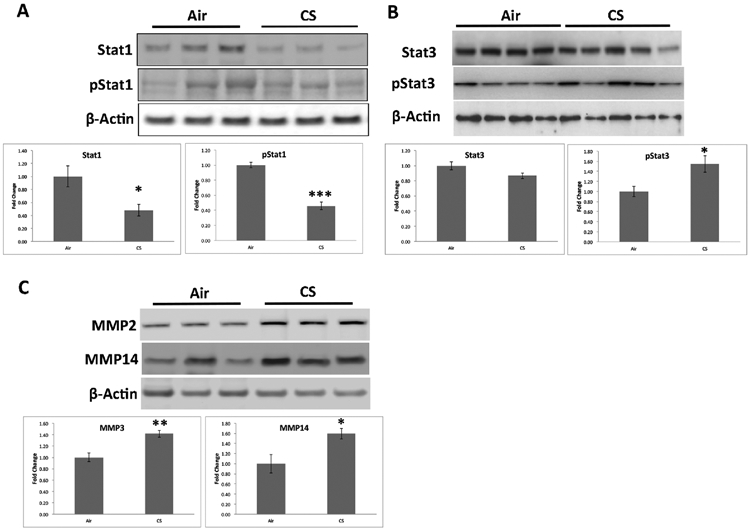
Reciprocal STAT1 and STAT3 signaling in the RPE/choroid after chronic smoking. A. Western blots of STAT1 and pSTAT1 from the RPE/choroid of air and smoke exposed mice. STAT1 and p-STAT1 were plotted as fold change of smoke to air treated mice. B. Western blots of STAT3 and pSTAT3 from the RPE/choroid of air and smoke exposed mice. STAT3 and p-STAT3 were plotted as fold change of smoke to air treated mice. C. Western blots of MMP3 and MMP14 from the RPE/choroid of air and smoke exposed mice. MMP3 and MMP14 were plotted as fold change of smoke to air treated mice. Data were normalized to β-actin. *p<0.05. **p<0.01, ***p<0.001.
STAT1 and STAT3 signaling responses were also similar in human RPE cells treated with cigarette smoke extract (CSE). Basal STAT1 activity is low in RPE cells and was enhanced with 100 ng/ml IFNα. Under these conditions, STAT1 signaling was reduced by CSE relative to controls (Figure 7A).
Figure 7.
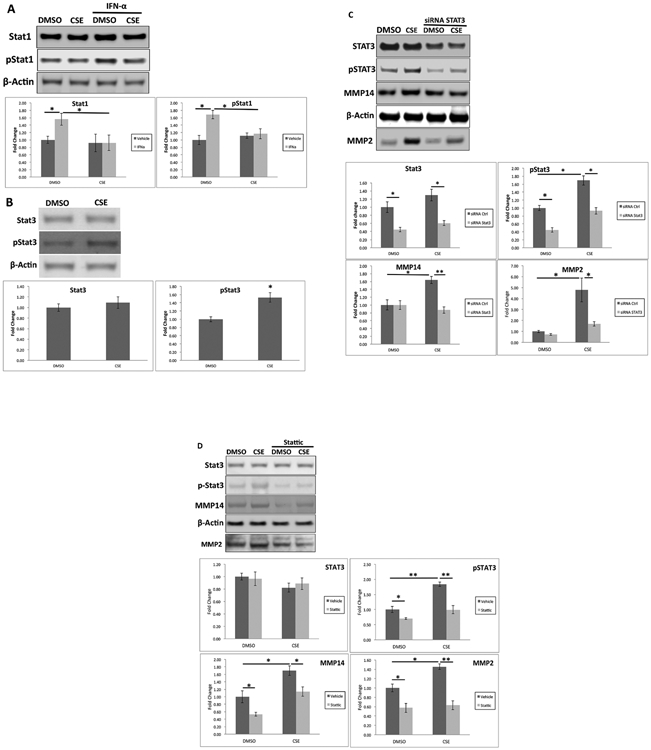
STAT1 is decreased by cigarette smoke extract (CSE). A. RPE cells were treated with DMSO or 125 μg/ml CSE in the presence of 100 ng/ml IFN- α for 6 hrs. Western blots of STAT1 and pSTAT1 and their abundances were plotted as fold change. B. Cells were treated with DMSO or CSE for 24 hrs. Western blots of STAT3 and pSTAT3, and their abundances were plotted as fold change. *p<0.05. STAT3 silencing abrogates CSE-induced MMP14 production and MMP2 secretion. C. ARPE-19 cells were transfected with STAT3 siRNA and treated with 125 μg/ml CSE for 24 h. Western blot shows total STAT3, p-STAT3, and MMP14, and their abundances were plotted as fold change relative to DMSO-treated control siRNA treated cells. Data were normalized to β-actin. D. RPE cells were transfected with STAT3 siRNA, and treated with 125 μg/ml CSE for 24 h. Western blot shows total STAT3, p-STAT3, and MMP14 from cell lysates and MMP2 from the supernatant, and their abundances were plotted as fold change relative to DMSO-treated control siRNA treated cells. Data were normalized to β-actin. *p<0.05; **p<0.01.
As in the RPE/choroid from mice exposed to chronic smoke, STAT3 was unchanged, and pSTAT3 was increased with CSE treatment compared to vehicle controls (Figure 7B). The pSTAT3 increase by CSE induced MMP3 and MMP14 production compared to vehicle control cells. This induction could be abrogated by silencing of STAT3 with either an siRNA against STAT3 (Figure 7C) or 1 uM of the STAT3 inhibitor Stattic (Figure 7D).
Discussion
The relative contribution of the dysfunctional pathways that are influenced by smoking to early AMD is not clear. Here, we report the global expression profile of the RPE/choroid from mice exposed to chronic smoking, which develop features of early AMD. We show that the overall RPE transcriptome after smoking exhibits marked suppression of the innate immune response including the antiviral response with type I and II interferons and an upregulation of cell differentiation and morphogenesis gene clusters, which indicates an attempt by the RPE to maintain its function despite smoke-induced injury. These novel changes are distinct from the previously acknowledged pathways involved in AMD pathobiology, such as mitochondrial dysfunction, autophagy, antioxidant response, and complement activation, and may represent events preceding the onset of established dysfunctions. These transcriptome changes are relevant because they are both in response to chronic smoking, and in a model that simulates RPE changes seen in early AMD. Since mice have a similar RPE and rod/cone density as the human perifovea[19], a site of early AMD changes, these alterations may be relevant to the perifovea.
Smoking induces the formation of danger associated molecular patterns (DAMPS), including oxidation-specific epitopes (OSEs), or oxidatively modified nucleic acids, proteins, and lipids that form when antioxidant systems inadequately neutralize reactive oxygen species[62]. DAMPS are recognized by pattern recognition receptors (PRRs) that activate the innate immune response. Relevant to AMD, we previously identified complement factor H and lipoprotein(a) as PRRs that bind to malondialdehyde and oxidized phospholipids, respectively, to induce an innate immune response[63, 64]. PRRs can activate type I and II IFNs in order to neutralize these potentially disease-causing molecules[65, 66]. The suppressed response by the RPE/choroid suggests that the immune response to DAMPs that are generated from smoking was inadequate and could lead to RPE dysfunction. The correlation of this impaired IFN response with an early AMD phenotype suggests that this failed arm of the innate immune response might be an early event in AMD pathobiology. A suppressed immune response is in contrast to work suggesting an overactive innate immune response, especially complement and the inflammasome. We note that our findings are from a model of early AMD, whereas the genetic link of complement factors with AMD risk was conducted in patients with advanced AMD[67-70]. In addition, our findings are at the transcriptional level. Since a pathogenic role for complement is best studied by determining both its activity and the extent that this enhanced activity damages tissue, we are not able to rule out a role for complement despite minimal transcriptional changes. Likewise, we note that the best evidence of a role for the inflammasome is in late disease, such as in geographic atrophy or neovascular AMD[71, 72]. The innate immune response is complex, cell type specific, and situational. The AMD stage and the specific arm of immunity are fundamental requirements needed in any study design to fully decipher the role of innate immunity on AMD pathobiology. Future investigations will focus on the IFN response on RPE function and the degree of tissue injury induced by smoking induced suppression of this response in early AMD.
The over-representation of upregulated genes involved in morphogenesis, differentiation, and development suggest a transcriptional response by the RPE intended to maintain or re-establish its epithelial state to compensate for injury caused by chronic smoke exposure. We had previously observed that some RPE cells enter EMT in human AMD samples[51]. Our RNA-seq analysis uncovered DEGs related to EMT, but the expression pattern was indicative of mesenchymal epithelial transition, which is consistent with the cell’s attempt to differentiate. The RPE has a heterogeneous morphology with a spectrum of normal appearing RPE to marked mesenchymal morphology in both AMD and this model as suggested in Fig. 1[20-22, 73, 74]. It is likely that each RPE cell’s attempt to recover from smoking related injury is mosaic. Future studies might benefit from single cell RNA-sequencing to enable characterization of the RPE’s heterogeneous response to smoking.
STAT1 and STAT3 are essential signaling components of type I and II IFN responses, which was verified by String analysis. In response to cigarette smoke, STAT1 was decreased while STAT3 was increased. Smoking is known to suppress type I and II IFN responses through several mechanisms. Upon interaction of IFNα with its IFN receptor, JAK1 and TYK2 are phosphorylated, which then phosphorylate STAT1 and STAT2 to enable heterodimerization. The heterodimer translocates into the nucleus and binds with nuclear p48/IRF-9 proteins to form the ISGF-3 complex, which binds to interferon stimulated response element in the promoters of IFNα-stimulated genes to induce their expression[75-77]. Cigarette smoke can up-regulate the catalytic activity of serine/threonine phosphatases[78], inhibit IRF-3[79], or reduce STAT1 phosphorylation[80]. With type II IFN signaling, smoking can decrease the expression of IFN-γR, which decreases STAT1 phosphorylation[81].
In contrast, STAT3 is induced by cigarette smoke as a protective response. In the lung, STAT3 is activated by cigarette smoke to regulate key inflammatory, proteolytic and apoptotic responses. At the same time, STAT3 modulates the anti-inflammatory response by increasing SOCS3 and IL-10 expression to prevent tissue injury[82]. When STAT3 is lacking, as in Stat3−/− mice, smoke exposure enhances inflammatory, proteolytic, and apoptotic responses, but with a deficient anti-inflammatory response that results in tissue injury. Likewise, STAT3 suppression by siRNA severely damages DNA to induce cell death after cigarette smoke exposure[83]. Consistent with these studies, the STAT3 induction is a protective response.
