Abstract
During lung development, the mesenchyme and epithelium are dependent on each other for instructive morphogenic cues that direct proliferation, cellular differentiation and organogenesis. Specification of epithelial and mesenchymal cell lineages occur in parallel, forming cellular subtypes that guide the formation of both transitional developmental structures and the permanent architecture of the adult lung. While epithelial cell types and lineages have been relatively well defined in recent years, the definition of mesenchymal cell types and lineage relationships has been more challenging. Transgenic mouse lines with permanent and inducible lineage tracers have been instrumental in identifying lineage relationships among epithelial progenitor cells and their differentiation into distinct airway and alveolar epithelial cells. Lineage tracing with reporter mice used to identify fibroblast progenitors and their lineage trajectories have been limited by the number of cell specific genes and the developmental timepoint when the lineage trace was activated. In this review, we discuss major developmental mesenchymal lineages, focusing on time of origin, major cell type, and other lineage derivatives, as well as the transgenic tools used to find and define them. We describe lung fibroblasts using function, location, and molecular markers in order to compare and contrast cells with similar functions. The temporal and cell-type specific expression of thirteen “fibroblast lineage” genes were identified in single-cell RNA-sequencing data from LungMAP in the LGEA database. Using these lineage signature genes as guides, we clustered murine lung fibroblast populations from embryonic day 16.5 to postnatal day 28 (E16.5-PN28) and generated heatmaps to illustrate expression of transcription factors, signaling receptors and ligands in a temporal and population specific manner.
Introduction:
Recombination experiments with fetal lung and tracheal cells in the early 1990s revealed that inductive signals from distal lung mesenchyme specified distal epithelial differentiation and proximal mesenchyme specified proximal epithelial differentiation; a process which was independent of whether the epithelium was of proximal or distal origin. These studies demonstrated that lung mesenchyme directed proximal and distal epithelial specification rather than a process intrinsic to or regulated by the epithelium [1]. Developmental studies identified that splanchnic mesenchyme signals to the epithelium through Fgf, WNT, BMP, RA, and TGFβ in a temporal and spatial context-dependent manner to direct endoderm specification towards pulmonary epithelium [2–12]. While mesenchymal cells play a critical role in programming the epithelial cell fate, the mesenchyme is a homogenous and poorly defined group of cells with poor functional and structural annotations. The idea of a cell “lineage” is widely used for hematopoietic and epithelial populations and some attempts have been made to extend “lineage” to lung mesenchyme. However, sorting mesenchymal cells into “lineage” or cell populations has been difficult due to overlapping markers, spatial separation of similar functional roles and the understanding of cellular plasticity of mesenchymal populations. The goal of this review is to summarize current knowledge of the diverse pulmonary mesenchymal cell types and to facilitate comparison of fibroblast subpopulations and functions in normal and diseased tissue. We will limit the use of lineage, where lineage tracing experiments have been used to define cells and their progeny and use the term population or cell stage for studies without lineage trace.
Mesenchymal lineages and other classifications of activation
Early lung lineage labeling experiments took advantage of epithelial cell type specific reporter genes and demonstrated that proximal and distal lung epithelial lineages are specified before E10.5 [13]. Subsequent lineage tracing experiments identified proximal and distal progenitor cells and their cell type specific progenies [14–16]. The role of lung epithelial progenitor cells and their potential to regenerate lung epithelium after injury is an area of active study, using cell specific markers identified by single cell-seq analysis to “predict” small progenitor populations in the adult lung [16–18]. Transgenic tools for lineage labeling in the epithelium take advantage of epithelial specific gene expression. For example, the Surfactant protein C locus is used to drive gene expression in alveolar type 2 (AT2) cells, and key transcription factors that regulate differentiation of very specific epithelial differentiation programs like NKX2.1, SPDEF, or Foxj1 are used to target their respective cell types. Three major factors have made lineage tracing experiments in lung fibroblasts more difficult than in the epithelium: 1) There are very few cell type specific markers that label mesenchymal progenitor cells rather than terminally differentiated cells (eg. Sm22 for differentiated smooth muscle [19]), 2) Key transcription factors that govern fibroblast differentiation programs often label spatially unrelated cells (proximal-distal, peribronchiolar, perivascular) at different times during mesenchymal maturation (eg. TBX4 driving smooth muscle maturation [20]), and 3) Key ligands and receptors of major pathways like Pdgfrs, Glis, Wnts, and Fgfs used to trace lineage in the mesenchyme are unreliable because of their context and time dependent activation. Cells that are neither spatially nor temporally related activate these common signaling pathways multiple times during lung development, injury and repair, and may function as very distinct lineages. Thus, the timing of activation of the lineage trace plays an important role in identifying progenitors and determining their lineage trajectory.
The diverse populations of the pulmonary mesenchyme
The lung mesenchyme can be separated into distinct spatial and functional populations, each with their own developmental lineage histories. Peribronchiolar and perivascular smooth muscle cells; pericytes associated with vessels; parenchymal myo-, matrix -, and lipo-fibroblasts; a newly defined alveolar niche cell, suited for supporting epithelial proliferation and differentiation, after injury; mesothelial cells lining the lung pleura; and mesenchymal stem cells (MSC) a rare poorly described progenitor populations that has proliferative potential after injury and can differentiate into multiple mesenchymal cell types. We will discuss each of these populations in further detail, with a focus on their developmental lineages using both genetic lineage labeling experiments and scRNA-seq time course analysis. Recent single cell analyses of normal and diseased lungs have identified a plethora of “new” cell populations with cell type specific functions, but again have complicated the definition of lung stromal cell populations by introducing the concept of an activation state in response to developmental or injury stimuli [21]. At the end of this review we will briefly discuss the potential of single cell seq data analysis and spatial integration to categorize and “name” lung fibroblasts in the future.
Early lung mesenchyme and tracheal patterning:
The embryonic trachea and esophagus share a common lumen until a longitudinal tracheoesophageal septum divides them into two distinct tubes. As the trachea and esophagus grow apart, they are surrounded by independently organized populations of mesenchymal cells. The cellular and molecular processes mediating morphogenesis of tracheal cartilage and smooth muscle development has been recently reviewed and will not be further discussed [22, 23] During embryonic development in the mouse, the foregut endoderm buds ventrally and the lung buds elongate into the splanchnic mesoderm before E8–9. At this time the splanchnic mesoderm adjacent to the ventral foregut contains multipotent cardiopulmonary Gli1+Wnt2+Isl1+ progenitors that form the myocardium, pulmonary airway, vascular smooth muscle, pericytes, and proximal endothelium in the lung [24]. Thereafter, mesenchymal cell subtype differentiation depends on their location along the cephalo-caudal axis, association with pulmonary structures, and functional phenotypes. Peribronchiolar smooth muscle, perivascular smooth muscle, pericyte, mesothelium, alveolar myofibroblast and alveolar lipofibroblast have been identified decades ago and described by their function and location. However, matrix fibroblasts, adult mesenchymal stem cells and alveolar niche cells have been discovered more recently and are the focus of many regeneration studies. Research on early mesoderm differentiation and functional differentiation of mesenchymal subtypes identified about 16 fibroblast markers that define fibroblast populations [25–38]. For this review we summarized current literature about these fibroblast markers and presumptive lineages with respect to spatio-temporal location and functional phenotype (Table 1)(Fig.1).
Table 1: “What happened?”.
Summary of transgenic tools used to lineage trace lung fibroblasts, combined with literature references and brief description of the finding. List of major mesenchymal lineages during lung development, repair, and homeostasis. Reports of similar and overlapping populations are grouped by lineage, transgenic tool and publication. A short description of when cells were labeled, and progenies documented.
| Lineage | Cell Type | Transgenic Mouse | Reference | Start | End |
|---|---|---|---|---|---|
| TBX4 | Myofibroblast | Tbx4LME-Cre JAX # 033331 (Greif et al., 2012) | (Xie et al., 2016) (Kumar et al., 2014) (Zhang et al., 2013) |
E9.25 | E15.5 |
| Smooth Muscle Cell | Tbx4LME-CreER JAX # N/A (Kumar et al., 2014) | (Kumar et al., 2014) | E10.5 | E13.5 | |
| Pericyte | Tbx4LME-Cre JAX # 033331 (Greif et al., 2012) | (Xie et al., 2016) | E9.25 | E15.5 | |
| Endothelium | Tbx4LME -Cre JAX # 033331 (Greif et al., 2012) Tbx4-rtTA/TetO-Cre JAX # N/A (Zhang et al., 2013) |
(Xie et al., 2016) (Zhang et al., 2013) |
E9.25 | E15.5 | |
| (Zhang et al., 2013) | E11.5 | E15.5 | |||
| Mesothelium | Tbx4LME-CreER JAX # N/A (Kumar et al., 2014) | (Kumar et al., 2014) | E10.5 | E13.5 | |
| Lipofibroblast | Tbx4LME-Cre JAX # 033331 (Greif et al., 2012) | (Xie et al., 2016) | E9.25 | E15.5 | |
| GLI1 | Myofibroblast | Gli1-CreERt2 JAX #007913 (Ahn and Joyner, 2004) | (Li et al., 2015) | E10.5–11.5 | E18.5 |
| P5–6 |
P11 P14 |
||||
| (Kugler et al., 2017) | P1 | P6, P10 | |||
| Mesenchymal Stem Cell | Gli1-CreERt2 JAX #007913 (Ahn and Joyner, 2004) | (Kramann et al., 2015) | 8-Week- | Two days | |
| Pericyte | Gli1-CreERt2 JAX #007913 (Ahn and Joyner, 2004) | (Li et al., 2015) | P5–6 | Adult | |
| Smooth Muscle Cell | Gli1-CreERt2JAX #007913 (Ahn and Joyner, 2004) | (Li et al., 2015) | E10.5–11.5 | E12.5, P11 | |
| (Li et al., 2015) | P5–6 | P11 | |||
| (Kugler et al., 2017) | P1 |
P6 P10 |
|||
| Mesothelium | Gli1-CreERt2 JAX #007913 (Ahn and Joyner, 2004) | (Li et al., 2015) | E10.5–11.5 |
P11 P14 |
|
| (Li et al., 2015) | P5–6 |
P11 P14 |
|||
| (Kugler et al., 2017) | P1 |
P6, P10 |
|||
| PDGFRa | Myofibroblast | C57/BL6 | (Bostrom et al., 2002; Bostrom et al., 1996) | N/A | E15.5 |
| PdgfrartTA;tetO-cre JAX # N/A (Li et al., 2018) | (Li et al., 2018) | E9.5–P7 | P7 | ||
| P0–P7 | P7 | ||||
| P1–P20 | P40 | ||||
| Pdgfra-creER™ JAX # 018280 (De Biase et al., 2011) | (Ntokou et al., 2015) | E9.5 |
E18.5, P5 P7 |
||
|
P2 P5 |
P7 P9 P14 |
||||
| PDGFRαEGFP JAX # 007669 (Hamilton et al., 2003) | (Endale et al., 2017) | N/A |
E16.5 E18.5 P7 |
||
| Matrix fibroblast | Tbx4LME-Cre JAX # 033331 (Greif et al., 2012) | (Xie et al., 2018) | E9.25 | Adult | |
| PDGFRαEGFP JAX # 007669 (Hamilton et al., 2003) | (Endale et al., 2017) (Green et al., 2016) | PNX | Adult | ||
| Lipofibroblast | PdgfrartTA;tetO-cre JAX # N/A (Li et al., 2018) | (Li et al., 2018) | E9.5–P7 | P7 | |
| P0–P7 | P7 | ||||
| P1–P20 | P40 | ||||
| Pdgfra-creER™ JAX # 018280 (De Biase et al., 2011) | (Ntokou et al., 2015) |
E9.5 P2 P5 |
E18.5 P5 P7 P7 to P14 |
||
| Smooth muscle cell | PDGFRαEGFP JAX # 007669 (Hamilton et al., 2003) | (Ntokou et al., 2015) (El Agha et al., 2017) |
E9.5 | E14.5 to P3 | |
| FGF18 | Myofibroblast | FGF18CreERT2 JAX # N/A (Hagan et al., 2019a) Gli1LacZ JAX # 008211 (Bai et al., 2002) Gli1CreERT2 JAX # 007913 (Ahn and Joyner, 2004) |
(Hagan et al., 2019b) | PN5–8 | PN9, PN21 |