We identified ECM regulation by STAT3 signaling, and that smoking induced MMPs. The RPE is attached to Bruch’s membrane, a pentalaminar matrix. Normally, Bruch’s membrane undergoes constant remodeling by MMPs that are modulated by TIMPs, which are in part, produced by the RPE[84-86]. With aging, Bruch’s membrane thickens due to matrix protein accumulation, lipid deposition, and oxidative modification including advanced glycation end product formation that decreases hydraulic conductivity and nutrient transport across Bruch’s membrane[74, 87-92]. These changes can enable the accumulation of cellular fragments, lipoproteins, and inflammatory debris during the formation of basal deposits and drusen, hallmark lesions of AMD. As an early event, altered MMP activity by the RPE may contribute to age-related Bruch’s membrane thickening[86, 93]. The induction of MMPs through STAT3 by smoking both in vitro and in vivo suggests that the normal remodeling function of MMPs is intact. With further smoke exposure, it is possible that this process fails and could contribute to basal deposit formation.
Some limitations of this study are recognized. The transcriptomes that we identified were from bulk RNA-seq. While the physiological impact may be similar, we acknowledge that some of the expression profiles could originate from choroidal cells. Single cell RNA-seq would be a valuable approach to separate the transcriptional response of the RPE from choroidal cells. ARPE-19 cells were chosen to study the effects in human cells and did confirm the findings of our mouse studies. However, ARPE-19 cells have their shortcomings, and do other in vitro systems such as human fetal RPE cells or RPE cells from donor globes.
Conclusions
The unbiased global RNA-seq analysis of the RPE/choroid after chronic smoking uncovered an unexpected decline in the innate immune response that coincided with a transcriptional attempt to maintain its epithelial state in a model that simulates early AMD. This work provides insight into the early events caused by smoking that could lead to the conversion to early AMD. While valuable, the RPE response is unlikely to be uniform given its known heterogeneity in AMD. The role of the processes identified in this investigation might be enhanced by implementing single cell RNA-sequencing to identify the subgroups of RPE that may be expressing these potentially pathogenic and protective signals identified in this investigation. Future investigations to evaluate the relationship of smoking-induced transcriptional changes in RPE (reported here) to epigenomic alterations in aging (Corso Diaz et al. Cell Reports in press) and AMD-associated expression quantitative trait loci from human retina and RPE[94, 95] will be helpful in formulating a coherent and comprehensive platform for analyses of interactions among distinct susceptibility factors leading to AMD pathology.
Supplementary Material
Figure S1. A. RNA-Seq quality control graph. Total RNA from mouse RPE/choroid (air vs 6-mo smoking) was used for library construction and RNASeq reading. The ratio of aligned targets among total reading is calculated for each sample and falls between 84-88%, suggesting good reading quality. B. Principal component analysis showing Air.3 as an outlier. C. High level of retinal gene expression from Air.3 indicates retinal contamination. This sample was eliminated from the analysis.
Highlights.
Chronic smoking suppressed the immune response including interferons in the RPE
Cell differentiation genes were also upregulated as a compensatory response
STAT1 and STAT3 were reciprocally activated to regulate the extracellular matrix
Matrix alterations to Bruch’s membrane are early events in AMD
The transcriptome to smoke is complex with both impaired and compensatory responses
Acknowledgements
This work was supported by the National Institutes of Health [NIH NEI R01 EY027691 to JTH], NEI Core grant [EY001765], Macular Degeneration Foundation to JTH, RPB (Wilmer Eye Institute), NEI-Intramural Research Program [ZIAEY000450 and ZIAEY000546 to AS]. JTH is the Robert Bond Welch Professor. We thank Kathleen Boesze-Battaglia, PhD for providing the RPE cell template for the Graphic Abstract.
Footnotes
Conflict of Interest Statement: JTH received grant funding and royalties, and MC grant funding from Bayer Pharmaceutical, Inc.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Smith CJ, Hansch C, The relative toxicity of compounds in mainstream cigarette smoke condensate, Food Chem Toxicol 38(7) (2000) 637–46. [DOI] [PubMed] [Google Scholar]
- [2].The Health Consequences of Smoking-50 Years of Progress: A Report of the Surgeon General, Atlanta (GA), 2014. [Google Scholar]
- [3].Wong WL, Su X, Li X, Cheung CM, Klein R, Cheng CY, Wong TY, Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis, Lancet Glob Health 2(2) (2014) e106–16. [DOI] [PubMed] [Google Scholar]
- [4].Pennington KL, DeAngelis MM, Epidemiology of age-related macular degeneration (AMD): associations with cardiovascular disease phenotypes and lipid factors, Eye Vis (Lond) 3 (2016) 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Handa JT, Bowes Rickman C, Dick AD, Gorin MB, Miller JW, Toth CA, Ueffing M, Zarbin M, Farrer LA, A systems biology approach towards understanding and treating non-neovascular age-related macular degeneration, Nature communications 10(1) (2019) 3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lutein + zeaxanthin and omega-3 fatty acids for age-related macular degeneration: the Age-Related Eye Disease Study 2 (AREDS2) randomized clinical trial, JAMA 309(19) (2013) 2005–15. [DOI] [PubMed] [Google Scholar]
- [7].Rosenfeld PJ, Brown DM, Heier JS, Boyer DS, Kaiser PK, Chung CY, Kim RY, Group MS, Ranibizumab for neovascular age-related macular degeneration, N Engl J Med 355(14) (2006) 1419–31. [DOI] [PubMed] [Google Scholar]
- [8].Brown DM, Kaiser PK, Michels M, Soubrane G, Heier JS, Kim RY, Sy JP, Schneider S, Group AS, Ranibizumab versus verteporfin for neovascular age-related macular degeneration, N Engl J Med 355(14) (2006) 1432–44. [DOI] [PubMed] [Google Scholar]
- [9].Swaroop A, Branham KE, Chen W, Abecasis G, Genetic susceptibility to age-related macular degeneration: a paradigm for dissecting complex disease traits, Hum Mol Genet 16 Spec No. 2 (2007) R174–82. [DOI] [PubMed] [Google Scholar]
- [10].Fritsche LG, Fariss RN, Stambolian D, Abecasis GR, Curcio CA, Swaroop A, Age-related macular degeneration: genetics and biology coming together, Annu Rev Genomics Hum Genet 15 (2014) 151–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Fritsche LG, Igl W, Bailey JN, Grassmann F, Sengupta S, Bragg-Gresham JL, Burdon KP, Hebbring SJ, Wen C, Gorski M, Kim IK, Cho D, Zack D, Souied E, Scholl HP, Bala E, Lee KE, Hunter DJ, Sardell RJ, Mitchell P, Merriam JE, Cipriani V, Hoffman JD, Schick T, Lechanteur YT, Guymer RH, Johnson MP, Jiang Y, Stanton CM, Buitendijk GH, Zhan X, Kwong AM, Boleda A, Brooks M, Gieser L, Ratnapriya R, Branham KE, Foerster JR, Heckenlively JR, Othman MI, Vote BJ, Liang HH, Souzeau E, McAllister IL, Isaacs T, Hall J, Lake S, Mackey DA, Constable IJ, Craig JE, Kitchner TE, Yang Z, Su Z, Luo H, Chen D, Ouyang H, Flagg K, Lin D, Mao G, Ferreyra H, Stark K, von Strachwitz CN, Wolf A, Brandl C, Rudolph G, Olden M, Morrison MA, Morgan DJ, Schu M, Ahn J, Silvestri G, Tsironi EE, Park KH, Farrer LA, Orlin A, Brucker A, Li M, Curcio CA, Mohand-Said S, Sahel JA, Audo I, Benchaboune M, Cree AJ, Rennie CA, Goverdhan SV, Grunin M, Hagbi-Levi S, Campochiaro P, Katsanis N, Holz FG, Blond F, Blanche H, Deleuze JF, Igo RP Jr., Truitt B, Peachey NS, Meuer SM, Myers CE, Moore EL, Klein R, Hauser MA, Postel EA, Courtenay MD, Schwartz SG, Kovach JL, Scott WK, Liew G, Tan AG, Gopinath B, Merriam JC, Smith RT, Khan JC, Shahid H, Moore AT, McGrath JA, Laux R, Brantley MA Jr., Agarwal A, Ersoy L, Caramoy A, Langmann T, Saksens NT, de Jong EK, Hoyng CB, Cain MS, Richardson AJ, Martin TM, Blangero J, Weeks DE, Dhillon B, van Duijn CM, Doheny KF, Romm J, Klaver CC, Hayward C, Gorin MB, Klein ML, Baird PN, den Hollander AI, Fauser S, Yates JR, Allikmets R, Wang JJ, Schaumberg DA, Klein BE, Hagstrom SA, Chowers I, Lotery AJ, Leveillard T, Zhang K, Brilliant MH, Hewitt AW, Swaroop A, Chew EY, Pericak-Vance MA, DeAngelis M, Stambolian D, Haines JL, Iyengar SK, Weber BH, Abecasis GR, Heid IM, A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants, Nat Genet 48(2) (2016) 134–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Smith W, Assink J, Klein R, Mitchell P, Klaver CC, Klein BE, Hofman A, Jensen S, Wang JJ, de Jong PT, Risk factors for age-related macular degeneration: Pooled findings from three continents, Ophthalmology 108(4) (2001) 697–704. [DOI] [PubMed] [Google Scholar]
- [13].Tan JS, Mitchell P, Kifley A, Flood V, Smith W, Wang JJ, Smoking and the long-term incidence of age-related macular degeneration: the Blue Mountains Eye Study, Arch Ophthalmol 125(8) (2007) 1089–95. [DOI] [PubMed] [Google Scholar]
- [14].Mitchell P, Wang JJ, Smith W, Leeder SR, Smoking and the 5-year incidence of age-related maculopathy: the Blue Mountains Eye Study, Arch Ophthalmol 120(10) (2002) 1357–63. [DOI] [PubMed] [Google Scholar]
- [15].Curcio CA, Millican CL, Allen KA, Kalina RE, Aging of the human photoreceptor mosaic: evidence for selective vulnerability of rods in central retina, Invest Ophthalmol Vis Sci 34(12) (1993) 3278–96. [PubMed] [Google Scholar]
- [16].Curcio CA, Medeiros NE, Millican CL, Photoreceptor loss in age-related macular degeneration, Invest Ophthalmol Vis Sci 37(7) (1996) 1236–49. [PubMed] [Google Scholar]
- [17].Starnes AC, Huisingh C, McGwin G Jr., Sloan KR, Ablonczy Z, Smith RT, Curcio CA, Ach T, Multi-nucleate retinal pigment epithelium cells of the human macula exhibit a characteristic and highly specific distribution, Vis Neurosci 33 (2016) e001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chen M, Rajapakse D, Fraczek M, Luo C, Forrester JV, Xu H, Retinal pigment epithelial cell multinucleation in the aging eye - a mechanism to repair damage and maintain homoeostasis, Aging Cell 15(3) (2016) 436–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Volland S, Esteve-Rudd J, Hoo J, Yee C, Williams DS, A comparison of some organizational characteristics of the mouse central retina and the human macula, PLoS One 10(4) (2015) e0125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Fujihara M, Nagai N, Sussan TE, Biswal S, Handa JT, Chronic cigarette smoke causes oxidative damage and apoptosis to retinal pigmented epithelial cells in mice, PLoS ONE 3(9) (2008) e3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Cano M, Thimmalappula R, Fujihara M, Nagai N, Sporn M, Wang AL, Neufeld AH, Biswal S, Handa JT, Cigarette smoking, oxidative stress, the anti-oxidant response through Nrf2 signaling, and Age-related Macular Degeneration, Vision Res 50(7) (2010) 652–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ebrahimi K, Cano M, Rhee J, Datta S, Wang L, Handa J, Oxidative stress induces an interactive decline in Wnt and Nrf2 signaling in degenerating retinal pigment epithelium, Antioxid Redox Signal (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Brooks MJ, Chen HY, Kelley RA, Mondal AK, Nagashima K, De Val N, Li T, Chaitankar V, Swaroop A, Improved Retinal Organoid Differentiation by Modulating Signaling Pathways Revealed by Comparative Transcriptome Analyses with Development In Vivo, Stem Cell Reports 13(5) (2019) 891–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kaya KD, Chen HY, Brooks MJ, Kelley RA, Shimada H, Nagashima K, de Val N, Drinnan CT, Gieser L, Kruczek K, Erceg S, Li T, Lukovic D, Adlakha YK, Welby E, Swaroop A, Transcriptome-based molecular staging of human stem cell-derived retinal organoids uncovers accelerated photoreceptor differentiation by 9-cis retinal, Mol Vis 25 (2019) 663–678. [PMC free article] [PubMed] [Google Scholar]
- [25].Robinson MD, McCarthy DJ, Smyth GK, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data, Bioinformatics 26(1) (2010) 139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, Vilo J, g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update), Nucleic Acids Res 47(W1) (2019) W191–W198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].C. The Gene Ontology, The Gene Ontology Resource: 20 years and still GOing strong, Nucleic Acids Res 47(D1) (2019) D330–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles, Proc Natl Acad Sci U S A 102(43) (2005) 15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, Bork P, von Mering C, STRING 8--a global view on proteins and their functional interactions in 630 organisms, Nucleic Acids Res 37(Database issue) (2009) D412–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ, STRING v9.1: protein-protein interaction networks, with increased coverage and integration, Nucleic Acids Res 41(Database issue) (2013) D808–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Yu G, Wang LG, Yan GR, He QY, DOSE: an R/Bioconductor package for disease ontology semantic and enrichment analysis, Bioinformatics 31(4) (2015) 608–9. [DOI] [PubMed] [Google Scholar]
- [32].Dunn KC, Aotaki-Keen AE, Putkey FR, Hjelmeland LM, ARPE-19, a human retinal pigment epithelial cell line with differentiated properties, Exp Eye Res 62(2) (1996) 155–69. [DOI] [PubMed] [Google Scholar]
- [33].Wang L, Cano M, Handa JT, p62 provides dual cytoprotection against oxidative stress in the retinal pigment epithelium, Biochim Biophys Acta 1843(7) (2014) 1248–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Olsen TS, Wassef NF, Olsen HS, Hansen HE, Ultrastructure of the kidney in acute interstitial nephritis, Ultrastruct Pathol 10(1) (1986) 1–16. [DOI] [PubMed] [Google Scholar]
- [35].Olsen S, Burdick JF, Keown PA, Wallace AC, Racusen LC, Solez K, Primary acute renal failure ("acute tubular necrosis") in the transplanted kidney: morphology and pathogenesis, Medicine (Baltimore) 68(3) (1989) 173–87. [DOI] [PubMed] [Google Scholar]
- [36].Drueke T, Hennessen U, Nabarra B, Ben Nasr L, Lucas PA, Dang P, Thomasset M, Lacour B, Coudrier E, McCarron DA, Ultrastructural and functional abnormalities of intestinal and renal epithelium in the SHR, Kidney Int 37(6) (1990) 1438–48. [DOI] [PubMed] [Google Scholar]
- [37].Anderson DH, Mullins RF, Hageman GS, Johnson LV, A role for local inflammation in the formation of drusen in the aging eye, Am J Ophthalmol 134(3) (2002) 411–31. [DOI] [PubMed] [Google Scholar]
- [38].Iba MM, Scholl H, Fung J, Thomas PE, Alam J, Induction of pulmonary CYP1A1 by nicotine, Xenobiotica 28(9) (1998) 827–43. [DOI] [PubMed] [Google Scholar]
- [39].Stejskalova L, Pavek P, The function of cytochrome P450 1A1 enzyme (CYP1A1) and aryl hydrocarbon receptor (AhR) in the placenta, Curr Pharm Biotechnol 12(5) (2011) 715–30. [DOI] [PubMed] [Google Scholar]
- [40].Nickel AG, von Hardenberg A, Hohl M, Loffler JR, Kohlhaas M, Becker J, Reil JC, Kazakov A, Bonnekoh J, Stadelmaier M, Puhl SL, Wagner M, Bogeski I, Cortassa S, Kappl R, Pasieka B, Lafontaine M, Lancaster CR, Blacker TS, Hall AR, Duchen MR, Kastner L, Lipp P, Zeller T, Muller C, Knopp A, Laufs U, Bohm M, Hoth M, Maack C, Reversal of Mitochondrial Transhydrogenase Causes Oxidative Stress in Heart Failure, Cell Metab 22(3) (2015) 472–84. [DOI] [PubMed] [Google Scholar]
- [41].Ren P, Hughes M, Krishnamoorthy S, Zou S, Zhang L, Wu D, Zhang C, Curci JA, Coselli JS, Milewicz DM, LeMaire SA, Shen YH, Critical Role of ADAMTS-4 in the Development of Sporadic Aortic Aneurysm and Dissection in Mice, Sci Rep 7(1) (2017) 12351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Collin GB, Hubmacher D, Charette JR, Hicks WL, Stone L, Yu M, Naggert JK, Krebs MP, Peachey NS, Apte SS, Nishina PM, Disruption of murine Adamtsl4 results in zonular fiber detachment from the lens and in retinal pigment epithelium dedifferentiation, Hum Mol Genet 24(24) (2015) 6958–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Liu K, Tian LX, Tang X, Wang J, Tang WQ, Ma ZF, Chen T, Liang HP, Neutrophilic granule protein (NGP) attenuates lipopolysaccharide-induced inflammatory responses and enhances phagocytosis of bacteria by macrophages, Cytokine 128 (2020) 155001. [DOI] [PubMed] [Google Scholar]