| PN1 |
PN2, PN7,
PN21 |
||||
| Gli1CreERT2 JAX # 007913 (Ahn and Joyner, 2004) Gli1LacZ JAX # 008211 (Bai et al., 2002) |
(Hagan et al., 2019b) | PN5–8 | PN9, PN21 | ||
| Lipofibroblast | FGF18CreERT2 JAX # N/A (Hagan et al., 2019a) Gli1LacZ JAX # 008211 (Bai et al., 2002) Gli1CreERT2 JAX # 007913 (Ahn and Joyner, 2004) |
(Hagan et al., 2019b) | PN5–8 | PN9, PN21 | |
| PN1 | PN2, PN7, PN21 | ||||
| Gli1CreERT2 JAX # 007913 (Ahn and Joyner, 2004) Gli1LacZ JAX # 008211 (Bai et al., 2002) |
(Hagan et al., 2019b) | PN5–8 | PN9, PN21 | ||
| Mesothelium | FGF18CreERT2 JAX # N/A (Hagan et al., 2019a) Gli1LacZ JAX # 008211 (Bai et al., 2002) |
(Hagan et al., 2019b) | PN5–8 | PN9, PN21 | |
| THY1 | Lipofibroblast | C57/B6; Thy1−/− JAX # N/A (Dr. Koger Morris, King’s College, London, UK) | (Varisco et al., 2012) | N/A | E18.5 to Adult |
| Mesenchymal Stem Cell | C57BL/6 | (McQualter et al., 2009) | Adult | Adult | |
| FGF10 | Lipofibroblast | Fgf10iCre JAX # 033807 (El Agha et al., 2012) | (El Agha et al., 2014) |
E11.5 E15.5 |
E15.5 E18.5 |
| Myofibroblast | Fgf10iCre JAX # 033807 (El Agha et al., 2012) | (El Agha et al., 2017) | Bleo Injury | Adult | |
| Smooth Muscle Cell | Fgf10iCre JAX # 033807 (El Agha et al., 2012) (Fgf10)-lacZ MGI:3629660 (Kelly et al., 2001) | (El Agha et al., 2014) | E11.5 | E13.5 to E18.5 | |
| (Mailleux et al., 2005) | E10.5 | E11.5 to E14.5 | |||
| Mesenchymal Stem Cell | Fgf10iCre JAX # 033807 (El Agha et al., 2012) | (El Agha et al., 2014) | E11.5 |
P30 Adult |
|
| TCF21 | Lipofibroblast | TCF21-creERT2 JAX # N/A (Park et al., 2019) | (Park et al., 2019) |
E9.5 11.5 15.5 |
E18.5 |
| Matrix Fibroblast | Tbx4LME-Cre JAX # 033331 (Greif et al., 2012) | (Xie et al., 2018) | E9.25 | Adult | |
| Myofibroblast | TCF21-creERT2 JAX # N/A (Park et al., 2019) | (Park et al., 2019) | P2 | P7 | |
| Smooth Muscle Cell | TCF21-creERT2 JAX # N/A (Park et al., 2019) | (Park et al., 2019) |
E9.5 11.5 |
E18.5 | |
| PDGFKb | Pericyte | PDGFKb−/+ JAX # 007846 (Soriano, 1994) | (Greif et al., 2012; Hellstrom et al., 1999) | E11.5 |
E13.5, E18.5 |
| Smooth Muscle Cell | PDGFR-β-Cre JAX # N/A (Foo et al., 2006) | (Greif et al., 2012) | E11.5 | E13.5, E18.5 | |
| Lipofibroblast | Tbx4LME-Cre JAX # 033331 (Greif et al., 2012) | (Xie et al., 2018) | Adult | Adult After Injury | |
| Myofibroblast | Tbx4LME-Cre JAX # 033331 (Greif et al., 2012) | (Xie et al., 2018) | E9.25 | Adult After Injury | |
| (Henderson et al., 2013) | Adult | Adult After Injury | |||
| NG2 (CSPG4) | Pericyte | NG2-CreEK™ JAX # 008538 (Zhu et al., 2011) | (Rock et al., 2011) | Adult | Adult after injury |
| Myofibroblast | NG2-CreEK™ JAX # 008538 (Zhu et al., 2011) | (Rock et al., 2011) | Adult | Adult after injury | |
| Lipofibroblast | NG2-CreEK™ JAX # 008538 (Zhu et al., 2011) | (Rock et al., 2011) | Adult | Adult after injury | |
| FOXD1 | Pericyte | Foxd1+/GFPCreER JAX # 012464 (Humphreys et al., 2010) | (Hung et al., 2013) | E11.5 | E14.5, Adult |
| Myofibroblast | Foxd1+/GFPCreER JAX # 012464 (Humphreys et al., 2010) | (Hung et al., 2013) | Adult | Adult 7 days after injury | |
| Lipofibroblast | Foxd1-Cre JAX # 012463 (Humphreys et al., 2010) | (Hung et al., 2013) | E11.5 | Adult | |
| COL13A1 | Matrix Fibroblast | Tbx4LME-Cre JAX # 033331 (Greif et al., 2012) | (Xie et al., 2018) | E9.25 | E16.5-Adult |
| Myofibroblast | Tbx4LME-Cre JAX # 033331 (Greif et al., 2012) | (Xie et al., 2018) | E9.25 | E16.5-Adult | |
| Lipofibroblast | Tbx4LME-Cre JAX # 033331 (Greif et al., 2012) | (Xie et al., 2018) | E9.25 | E16.5-Adult | |
| COL14A1 | Matrix Fibroblast | Tbx4LME-Cre JAX # 033331 (Greif et al., 2012) | (Xie et al., 2018) | E9.25 | E16.5-Adult |
| Myofibroblast | Tbx4LME-Cre JAX # 033331 (Greif et al., 2012) | (Xie et al., 2018) | E9.25 | E16.5-Adult | |
| Lipofibroblast | Tbx4LME-Cre JAX # 033331 (Greif et al., 2012) | (Xie et al., 2018) | E9.25 | E16.5-Adult | |
| SCA-1 | Mesenchymal Stem Cell | Ly6aCre (Sca-1Cre) JAX #032621 (Vagnozzi et al., 2018) | (McQualter et al., 2009) | E18.5 | Adult |
| Myofibroblast | C57/BL6 | (Cao et al., 2018) | Adult | In Vitro After Injury | |
| Lipofibroblast | Ly6aCre (Sca-1Cre) JAX #032621 (Vagnozzi et al., 2018) | (McQualter et al., 2009) | E18.5 | Adult | |
| (Xie et al., 2018) | Adult | Adult | |||
| Matrix Fibroblast | PDGFRαEGFP JAX # 007669 (Hamilton et al., 2003) | (Green et al., 2016) | Adult | Adult after PNX | |
| (Xie et al., 2018) | Adult | Adult | |||
| Pericyte | Ly6aCre (Sca-1Cre) JAX #032621 (Vagnozzi et al., 2018) | (McQualter et al., 2009) | E18.5 | Adult | |
| Smooth Muscle Cell | Ly6aCre (Sca-1Cre) JAX #032621 (Vagnozzi et al., 2018) | (McQualter et al., 2009) | E18.5 | Adult | |
| ABCG2 | Mesenchymal Stem Cell | Abcg2CreERT2 JAX # 021961 (Fatima et al., 2012) | (Gaskill et al., 2017) | Adult | Adult |
| Pericyte | Abcg2CreERT2 JAX # 021961 (Fatima et al., 2012) | (Gaskill et al., 2017) | Adult | Adult, Adult after injury | |
| LGR5 | Alveolar Niche Cell | Lgr5EGFP-IRES-creERT2 JAX # 008875 (Barker et al., 2007) | (Lee et al., 2017) | Adult | Adult |
| Myofibroblast | Lgr5EGFP-IRES-creERT2 JAX # 008875 (Barker et al., 2007) | (Lee et al., 2017) | Adult | Adult, Adult After Injury | |
| AXIN2/WNT2 | Alveolar Niche Cell | Axin2CreERT2 JAX # 018867 (van Amerongen et al., 2012) Wnt2CreERT2 JAX # N/A (Peng et al., 2013) PDGFRαEGFP JAX # 007669 (Hamilton et al., 2003) |
(Zepp et al., 2017) | Adult | Adult |
| Myofibroblast | Axin2CreERT2 JAX # 018867 (van Amerongen et al., 2012) Wnt2CreERT2 JAX # N/A (Peng et al., 2013) PDGFRαEGFP JAX # 007669 (Hamilton et al., 2003) |
(Zepp et al., 2017) | Adult | Adult, Adult after injury | |
| LGR6 | Smooth Muscle Cell | Lgr6GFP-ires-CreERT2 JAX #016934 (Snippert et al., 2010) | (Lee et al., 2017) | Adult | Adult |
| Myofibroblast | Lgr6GFP-ires-CreERT2 JAX # 016934 (Snippert et al., 2010) | (Lee et al., 2017) | Adult | Adult | |
| WT1 | Mesothelium | Wt1CreERT2 JAX # 010912 (Zhou et al., 2008) | (Que et al., 2008) (Colvin et al., 2001) |
E10.5 | E11.5, E15.5, P10 |
| Smooth Muscle Cell | Wt1CreERT2 JAX # 010912 (Zhou et al., 2008) | (Que et al., 2008) | E10.5 | E15.5, P10 | |
| Myofibroblast | Wt1CreERT2 JAX # 010912 (Zhou et al., 2008) | (Que et al., 2008) | E10.5 | E15.5, P10 |
Ahn, S., and Joyner, A.L. (2004). Dynamic changes in the response of cells to positive hedgehog signaling during mouse limb patterning. Cell 118, 505–516.
Bai, C.B., Auerbach, W., Lee, J.S., Stephen, D., and Joyner, A.L. (2002). Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development 129, 4753–4761.
Barker, N., van Es, J.H., Kuipers, J., Kujala, P., van den Born, M., Cozijnsen, M., Haegebarth, A., Korving, J., Begthel, H., Peters, P.J., et al. (2007). Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007.
Bostrom, H., Gritli-Linde, A., and Betsholtz, C. (2002). PDGF-A/PDGF alpha-receptor signaling is required for lung growth and the formation of alveoli but not for early lung branching morphogenesis. Dev Dyn 223, 155–162.
Bostrom, H., Willetts, K., Pekny, M., Leveen, P., Lindahl, P., Hedstrand, H., Pekna, M., Hellstrom, M., Gebre-Medhin, S., Schalling, M., et al. (1996). PDGF-A signaling is a critical event in lung alveolar myofibroblast development and alveogenesis. Cell 85, 863–873.
Cao, H., Wang, C., Chen, X., Hou, J., Xiang, Z., Shen, Y., and Han, X. (2018). Inhibition of Wnt/beta-catenin signaling suppresses myofibroblast differentiation of lung resident mesenchymal stem cells and pulmonary fibrosis. Sci Rep 8, 13644.
Colvin, J.S., White, A.C., Pratt, S.J., and Ornitz, D.M. (2001). Lung hypoplasia and neonatal death in Fgf9-null mice identify this gene as an essential regulator of lung mesenchyme. Development 128, 2095–2106.
De Biase, L.M., Kang, S.H., Baxi, E.G., Fukaya, M., Pucak, M.L., Mishina, M., Calabresi, P.A., and Bergles, D.E. (2011). NMDA receptor signaling in oligodendrocyte progenitors is not required for oligodendrogenesis and myelination. J Neurosci 31, 12650–12662.
El Agha, E., Al Alam, D., Carraro, G., MacKenzie, B., Goth, K., De Langhe, S.P., Voswinckel, R., Hajihosseini, M.K., Rehan, V.K., and Bellusci, S. (2012). Characterization of a novel fibroblast growth factor 10 (Fgf10) knock-in mouse line to target mesenchymal progenitors during embryonic development. PLoS One 7, e38452.
El Agha, E., Herold, S., Al Alam, D., Quantius, J., MacKenzie, B., Carraro, G., Moiseenko, A., Chao, C.M., Minoo, P., Seeger, W., et al. (2014). Fgf10-positive cells represent a progenitor cell population during lung development and postnatally. Development 141, 296–306.
El Agha, E., Moiseenko, A., Kheirollahi, V., De Langhe, S., Crnkovic, S., Kwapiszewska, G., Szibor, M., Kosanovic, D., Schwind, F., Schermuly, R.T., et al. (2017). Two-Way Conversion between Lipogenic and Myogenic Fibroblastic Phenotypes Marks the Progression and Resolution of Lung Fibrosis. Cell Stem Cell 20, 571.
Endale, M., Ahlfeld, S., Bao, E., Chen, X., Green, J., Bess, Z., Weirauch, M.T., Xu, Y., and Perl, A.K. (2017). Temporal, spatial, and phenotypical changes of PDGFRalpha expressing fibroblasts during late lung development. Dev Biol 425, 161–175.
Fatima, S., Zhou, S., and Sorrentino, B.P. (2012). Abcg2 expression marks tissue-specific stem cells in multiple organs in a mouse progeny tracking model. Stem Cells 30, 210–221.
Foo, S.S., Turner, C.J., Adams, S., Compagni, A., Aubyn, D., Kogata, N., Lindblom, P., Shani, M., Zicha, D., and Adams, R.H. (2006). Ephrin-B2 controls cell motility and adhesion during blood-vessel-wall assembly. Cell 124, 161–173.
Gaskill, C.F., Carrier, E.J., Kropski, J.A., Bloodworth, N.C., Menon, S., Foronjy, R.F., Taketo, M.M., Hong, C.C., Austin, E.D., West, J.D., et al. (2017). Disruption of lineage specification in adult pulmonary mesenchymal progenitor cells promotes microvascular dysfunction. J Clin Invest 127, 2262–2276.
Green, J., Endale, M., Auer, H., and Perl, A.K. (2016). Diversity of Interstitial Lung Fibroblasts Is Regulated by Platelet-Derived Growth Factor Receptor alpha Kinase Activity. Am J Respir Cell Mol Biol 54, 532–545.
Greif, D.M., Kumar, M., Lighthouse, J.K., Hum, J., An, A., Ding, L., Red-Horse, K., Espinoza, F.H., Olson, L., Offermanns, S., et al. (2012). Radial construction of an arterial wall. Dev Cell 23, 482–493.
Hagan, A.S., Boylan, M., Smith, C., Perez-Santamarina, E., Kowalska, K., Hung, I.H., Lewis, R.M., Hajihosseini, M.K., Lewandoski, M., and Ornitz, D.M. (2019a). Generation and validation of novel conditional flox and inducible Cre alleles targeting fibroblast growth factor 18 (Fgf18). Dev Dyn 248, 882–893.
Hagan, A.S., Zhang, B., and Ornitz, D.M. (2019b). Identification of an FGF18-expressing alveolar myofibroblast that is developmentally cleared during alveologenesis. Development.
Hamilton, T.G., Klinghoffer, R.A., Corrin, P.D., and Soriano, P. (2003). Evolutionary divergence of platelet-derived growth factor alpha receptor signaling mechanisms. Mol Cell Biol 23, 4013–4025.
Hellstrom, M., Kalen, M., Lindahl, P., Abramsson, A., and Betsholtz, C. (1999). Role of PDGF-B and PDGFR-beta in recruitment of vascular smooth muscle cells and pericytes during embryonic blood vessel formation in the mouse. Development 126, 3047–3055.
Henderson, N.C., Arnold, T.D., Katamura, Y., Giacomini, M.M., Rodriguez, J.D., McCarty, J.H., Pellicoro, A., Raschperger, E., Betsholtz, C., Ruminski, P.G., et al. (2013). Targeting of alphav integrin identifies a core molecular pathway that regulates fibrosis in several organs. Nat Med 19, 1617–1624.
Humphreys, B.D., Lin, S.L., Kobayashi, A., Hudson, T.E., Nowlin, B.T., Bonventre, J.V., Valerius, M.T., McMahon, A.P., and Duffield, J.S. (2010). Fate tracing reveals the pericyte and not epithelial origin of myofibroblasts in kidney fibrosis. Am J Pathol 176, 85–97.
Hung, C., Linn, G., Chow, Y.H., Kobayashi, A., Mittelsteadt, K., Altemeier, W.A., Gharib, S.A., Schnapp, L.M., and Duffield, J.S. (2013). Role of lung pericytes and resident fibroblasts in the pathogenesis of pulmonary fibrosis. Am J Respir Crit Care Med 188, 820–830.
Kelly, R.G., Brown, N.A., and Buckingham, M.E. (2001). The arterial pole of the mouse heart forms from Fgf10-expressing cells in pharyngeal mesoderm. Dev Cell 1, 435–440.
Kramann, R., Schneider, R.K., DiRocco, D.P., Machado, F., Fleig, S., Bondzie, P.A., Henderson, J.M., Ebert, B.L., and Humphreys, B.D. (2015). Perivascular Gli1+ progenitors are key contributors to injury-induced organ fibrosis. Cell Stem Cell 16, 51–66.
Kugler, M.C., Loomis, C.A., Zhao, Z., Cushman, J.C., Liu, L., and Munger, J.S. (2017). Sonic Hedgehog Signaling Regulates Myofibroblast Function During Alveolar Septum Formation in Murine Postnatal Lung. Am J Respir Cell Mol Biol.
Kumar, M.E., Bogard, P.E., Espinoza, F.H., Menke, D.B., Kingsley, D.M., and Krasnow, M.A. (2014). Mesenchymal cells. Defining a mesenchymal progenitor niche at single-cell resolution. Science 346, 1258810.
Lee, J.H., Tammela, T., Hofree, M., Choi, J., Marjanovic, N.D., Han, S., Canner, D., Wu, K., Paschini, M., Bhang, D.H., et al. (2017). Anatomically and Functionally Distinct Lung Mesenchymal Populations Marked by Lgr5 and Lgr6. Cell 170, 1149–1163 e1112.
Li, C., Li, M., Li, S., Xing, Y., Yang, C.Y., Li, A., Borok, Z., De Langhe, S., and Minoo, P. (2015). Progenitors of secondary crest myofibroblasts are developmentally committed in early lung mesoderm. Stem Cells 33, 999–1012.
Li, R., Bernau, K., Sandbo, N., Gu, J., Preissl, S., and Sun, X. (2018). Pdgfra marks a cellular lineage with distinct contributions to myofibroblasts in lung maturation and injury response. Elife 7.
Mailleux, A.A., Kelly, R., Veltmaat, J.M., De Langhe, S.P., Zaffran, S., Thiery, J.P., and Bellusci, S. (2005). Fgf10 expression identifies parabronchial smooth muscle cell progenitors and is required for their entry into the smooth muscle cell lineage. Development 132, 2157–2166.
McQualter, J.L., Brouard, N., Williams, B., Baird, B.N., Sims-Lucas, S., Yuen, K., Nilsson, S.K., Simmons, P.J., and Bertoncello, I. (2009). Endogenous fibroblastic progenitor cells in the adult mouse lung are highly enriched in the sca-1 positive cell fraction. Stem Cells 27, 623–633.
Ntokou, A., Klein, F., Dontireddy, D., Becker, S., Bellusci, S., Richardson, W.D., Szibor, M., Braun, T., Morty, R.E., Seeger, W., et al. (2015). Characterization of the platelet-derived growth factor receptor-alpha-positive cell lineage during murine late lung development. Am J Physiol Lung Cell Mol Physiol 309, L942–958.
Park, J., Ivey, M.J., Deana, Y., Riggsbee, K.L., Sorensen, E., Schwabl, V., Sjoberg, C., Hjertberg, T., Park, G.Y., Swonger, J.M., et al. (2019). The Tcf21 lineage constitutes the lung lipofibroblast population. Am J Physiol Lung Cell Mol Physiol 316, L872-L885.
Peng, T., Tian, Y., Boogerd, C.J., Lu, M.M., Kadzik, R.S., Stewart, K.M., Evans, S.M., and Morrisey, E.E. (2013). Coordination of heart and lung co-development by a multipotent cardiopulmonary progenitor. Nature 500, 589–592.
Que, J., Wilm, B., Hasegawa, H., Wang, F., Bader, D., and Hogan, B.L. (2008). Mesothelium contributes to vascular smooth muscle and mesenchyme during lung development. Proc Natl Acad Sci U S A 105, 16626–16630.
Rock, J.R., Barkauskas, C.E., Cronce, M.J., Xue, Y., Harris, J.R., Liang, J., Noble, P.W., and Hogan, B.L. (2011). Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc Natl Acad Sci U S A 108, E1475–1483.
Snippert, H.J., Haegebarth, A., Kasper, M., Jaks, V., van Es, J.H., Barker, N., van de Wetering, M., van den Born, M., Begthel, H., Vries, R.G., et al. (2010). Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science 327, 1385–1389.
Soriano, P. (1994). Abnormal kidney development and hematological disorders in PDGF beta-receptor mutant mice. Genes Dev 8, 1888–1896.
Vagnozzi, R.J., Sargent, M.A., Lin, S.J., Palpant, N.J., Murry, C.E., and Molkentin, J.D. (2018). Genetic Lineage Tracing of Sca-1(+) Cells Reveals Endothelial but Not Myogenic Contribution to the Murine Heart. Circulation 138, 2931–2939.
van Amerongen, R., Bowman, A.N., and Nusse, R. (2012). Developmental stage and time dictate the fate of Wnt/beta-catenin-responsive stem cells in the mammary gland. Cell Stem Cell 11, 387–400.
Varisco, B.M., Ambalavanan, N., Whitsett, J.A., and Hagood, J.S. (2012). Thy-1 signals through PPARgamma to promote lipofibroblast differentiation in the developing lung. Am J Respir Cell Mol Biol 46, 765–772.
Xie, T., Liang, J., Liu, N., Huan, C., Zhang, Y., Liu, W., Kumar, M., Xiao, R., D’Armiento, J., Metzger, D., et al. (2016). Transcription factor TBX4 regulates myofibroblast accumulation and lung fibrosis. J Clin Invest 126, 3626.
Xie, T., Wang, Y., Deng, N., Huang, G., Taghavifar, F., Geng, Y., Liu, N., Kulur, V., Yao, C., Chen, P., et al. (2018). Single-Cell Deconvolution of Fibroblast Heterogeneity in Mouse Pulmonary Fibrosis. Cell Rep 22, 3625–3640.
Zepp, J.A., Zacharias, W.J., Frank, D.B., Cavanaugh, C.A., Zhou, S., Morley, M.P., and Morrisey, E.E. (2017). Distinct Mesenchymal Lineages and Niches Promote Epithelial Self-Renewal and Myofibrogenesis in the Lung. Cell 170, 1134–1148 e1110.
Zhang, W., Menke, D.B., Jiang, M., Chen, H., Warburton, D., Turcatel, G., Lu, C.H., Xu, W., Luo, Y., and Shi, W. (2013). Spatial-temporal targeting of lung-specific mesenchyme by a Tbx4 enhancer. BMC Biol 11, 111.
Zhou, B., Ma, Q., Rajagopal, S., Wu, S.M., Domian, I., Rivera-Feliciano, J., Jiang, D., von Gise, A., Ikeda, S., Chien, K.R., et al. (2008). Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature 454, 109–113.
Zhu, X., Hill, R.A., Dietrich, D., Komitova, M., Suzuki, R., and Nishiyama, A. (2011). Age-dependent fate and lineage restriction of single NG2 cells. Development 138, 745–753.
Figure 1-. Location, Location, Location.
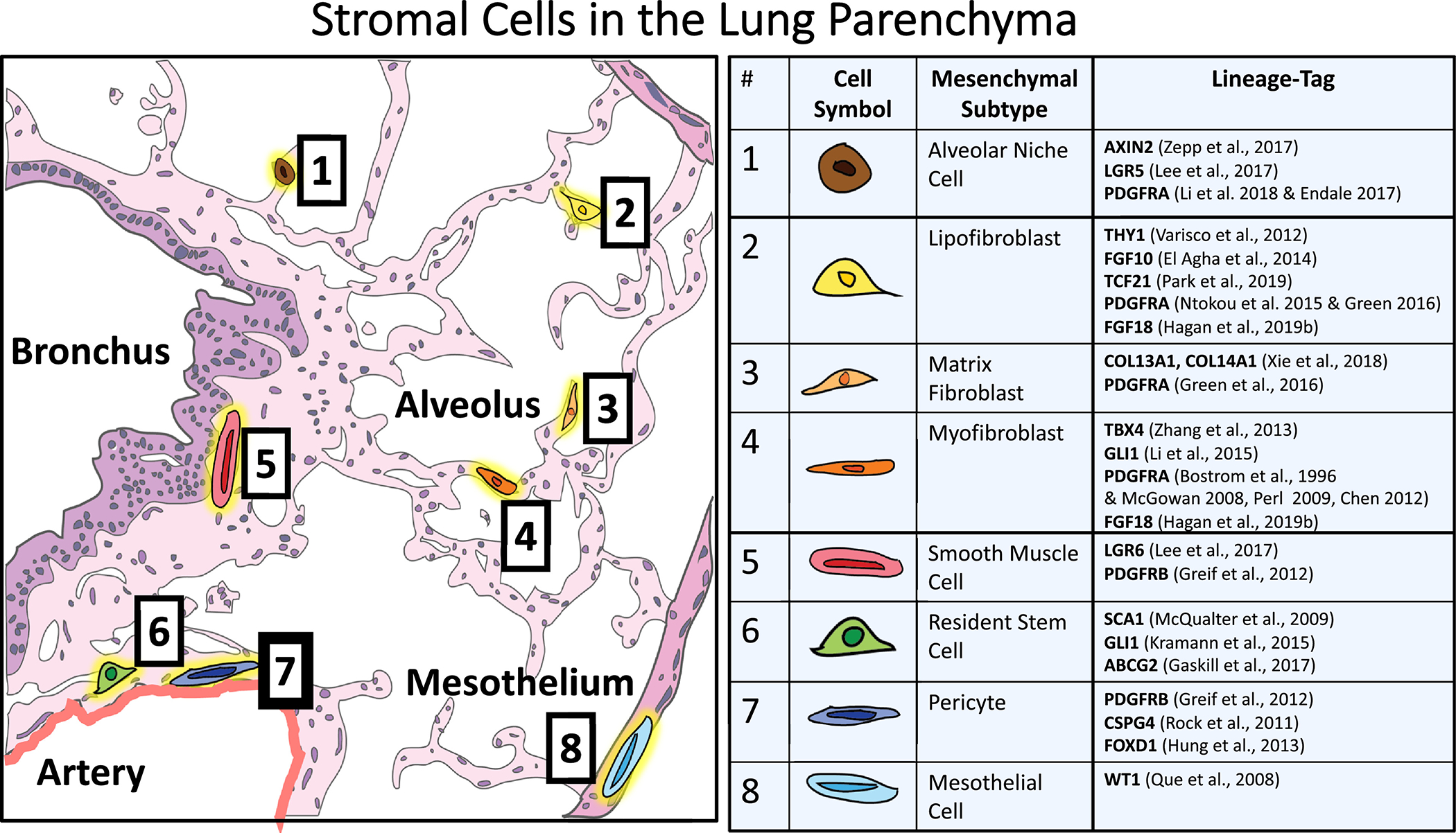
Representational drawing of an adult donor lung histology section. Each of the eight functional mesenchymal subtypes found in the adult lung are located to their niche based on their lineage definition in the literature. The table includes accepted lineage labels currently available in the field.
The TBX4 Lineage traced cells- Cells at the right place and the right time:
T-box gene 4 (Tbx4) is a mesenchyme-specific transcription factor that plays an important role in airway morphogenesis and is expressed in the early splanchnic mesoderm that surrounds the lung buds as early as E9.5 [39]. Using a Tbx4-rtTA/tetOCre and doxycycline activation from E6.5 to E10.5 targets almost all splanchnic mesoderm surrounding the lung buds at E10.5, the same region where the Gli1+Wnt2+Isl1+ cardiopulmonary progenitors reside [25]. Activating Tbx4-rtTA lineage trace before E11.5 and E15.5 traces pulmonary endothelial cells and vascular smooth muscle, while other undefined fibroblasts, smooth muscle cells, and pericytes are labeled throughout embryonic development [25]. The constitutive Tbx4LME-Cre; R26tdT/tdT, lineage tracer tracks Tbx4 expressing cells starting as early as E9.5. These Tbx4 traced cells give raise to embryonic (at E15.5) and adult-stage (PN56) fibroblasts along blood vessels, underneath the bronchioles, and within the distal alveolar (interstitial) mesenchyme [20]. Multiple groups showed that Tbx4 expressing cells give rise to nearly all mesenchymal cell types, including mesothelium, vascular and bronchiolar smooth muscle, pericytes, endothelium, lipofibroblasts, matrix fibroblasts, and myofibroblasts [20, 25, 40]. Combining the constitutive Tbx4-lineage trace with a smooth muscle cell reporter (ACTA2-GFP) and a collagen producing fibroblast reporter (COL1α1-GFP) revealed Tbx4 lineage contribution to smooth muscle cells and collagen producing cells by flow cytometry before and after bleomycin injury [20]. In uninjured mice, 90.6% of the αSma-GFP+ cells and 87% of all COL1α1-GFP cells were Tbx4 derived. After bleomycin injury 97.6% of the αSma-GFP+ cells and 95% of all COL1α1-GFP cells were Tbx4 derived. These data demonstrate that the vast majority of fibroblasts activated during repair are derived from resident Tbx4 lineage-labeled fibroblasts or fibroblasts that activated Tbx4 during injury/repair. Conditionally ablating Tbx4-expressing cells in adults did not disturb lung homeostasis. Inactivation of Tbx4 gene expression in COL1A2- or ACTA2- expressing cells attenuated lung fibrosis after bleomycin-induced injury, demonstrating that fibrotic fibroblast activation is dependent on Tbx4-mediated gene activation [20]. TBX4 is thus a critical transcription factor for mesenchymal differentiation during development and injury response, as it most likely induces expression of genes associated with smooth muscle contraction and mesenchymal cell structure. To compare TBX4 expression within the mesenchymal cells over time please refer to the “lineage marker” and “Transcription factor” Heatmap (Fig 2, Fig.3).