- [44].Sundman L, Saarialho-Kere U, Vendelin J, Lindfors K, Assadi G, Kaukinen K, Westerholm-Ormio M, Savilahti E, Maki M, Alenius H, D'Amato M, Pulkkinen V, Kere J, Saavalainen P, Neuropeptide S receptor 1 expression in the intestine and skin--putative role in peptide hormone secretion, Neurogastroenterol Motil 22(1) (2010) 79–87, e30. [DOI] [PubMed] [Google Scholar]
- [45].Yang K, Xiao Y, Xu T, Yu W, Ruan Y, Luo P, Cheng F, Integrative analysis reveals CRHBP inhibits renal cell carcinoma progression by regulating inflammation and apoptosis, Cancer Gene Ther (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Moschen AR, Adolph TE, Gerner RR, Wieser V, Tilg H, Lipocalin-2: A Master Mediator of Intestinal and Metabolic Inflammation, Trends Endocrinol Metab 28(5) (2017) 388–397. [DOI] [PubMed] [Google Scholar]
- [47].Wang S, Song R, Wang Z, Jing Z, Wang S, Ma J, S100A8/A9 in Inflammation, Front Immunol 9 (2018) 1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kalluri R, Neilson EG, Epithelial-mesenchymal transition and its implications for fibrosis, J Clin Invest 112(12) (2003) 1776–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Radeke MJ, Radeke CM, Shih YH, Hu J, Bok D, Johnson LV, Coffey PJ, Restoration of mesenchymal retinal pigmented epithelial cells by TGFbeta pathway inhibitors: implications for age-related macular degeneration, Genome Med 7(1) (2015) 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kimura K, Orita T, Liu Y, Yang Y, Tokuda K, Kurakazu T, Noda T, Yanai R, Morishige N, Takeda A, Ishibashi T, Sonoda KH, Attenuation of EMT in RPE cells and subretinal fibrosis by an RAR-gamma agonist, J Mol Med (Berl) 93(7) (2015) 749–58. [DOI] [PubMed] [Google Scholar]
- [51].Ghosh S, Shang P, Terasaki H, Stepicheva N, Hose S, Yazdankhah M, Weiss J, Sakamoto T, Bhutto IA, Xia S, Zigler JS Jr., Kannan R, Qian J, Handa JT, Sinha D, A Role for betaA3/A1-Crystallin in Type 2 EMT of RPE Cells Occurring in Dry Age-Related Macular Degeneration, Invest Ophthalmol Vis Sci 59(4) (2018) AMD104–AMD113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Duerr CU, McCarthy CD, Mindt BC, Rubio M, Meli AP, Pothlichet J, Eva MM, Gauchat JF, Qureshi ST, Mazer BD, Mossman KL, Malo D, Gamero AM, Vidal SM, King IL, Sarfati M, Fritz JH, Type I interferon restricts type 2 immunopathology through the regulation of group 2 innate lymphoid cells, Nat Immunol 17(1) (2016) 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Regis G, Pensa S, Boselli D, Novelli F, Poli V, Ups and downs: the STAT1:STAT3 seesaw of Interferon and gp130 receptor signalling, Semin Cell Dev Biol 19(4) (2008) 351–9. [DOI] [PubMed] [Google Scholar]
- [54].Takagi Y, Harada J, Chiarugi A, Moskowitz MA, STAT1 is activated in neurons after ischemia and contributes to ischemic brain injury, J Cereb Blood Flow Metab 22(11) (2002) 1311–8. [DOI] [PubMed] [Google Scholar]
- [55].Hsu WL, Ma YL, Hsieh DY, Liu YC, Lee EH, STAT1 negatively regulates spatial memory formation and mediates the memory-impairing effect of Abeta, Neuropsychopharmacology 39(3) (2014) 746–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Chiba T, Yamada M, Sasabe J, Terashita K, Shimoda M, Matsuoka M, Aiso S, Amyloid-beta causes memory impairment by disturbing the JAK2/STAT3 axis in hippocampal neurons, Mol Psychiatry 14(2) (2009) 206–22. [DOI] [PubMed] [Google Scholar]
- [57].von Mering C, Jensen LJ, Snel B, Hooper SD, Krupp M, Foglierini M, Jouffre N, Huynen MA, Bork P, STRING: known and predicted protein-protein associations, integrated and transferred across organisms, Nucleic Acids Res 33(Database issue) (2005) D433–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Jia ZH, Jia Y, Guo FJ, Chen J, Zhang XW, Cui MH, Phosphorylation of STAT3 at Tyr705 regulates MMP-9 production in epithelial ovarian cancer, PLoS One 12(8) (2017) e0183622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Xie TX, Wei D, Liu M, Gao AC, Ali-Osman F, Sawaya R, Huang S, Stat3 activation regulates the expression of matrix metalloproteinase-2 and tumor invasion and metastasis, Oncogene 23(20) (2004) 3550–60. [DOI] [PubMed] [Google Scholar]
- [60].Joo A, Aburatani H, Morii E, Iba H, Yoshimura A, STAT3 and MITF cooperatively induce cellular transformation through upregulation of c-fos expression, Oncogene 23(3) (2004) 726–34. [DOI] [PubMed] [Google Scholar]
- [61].Sonnenblick A, Levy C, Razin E, Interplay between MITF, PIAS3, and STAT3 in mast cells and melanocytes, Molecular and cellular biology 24(24) (2004) 10584–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Miller YI, Choi SH, Wiesner P, Fang L, Harkewicz R, Hartvigsen K, Boullier A, Gonen A, Diehl CJ, Que X, Montano E, Shaw PX, Tsimikas S, Binder CJ, Witztum JL, Oxidation-specific epitopes are danger-associated molecular patterns recognized by pattern recognition receptors of innate immunity, Circ Res 108(2) (2011) 235–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Weismann D, Hartvigsen K, Lauer N, Bennett KL, Scholl HP, Charbel Issa P, Cano M, Brandstatter H, Tsimikas S, Skerka C, Superti-Furga G, Handa JT, Zipfel PF, Witztum JL, Binder CJ, Complement factor H binds malondialdehyde epitopes and protects from oxidative stress, Nature 478(7367) (2011) 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Handa JT, Tagami M, Ebrahimi K, Leibundgut G, Janiak A, Witztum JL, Tsimikas S, Lipoprotein(A) with An Intact Lysine Binding Site Protects the Retina From an Age-Related Macular Degeneration Phenotype in Mice (An American Ophthalmological Society Thesis), Trans Am Ophthalmol Soc 113 (2015) T51–T522. [PMC free article] [PubMed] [Google Scholar]
- [65].Saxena AR, Gao LY, Srivatsa S, Bobersky EZ, Periasamy S, Hunt DT, Altman KE, Crawford DR, Oxidized and Original article degraded mitochondrial polynucleotides (DeMPs), especially RNA, are potent immunogenic regulators in primary mouse macrophages, Free Radic Biol Med 104 (2017) 371–379. [DOI] [PubMed] [Google Scholar]
- [66].Little JP, Simtchouk S, Schindler SM, Villanueva EB, Gill NE, Walker DG, Wolthers KR, Klegeris A, Mitochondrial transcription factor A (Tfam) is a pro-inflammatory extracellular signaling molecule recognized by brain microglia, Mol Cell Neurosci 60 (2014) 88–96. [DOI] [PubMed] [Google Scholar]
- [67].Edwards AO, Ritter Iii R, Abel KJ, Manning A, Panhuysen C, Farrer LA, Complement Factor H Polymorphism and Age-Related Macular Degeneration, Science 308(5720) (2005) 421–424. [DOI] [PubMed] [Google Scholar]
- [68].Hageman GS, Anderson DH, Johnson LV, Hancox LS, Taiber AJ, Hardisty LI, Hageman JL, Stockman HA, Borchardt JD, Gehrs KM, Smith RJ, Silvestri G, Russell SR, Klaver CC, Barbazetto I, Chang S, Yannuzzi LA, Barile GR, Merriam JC, Smith RT, Olsh AK, Bergeron J, Zernant J, Merriam JE, Gold B, Dean M, Allikmets R, A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration, Proc Natl Acad Sci U S A 102(20) (2005) 7227–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, Gallins P, Spencer KL, Kwan SY, Noureddine M, Gilbert JR, Schnetz-Boutaud N, Agarwal A, Postel EA, Pericak-Vance MA, Complement Factor H Variant Increases the Risk of Age-Related Macular Degeneration, Science (2005). [DOI] [PubMed] [Google Scholar]
- [70].Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, Sangiovanni JP, Mane SM, Mayne ST, Bracken MB, Ferris FL, Ott J, Barnstable C, Hoh J, Complement Factor H Polymorphism in Age-Related Macular Degeneration, Science (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Tarallo V, Hirano Y, Gelfand BD, Dridi S, Kerur N, Kim Y, Cho WG, Kaneko H, Fowler BJ, Bogdanovich S, Albuquerque RJ, Hauswirth WW, Chiodo VA, Kugel JF, Goodrich JA, Ponicsan SL, Chaudhuri G, Murphy MP, Dunaief JL, Ambati BK, Ogura Y, Yoo JW, Lee DK, Provost P, Hinton DR, Nunez G, Baffi JZ, Kleinman ME, Ambati J, DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88, Cell 149(4) (2012) 847–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Doyle SL, Campbell M, Ozaki E, Salomon RG, Mori A, Kenna PF, Farrar GJ, Kiang AS, Humphries MM, Lavelle EC, O'Neill LA, Hollyfield JG, Humphries P, NLRP3 has a protective role in age-related macular degeneration through the induction of IL-18 by drusen components, Nat Med (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Green WR, Enger C, Age-related macular degeneration histopathologic studies. The 1992 Lorenz E. Zimmerman Lecture, Ophthalmology 100(10) (1993) 1519–35. [DOI] [PubMed] [Google Scholar]