Figure 2. Heatmap “Lineage Markers”:
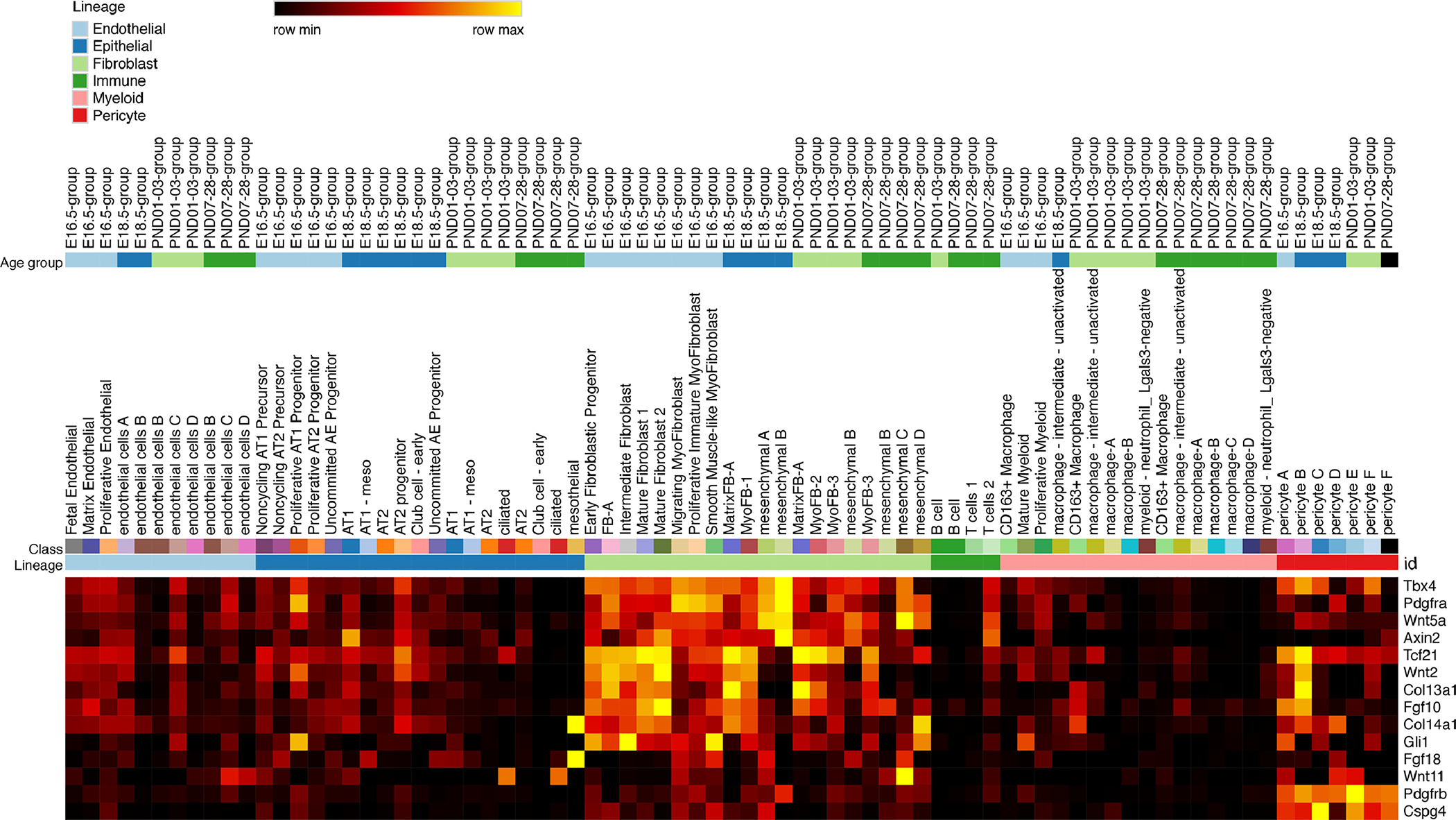
Expression of established lineage markers in mesenchymal populations identified by single cell RNA-Seq heatmap: 14 of the 16 lineage markers were identified in the scRNA-seq dataset and displayed in a heatmap (Wt1, Ng2, Thy1, Lgr5 and Lgr6 were expressed at very low levels). The x-axis was arranged by cell type, cell subtype, and age group, respectively. Yellow represents high expression and black represents little to no expression.
Figure 3: Heatmap “Transcription factors”.
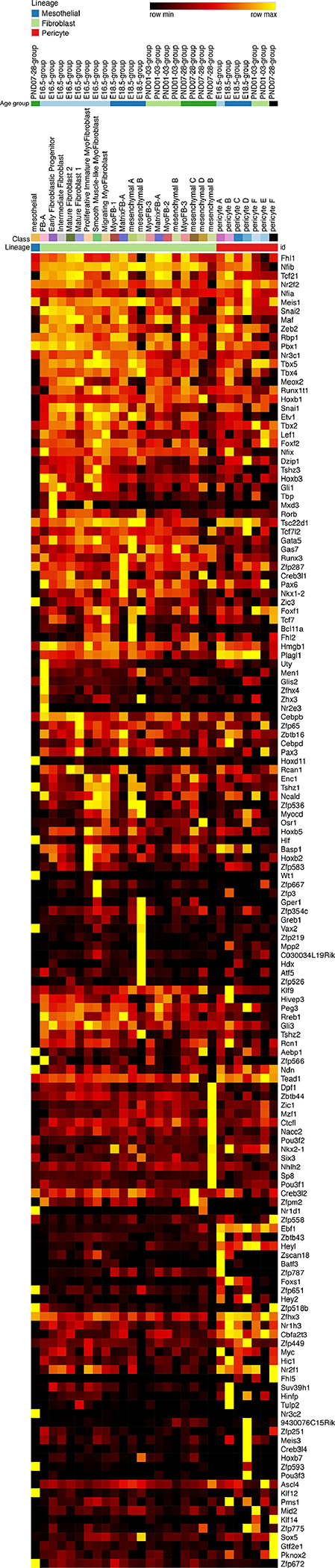
Expression of transcription factors in mesenchymal populations identified by single cell RNA-Seq heatmap. These transcription factors are the highest expressed (TPM>2.5) and most specifically expressed within the mesenchymal population. Transcription factors were placed on the y-axis of a single cell RNA-seq heatmap. The x-axis was arranged by cell type, cell subtype, and age group, respectively. Yellow represents highest expression.
Gli1 Lineage-traced cells: Vote early and often:
During embryonic lung growth, paracrine Shh signal from the epithelium regulates proliferation and differentiation of the responding Gli1 expressing mesenchyme. Lineage tracing of the Gli1+ cells at E8–9 shows that the cardiac progenitors give rise to pulmonary vasculature and subsets of cells in the lung mesenchyme [24]. Labeling Gli1+ cells during E10.5–11.5 gives rise to mesothelial cells, interstitial myofibroblast, perivascular and peribronchiolar smooth muscle in post-natal day 14 lungs, but not lipofibroblasts [41]. From PN5–7, the Gli1 reporter labels myofibroblasts at the septal tips where these hedgehog-responsive fibroblasts play a critical role in alveolar septation [41, 42]. At PN11, PN5-PN7 lineage traced Gli+ fibroblasts differentiate into myofibroblasts but not to lipofibroblasts [41]. These data support a clear lineage distinction between interstitial myo- and lipofibroblasts, in which alveolar myofibroblasts are responsive to Shh and alveolar lipofibroblasts are modulators of other major signaling pathways, such as FGF [29, 43]. PDGFA signaling is also essential for alveolar septation and alveolar myofibroblast differentiation [27, 44–48]. At the end of the saccular phase (E18.5) 60% of the Gli1+ lineage cells expressed Pdgfra, whereas 97% of the Gli1+ lineage cells expressed Pdgfra toward the end of alveolarization at PN14, supporting the concept that myofibroblasts in the Gli1 lineage activate Pdgfra expression during alveolar septation.
Gli1-LacZ-labeled perivascular pericytes and peribronchiolar SMCs are responsive to hedgehog well into adulthood [42]. The number of alveolar interstitial Gli1-expressing cells is decreased by PN14 consistent with reduced Shh signaling and slowing of alveologenesis. Approximately 30% of alveolar PN1 labeled Gli1+ cells give raise to PN6 crest myofibroblasts that co-express αSma. Approximately 20–25% of peribronchial and 10–15% of alveolar Gli1+ cells co-express Pdgfra these dual positive cells are located at the septal tips, increase in number during alveolarization and undergo apoptosis during septal wall thinning after PN10. Genetic loss of hedgehog signaling in the epithelium decreases the number of septal tips and inhibits lineage traced Gli1+ cell proliferation and differentiation into myofibroblasts by P7 [42]. Since elastin expression is unchanged when Gli1+ myofibroblasts are lost, it is assumed that there is a population of cells that undergo Gli1-independent matrix fibroblast differentiation. Very few Gli1+ cells remain In the adult lung and are mainly found in perivascular mesenchyme, functioning as progenitor cells during injury repair [26]. During lung fibrosis both Gli1 lineage traced cells and cells actively expressing Gli1 expand and contribute significantly to the pool of myofibroblasts in fibrotic areas [49] and have a significant paracrine function in promoting epithelial differentiation in IPF [50]. Gli1+ mesenchymal stromal cells (MSCs) integrate hedgehog signaling, upregulate BMP antagonist expression in response, and promote metaplastic differentiation of airway progenitors into KRT5+ basal cells. If bypassed by exogenously upregulating BMP signaling with recombinant hBMP2, this effect is blunted, and proper alveolarization occurs. These data demonstrate that the Gli1 population is a myofibroblast that integrates epithelial shh signaling and reciprocally induces epithelial differentiation. To compare Gli expression within the stromal cells during development please refer to the “Transcription factor” Heatmap (Fig.3).
Pdgfra lineage traced cells and Pdgfra expressing populations: Jack of all trades, master of none
Platelet-derived growth factor-A (PDGFA) and its receptor, platelet-derived growth factor receptor-alpha (Pdgfra), are required for formation of the secondary alveolar septa in mice [27, 44–48]. Pdgfra+ cells are distinct from Pdgfrb+ pericytes [44, 51]. A transgenic reporter mouse with a green fluorescent protein (GFP) knocked into the Pdgfra gene locus Pdgfra-GFP+ (Pdgfratm11(EGFP)Sor [52]) has widely been used to track real-time expression of Pdgfra during both lung development and alveolar regeneration [44, 53–62]. Three populations of fibroblasts were identified on the basis of absent, dim, or bright Pdgfra-GFP expression. During alveolarizarization bright GFP+ cells were more proliferative at PN4 and non-proliferative by PN12. These GFP bright Pdgfra+ fibroblasts are primarily located in the alveolar entry ring where alpha smooth muscle actin (aSMA) and elastin fibers accumulate [53]. In contrast, Pdgfra-expression is not associated with neutral lipid accumulation in lipofibroblasts, which are located at the alveolar base [53–55]. During alveolar regeneration Pdgfra-GFP+ dim cells give raise to interstitial myofibroblast. Inhibition of Fgfr2 inhibited myofibroblast differentiation and realveolarization but increased the number of Pdgfra-GFP+ bright cells. Loss of αSma expression and “brighter” Pdgfra-GFP expression was accompanied with increased Wnt2a and BMP4 RNA in Pdgfra+ fibroblasts [56, 58]. Two GFP reporter mice (Pdgfratm11(EGFP)Sor and Pdgfra-CreERT2 BAC transgenic line), were used to track expression and permanent lineage traced Pdgfra cells. Pdgfra-GFP+ cells were primarily located around the proximal airways during the pseudoglandular period of development and more distally within the peripheral airways in the canalicular stage. Upon lineage tracing, neither of these cells contributed to alveolar Pdgfra+ fibroblasts. In contrast, E18.5 Pdgfra-GFP labeled fibroblasts contributed to myo and lipofibroblasts located postnatally in the primary septae during alveolarization and to a sparse set of fibroblasts in the adult lung [60]. These data demonstrate that lineage traced Pdgfra fibroblasts are associated with conducting airways, do not migrate into the alveolar saccules, and are a distinct cell population from other distal fibrobalsts. At E18.5 fibroblasts that are associated with peripheral lung saccules express Pdgfra de novo, which was an underappreciated finding in the early studies that suggested migration of proximal fibroblasts to the distal saccules [27, 44–48]. In contrast to the Pdgfra-CreERT2 BAC transgenic line which labels lipofibroblast progenitors, the PdgfrartTA knockin mice labels (E9.5-PN7) progenitor cells that give raise to ~95% of myofibroblasts and not lipofibroblasts [63]. These different transgenic mouse lines may target overlapping but non-identical cell populations which should always be considered when interpreting results obtained from different transgenic mouse lines [15]. To compare Pdgfra RNA expression within various mesenchymal cells during development please refer to the “Receptor” Heatmap (Fig.4).
Figure 4: Heatmap “Sender-Ligands”.
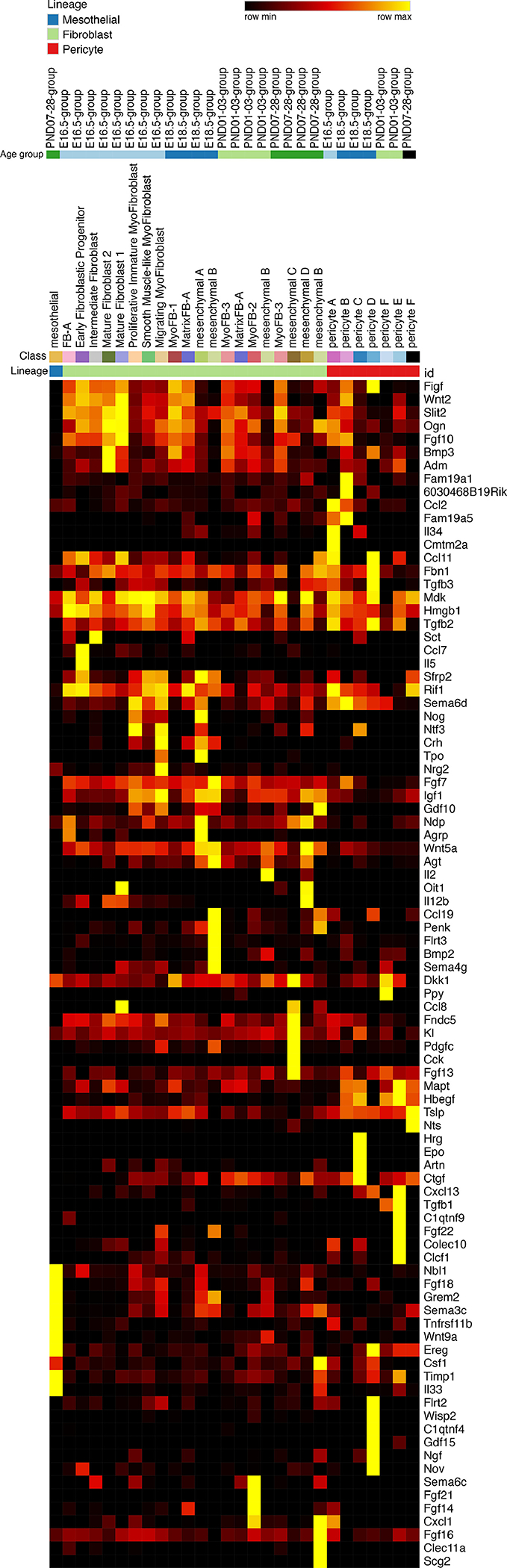
Expression of signaling pathway ligands in mesenchymal populations identified by single cell RNA-Seq heatmap: These ligands are the highest and most specifically expressed in the mesenchyme compared to all other cell types. Ligands were placed on the y-axis of a single cell RNA-seq heatmap. The x-axis was arranged by cell type, cell subtype, and age group, respectively. Yellow represents highest expression.
In adult lungs, bright and dim Pdgfra-GFP expressing fibroblasts are located in the alveolar iterstitium, and therefore called interstitial resident fibroblasts (iReFs). To distinguish them from circulating mesenchymal stem cells or other mesenchymal stromal cells (MSC) we profiled the bright and dim Pdgfra-GFP+ cells during alveolar regeneration after partial pneumonectomy by flow cytometry and by gene expression microarrays [59]. We identified that dim Pdgfra-GFP fibroblasts express CD29 (integrin beta 1) and aSma during septal regeneration and are characterized by a “contractile” gene signature (myo-iReF). Bright Pdgfra-GFP fibroblasts express CD34, a stem cell associated marker, and are characterized by a “matrix remodeling” gene signature (matrix/lipo-iReF). Other known fibroblast and MSC surface markers (SCA-1, Thy1 (CD90), CD44, and CD49, and CD40) were not selectively expressed in GFP-bright or GFP-dim fibroblasts, nor were they changed during alveolar regeneration [59]. The spatial and temporal location of Pdgfra-GFP expressing cells was mapped throughout lung development, and is available in the LungMap database. In the saccular and canalicular phase, peribronchiolar Pdgfra-GFP+ smooth muscle cells are localized in close proximity to Sox2 bronchiolar epithelial cells. Pdgfra-GFP+/ αSma neg fibroblasts are located near HOPX-positive cells in the bronchiolar-alveolar transition zone. Pdgfra-GFP+ cells were never found in close proximity to Sox9-positive alveolar epithelial progenitor cells. Gene expression profiling was performed on key developmental stages in Pdgfra expressing fibroblasts, isolated with the CD140 (Pdgfra) antibody. Pdgfra (CD140) cells dynamically change gene expression between the four developmental timepoints that were measured [44, 57]. This gene expression data can be accessed and interrogated on the Lung Gene Expression Analysis open source Web Portal [64]
These studies support that Pdgfra expressing fibroblasts are neither a single homogenous population nor a distinct lineage, but are a diverse range of fibroblastic subtypes, including myo, matrix and lipo interstitial resident fibroblasts. During alveolar septation, the contribution of myo, matrix and lipo fibroblasts to the Pdgfra expressing fibroblast pool changes dynamically, reflecting the functional changes fibroblasts have to undergo to support 1) elongating septal tips (myo-FB), 2) building structural support (matrix-FB), and 3) supporting alveolar epithelial cells (lipo-FB).
The role of Pdgfra-GFP+ lung fibroblasts in alveolar development was assessed in organoid culture with primary lung epithelial cells. A mixed population of dim and bright Pdgfra-GFP+ fibroblasts support alveolar organoid formation and epithelial differentiation, while Pdgfraneg lung fibroblasts supported bronchiolar organoids and epithelial differentiation [65]. These findings may help explain unresolved pathologies in neonatal diseases where alveolarization is disturbed. Premature birth and perinatal ventilation causes an arrest and/or delay of alveologenesis, resulting in bronchopulmonary dysplasia (BPD)[66]. PDGFRA expression is reduced in BPD patients in whole lung homogenate and isolated mesenchyme [44, 67]. Expectedly, In a neonatal murine hyperoxia model of BPD, Pdgfra lineage-derived cells decreased and did not substantially contribute to pathological myofibroblasts [63]. Alternatively, in adult mouse models of lung injury, Pdgfra lineage labeled cells contribute to pathological myofibroblasts and fibrotic scars after bleomycin injury [31, 36, 63]. The role of Pdgfr signaling in various fibrotic lung disease is currently being investigated by several groups, and we expect to learn more about the role of this fibroblast population in lung disease in the near future.
Bronchiolar and alveolar niche cells: The way the “WNT” blows
Recent studies using genetic lineage tracing, single-cell RNA sequencing, and organoid culture approaches defined bronchiolar and alveolar niche fibroblasts and their role in regeneration [35, 36].
Lgr5/Lgr6:
Leucine Rich Repeat Containing G Protein-Coupled Receptor 5 and 6 (Lgr5, Lgr6) are well known markers of epithelial stem cells in other organs but have otherwise not been described to have any function in mesenchymal cell development or maintenance [68]. Lgr5 and 6 bind R-spondin, allowing for b-catenin to be released from the membrane, enter the nucleus, and activate Wnt responsive genes [69]. In adult lungs, Wnt-responsive Lgr5 lineage-labeled cells are mostly aSmaneg and constitute only 1.24% ± 0.42% of resident alveolar fibroblasts [35]. Combining FACS isolated Lgr5 lung fibroblasts with primary murine lung epithelium directs differentiation of alveolar organoids through secretion of Wnt3a [35]. However, Lgr5 is not exclusive to adult alveolar fibroblasts. Based on single-cell RNA-seq datasets from developing murine lungs, Lgr5 is expressed in many cell types during early lung development and only highly expressed in transient myofibroblasts at PN7. Lgr5 expression levels go undetected by PN28, with the exception of expression in macrophages. Bulk RNA-seq on sorted mesenchyme shows peak expression of Lgr5 at E18.5, which remains higher than in other cell types after birth [64]. In adult lungs, Wnt-responsive Lgr6 lineage-labeled cells are bronchiolar smooth muscle cells that underlie SCGB1A1+ Club cells [35]. Combining FACS isolated Lgr6 lung fibroblasts with primary murine lung epithelium directs differentiation of bronchiolar epithelium through secretion of Wnt7b [35]. During lung development Lgr6 is expressed in myofibroblasts and SMC progenitors at E16.5, is turned off by E18.5, and expression peaks in myofibroblasts postnatally [64]. These Lgr6 bronchiolar fibroblasts very likely are the same “niche cells” that support bronchiolar club cell regeneration after naphthalene injury via reactivation of a Wnt/Fgf10 embryonic signaling cascade [70]. To compare Lgr5 and Lgr6 expression within the stromal cells over time please refer to the “ligand” Heatmap (Fig.4).
MANCs and AMPs:
Two other populations of adult alveolar niche cells, mesenchymal alveolar niche cells (MANCs) and axin2+ myofibrogenic progenitor (AMPs), were identified using single-cell RNA-seq, signaling lineage reporters and organoid cultures [36]. MANCs support alveolar growth and regeneration, and AMPs contribute to peribronchiolar smooth muscle and pathological myofibroblast response after injury. An Axin2CreERT2:tdT reporter line [71], a Wnt2CreERT2 line [24], and the Pdgfra-GFP reporter line [52] were used to lineage label and FACS isolate these specific cell populations. Wnt-responsive Axin2+ fibroblasts were located in the alveolar regions, and surrounding the conducting airways and blood vessels. Axin2 lineage-traced cells around airways primarily expressed Pdgfrb, identifying them as pericytes. In the alveolar cells Pdgfra-GFP was co-expressed in 74% of the Axin2 lineage-traced and 90% of the Wnt2 lineage-traced cells. Based on these studies, the previously described iRefs can be subdivided into three subpopulations: Axin2+/Pdgfra-GFP+, Wnt2+/Pdgfra-GFP+ and Pdgfra-GFP+. Each of these linage traced fibroblast populations were isolated by flow cytometry and were bulk and scRNA-sequenced. The Axin2+ single positive peribronchiolar cells expressed genes associated with “smooth muscle”, “myofibroblast” or “pericyte-like cells”, but not “contractile” proteins, suggesting that these cells are primed but not yet committed to these differentiated lineages. In contrast, Axin2+/Pdgfra-GFP+, Wnt2+ and Pdgfra-GFP+ cells expressed genes associated with a matrix fibroblast phenotype.
When combined with alveolar epithelial cells the Axin2+/Pdgfra-GFP+ fibroblasts generated the most and largest organoids, which also contained the highest AT1/AT2 ratio. To identify which of the fibroblast populations contribute to fibrosis, Axin2CreERT2:tdT, PdgfraCreERT2 and Wnt2CreERT2 were crossed to R26REYFP and lineage labeled prior to bleomycin injury. All three lineages generated αSma+ cells; the Axin2 lineage contributed 50% of the myofibroblasts, and less than 20% were derived from either the Wnt2 or Pdgfra lineages. Many myofibroblasts maintained expression of Axin2 but expressed little or no Pdgfra. After naphthalene injury and repair, 50% of peribronchial Axin2+ cells acquired αSma expression. These data demonstrate that the Axin2+/Pdgfra-GFP+ cells support the alveolar niche and transcriptionally overlap with the Lgr5+ population, and the Axin2+ peribronchiolar cells contain smooth muscle progenitors that contribute to pathological injury repair and overlap with the Lgr6+ population. A similar bleomycine injury study associated additional markers of the canonical Wnt signaling with these fibroblasts [33].
While these studies provided evidence for the alveolar niche cell and various functional fibroblast stages, they did not give insight into the developmental origin of these cells. Crossing multiple inducible driver lines Fgf10CreERT2, Wt1CreERT2, Gli1CreERT2, with a tdTomatoflox reporter line lineage revealed thatFgf10+ and WT1+ cells show a minor contribution to the smooth muscle cells, while GLI1+ and AXIN2+ cells significantly contribute to smooth muscle cells and interstitial myofibroblasts [43]. taken together, Wnt-responsive cells labeled during development or in the adult contribute to smooth muscle populations that support the bronchiolar epithelial niche, and resident interstitial fibroblasts that support the alveolar epithelial niche.
Lipofibroblasts:
Lipofibroblasts are a specific subtype of mesenchymal cells that contain lipid vesicles and support the alveolar epithelium during development, regeneration, and homeostasis [65, 72–74]. Expression of adipose differentiation-related protein (ADRP, encoded by Plin2) enables fibroblasts to take up, store, and subsequently transfer Triglycerides to neighboring alveolar type 2 cells to be incorporated into surfactant phospholipids[75, 76]. The previously discussed alveolar niche cells do not necessarily have these lipid inclusions [17, 36]. Activation of peroxisome proliferator-activated receptor-γ (PPARγ) by rosiglitazone and metformin [77, 78] or stimulation by Fgf2 [34] induced lipofibroblast differentiation. The lineage origin of lipofibroblasts is still poorly defined, as only common adipocyte genes were used to label and trace them, these markers include Pparg, Plin2, Fabp1, Fabp4, Fabp5, Lpl, and Lipa, [28, 29, 79, 80].
Thy1 expression and lipofibroblasts in fibrosis
Thymocyte differentiation antigen-1 (THY1), also known as CD90, expressing fibroblasts were characterized as 1) having increased expression of αSma and Collagen, 2) responding differently to cytokines and growth factors, and 3) showing increased migration patterns compared to other pulmonary mesenchymal cells [28, 81–85]. Loss of Thy1 impairs postnatal alveolar septation [86]. Further phenotyping revealed that Thy-1+ fibroblasts cells expresse Adrp, produce lipid vesicles, and are located next to AT2 cells. Moreover, transfection of Thy-1 into Thy-1neg fibroblasts induced expression of the transcription factor PPARγ, which directly induces lipofibroblast differentiation, inhibits collagen matrix contraction, and reduces cell survival [28, 87]. scRNA-seq confirmed that Thy-1 is expressed in lipofibroblasts during alveolarization, but lost in adult lipofibroblasts [33].
FACS analysis revealed that Thy-1+ and Thy-1neg fibroblasts exist as stable populations in the adult rat lung [87]. Thy-1neg cells have significantly higher myofibroblast and myogenic regulatory factor gene and protein expression compared with Thy-1+ cells [87]. Thy-1neg myofibroblasts accumulate in lungs with idiopathic pulmonary fibrosis (IPF) and expand in bleomycin-induced lung injury [83, 85].Therefore, the absence of Thy1 was associated with a higher susceptibility for lung fibrosis [83]. However, after bleomycin injury, chimeric Thy1−/− mice with Thy-1+ lymphocytes and Thy-1neg myofibroblasts showed fibrosis similar to wild-type mice, but decreased inflammation [83]. These data demonstrate that inflammation is not essential for evolution of fibrosis and lack of Thy-1 in fibroblasts is not protective [83]. However, regions of active fibrosis in IPF lose expression of Thy-1 [83]. Thy 1 was determined to be the control of mechanotransduction, via inactivating αvβ3 integrin, controlling cell contraction and force-induced Rho signaling [88, 89]. Targeting this integrin with a soluble Thy-1 recombinant protein is sufficient to halt initiation of fibrosis [85]. These data suggest that adult Thy-1+ cells may be better described as mechanosensory niche cells, as their role in IPF progression suggests [83, 85, 88, 89]
FGF signaling and fibroblasts
Fibroblast growth factor 10 (Fgf10) is expressed in the early splanchnic mesoderm surrounding the foregut around E9.5 when the primary lung buds start to emerge. The splanchnic mesoderm expresses the transcription factor Tbx4 and also contains the multipotent cardiopulmonary progenitors Gli1+Wnt2+Isl1+ [24, 90, 91]. The Fgf10 expressing fibroblasts are critical for maintaining epithelial progenitor cell proliferation during pseudoglandular and canalicular stages (E9.5-E16.5), as Fgf10 knockout phenocopies Fgfr2b KO, halting lung development entirely [92–94]. Fgf10 remains expressed in the distal tip mesenchyme, initiating proliferation and halting lineage commitment in the distal-most epithelium [95]. During homeostasis, Fgf10 is expressed in mesenchymal stromal niches, between cartilage rings in the upper conducting airway, driving submucosal gland (SMG) and basal cell development and maintenance [29, 95–98]. Fgf10+ fibroblasts give rise to and coordinate formation of peribronchiolar and perivascular smooth muscle. Later in development, they give rise to alveolar myofibroblasts and lipofibroblasts [29, 43, 94, 99]. Alveolar fibroblasts that express Fgf10 are a heterogenous mixture of Pdgfra+ and Gli1+ cells, but most Fgf10+ fibroblasts are lipofibroblasts adjacent to AT2 cells [29, 100]. These cells support AT2 self-renewal in homeostasis and injury, and may overlap with the MANCs and Lgr5+ niche cells [17, 35, 36]. Lineage-labeled Fgf10+ lipofibroblasts differentiate into myofibroblasts and upregulate Fgf10 during injury, and then de-differentiate back into lipofibroblasts during resolution. These data propose that Fgf10 fibroblasts can switch both ways between myo- and lipo- phenotypes, and support the concept that Fgf10+ fibroblasts are niche-specific cells that provide a proper paracrine environment to the epithelium during injury and repair [78, 101]. Fgf10 expression is reduced in biopsies from BPD patients [102], and in a hyperoxia model of BPD in mice, expression of dnFgfr2 during repair inhibited epithelial repair [103, 104]. During realveolarization after perinatal dexamethasone exposure or partial pneumonectomy, and in the context of RA-induced repair, expression of dnFgfR2 inhibited myofibroblast differentiation and increased expression of Pdgfra [56, 58].