- [74].Sarks SH, Ageing and degeneration in the macular region: a clinico-pathological study, Br J Ophthalmol 60(5) (1976) 324–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Schindler C, Levy DE, Decker T, JAK-STAT signaling: from interferons to cytokines, J Biol Chem 282(28) (2007) 20059–63. [DOI] [PubMed] [Google Scholar]
- [76].Feld JJ, Hoofnagle JH, Mechanism of action of interferon and ribavirin in treatment of hepatitis C, Nature 436(7053) (2005) 967–72. [DOI] [PubMed] [Google Scholar]
- [77].Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD, How cells respond to interferons, Annu Rev Biochem 67 (1998) 227–64. [DOI] [PubMed] [Google Scholar]
- [78].Joshi-Barve S, Amancherla K, Patil M, Bhatnagar A, Mathews S, Gobejishvili L, Cave M, McClain C, Barve S, Acrolein, a ubiquitous pollutant and lipid hydroperoxide product, inhibits antiviral activity of interferon-alpha: relevance to hepatitis C, Free Radic Biol Med 47(1) (2009) 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Mian MF, Stampfli MR, Mossman KL, Ashkar AA, Cigarette smoke attenuation of poly I:C-induced innate antiviral responses in human PBMC is mainly due to inhibition of IFN-beta production, Mol Immunol 46(5) (2009) 821–9. [DOI] [PubMed] [Google Scholar]
- [80].Chaudhary V, Zhang S, Yuen KS, Li C, Lui PY, Fung SY, Wang PH, Chan CP, Li D, Kok KH, Liang M, Jin DY, Suppression of type I and type III IFN signalling by NSs protein of severe fever with thrombocytopenia syndrome virus through inhibition of STAT1 phosphorylation and activation, J Gen Virol 96(11) (2015) 3204–11. [DOI] [PubMed] [Google Scholar]
- [81].Dhillon NK, Murphy WJ, Filla MB, Crespo AJ, Latham HA, O'Brien-Ladner A, Down modulation of IFN-gamma signaling in alveolar macrophages isolated from smokers, Toxicology and applied pharmacology 237(1) (2009) 22–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Geraghty P, Wyman AE, Garcia-Arcos I, Dabo AJ, Gadhvi S, Foronjy R, STAT3 modulates cigarette smoke-induced inflammation and protease expression, Front Physiol 4 (2013) 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Liu X, STAT3 activation inhibits human bronchial epithelial cell apoptosis in response to cigarette smoke exposure, Biochem Biophys Res Commun 353(1) (2007) 121–6. [DOI] [PubMed] [Google Scholar]
- [84].Ruiz A, Brett P, Bok D, TIMP-3 is expressed in the human retinal pigment epithelium, Biochem Biophys Res Commun 226(2) (1996) 467–74. [DOI] [PubMed] [Google Scholar]
- [85].Alexander JP, Bradley JM, Gabourel JD, Acott TS, Expression of matrix metalloproteinases and inhibitor by human retinal pigment epithelium, Invest Ophthalmol Vis Sci 31(12) (1990) 2520–8. [PubMed] [Google Scholar]
- [86].Leu ST, Batni S, Radeke MJ, Johnson LV, Anderson DH, Clegg DO, Drusen are Cold Spots for Proteolysis: Expression of Matrix Metalloproteinases and Their Tissue Inhibitor Proteins in Age-related Macular Degeneration, Exp Eye Res 74(1) (2002) 141–54. [DOI] [PubMed] [Google Scholar]
- [87].Moore DJ, Hussain AA, Marshall J, Age-related variation in the hydraulic conductivity of Bruch's membrane, Invest Ophthalmol Vis Sci 36(7) (1995) 1290–7. [PubMed] [Google Scholar]
- [88].Starita C, Hussain AA, Pagliarini S, Marshall J, Hydrodynamics of ageing Bruch's membrane: implications for macular disease, Exp Eye Res 62(5) (1996) 565–72. [DOI] [PubMed] [Google Scholar]
- [89].Handa JT, Verzijl N, Matsunaga H, Aotaki-Keen A, Lutty GA, te Koppele JM, Miyata T, Hjelmeland LM, Increase in the advanced glycation end product pentosidine in Bruch's membrane with age, Invest Ophthalmol Vis Sci 40(3) (1999) 775–9. [PubMed] [Google Scholar]
- [90].Holz FG, Sheraidah G, Pauleikhoff D, Bird AC, Analysis of lipid deposits extracted from human macular and peripheral Bruch's membrane, Arch Ophthalmol 112(3) (1994) 402–6. [DOI] [PubMed] [Google Scholar]
- [91].Pauleikhoff D, Chen JC, Chisholm IH, Bird AC, Choroidal perfusion abnormality with age-related Bruch's membrane change, Am J Ophthalmol 109(2) (1990) 211–7. [DOI] [PubMed] [Google Scholar]
- [92].Hussain AA, Rowe L, Marshall J, Age-related alterations in the diffusional transport of amino acids across the human Bruch's-choroid complex, J Opt Soc Am A Opt Image Sci Vis 19(1) (2002) 166–72. [DOI] [PubMed] [Google Scholar]
- [93].Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, Adhihetty PJ, Adler SG, Agam G, Agarwal R, Aghi MK, Agnello M, Agostinis P, Aguilar PV, Aguirre-Ghiso J, Airoldi EM, Ait-Si-Ali S, Akematsu T, Akporiaye ET, Al-Rubeai M, Albaiceta GM, Albanese C, Albani D, Albert ML, Aldudo J, Algul H, Alirezaei M, Alloza I, Almasan A, Almonte-Beceril M, Alnemri ES, Alonso C, Altan-Bonnet N, Altieri DC, Alvarez S, Alvarez-Erviti L, Alves S, Amadoro G, Amano A, Amantini C, Ambrosio S, Amelio I, Amer AO, Amessou M, Amon A, An Z, Anania FA, Andersen SU, Andley UP, Andreadi CK, Andrieu-Abadie N, Anel A, Ann DK, Anoopkumar-Dukie S, Antonioli M, Aoki H, Apostolova N, Aquila S, Aquilano K, Araki K, Arama E, Aranda A, Araya J, Arcaro A, Arias E, Arimoto H, Ariosa AR, Armstrong JL, Arnould T, Arsov I, Asanuma K, Askanas V, Asselin E, Atarashi R, Atherton SS, Atkin JD, Attardi LD, Auberger P, Auburger G, Aurelian L, Autelli R, Avagliano L, Avantaggiati ML, Avrahami L, Awale S, Azad N, Bachetti T, Backer JM, Bae DH, Bae JS, Bae ON, Bae SH, Baehrecke EH, Baek SH, Baghdiguian S, Bagniewska-Zadworna A, Bai H, Bai J, Bai XY, Bailly Y, Balaji KN, Balduini W, Ballabio A, Balzan R, Banerjee R, Banhegyi G, Bao H, Barbeau B, Barrachina MD, Barreiro E, Bartel B, Bartolome A, Bassham DC, Bassi MT, Bast RC Jr., Basu A, Batista MT, Batoko H, Battino M, Bauckman K, Baumgarner BL, Bayer KU, Beale R, Beaulieu JF, Beck GR Jr., Becker C, Beckham JD, Bedard PA, Bednarski PJ, Begley TJ, Behl C, Behrends C, Behrens GM, Behrns KE, Bejarano E, Belaid A, Belleudi F, Benard G, Berchem G, Bergamaschi D, Bergami M, Berkhout B, Berliocchi L, Bernard A, Bernard M, Bernassola F, Bertolotti A, Bess AS, Besteiro S, Bettuzzi S, Bhalla S, Bhattacharyya S, Bhutia SK, Biagosch C, Bianchi MW, Biard-Piechaczyk M, Billes V, Bincoletto C, Bingol B, Bird SW, Bitoun M, Bjedov I, Blackstone C, Blanc L, Blanco GA, Blomhoff HK, Boada-Romero E, Bockler S, Boes M, Boesze-Battaglia K, Boise LH, Bolino A, Boman A, Bonaldo P, Bordi M, Bosch J, Botana LM, Botti J, Bou G, Bouche M, Bouchecareilh M, Boucher MJ, Boulton ME, Bouret SG, Boya P, Boyer-Guittaut M, Bozhkov PV, Brady N, Braga VM, Brancolini C, Braus GH, Bravo-San Pedro JM, Brennan LA, Bresnick EH, Brest P, Bridges D, Bringer MA, Brini M, Brito GC, Brodin B, Brookes PS, Brown EJ, Brown K, Broxmeyer HE, Bruhat A, Brum PC, Brumell JH, Brunetti-Pierri N, Bryson-Richardson RJ, Buch S, Buchan AM, Budak H, Bulavin DV, Bultman SJ, Bultynck G, Bumbasirevic V, Burelle Y, Burke RE, Burmeister M, Butikofer P, Caberlotto L, Cadwell K, Cahova M, Cai D, Cai J, Cai Q, Calatayud S, Camougrand N, Campanella M, Campbell GR, Campbell M, Campello S, Candau R, Caniggia I, Cantoni L, Cao L, Caplan AB, Caraglia M, Cardinali C, Cardoso SM, Carew JS, Carleton LA, Carlin CR, Carloni S, Carlsson SR, Carmona-Gutierrez D, Carneiro LA, Carnevali O, Carra S, Carrier A, Carroll B, Casas C, Casas J, Cassinelli G, Castets P, Castro-Obregon S, Cavallini G, Ceccherini I, Cecconi F, Cederbaum AI, Cena V, Cenci S, Cerella C, Cervia D, Cetrullo S, Chaachouay H, Chae HJ, Chagin AS, Chai CY, Chakrabarti G, Chamilos G, Chan EY, Chan MT, Chandra D, Chandra P, Chang CP, Chang RC, Chang TY, Chatham JC, Chatterjee S, Chauhan S, Che Y, Cheetham ME, Cheluvappa R, Chen CJ, Chen G, Chen GC, Chen G, Chen H, Chen JW, Chen JK, Chen M, Chen M, Chen P, Chen Q, Chen Q, Chen SD, Chen S, Chen SS, Chen W, Chen WJ, Chen WQ, Chen W, Chen X, Chen YH, Chen YG, Chen Y, Chen Y, Chen Y, Chen YJ, Chen YQ, Chen Y, Chen Z, Chen Z, Cheng A, Cheng CH, Cheng H, Cheong H, Cherry S, Chesney J, Cheung CH, Chevet E, Chi HC, Chi