Global or mesenchyme-specific knockouts of FGFR3/4 during alveolarization lead to extensive alveolar simplification [105–107], and FGF18 is specifically upregulated during rat and human alveologenesis [108–110]. In a recent study, labeling FGF18-expressing cells from PN5 to PN8 in the lung using an FGF18CreERT2 mouse revealed that 14% of alveolar myofibroblasts express FGF18 during alveologenesis (PN9) [111, 112]. Amongst this population, ~72% also expressed aSMA and PDGFRa. By scRNA-seq, high FGF18 expression is correlated with high ACTA2-expression in myofibroblasts. At the end of alveolarization at P21, ~88% of these myofibroblasts are lost. Some fluorescent particles were found in immune cells, suggesting that myofibroblasts are cleared by immune-mediated phagocytosis. Notably, ~23% of HOPX+ AT1 cells and 65% of WT1+ mesothelial cells were labeled with FGF18 at PN9, suggesting the FGF18 expression is not exclusive in interstitial lung fibroblasts.
A combination of GLI1-LacZ with FGF18CreERT2 and lineage labeling from PN5–8 revealed that the majority (77%) of PFGF18-labeled cells co-expressed Gli-LacZ, indicating that these populations largely overlap. However, lineage tracing using Gli1CreERT2 revealed that less Gli1 cells than FGF18 cells were lost by PN21, and many that the remaining Gli1 cells were alveolar lipofibroblasts. These data suggest that lipofibroblasts labeled early in development are more permanent than previously believed and demonstrate an active role of myofibroblast-directed FGF18 signaling. Important basic developmental experiments determining the autocrine vs. paracrine effects of FGF18 on the various cells involved in alveologenesis remains as an exciting future direction [111].
Taken together Fgf signaling in combination with wnt, gli and pdgfra signaling is very context dependent and critical for proper differentiation and proliferation of both myofibroblasts and lipofibroblasts during development, disease, injury and repair.
Tcf21 lineage: lipofibroblast or matrix? (the squeaky wheel gets the grease)
Transcription Factor 21 (Tcf21) is expressed in the early embryonic lung, and embryonic Tcf21 gene inactivation results in hypoplastic lungs [113]. Genetic lineage tracing using Tcf21mCrem/+ ;R26tdT/tdT transgenic mice during the embryonic phase at E11.5 in Tcf21mCrem/+ ;R26tdT/tdT mice revealed labeled lineage-traced progenies (at E18.5) in peribronchiolar and perivascular smooth muscle, Pdgfra+ fibroblasts and LipidTOX+ fibroblasts [30]. Tracing Tcf21 expressing cells during the pseudoglandular stage at E15.5 traced less than 1% αSma+ but 89.4% Adrp+ distal fibroblasts, suggesting that lipofibroblast specification occurs during the saccular phase in TCF21 expressing cells, and these cells are no longer precursors to myofibroblasts or smooth muscle cells. In adult lungs, Tcf21 expression is detected in Pdgfra+ lipofibroblasts [114]. Pdgfra expressing cells were analyzed to determine the overlap lipofibroblasts and myofibroblasts within the Tcf21 expressing population during development. Tracing Tcf21 expressing cells during the pseudoglandular stage at E15.5 showed that in 25% of the Tcf21+ traced cells express Pdgfra-GFP+ and that 30% of the Pdgfra-GFP+ expressing cells were traced by Tcf21+ [30]. These data demonstrate that at E18.5 there are three non-overlapping types of cells Tcf21/Pdgfra-GFP, Tcf21 and Pdgfra-GFP. As the majority of progeny of the Tcf21 cells expressed the lipofibroblast marker ADRP, these progenitor cells captured at E15.5 give raise to lipofibroblasts. These data predict that the Pdgfra-GFP+/Tcf21neg cells are the progenitors for the interstitial myofibroblasts, which has not been tested. Nonetheless, this clearly demonstrates that some Tcf21+ cells become a fibroblast population distinct from smooth muscle and Pdgfra expressing cells.
Recent single cell transcriptome analysis suggest that active Tcf21 expression is strongly associated with Col13a1 expression in the adult lung [33]. Tcf21neg lineage lipofibroblasts but not Tcf21+ lipofibroblast expressed high levels of Col13a1 at PN7. However, using a Ribotag mouse to isolate ribosomal-associated transcripts from the Tcf21 lineage, they demonstrated that Col13a1 and Plin2 were enriched in perinatal and adult Tcf21 lineages, while Fgf10 and Zfp423 were only enriched perinatally and Thy1 and desmin were only enriched in postnatal fibroblasts. These data suggest that the prenatal (E15.5) and postnatal (PN2) Tcf21-labeled cells are progenitor cells of two distinct lipofibroblasts, one with a more matrix function than the other. To compare Tcf21 expression within the stromal cells over time please refer to the “Transcription factor” Heatmap (Fig.3).
Heterogeneity of fibroblasts in bleomycin induced fibrosis was assessed and two distinct matrix fibroblast populations were characterized based on gene signatures associated with ECM and adhesion and discerned by Col13a1 and Col14a1 expression [33]. The Col13a1 matrix fibroblasts also expressed Itga8, Cxcl14, Npnt, and Tcf21, bringing them in close relationship to lipofibroblasts. As lipofibroblasts are associated with alveolar maintenance and repair [35, 36], the increase in Col13a1+ fibroblasts during bleomycin injury and repair may represent a regenerative population that is present 21 days after injury. Developmental studies on P7 murine lungs validated expression of Col13A1 in the lipofibroblast population, in both lineage traced TCF21 progeny and cells that co-expressed TCF21 [30]. At PN7, the peak of alveolarization is a timepoint when fibroblast would have to switch to a matrix phenotype to lay down the scaffold in the newly forming septae [30]. Developmentally, Col13a1+ and Col14a1+ matrix fibroblasts have yet to be further defined, but according to the LGEA scRNA-seq dataset, they are expressed in distinct subsets of fibroblasts from E16.5-P28.
Matrix Fibroblasts:
Until recently, matrix fibroblasts have not gained much attention and were always grouped in with myofibroblasts and “activated”-pathological fibroblasts. Recent single cell seq analysis clearly identified several lung matrix fibroblasts, but their developmental and/org regenerative role and spatial location still remain elusive [33, 63, 64, 115–117]. During lung development, matrix producing and modulating cells are important to create the gracile scaffold that holds up the lung parenchyma, like poles and beams hold up the circus tent or Renaissance-age dome structures. Based on transcriptional profiles, matrix fibroblasts are more specialized to form and modulate the ECM during development and repair, while myofibroblasts still produce matrix but also contract and provide tensile strength to the alveolus [33, 59]. PDGFRa-GFP transgenic mice have been used to characterize determine interstitial myo, lipo and matrix fibroblasts all of which express PDGFRa-GFP but whether these are independent cell populations cell stages of the same populations remains to be determined [55, 59, 114, 118–121]. Intensity of the GFP signal has been used to distinguish different populations [43, 44, 56, 58, 59]. Developmental studies demonstrate that the GFP-dim cells become myofibroblast during active septation [55, 114, 118–121]. However, PDGFRa-GFP+ bright fibroblasts from adult mice expressed a matrix remodeling and matrix synthesizing gene expression profile, while PDGFRa-GFP+ dim fibroblasts expressed a contractile and smooth muscle phenotype [58, 59]. Further analysis of the dataset revealed that Pdgfra+/CD29+ dual positive cells were myofibroblasts, while Pdgfra+/CD34+ dual positive cells were matrix fibroblasts [59]. The same study showed, that increased Pdgfra kinase activity promotes the matrix fibroblast phenotype. In IPF, PDGFRa-bright matrix fibroblasts are lost, and when returned to recombinant mouse-human organoids, can direct regeneration of aged epithelium into healthy epithelium [122]. Thus, the difference between a myofibroblast and a matrix fibroblast during lung regeneration might be a cell stage regulated by the amount of Pdgfra signaling input on the fibroblast.
Pericytes: Free hugs, nothing is free
Pericytes are vascular smooth muscle (VSMC) related mesenchymal cells that lie within the capillary basement membrane and closely neighbor endothelial cells [51, 123]. In the lung, pericytes adhere to the basement membrane and tightly to the endothelium through gap, tight, and adherence junctions, correlating closely with the strong barrier and low turnover rate of pulmonary endothelium [124]. Pericytes are generally defined by surface expression of Pdgfrb, Pdgfra, CD146, ABCG2, Ng2, αSma, Collagen1a1, vimentin, and desmin; the most specific marker is Pdgfrb [31, 32, 124–127]. Multiple reports suggest that NG2+ pericytes arise from a transient mesenchymal pool present in the distal lung buds from E9.5 TBX4+ progenitors [25] or E11-E13, expressing transcription factor FoxD1 [32]. Lineage tracing using Tbx4, by either labeling throughout embryogenesis or only after embryonic day 15.5, gave raise to Ng2+/ αSma neg pericytes, smooth muscle cells, rare endothelial cells, myofibroblasts, and alveolar lipofibroblasts [20]. Less than 60% of pericytes express NG2 [32]. Approximately 80% of the FoxD1-traced cells did not express Col1a1 or αSma, but fit the definition of pericytes: stellate-shaped with long cell processes, attached to endothelial cells, separated from the airways, and expressing Pdgfr-b [32, 51]. Also, a Pdgfrβ+/Ng2+/ αSma neg perivascular population arose from NG2-traced cells [31]. Foxd1-expressing progenitors also labeled Ng2+/ αSma neg pericyte. Pdgfra-GFP tracing identified two Pdgfra-GFP expressing “pericyte” populations, which are located perivascular or peribronchiolar and co-express Pdgfrβ but not Ng2 (CSPG4) and stained variably for αSma [44]. Lineage-traced Gli1+ perivascular fibroblasts express Pdgfrβ but not Ng2, and can contribute to fibrotic myofibroblasts and Ng2+ cells (pericytes) after injury [26].
Mesenchymal Stem Cells, Mesenchymal Stromal Cells, Mesenchymal progenitor cells: More of the Same?
Stem cells antigen 1 (Sca-1) is expressed on many different cell types in the lung. Excluding CD45+, CD31+, and CD326+ cells the role of Sca1- positive fibroblasts in lung development and regeneration was assessed [128–130]. Multiple groups reported that CD45negCD31negSca-1+ cells had mesenchymal characteristics in the adult lung but their origin and function remained unclear [131, 132]. A systematic analysis of the cellular identity and composition of CD45negCD31negSca-1+CD34+ cells in neonatal and adult lungs supported that these postnatally-arriving cells were MSCs [34]. The CD45negCD31negSca-1+ fraction uniformly expressed Pdgfra and CD34, while a portion of these cells expressed Thy-1. Isolated and plated CD45negCD31negSca-1+Thy-1high cells preferentially differentiated into lipofibroblasts, while CD45negCD31negSca-1+Thy-1low cells differentiated into microtubule-forming non-lipofibroblasts. During alveolar regeneration no changes of proliferation, differentiation or expression of CD34 or CD29 was detected in the Sca-1 populations [58, 59]. In the adult a portion of Fgf10+ cells are Sca-1+, suggesting that MSCs fit the descriptions of a lipofibroblast [29]. SCA-1 immunoreactivity in the distal lung is predominantly restricted to endothelial and perivascular cells.
A “mesenchymal progenitor cell” (MPC), labeled by ABCG2 in adult lungs falls into the MSC category due to its bipotency in pulmonary angiogenesis [127]. These cells contributed to vascular homeostasis, and when challenged with fibrosis-inducing bleomycin or depletion of BMP/stabilization of β catenin (both increasing WNT signaling input), these cells proliferated and improperly differentiated. Bleomycin caused the cells to upregulate aSMA and differentiate into a perivascular myofibroblast. Increased WNT caused the cells to downregulate aSMA and proliferate, expanding into immature pericytes that contributed to vascular leak and reduced vascular contractility. ABCG2+ cells did not directly contribute to fibrotic lesions, but contributed to abnormally stiff, aberrant vessels nearby. These results suggest that ABCG2+ cells have, bipotency, and contribute to proper perivascular maintenance [127]. Further studies are required to determine if these cells overlap with the aforementioned Gli1+ perivascular stem cells and Sca-1 stem cells [26, 133].
Mesothelium; the thin lining
Wilm’s Tumor 1 (wt1) is a tumor suppressor gene that, when mutated, contributes to the development of Wilm’s Tumor disease progression in the kidney of children [134]. Wt1 is highly expressed in the mesothelium lining the lung pleurae, peritoneum, and pericardium, and increases in mesothelioma [135]. During development, Wt1 regulates the development of the mesothelium. The mesothelium releases Fgf9, stimulating Fgf10 expression in the mesenchyme and subsequent epithelial proliferation and lung branching [10, 136]. Deletion of Fgf9 in the Wt1 lineage resulted in loss of Wnt2a expression in the mesenchyme and decreased airway branching [137]. Lineage tracing using a Wt1-Cre; LacZ revealed that mesothelial WT1+ cells contribute to smooth muscle and other undefined fibroblasts surrounding pulmonary vasculature [38]. In IPF, sub-pleural fibrosis is an early indication of disease progression, and studies have shown that both WT1+ mesothelial cells and WT1− mesenchyme contribute to lesions [138, 139]. Recent scRNA-seq and validating RNA in-situ hybridization studies demonstrate that HAS1hi fibroblasts (hyaluronan synthase) exist in the subpleural regions of the lung, and express WT1. [140]. In IPF these HAS1hi fibroblasts also express COL1A1, and markers of epithelial mesenchymal transition (TWIST, SNAI1) and are immune responsive (IL4/IL13 signaling). It remains to be seen if HAS1hi cells arose from lineage-negative or lineage-positive WT1 cells, and whether the WT1 transcription factor regulates expression of HAS1. Future lineage tracing and genetic epistasis studies could reveal how these subpleural and mesothelial cell subsets contribute to IPF.
Smooth Muscle Cells
In the lung, smooth muscle cells (SMC) form circular peribronchiolar structures and mesh like perivascular structures, reflecting different mechanical requirements in the lumen of the bronchioles and vessels [44, 57, 141, 142]. During lung development, airway SMCs form adjacent to the proximal Sox2 positive epithelial cells but not next to Hopx+ epithelium in the bronchio-alveolar transition zone [44, 57, 143, 144]. Airway smooth muscle progenitors map exclusively to mesenchyme ahead of budding airways. Progenitors recruited from these tip pools differentiate into SMC around airway stalks. Mesenchyme flanking the bronchiolar stalk can be induced to form SMC by focal Wnt signal from epithelial lung tips and lateral buds [40]. The earliest lineage labels of SMCs are TCF21 and PDGFRa at E9.5 [30, 60, 101]; Gli1, WT1, FGF10, and TBX4 at E10.5 [38, 40, 41, 99]; and FGF10 and PDGFRb at E11.5 [29, 37], suggesting that SMCs are committed early during development. Later developmental lineage traces show that SMCs arise from E18.5 SCA-1 cells [34] and P1–6 Gli1 cells [41, 42]. In the adult lung, the smooth muscle layer contains the Axin2+ peribronchiolar AMPs and PDGFRb+ Lgr6 cells, which are still Wnt responsive and can contribute to pathological repair after injury [35, 36]. However, Wnt2 is not required for the differentiation of early mesenchyme into vascular SMCs, supporting lineage restrictions in SMC differentiation early in lung development [5].