SG, Chiacchiera F, Chiang HL, Chiarelli R, Chiariello M, Chieppa M, Chin LS, Chiong M, Chiu GN, Cho DH, Cho SG, Cho WC, Cho YY, Cho YS, Choi AM, Choi EJ, Choi EK, Choi J, Choi ME, Choi SI, Chou TF, Chouaib S, Choubey D, Choubey V, Chow KC, Chowdhury K, Chu CT, Chuang TH, Chun T, Chung H, Chung T, Chung YL, Chwae YJ, Cianfanelli V, Ciarcia R, Ciechomska IA, Ciriolo MR, Cirone M, Claerhout S, Clague MJ, Claria J, Clarke PG, Clarke R, Clementi E, Cleyrat C, Cnop M, Coccia EM, Cocco T, Codogno P, Coers J, Cohen EE, Colecchia D, Coletto L, Coll NS, Colucci-Guyon E, Comincini S, Condello M, Cook KL, Coombs GH, Cooper CD, Cooper JM, Coppens I, Corasaniti MT, Corazzari M, Corbalan R, Corcelle-Termeau E, Cordero MD, Corral-Ramos C, Corti O, Cossarizza A, Costelli P, Costes S, Cotman SL, Coto-Montes A, Cottet S, Couve E, Covey LR, Cowart LA, Cox JS, Coxon FP, Coyne CB, Cragg MS, Craven RJ, Crepaldi T, Crespo JL, Criollo A, Crippa V, Cruz MT, Cuervo AM, Cuezva JM, Cui T, Cutillas PR, Czaja MJ, Czyzyk-Krzeska MF, Dagda RK, Dahmen U, Dai C, Dai W, Dai Y, Dalby KN, Dalla Valle L, Dalmasso G, D'Amelio M, Damme M, Darfeuille-Michaud A, Dargemont C, Darley-Usmar VM, Dasarathy S, Dasgupta B, Dash S, Dass CR, Davey HM, Davids LM, Davila D, Davis RJ, Dawson TM, Dawson VL, Daza P, de Belleroche J, de Figueiredo P, de Figueiredo RC, de la Fuente J, De Martino L, De Matteis A, De Meyer GR, De Milito A, De Santi M, de Souza W, De Tata V, De Zio D, Debnath J, Dechant R, Decuypere JP, Deegan S, Dehay B, Del Bello B, Del Re DP, Delage-Mourroux R, Delbridge LM, Deldicque L, Delorme-Axford E, Deng Y, Dengjel J, Denizot M, Dent P, Der CJ, Deretic V, Derrien B, Deutsch E, Devarenne TP, Devenish RJ, Di Bartolomeo S, Di Daniele N, Di Domenico F, Di Nardo A, Di Paola S, Di Pietro A, Di Renzo L, DiAntonio A, Diaz-Araya G, Diaz-Laviada I, Diaz-Meco MT, Diaz-Nido J, Dickey CA, Dickson RC, Diederich M, Digard P, Dikic I, Dinesh-Kumar SP, Ding C, Ding WX, Ding Z, Dini L, Distler JH, Diwan A, Djavaheri-Mergny M, Dmytruk K, Dobson RC, Doetsch V, Dokladny K, Dokudovskaya S, Donadelli M, Dong XC, Dong X, Dong Z, Donohue TM Jr., Doran KS, D'Orazi G, Dorn GW 2nd, Dosenko V, Dridi S, Drucker L, Du J, Du LL, Du L, du Toit A, Dua P, Duan L, Duann P, Dubey VK, Duchen MR, Duchosal MA, Duez H, Dugail I, Dumit VI, Duncan MC, Dunlop EA, Dunn WA Jr., Dupont N, Dupuis L, Duran RV, Durcan TM, Duvezin-Caubet S, Duvvuri U, Eapen V, Ebrahimi-Fakhari D, Echard A, Eckhart L, Edelstein CL, Edinger AL, Eichinger L, Eisenberg T, Eisenberg-Lerner A, Eissa NT, El-Deiry WS, El-Khoury V, Elazar Z, Eldar-Finkelman H, Elliott CJ, Emanuele E, Emmenegger U, Engedal N, Engelbrecht AM, Engelender S, Enserink JM, Erdmann R, Erenpreisa J, Eri R, Eriksen JL, Erman A, Escalante R, Eskelinen EL, Espert L, Esteban-Martinez L, Evans TJ, Fabri M, Fabrias G, Fabrizi C, Facchiano A, Faergeman NJ, Faggioni A, Fairlie WD, Fan C, Fan D, Fan J, Fang S, Fanto M, Fanzani A, Farkas T, Faure M, Favier FB, Fearnhead H, Federici M, Fei E, Felizardo TC, Feng H, Feng Y, Feng Y, Ferguson TA, Fernandez AF, Fernandez-Barrena MG, Fernandez-Checa JC, Fernandez-Lopez A, Fernandez-Zapico ME, Feron O, Ferraro E, Ferreira-Halder CV, Fesus L, Feuer R, Fiesel FC, Filippi-Chiela EC, Filomeni G, Fimia GM, Fingert JH, Finkbeiner S, Finkel T, Fiorito F, Fisher PB, Flajolet M, Flamigni F, Florey O, Florio S, Floto RA, Folini M, Follo C, Fon EA, Fornai F, Fortunato F, Fraldi A, Franco R, Francois A, Francois A, Frankel LB, Fraser ID, Frey N, Freyssenet DG, Frezza C, Friedman SL, Frigo DE, Fu D, Fuentes JM, Fueyo J, Fujitani Y, Fujiwara Y, Fujiya M, Fukuda M, Fulda S, Fusco C, Gabryel B, Gaestel M, Gailly P, Gajewska M, Galadari S, Galili G, Galindo I, Galindo MF, Galliciotti G, Galluzzi L, Galluzzi L, Galy V, Gammoh N, Gandy S, Ganesan AK, Ganesan S, Ganley IG, Gannage M, Gao FB, Gao F, Gao JX, Garcia Nannig L, Garcia Vescovi E, Garcia-Macia M, Garcia-Ruiz C, Garg AD, Garg PK, Gargini R, Gassen NC, Gatica D, Gatti E, Gavard J, Gavathiotis E, Ge L, Ge P, Ge S, Gean PW, Gelmetti V, Genazzani AA, Geng J, Genschik P, Gerner L, Gestwicki JE, Gewirtz DA, Ghavami S, Ghigo E, Ghosh D, Giammarioli AM, Giampieri F, Giampietri C, Giatromanolaki A, Gibbings DJ, Gibellini L, Gibson SB, Ginet V, Giordano A, Giorgini F, Giovannetti E, Girardin SE, Gispert S, Giuliano S, Gladson CL, Glavic A, Gleave M, Godefroy N, Gogal RM Jr., Gokulan K, Goldman GH, Goletti D, Goligorsky MS, Gomes AV, Gomes LC, Gomez H, Gomez-Manzano C, Gomez-Sanchez R, Goncalves DA, Goncu E, Gong Q, Gongora C, Gonzalez CB, Gonzalez-Alegre P, Gonzalez-Cabo P, Gonzalez-Polo RA, Goping IS, Gorbea C, Gorbunov NV, Goring DR, Gorman AM, Gorski SM, Goruppi S, Goto-Yamada S, Gotor C, Gottlieb RA, Gozes I, Gozuacik D, Graba Y, Graef M, Granato GE, Grant GD, Grant S, Gravina GL, Green DR, Greenhough A, Greenwood MT, Grimaldi B, Gros F, Grose C, Groulx JF, Gruber F, Grumati P, Grune T, Guan JL, Guan KL, Guerra B, Guillen C, Gulshan K, Gunst J, Guo C, Guo L, Guo M, Guo W, Guo XG, Gust AA, Gustafsson AB, Gutierrez E, Gutierrez MG, Gwak HS, Haas A, Haber JE, Hadano S, Hagedorn M, Hahn DR, Halayko AJ, Hamacher-Brady A, Hamada K, Hamai A, Hamann A, Hamasaki M, Hamer I, Hamid Q, Hammond EM, Han F, Han W, Handa JT, Hanover JA, Hansen M, Harada M, Harhaji-Trajkovic L, Harper JW, Harrath AH, Harris AL, Harris J, Hasler U, Hasselblatt P, Hasui K, Hawley RG, Hawley TS, He C, He CY, He F, He G, He RR, He XH, He YW, He YY, Heath JK, Hebert MJ, Heinzen RA, Helgason GV, Hensel M, Henske EP, Her C, Herman PK, Hernandez A, Hernandez C, Hernandez-Tiedra S, Hetz C, Hiesinger PR, Higaki K, Hilfiker S, Hill BG, Hill JA, Hill WD, Hino K, Hofius D, Hofman P, Hoglinger GU, Hohfeld J, Holz MK, Hong Y, Hood DA, Hoozemans JJ, Hoppe T, Hsu C, Hsu CY, Hsu LC, Hu D, Hu G, Hu HM, Hu H, Hu MC, Hu YC, Hu ZW, Hua F, Hua Y, Huang C, Huang HL, Huang KH, Huang KY, Huang S, Huang S, Huang WP, Huang YR, Huang Y, Huang Y, Huber TB, Huebbe P, Huh WK, Hulmi JJ, Hur GM, Hurley JH, Husak Z, Hussain SN, Hussain S, Hwang JJ, Hwang S, Hwang TI, Ichihara A, Imai Y, Imbriano C, Inomata M, Into T, Iovane V, Iovanna JL, Iozzo RV, Ip NY, Irazoqui JE, Iribarren P, Isaka Y, Isakovic AJ, Ischiropoulos H, Isenberg JS, Ishaq M, Ishida H, Ishii I, Ishmael JE, Isidoro C, Isobe K, Isono E, Issazadeh-Navikas S, Itahana K, Itakura E, Ivanov AI, Iyer AK, Izquierdo JM, Izumi Y, Izzo V, Jaattela M, Jaber N, Jackson DJ, Jackson WT, Jacob TG, Jacques TS, Jagannath C, Jain A, Jana NR, Jang BK, Jani A, Janji B, Jannig PR, Jansson PJ, Jean S, Jendrach M, Jeon JH, Jessen N, Jeung EB, Jia K, Jia L, Jiang H, Jiang H, Jiang L, Jiang T, Jiang X, Jiang X, Jiang X, Jiang Y, Jiang Y, Jimenez A, Jin C, Jin H, Jin L, Jin M, Jin S, Jinwal UK, Jo EK, Johansen T, Johnson DE, Johnson GV, Johnson JD, Jonasch E, Jones C, Joosten LA, Jordan J, Joseph AM, Joseph B, Joubert AM, Ju D, Ju J, Juan HF, Juenemann K, Juhasz G, Jung HS, Jung JU, Jung YK, Jungbluth H, Justice MJ, Jutten B, Kaakoush NO, Kaarniranta K, Kaasik A, Kabuta T, Kaeffer B, Kagedal K, Kahana A, Kajimura S, Kakhlon O, Kalia M, Kalvakolanu DV, Kamada Y, Kambas K, Kaminskyy VO, Kampinga HH, Kandouz M, Kang C, Kang R, Kang TC, Kanki T, Kanneganti TD, Kanno H, Kanthasamy AG, Kantorow M, Kaparakis-Liaskos M, Kapuy O, Karantza V, Karim MR, Karmakar P, Kaser A, Kaushik S, Kawula T, Kaynar AM, Ke PY, Ke ZJ, Kehrl JH, Keller KE, Kemper JK, Kenworthy AK, Kepp O, Kern A, Kesari S, Kessel D, Ketteler R, Kettelhut Ido C, Khambu B, Khan MM, Khandelwal VK, Khare S, Kiang JG, Kiger AA, Kihara A, Kim AL, Kim CH, Kim DR, Kim DH, Kim EK, Kim HY, Kim HR, Kim JS, Kim JH, Kim JC, Kim JH, Kim KW, Kim MD, Kim MM, Kim PK, Kim SW, Kim SY, Kim YS, Kim Y, Kimchi A, Kimmelman AC, Kimura T, King JS, Kirkegaard K, Kirkin V, Kirshenbaum LA, Kishi S, Kitajima Y, Kitamoto K, Kitaoka Y, Kitazato K, Kley RA, Klimecki WT, Klinkenberg M, Klucken J, Knaevelsrud H, Knecht E, Knuppertz L, Ko JL, Kobayashi S, Koch JC, Koechlin-Ramonatxo C, Koenig U, Koh YH, Kohler K, Kohlwein SD, Koike M, Komatsu M, Kominami E, Kong D, Kong HJ, Konstantakou EG, Kopp BT, Korcsmaros T, Korhonen L, Korolchuk VI, Koshkina NV, Kou Y, Koukourakis MI, Koumenis C, Kovacs AL, Kovacs T, Kovacs WJ, Koya D, Kraft C, Krainc D, Kramer H, Kravic-Stevovic T, Krek W, Kretz-Remy C, Krick R, Krishnamurthy M, Kriston-Vizi J, Kroemer G, Kruer MC, Kruger R, Ktistakis NT, Kuchitsu K, Kuhn C, Kumar AP, Kumar A, Kumar A, Kumar D, Kumar D, Kumar R, Kumar S, Kundu M, Kung HJ, Kuno A, Kuo SH, Kuret J, Kurz T, Kwok T, Kwon TK, Kwon YT, Kyrmizi I, La Spada AR, Lafont F, Lahm T, Lakkaraju A, Lam T, Lamark T, Lancel S, Landowski TH, Lane DJ, Lane JD, Lanzi C, Lapaquette P, Lapierre LR, Laporte J, Laukkarinen J, Laurie GW, Lavandero S, Lavie L, LaVoie MJ, Law BY, Law HK, Law KB, Layfield R, Lazo PA, Le Cam L, Le Roch KG, Le Stunff H, Leardkamolkarn V, Lecuit M, Lee BH, Lee CH, Lee EF, Lee GM, Lee HJ, Lee H, Lee JK, Lee J, Lee JH, Lee JH, Lee M, Lee MS, Lee PJ, Lee SW, Lee SJ, Lee SJ, Lee SY, Lee SH, Lee SS, Lee SJ, Lee S, Lee YR, Lee YJ, Lee YH, Leeuwenburgh C, Lefort S, Legouis R, Lei J, Lei QY, Leib DA, Leibowitz G, Lekli I, Lemaire SD, Lemasters JJ, Lemberg MK, Lemoine A, Leng S, Lenz G, Lenzi P, Lerman LO, Lettieri Barbato D, Leu JI, Leung HY, Levine B, Lewis PA, Lezoualc'h F, Li C, Li F, Li FJ, Li J, Li K, Li L, Li M, Li M, Li Q, Li R, Li S, Li W, Li W, Li X, Li Y, Lian J, Liang C, Liang Q, Liao Y, Liberal J, Liberski PP, Lie P, Lieberman AP, Lim HJ, Lim KL, Lim K, Lima RT, Lin CS, Lin CF, Lin F, Lin F, Lin FC, Lin K, Lin KH, Lin PH, Lin T, Lin WW, Lin YS, Lin Y, Linden R, Lindholm D, Lindqvist LM, Lingor P, Linkermann A, Liotta LA, Lipinski MM, Lira VA, Lisanti MP, Liton PB, Liu B, Liu C, Liu CF, Liu F, Liu HJ, Liu J, Liu JJ, Liu JL, Liu K, Liu L, Liu L, Liu Q, Liu RY, Liu S, Liu S, Liu W, Liu XD, Liu X, Liu XH, Liu X, Liu X, Liu X, Liu Y, Liu Y, Liu Z, Liu Z, Liuzzi JP, Lizard G, Ljujic M, Lodhi IJ, Logue SE, Lokeshwar BL, Long YC, Lonial S, Loos B, Lopez-Otin C, Lopez-Vicario C, Lorente M, Lorenzi PL, Lorincz P, Los M, Lotze MT, Lovat PE, Lu B, Lu B, Lu J, Lu Q, Lu SM, Lu S, Lu Y, Luciano F, Luckhart S, Lucocq JM, Ludovico P, Lugea A, Lukacs NW, Lum JJ, Lund AH, Luo H, Luo J, Luo S, Luparello C, Lyons T, Ma J, Ma Y, Ma Y, Ma Z, Machado J, Machado-Santelli GM, Macian F, MacIntosh GC, MacKeigan JP, Macleod KF, MacMicking JD, MacMillan-Crow LA, Madeo F, Madesh M, Madrigal-Matute J, Maeda A, Maeda T, Maegawa G, Maellaro E, Maes H, Magarinos M, Maiese K, Maiti TK, Maiuri L, Maiuri MC, Maki CG, Malli R, Malorni W, Maloyan A, Mami-Chouaib F, Man N, Mancias JD, Mandelkow EM, Mandell MA, Manfredi AA, Manie SN, Manzoni C, Mao K, Mao Z, Mao ZW, Marambaud P, Marconi AM, Marelja Z, Marfe G, Margeta M, Margittai E, Mari M, Mariani FV, Marin C, Marinelli S, Marino G, Markovic I, Marquez R, Martelli AM, Martens S, Martin KR, Martin SJ, Martin S, Martin-Acebes MA, Martin-Sanz P, Martinand-Mari C, Martinet W, Martinez J, Martinez-Lopez N, Martinez-Outschoorn U, Martinez-Velazquez M, Martinez-Vicente M, Martins WK, Mashima H, Mastrianni JA, Matarese G, Matarrese P, Mateo R, Matoba S, Matsumoto N, Matsushita T, Matsuura A, Matsuzawa T, Mattson MP, Matus S, Maugeri N, Mauvezin C, Mayer A, Maysinger D, Mazzolini GD, McBrayer MK, McCall K, McCormick C, McInerney GM, McIver SC, McKenna S, McMahon JJ, McNeish IA, Mechta-Grigoriou F, Medema JP, Medina DL, Megyeri K, Mehrpour M, Mehta JL, Mei Y, Meier UC, Meijer AJ, Melendez A, Melino G, Melino S, de Melo EJ, Mena MA, Meneghini MD, Menendez JA, Menezes R, Meng L, Meng LH, Meng S, Menghini R, Menko AS, Menna-Barreto RF, Menon MB, Meraz-Rios MA, Merla G, Merlini L, Merlot AM, Meryk A, Meschini S, Meyer JN, Mi MT, Miao CY, Micale L, Michaeli S, Michiels C, Migliaccio AR, Mihailidou AS, Mijaljica D, Mikoshiba K, Milan E, Miller-Fleming L, Mills GB, Mills IG, Minakaki G, Minassian BA, Ming XF, Minibayeva F, Minina EA, Mintern JD, Minucci S, Miranda-Vizuete A, Mitchell CH, Miyamoto S, Miyazawa K, Mizushima N, Mnich K, Mograbi B, Mohseni S, Moita LF, Molinari M, Molinari M, Moller AB, Mollereau B, Mollinedo F, Mongillo M, Monick MM, Montagnaro S, Montell C, Moore DJ, Moore MN, Mora-Rodriguez R, Moreira PI, Morel E, Morelli MB, Moreno S, Morgan MJ, Moris A, Moriyasu Y, Morrison JL, Morrison LA, Morselli E, Moscat J, Moseley PL, Mostowy S, Motori E, Mottet D, Mottram JC, Moussa CE, Mpakou VE, Mukhtar H, Mulcahy Levy JM, Muller S, Munoz-Moreno R, Munoz-Pinedo C, Munz C, Murphy ME, Murray JT, Murthy A, Mysorekar IU, Nabi IR, Nabissi M, Nader GA, Nagahara Y, Nagai Y, Nagata K, Nagelkerke A, Nagy P, Naidu SR, Nair S, Nakano H, Nakatogawa H, Nanjundan M, Napolitano G, Naqvi NI, Nardacci R, Narendra DP, Narita M, Nascimbeni AC, Natarajan R, Navegantes LC, Nawrocki ST, Nazarko TY, Nazarko VY, Neill T, Neri LM, Netea MG, Netea-Maier RT, Neves BM, Ney PA, Nezis IP, Nguyen HT, Nguyen HP, Nicot AS, Nilsen H, Nilsson P, Nishimura M, Nishino I, Niso-Santano M, Niu H, Nixon RA, Njar VC, Noda T, Noegel AA, Nolte EM, Norberg E, Norga KK, Noureini SK, Notomi S, Notterpek L, Nowikovsky K, Nukina N, Nurnberger T, O'Donnell VB, O'Donovan T, O'Dwyer PJ, Oehme I, Oeste CL, Ogawa M, Ogretmen B, Ogura Y, Oh YJ, Ohmuraya M, Ohshima T, Ojha R, Okamoto K, Okazaki T, Oliver FJ, Ollinger K, Olsson S, Orban DP, Ordonez P, Orhon I, Orosz L, O'Rourke EJ, Orozco H, Ortega AL, Ortona E, Osellame LD, Oshima J, Oshima S, Osiewacz HD, Otomo T, Otsu K, Ou JH, Outeiro TF, Ouyang DY, Ouyang H, Overholtzer M, Ozbun MA, Ozdinler PH, Ozpolat B, Pacelli C, Paganetti P, Page G, Pages G, Pagnini U, Pajak B, Pak SC, Pakos-Zebrucka K, Pakpour N, Palkova Z, Palladino F, Pallauf K, Pallet N, Palmieri M, Paludan SR, Palumbo C, Palumbo S, Pampliega O, Pan H, Pan W, Panaretakis T, Pandey A, Pantazopoulou A, Papackova Z, Papademetrio DL, Papassideri I, Papini A, Parajuli N, Pardo J, Parekh VV, Parenti G, Park JI, Park J, Park OK, Parker R, Parlato R, Parys JB, Parzych KR, Pasquet JM, Pasquier B, Pasumarthi KB, Patschan D, Patterson C, Pattingre S, Pattison S, Pause A, Pavenstadt H, Pavone F, Pedrozo Z, Pena FJ, Penalva MA, Pende M, Peng J, Penna F, Penninger JM, Pensalfini A, Pepe S, Pereira GJ, Pereira PC, Perez-de la Cruz V, Perez-Perez ME, Perez-Rodriguez D, Perez-Sala D, Perier C, Perl A, Perlmutter DH, Perrotta I, Pervaiz S, Pesonen M, Pessin JE, Peters GJ, Petersen M, Petrache I, Petrof BJ, Petrovski G, Phang JM, Piacentini M, Pierdominici M, Pierre P, Pierrefite-Carle V, Pietrocola F, Pimentel-Muinos FX, Pinar M, Pineda B, Pinkas-Kramarski R, Pinti M, Pinton P, Piperdi B, Piret JM, Platanias LC, Platta HW, Plowey ED, Poggeler S, Poirot M, Polcic P, Poletti A, Poon AH, Popelka H, Popova B, Poprawa I, Poulose SM, Poulton J, Powers SK, Powers T, Pozuelo-Rubio M, Prak K, Prange R, Prescott M, Priault M, Prince S, Proia RL, Proikas-Cezanne T, Prokisch H, Promponas VJ, Przyklenk K, Puertollano R, Pugazhenthi S, Puglielli L, Pujol A, Puyal J, Pyeon D, Qi X, Qian WB, Qin ZH, Qiu Y, Qu Z, Quadrilatero J, Quinn F, Raben N, Rabinowich H, Radogna F, Ragusa MJ, Rahmani M, Raina K, Ramanadham S, Ramesh R, Rami A, Randall-Demllo S, Randow F, Rao H, Rao VA, Rasmussen BB, Rasse TM, Ratovitski EA, Rautou PE, Ray SK, Razani B, Reed BH, Reggiori F, Rehm M, Reichert AS, Rein T, Reiner DJ, Reits E, Ren J, Ren X, Renna M, Reusch JE, Revuelta JL, Reyes L, Rezaie AR, Richards RI, Richardson DR, Richetta C, Riehle MA, Rihn BH, Rikihisa Y, Riley BE, Rimbach G, Rippo MR, Ritis K, Rizzi F, Rizzo E, Roach PJ, Robbins J, Roberge M, Roca G, Roccheri MC, Rocha S, Rodrigues CM, Rodriguez CI, de Cordoba SR, Rodriguez-Muela N, Roelofs J, Rogov VV, Rohn TT, Rohrer B, Romanelli D, Romani L, Romano PS, Roncero MI, Rosa JL, Rosello A, Rosen KV, Rosenstiel P, Rost-Roszkowska M, Roth KA, Roue G, Rouis M, Rouschop KM, Ruan DT, Ruano D, Rubinsztein DC, Rucker EB 3rd, Rudich A, Rudolf E, Rudolf R, Ruegg MA, Ruiz-Roldan C, Ruparelia AA, Rusmini P, Russ DW, Russo GL, Russo G, Russo R, Rusten TE, Ryabovol V, Ryan KM, Ryter SW, Sabatini DM, Sacher M, Sachse C, Sack MN, Sadoshima J, Saftig P, Sagi-Eisenberg R, Sahni S, Saikumar P, Saito T, Saitoh T, Sakakura K, Sakoh-Nakatogawa M, Sakuraba Y, Salazar-Roa M, Salomoni P, Saluja AK, Salvaterra PM, Salvioli R, Samali A, Sanchez AM, Sanchez-Alcazar JA, Sanchez-Prieto R, Sandri M, Sanjuan MA, Santaguida