The Predictive Power of Single Cell Sequencing in Lung Lineage
The field of lung fibroblast biology has benefited greatly from classical lineage tracing experiments but is now on the cusp of a new era of lineage information supplied by scRNA sequencing. Gene expression data from fibroblast populations inherently contain more noise than data sets from epithelial populations [64, 115, 145, 146]. Lung epithelial cells make fate decisions early and can be grouped by hallmark gene expression into relatively non-overlapping subpopulations, thus incredibly amenable to scRNA-seq analysis [64, 115, 145, 146]. Fibroblasts respond dynamically to environmental cues such as ECM stiffness, paracrine ligands, and pH, directly altering gene expression and function. Thus, fibroblasts exist more on a spectrum of gene expression rather than in individual populations, and are therefore perfectly situated for trajectory inference algorithms like pseudo time analysis [116]. Pseudo time analyses use scRNA-seq datasets to reconstruct continuous or branching cell-state transitions from cells that have similar gene expression profiles. Thus, variations in gene expression relating to gene-ontological-categorized cellular processes can be utilized to predict a cell population’s lineage. Some of these programs include Monocle 1, Waterfall, Wanderlust, and TSCAN [147]. More advanced programs like Monocle 2, Wishbone, DPT, SLICE, SLINGSHOT, and DPT can predict multiple branch points [147]. These advanced algorithms can only predict lineage relationships which still need to be validated by genetic lineage labeling studies. Moreover, identifying the spatial and also temporal location of these predicted populations will give valuable insight in their function and role in development, repair and fibrosis. Thus, we are not ready to leave behind lineage tracing and histology, but rather expand our toolbox and incorporate sequencing technology and prediction algorithms to better understand the elusive lung fibroblast.
Bring on the heat:
In order to compare and contrast lineages and identify potential overlap due to expression of the same lineage markers at different time points, we subset a single cell dataset from LungMAP with fourteen “lineage tracer” genes as guides. The resulting heatmap visualize the dynamic expression patterns of presumable lineage markers all through developmental timepoints (E16.5-PN28) and demonstrate the overlap of gene expression between fibroblast populations and expression in non-mesenchymal celltypes. We re-clustered all stromal cells by gene expression and sorted for expression of transcription factors, signaling pathway ligands and receptors (Fig. 2–5). These heatmaps, “lineage”, “transcription factor”, “sender/ligand” and “receiver/receptor” provide a resourceful list to correlate one’s own experimental data with fibroblast lineages and to identify any lineage’s temporal lineage mark, transcriptional activators and how they receive and respond in their microenvironmental niche. The whole dataset can be interactively interrogated in the “toppcell” suite using this hyperlink: https://toppcell.cchmc.org/biosystems/go/index3/shred/LungMap/Output%20by%20Lineage%20by%20Class%20by%20Age%20group%20by%20Age
Figure 5: Heatmap “receiver-Receptors”.
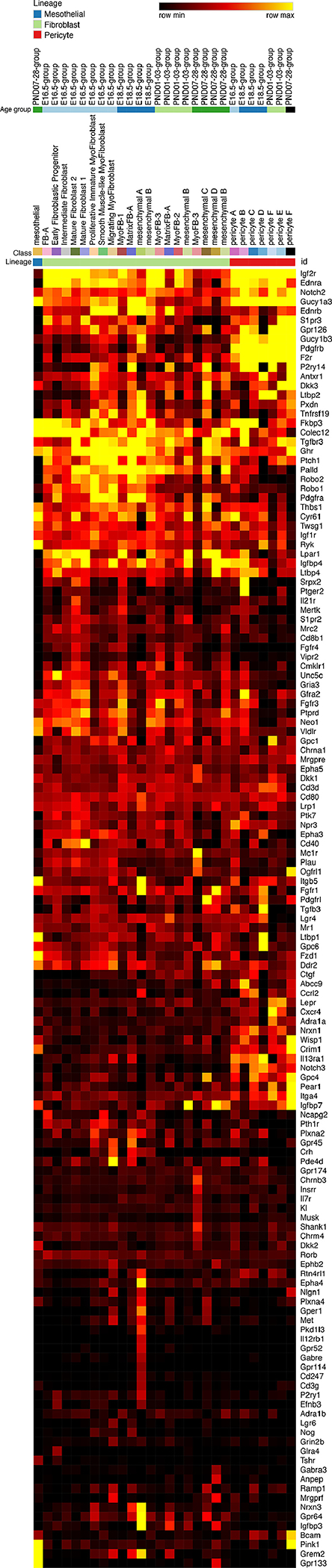
Expression of signaling pathway receptor in mesenchymal populations identified by single cell RNA-Seq heatmap: These receptors are the highest and most specifically expressed in the mesenchyme compared to all other cell types. Signaling pathway receptors were placed on the y-axis of a scRNA-seq heatmap. The x-axis was arranged by cell type, cell subtype, and age group, respectively. Yellow represents highest expression.
Conclusion
Fibroblasts are much more plastic than epithelial cells. Epithelial cells tend to make a fate decision and then go either forward or backwards on that track, whereas the fibroblast continuously senses the environment and adapts its role, function, and activation status accordingly. Being in a functional stage and not a defined lineage probably allows the fibroblast to quickly adapt to dynamically changing signals from the environment (paracrine signals, mechanical tension, loss of epithelial cells, stress of epithelial cells, inflammation). Epithelial cells for the most part have a well-defined role in their niche and fibroblasts support that niche. During epithelial stress or injury, fibroblasts become activated to support the epithelial regeneration [148, 149]. This activation for the most part is an augmentation of their function rather than a trans differentiation [21, 117, 150]. It is well accepted in the field that fibroblast activation is characterized by increase of proliferation, migration, αSma expression, matrix degradation and matrix synthesis [151–153]. Fibroblasts lose some of their plasticity with chronic stress, repeated injury and advanced age and become shunted toward myofibroblast differentiation and excessive ECM producing cells [82, 122, 154, 155]. This loss of the plasticity results in maladaptive repair and regeneration [82, 122, 156, 157]. A recently published single cell RNAseq dataset from normal and bleomycin injured lung fibroblast identified that bleomycin induced fibrosis activated fibroblasts by augmenting a signature found in normal lung fibroblasts and not inducing a new “fibrosis” signature [21]. These data imply that activated fibroblasts are more proliferative, migratory, invading, contractile, express more matrix, or store more lipid. A recent single cell RNAseq analysis from normal, IPF and CODP lungs the origins of the activated IPF myofibroblast was inferred to be the extreme pole of a continuum of a contractile fibroblast found in a normal lung [117]. So, the question remains: “Is there a fibroblast lineage?” The answer is that some lineages exist, but most of what we see is a functional stage with variable degree of plasticity.
Highlights.
lung fibroblast populations and lineages
temporal gene expression profile in fibroblast population
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- [1].Shannon JM, Induction of alveolar type II cell differentiation in fetal tracheal epithelium by grafted distal lung mesenchyme, Dev Biol 166(2) (1994) 600–14. [DOI] [PubMed] [Google Scholar]
- [2].Serls AE, Doherty S, Parvatiyar P, Wells JM, Deutsch GH, Different thresholds of fibroblast growth factors pattern the ventral foregut into liver and lung, Development 132(1) (2005) 35–47. [DOI] [PubMed] [Google Scholar]
- [3].McCulley D, Wienhold M, Sun X, The pulmonary mesenchyme directs lung development, Curr Opin Genet Dev 32 (2015) 98–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Que J, Okubo T, Goldenring JR, Nam KT, Kurotani R, Morrisey EE, Taranova O, Pevny LH, Hogan BL, Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm, Development 134(13) (2007) 2521–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Goss AM, Tian Y, Cheng L, Yang J, Zhou D, Cohen ED, Morrisey EE, Wnt2 signaling is necessary and sufficient to activate the airway smooth muscle program in the lung by regulating myocardin/Mrtf-B and Fgf10 expression, Dev Biol 356(2) (2011) 541–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Harris-Johnson KS, Domyan ET, Vezina CM, Sun X, beta-Catenin promotes respiratory progenitor identity in mouse foregut, Proc Natl Acad Sci U S A 106(38) (2009) 16287–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Okubo T, Hogan BL, Hyperactive Wnt signaling changes the developmental potential of embryonic lung endoderm, J Biol 3(3) (2004) 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Domyan ET, Sun X, Patterning and plasticity in development of the respiratory lineage, Dev Dyn 240(3) (2011) 477–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Que J, Luo X, Schwartz RJ, Hogan BL, Multiple roles for Sox2 in the developing and adult mouse trachea, Development 136(11) (2009) 1899–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Weaver M, Dunn NR, Hogan BL, Bmp4 and Fgf10 play opposing roles during lung bud morphogenesis, Development 127(12) (2000) 2695–704. [DOI] [PubMed] [Google Scholar]
- [11].Chen F, Desai TJ, Qian J, Niederreither K, Lu J, Cardoso WV, Inhibition of Tgf beta signaling by endogenous retinoic acid is essential for primary lung bud induction, Development 134(16) (2007) 2969–79. [DOI] [PubMed] [Google Scholar]
- [12].Tsao PN, Vasconcelos M, Izvolsky KI, Qian J, Lu J, Cardoso WV, Notch signaling controls the balance of ciliated and secretory cell fates in developing airways, Development 136(13) (2009) 2297–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Perl AK, Wert SE, Nagy A, Lobe CG, Whitsett JA, Early restriction of peripheral and proximal cell lineages during formation of the lung, Proc Natl Acad Sci U S A 99(16) (2002) 10482–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Rawlins EL, Clark CP, Xue Y, Hogan BL, The Id2+ distal tip lung epithelium contains individual multipotent embryonic progenitor cells, Development 136(22) (2009) 3741–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rawlins EL, Perl AK, The a”MAZE”ing world of lung-specific transgenic mice, Am J Respir Cell Mol Biol 46(3) (2012) 269–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Vaughan AE, Brumwell AN, Xi Y, Gotts JE, Brownfield DG, Treutlein B, Tan K, Tan V, Liu FC, Looney MR, Matthay MA, Rock JR, Chapman HA, Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury, Nature 517(7536) (2015) 621–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zacharias WJ, Frank DB, Zepp JA, Morley MP, Alkhaleel FA, Kong J, Zhou S, Cantu E, Morrisey EE, Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor, Nature 555(7695) (2018) 251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Jacob A, Morley M, Hawkins F, McCauley KB, Jean JC, Heins H, Na CL, Weaver TE, Vedaie M, Hurley K, Hinds A, Russo SJ, Kook S, Zacharias W, Ochs M, Traber K, Quinton LJ, Crane A, Davis BR, White FV, Wambach J, Whitsett JA, Cole FS, Morrisey EE, Guttentag SH, Beers MF, Kotton DN, Differentiation of Human Pluripotent Stem Cells into Functional Lung Alveolar Epithelial Cells, Cell Stem Cell 21(4) (2017) 472–488 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Li L, Miano JM, Cserjesi P, Olson EN, SM22 alpha, a marker of adult smooth muscle, is expressed in multiple myogenic lineages during embryogenesis, Circ Res 78(2) (1996) 188–95. [DOI] [PubMed] [Google Scholar]
- [20].Xie T, Liang J, Liu N, Huan C, Zhang Y, Liu W, Kumar M, Xiao R, D’Armiento J, Metzger D, Chambon P, Papaioannou VE, Stripp BR, Jiang D, Noble PW, Transcription factor TBX4 regulates myofibroblast accumulation and lung fibrosis, J Clin Invest 126(9) (2016) 3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Peyser R, MacDonnell S, Gao Y, Cheng L, Kim Y, Kaplan T, Ruan Q, Wei Y, Ni M, Adler C, Zhang W, Devalaraja-Narashimha K, Grindley J, Halasz G, Morton L, Defining the Activated Fibroblast Population in Lung Fibrosis Using Single-Cell Sequencing, Am J Respir Cell Mol Biol 61(1) (2019) 74–85. [DOI] [PubMed] [Google Scholar]
- [22].Lu Han TN, Aaron M Zorn, Mesodermal lineages in the developing respiratory system, Trends in Developmental Biology 9 (2016) 91–110. [PMC free article] [PubMed] [Google Scholar]
- [23].Nasr T, Mancini P, Rankin SA, Edwards NA, Agricola ZN, Kenny AP, Kinney JL, Daniels K, Vardanyan J, Han L, Trisno SL, Cha SW, Wells JM, Kofron MJ, Zorn AM, Endosome-Mediated Epithelial Remodeling Downstream of Hedgehog-Gli Is Required for Tracheoesophageal Separation, Dev Cell 51(6) (2019) 665–674 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Peng T, Tian Y, Boogerd CJ, Lu MM, Kadzik RS, Stewart KM, Evans SM, Morrisey EE, Coordination of heart and lung co-development by a multipotent cardiopulmonary progenitor, Nature 500(7464) (2013) 589–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zhang W, Menke DB, Jiang M, Chen H, Warburton D, Turcatel G, Lu CH, Xu W, Luo Y, Shi W, Spatial-temporal targeting of lung-specific mesenchyme by a Tbx4 enhancer, BMC Biol 11 (2013) 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kramann R, Schneider RK, DiRocco DP, Machado F, Fleig S, Bondzie PA, Henderson JM, Ebert BL, Humphreys BD, Perivascular Gli1+ progenitors are key contributors to injury-induced organ fibrosis, Cell Stem Cell 16(1) (2015) 51–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Bostrom H, Willetts K, Pekny M, Leveen P, Lindahl P, Hedstrand H, Pekna M, Hellstrom M, Gebre-Medhin S, Schalling M, Nilsson M, Kurland S, Tornell J, Heath JK, Betsholtz C, PDGF-A signaling is a critical event in lung alveolar myofibroblast development and alveogenesis, Cell 85(6) (1996) 863–73. [DOI] [PubMed] [Google Scholar]