S, Santambrogio L, Santoni G, Dos Santos CN, Saran S, Sardiello M, Sargent G, Sarkar P, Sarkar S, Sarrias MR, Sarwal MM, Sasakawa C, Sasaki M, Sass M, Sato K, Sato M, Satriano J, Savaraj N, Saveljeva S, Schaefer L, Schaible UE, Scharl M, Schatzl HM, Schekman R, Scheper W, Schiavi A, Schipper HM, Schmeisser H, Schmidt J, Schmitz I, Schneider BE, Schneider EM, Schneider JL, Schon EA, Schonenberger MJ, Schonthal AH, Schorderet DF, Schroder B, Schuck S, Schulze RJ, Schwarten M, Schwarz TL, Sciarretta S, Scotto K, Scovassi AI, Screaton RA, Screen M, Seca H, Sedej S, Segatori L, Segev N, Seglen PO, Segui-Simarro JM, Segura-Aguilar J, Seki E, Sell C, Seiliez I, Semenkovich CF, Semenza GL, Sen U, Serra AL, Serrano-Puebla A, Sesaki H, Setoguchi T, Settembre C, Shacka JJ, Shajahan-Haq AN, Shapiro IM, Sharma S, She H, Shen CK, Shen CC, Shen HM, Shen S, Shen W, Sheng R, Sheng X, Sheng ZH, Shepherd TG, Shi J, Shi Q, Shi Q, Shi Y, Shibutani S, Shibuya K, Shidoji Y, Shieh JJ, Shih CM, Shimada Y, Shimizu S, Shin DW, Shinohara ML, Shintani M, Shintani T, Shioi T, Shirabe K, Shiri-Sverdlov R, Shirihai O, Shore GC, Shu CW, Shukla D, Sibirny AA, Sica V, Sigurdson CJ, Sigurdsson EM, Sijwali PS, Sikorska B, Silveira WA, Silvente-Poirot S, Silverman GA, Simak J, Simmet T, Simon AK, Simon HU, Simone C, Simons M, Simonsen A, Singh R, Singh SV, Singh SK, Sinha D, Sinha S, Sinicrope FA, Sirko A, Sirohi K, Sishi BJ, Sittler A, Siu PM, Sivridis E, Skwarska A, Slack R, Slaninova I, Slavov N, Smaili SS, Smalley KS, Smith DR, Soenen SJ, Soleimanpour SA, Solhaug A, Somasundaram K, Son JH, Sonawane A, Song C, Song F, Song HK, Song JX, Song W, Soo KY, Sood AK, Soong TW, Soontornniyomkij V, Sorice M, Sotgia F, Soto-Pantoja DR, Sotthibundhu A, Sousa MJ, Spaink HP, Span PN, Spang A, Sparks JD, Speck PG, Spector SA, Spies CD, Springer W, Clair DS, Stacchiotti A, Staels B, Stang MT, Starczynowski DT, Starokadomskyy P, Steegborn C, Steele JW, Stefanis L, Steffan J, Stellrecht CM, Stenmark H, Stepkowski TM, Stern ST, Stevens C, Stockwell BR, Stoka V, Storchova Z, Stork B, Stratoulias V, Stravopodis DJ, Strnad P, Strohecker AM, Strom AL, Stromhaug P, Stulik J, Su YX, Su Z, Subauste CS, Subramaniam S, Sue CM, Suh SW, Sui X, Sukseree S, Sulzer D, Sun FL, Sun J, Sun J, Sun SY, Sun Y, Sun Y, Sun Y, Sundaramoorthy V, Sung J, Suzuki H, Suzuki K, Suzuki N, Suzuki T, Suzuki YJ, Swanson MS, Swanton C, Sward K, Swarup G, Sweeney ST, Sylvester PW, Szatmari Z, Szegezdi E, Szlosarek PW, Taegtmeyer H, Tafani M, Taillebourg E, Tait SW, Takacs-Vellai K, Takahashi Y, Takats S, Takemura G, Takigawa N, Talbot NJ, Tamagno E, Tamburini J, Tan CP, Tan L, Tan ML, Tan M, Tan YJ, Tanaka K, Tanaka M, Tang D, Tang D, Tang G, Tanida I, Tanji K, Tannous BA, Tapia JA, Tasset-Cuevas I, Tatar M, Tavassoly I, Tavernarakis N, Taylor A, Taylor GS, Taylor GA, Taylor JP, Taylor MJ, Tchetina EV, Tee AR, Teixeira-Clerc F, Telang S, Tencomnao T, Teng BB, Teng RJ, Terro F, Tettamanti G, Theiss AL, Theron AE, Thomas KJ, Thome MP, Thomes PG, Thorburn A, Thorner J, Thum T, Thumm M, Thurston TL, Tian L, Till A, Ting JP, Titorenko VI, Toker L, Toldo S, Tooze SA, Topisirovic I, Torgersen ML, Torosantucci L, Torriglia A, Torrisi MR, Tournier C, Towns R, Trajkovic V, Travassos LH, Triola G, Tripathi DN, Trisciuoglio D, Troncoso R, Trougakos IP, Truttmann AC, Tsai KJ, Tschan MP, Tseng YH, Tsukuba T, Tsung A, Tsvetkov AS, Tu S, Tuan HY, Tucci M, Tumbarello DA, Turk B, Turk V, Turner RF, Tveita AA, Tyagi SC, Ubukata M, Uchiyama Y, Udelnow A, Ueno T, Umekawa M, Umemiya-Shirafuji R, Underwood BR, Ungermann C, Ureshino RP, Ushioda R, Uversky VN, Uzcategui NL, Vaccari T, Vaccaro MI, Vachova L, Vakifahmetoglu-Norberg H, Valdor R, Valente EM, Vallette F, Valverde AM, Van den Berghe G, Van Den Bosch L, van den Brink GR, van der Goot FG, van der Klei IJ, van der Laan LJ, van Doorn WG, van Egmond M, van Golen KL, Van Kaer L, van Lookeren Campagne M, Vandenabeele P, Vandenberghe W, Vanhorebeek I, Varela-Nieto I, Vasconcelos MH, Vasko R, Vavvas DG, Vega-Naredo I, Velasco G, Velentzas AD, Velentzas PD, Vellai T, Vellenga E, Vendelbo MH, Venkatachalam K, Ventura N, Ventura S, Veras PS, Verdier M, Vertessy BG, Viale A, Vidal M, Vieira HL, Vierstra RD, Vigneswaran N, Vij N, Vila M, Villar M, Villar VH, Villarroya J, Vindis C, Viola G, Viscomi MT, Vitale G, Vogl DT, Voitsekhovskaja OV, von Haefen C, von Schwarzenberg K, Voth DE, Vouret-Craviari V, Vuori K, Vyas JM, Waeber C, Walker CL, Walker MJ, Walter J, Wan L, Wan X, Wang B, Wang C, Wang CY, Wang C, Wang C, Wang C, Wang D, Wang F, Wang F, Wang G, Wang HJ, Wang H, Wang HG, Wang H, Wang HD, Wang J, Wang J, Wang M, Wang MQ, Wang PY, Wang P, Wang RC, Wang S, Wang TF, Wang X, Wang XJ, Wang XW, Wang X, Wang X, Wang Y, Wang Y, Wang Y, Wang YJ, Wang Y, Wang Y, Wang YT, Wang Y, Wang ZN, Wappner P, Ward C, Ward DM, Warnes G, Watada H, Watanabe Y, Watase K, Weaver TE, Weekes CD, Wei J, Weide T, Weihl CC, Weindl G, Weis SN, Wen L, Wen X, Wen Y, Westermann B, Weyand CM, White AR, White E, Whitton JL, Whitworth AJ, Wiels J, Wild F, Wildenberg ME, Wileman T, Wilkinson DS, Wilkinson S, Willbold D, Williams C, Williams K, Williamson PR, Winklhofer KF, Witkin SS, Wohlgemuth SE, Wollert T, Wolvetang EJ, Wong E, Wong GW, Wong RW, Wong VK, Woodcock EA, Wright KL, Wu C, Wu D, Wu GS, Wu J, Wu J, Wu M, Wu M, Wu S, Wu WK, Wu Y, Wu Z, Xavier CP, Xavier RJ, Xia GX, Xia T, Xia W, Xia Y, Xiao H, Xiao J, Xiao S, Xiao W, Xie CM, Xie Z, Xie Z, Xilouri M, Xiong Y, Xu C, Xu C, Xu F, Xu H, Xu H, Xu J, Xu J, Xu J, Xu L, Xu X, Xu Y, Xu Y, Xu ZX, Xu Z, Xue Y, Yamada T, Yamamoto A, Yamanaka K, Yamashina S, Yamashiro S, Yan B, Yan B, Yan X, Yan Z, Yanagi Y, Yang DS, Yang JM, Yang L, Yang M, Yang PM, Yang P, Yang Q, Yang W, Yang WY, Yang X, Yang Y, Yang Y, Yang Z, Yang Z, Yao MC, Yao PJ, Yao X, Yao Z, Yao Z, Yasui LS, Ye M, Yedvobnick B, Yeganeh B, Yeh ES, Yeyati PL, Yi F, Yi L, Yin XM, Yip CK, Yoo YM, Yoo YH, Yoon SY, Yoshida K, Yoshimori T, Young KH, Yu H, Yu JJ, Yu JT, Yu J, Yu L, Yu WH, Yu XF, Yu Z, Yuan J, Yuan ZM, Yue BY, Yue J, Yue Z, Zacks DN, Zacksenhaus E, Zaffaroni N, Zaglia T, Zakeri Z, Zecchini V, Zeng J, Zeng M, Zeng Q, Zervos AS, Zhang DD, Zhang F, Zhang G, Zhang GC, Zhang H, Zhang H, Zhang H, Zhang H, Zhang J, Zhang J, Zhang J, Zhang J, Zhang JP, Zhang L, Zhang L, Zhang L, Zhang L, Zhang MY, Zhang X, Zhang XD, Zhang Y, Zhang Y, Zhang Y, Zhang Y, Zhang Y, Zhao M, Zhao WL, Zhao X, Zhao YG, Zhao Y, Zhao Y, Zhao YX, Zhao Z, Zhao ZJ, Zheng D, Zheng XL, Zheng X, Zhivotovsky B, Zhong Q, Zhou GZ, Zhou G, Zhou H, Zhou SF, Zhou XJ, Zhu H, Zhu H, Zhu WG, Zhu W, Zhu XF, Zhu Y, Zhuang SM, Zhuang X, Ziparo E, Zois CE, Zoladek T, Zong WX, Zorzano A, Zughaier SM, Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition), Autophagy 12(1) (2016) 1–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Ratnapriya R, Sosina OA, Starostik MR, Kwicklis M, Kapphahn RJ, Fritsche LG, Walton A, Arvanitis M, Gieser L, Pietraszkiewicz A, Montezuma SR, Chew EY, Battle A, Abecasis GR, Ferrington DA, Chatterjee N, Swaroop A, Author Correction: Retinal transcriptome and eQTL analyses identify genes associated with age-related macular degeneration, Nat Genet 51(6) (2019) 1067. [DOI] [PubMed] [Google Scholar]
- [95].Orozco LD, Chen HH, Cox C, Katschke KJ Jr., Arceo R, Espiritu C, Caplazi P, Nghiem SS, Chen YJ, Modrusan Z, Dressen A, Goldstein LD, Clarke C, Bhangale T, Yaspan B, Jeanne M, Townsend MJ, van Lookeren Campagne M, Hackney JA, Integration of eQTL and a Single-Cell Atlas in the Human Eye Identifies Causal Genes for Age-Related Macular Degeneration, Cell Rep 30(4) (2020) 1246–1259 e6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. A. RNA-Seq quality control graph. Total RNA from mouse RPE/choroid (air vs 6-mo smoking) was used for library construction and RNASeq reading. The ratio of aligned targets among total reading is calculated for each sample and falls between 84-88%, suggesting good reading quality. B. Principal component analysis showing Air.3 as an outlier. C. High level of retinal gene expression from Air.3 indicates retinal contamination. This sample was eliminated from the analysis.