- [28].Varisco BM, Ambalavanan N, Whitsett JA, Hagood JS, Thy-1 signals through PPARgamma to promote lipofibroblast differentiation in the developing lung, Am J Respir Cell Mol Biol 46(6) (2012) 765–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].El Agha E, Herold S, Al Alam D, Quantius J, MacKenzie B, Carraro G, Moiseenko A, Chao CM, Minoo P, Seeger W, Bellusci S, Fgf10-positive cells represent a progenitor cell population during lung development and postnatally, Development 141(2) (2014) 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Park J, Ivey MJ, Deana Y, Riggsbee KL, Sorensen E, Schwabl V, Sjoberg C, Hjertberg T, Park GY, Swonger JM, Rosengreen T, Morty RE, Ahlbrecht K, Tallquist MD, The Tcf21 lineage constitutes the lung lipofibroblast population, Am J Physiol Lung Cell Mol Physiol 316(5) (2019) L872–L885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Rock JR, Barkauskas CE, Cronce MJ, Xue Y, Harris JR, Liang J, Noble PW, Hogan BL, Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition, Proc Natl Acad Sci U S A 108(52) (2011) E1475–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hung C, Linn G, Chow YH, Kobayashi A, Mittelsteadt K, Altemeier WA, Gharib SA, Schnapp LM, Duffield JS, Role of lung pericytes and resident fibroblasts in the pathogenesis of pulmonary fibrosis, Am J Respir Crit Care Med 188(7) (2013) 820–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Xie T, Wang Y, Deng N, Huang G, Taghavifar F, Geng Y, Liu N, Kulur V, Yao C, Chen P, Liu Z, Stripp B, Tang J, Liang J, Noble PW, Jiang D, Single-Cell Deconvolution of Fibroblast Heterogeneity in Mouse Pulmonary Fibrosis, Cell Rep 22(13) (2018) 3625–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].McQualter JL, Brouard N, Williams B, Baird BN, Sims-Lucas S, Yuen K, Nilsson SK, Simmons PJ, Bertoncello I, Endogenous fibroblastic progenitor cells in the adult mouse lung are highly enriched in the sca-1 positive cell fraction, Stem Cells 27(3) (2009) 623–33. [DOI] [PubMed] [Google Scholar]
- [35].Lee JH, Tammela T, Hofree M, Choi J, Marjanovic ND, Han S, Canner D, Wu K, Paschini M, Bhang DH, Jacks T, Regev A, Kim CF, Anatomically and Functionally Distinct Lung Mesenchymal Populations Marked by Lgr5 and Lgr6, Cell 170(6) (2017) 1149–1163 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zepp JA, Zacharias WJ, Frank DB, Cavanaugh CA, Zhou S, Morley MP, Morrisey EE, Distinct Mesenchymal Lineages and Niches Promote Epithelial Self-Renewal and Myofibrogenesis in the Lung, Cell 170(6) (2017) 1134–1148 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Greif DM, Kumar M, Lighthouse JK, Hum J, An A, Ding L, Red-Horse K, Espinoza FH, Olson L, Offermanns S, Krasnow MA, Radial construction of an arterial wall, Dev Cell 23(3) (2012) 482–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Que J, Wilm B, Hasegawa H, Wang F, Bader D, Hogan BL, Mesothelium contributes to vascular smooth muscle and mesenchyme during lung development, Proc Natl Acad Sci U S A 105(43) (2008) 16626–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Arora R, Metzger RJ, Papaioannou VE, Multiple roles and interactions of Tbx4 and Tbx5 in development of the respiratory system, PLoS Genet 8(8) (2012) e1002866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kumar ME, Bogard PE, Espinoza FH, Menke DB, Kingsley DM, Krasnow MA, Mesenchymal cells. Defining a mesenchymal progenitor niche at single-cell resolution, Science 346(6211) (2014) 1258810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Li C, Li M, Li S, Xing Y, Yang CY, Li A, Borok Z, De Langhe S, Minoo P, Progenitors of secondary crest myofibroblasts are developmentally committed in early lung mesoderm, Stem Cells 33(3) (2015) 999–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kugler MC, Loomis CA, Zhao Z, Cushman JC, Liu L, Munger JS, Sonic Hedgehog Signaling Regulates Myofibroblast Function During Alveolar Septum Formation in Murine Postnatal Lung, Am J Respir Cell Mol Biol (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Moiseenko A, Kheirollahi V, Chao CM, Ahmadvand N, Quantius J, Wilhelm J, Herold S, Ahlbrecht K, Morty RE, Rizvanov AA, Minoo P, El Agha E, Bellusci S, Origin and characterization of alpha smooth muscle actin-positive cells during murine lung development, Stem Cells 35(6) (2017) 1566–1578. [DOI] [PubMed] [Google Scholar]
- [44].Endale M, Ahlfeld S, Bao E, Chen X, Green J, Bess Z, Weirauch MT, Xu Y, Perl AK, Temporal, spatial, and phenotypical changes of PDGFRalpha expressing fibroblasts during late lung development, Dev Biol 425(2) (2017) 161–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Bostrom H, Gritli-Linde A, Betsholtz C, PDGF-A/PDGF alpha-receptor signaling is required for lung growth and the formation of alveoli but not for early lung branching morphogenesis, Dev Dyn 223(1) (2002) 155–62. [DOI] [PubMed] [Google Scholar]
- [46].Lau M, Masood A, Yi M, Belcastro R, Li J, Tanswell AK, Long-term failure of alveologenesis after an early short-term exposure to a PDGF-receptor antagonist, Am J Physiol Lung Cell Mol Physiol 300(4) (2011) L534–47. [DOI] [PubMed] [Google Scholar]
- [47].Lindahl P, Karlsson L, Hellstrom M, Gebre-Medhin S, Willetts K, Heath JK, Betsholtz C, Alveogenesis failure in PDGF-A-deficient mice is coupled to lack of distal spreading of alveolar smooth muscle cell progenitors during lung development, Development 124(20) (1997) 3943–53. [DOI] [PubMed] [Google Scholar]
- [48].Li C, Lee MK, Gao F, Webster S, Di H, Duan J, Yang CY, Bhopal N, Peinado N, Pryhuber G, Smith SM, Borok Z, Bellusci S, Minoo P, Secondary crest myofibroblast PDGFRalpha controls the elastogenesis pathway via a secondary tier of signaling networks during alveologenesis, Development 146(15) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kugler MC, Yie TA, Cai Y, Berger JZ, Loomis CA, Munger JS, The Hedgehog target Gli1 is not required for bleomycin-induced lung fibrosis, Exp Lung Res 45(1–2) (2019) 22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Cassandras M, Wang C, Kathiriya J, Tsukui T, Matatia P, Matthay M, Wolters P, Molofsky A, Sheppard D, Chapman H, Peng T, Gli1+ mesenchymal stromal cells modulate epithelial metaplasia in lung fibrosis, bioRxiv (2019) 841957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Barron L, Gharib SA, Duffield JS, Lung Pericytes and Resident Fibroblasts: Busy Multitaskers, Am J Pathol 186(10) (2016) 2519–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Hamilton TG, Klinghoffer RA, Corrin PD, Soriano P, Evolutionary divergence of platelet-derived growth factor alpha receptor signaling mechanisms, Mol Cell Biol 23(11) (2003) 4013–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].McGowan SE, Grossmann RE, Kimani PW, Holmes AJ, Platelet-derived growth factor receptor-alpha-expressing cells localize to the alveolar entry ring and have characteristics of myofibroblasts during pulmonary alveolar septal formation, Anat Rec (Hoboken) 291(12) (2008) 1649–61. [DOI] [PubMed] [Google Scholar]
- [54].Kimani PW, Holmes AJ, Grossmann RE, McGowan SE, PDGF-Ralpha gene expression predicts proliferation, but PDGF-A suppresses transdifferentiation of neonatal mouse lung myofibroblasts, Respir Res 10 (2009) 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].McGowan SE, McCoy DM, Fibroblasts expressing PDGF-receptor-alpha diminish during alveolar septal thinning in mice, Pediatr Res 70(1) (2011) 44–9. [DOI] [PubMed] [Google Scholar]
- [56].Perl AK, Gale E, FGF signaling is required for myofibroblast differentiation during alveolar regeneration, Am J Physiol Lung Cell Mol Physiol 297(2) (2009) L299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Endale M, Ahlfeld S, Bao E, Chen X, Green J, Bess Z, Weirauch M, Xu Y, Perl AK, Dataset on transcriptional profiles and the developmental characteristics of PDGFRalpha expressing lung fibroblasts, Data Brief 13 (2017) 415–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Chen L, Acciani T, Le Cras T, Lutzko C, Perl AK, Dynamic regulation of platelet-derived growth factor receptor alpha expression in alveolar fibroblasts during realveolarization, Am J Respir Cell Mol Biol 47(4) (2012) 517–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Green J, Endale M, Auer H, Perl AK, Diversity of Interstitial Lung Fibroblasts Is Regulated by Platelet-Derived Growth Factor Receptor alpha Kinase Activity, Am J Respir Cell Mol Biol 54(4) (2016) 532–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Ntokou A, Klein F, Dontireddy D, Becker S, Bellusci S, Richardson WD, Szibor M, Braun T, Morty RE, Seeger W, Voswinckel R, Ahlbrecht K, Characterization of the platelet-derived growth factor receptor-alpha-positive cell lineage during murine late lung development, Am J Physiol Lung Cell Mol Physiol 309(9) (2015) L942–58. [DOI] [PubMed] [Google Scholar]
- [61].Gouveia L, Betsholtz C, Andrae J, PDGF-A signaling is required for secondary alveolar septation and controls epithelial proliferation in the developing lung, Development 145(7) (2018). [DOI] [PubMed] [Google Scholar]
- [62].Gouveia L, Betsholtz C, Andrae J, Expression analysis of platelet-derived growth factor receptor alpha and its ligands in the developing mouse lung, Physiol Rep 5(6) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Li R, Bernau K, Sandbo N, Gu J, Preissl S, Sun X, Pdgfra marks a cellular lineage with distinct contributions to myofibroblasts in lung maturation and injury response, Elife 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Du Y, Kitzmiller JA, Sridharan A, Perl AK, Bridges JP, Misra RS, Pryhuber GS, Mariani TJ, Bhattacharya S, Guo M, Potter SS, Dexheimer P, Aronow B, Jobe AH, Whitsett JA, Xu Y, Lung Gene Expression Analysis (LGEA): an integrative web portal for comprehensive gene expression data analysis in lung development, Thorax (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Barkauskas CE, Cronce MJ, Rackley CR, Bowie EJ, Keene DR, Stripp BR, Randell SH, Noble PW, Hogan BL, Type 2 alveolar cells are stem cells in adult lung, J Clin Invest 123(7) (2013) 3025–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Husain AN, Siddiqui NH, Stocker JT, Pathology of arrested acinar development in postsurfactant bronchopulmonary dysplasia, Hum Pathol 29(7) (1998) 710–7. [DOI] [PubMed] [Google Scholar]
- [67].Popova AP, Bentley JK, Cui TX, Richardson MN, Linn MJ, Lei J, Chen Q, Goldsmith AM, Pryhuber GS, Hershenson MB, Reduced platelet-derived growth factor receptor expression is a primary feature of human bronchopulmonary dysplasia, Am J Physiol Lung Cell Mol Physiol 307(3) (2014) L231–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H, Identification of stem cells in small intestine and colon by marker gene Lgr5, Nature 449(7165) (2007) 1003–7. [DOI] [PubMed] [Google Scholar]
- [69].de Lau WB, Snel B, Clevers HC, The R-spondin protein family, Genome Biol 13(3) (2012) 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Volckaert T, Dill E, Campbell A, Tiozzo C, Majka S, Bellusci S, De Langhe SP, Parabronchial smooth muscle constitutes an airway epithelial stem cell niche in the mouse lung after injury, J Clin Invest 121(11) (2011) 4409–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Frank DB, Peng T, Zepp JA, Snitow M, Vincent TL, Penkala IJ, Cui Z, Herriges MJ, Morley MP, Zhou S, Lu MM, Morrisey EE, Emergence of a Wave of Wnt Signaling that Regulates Lung Alveologenesis by Controlling Epithelial Self-Renewal and Differentiation, Cell Rep 17(9) (2016) 2312–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Tahedl D, Wirkes A, Tschanz SA, Ochs M, Muhlfeld C, How common is the lipid body-containing interstitial cell in the mammalian lung?, Am J Physiol Lung Cell Mol Physiol 307(5) (2014) L386–94. [DOI] [PubMed] [Google Scholar]
- [73].McGowan SE, Torday JS, The pulmonary lipofibroblast (lipid interstitial cell) and its contributions to alveolar development, Annu Rev Physiol 59 (1997) 43–62. [DOI] [PubMed] [Google Scholar]
- [74].O’Hare KH, Sheridan MN, Electron microscopic observations on the morphogenesis of the albino rat lung, with special reference to pulmonary epithelial cells, Am J Anat 127(2) (1970) 181–205. [DOI] [PubMed] [Google Scholar]
- [75].Tansey JT, Sztalryd C, Hlavin EM, Kimmel AR, Londos C, The central role of perilipin a in lipid metabolism and adipocyte lipolysis, IUBMB Life 56(7) (2004) 379–85. [DOI] [PubMed] [Google Scholar]
- [76].Torday JS, Torres E, Rehan VK, The role of fibroblast transdifferentiation in lung epithelial cell proliferation, differentiation, and repair in vitro, Pediatr Pathol Mol Med 22(3) (2003) 189–207. [DOI] [PubMed] [Google Scholar]
- [77].Rehan VK, Sakurai R, Wang Y, Santos J, Huynh K, Torday JS, Reversal of nicotine-induced alveolar lipofibroblast-to-myofibroblast transdifferentiation by stimulants of parathyroid hormone-related protein signaling, Lung 185(3) (2007) 151–9. [DOI] [PubMed] [Google Scholar]
- [78].Kheirollahi V, Wasnick RM, Biasin V, Vazquez-Armendariz AI, Chu X, Moiseenko A, Weiss A, Wilhelm J, Zhang JS, Kwapiszewska G, Herold S, Schermuly RT, Mari B, Li X, Seeger W, Gunther A, Bellusci S, El Agha E, Metformin induces lipogenic differentiation in myofibroblasts to reverse lung fibrosis, Nat Commun 10(1) (2019) 2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Chen H, Jackson S, Doro M, McGowan S, Perinatal expression of genes that may participate in lipid metabolism by lipid-laden lung fibroblasts, J Lipid Res 39(12) (1998) 2483–92. [PubMed] [Google Scholar]
- [80].Schultz CJ, Torres E, Londos C, Torday JS, Role of adipocyte differentiation-related protein in surfactant phospholipid synthesis by type II cells, Am J Physiol Lung Cell Mol Physiol 283(2) (2002) L288–96. [DOI] [PubMed] [Google Scholar]
- [81].McIntosh JC, Hagood JS, Richardson TL, Simecka JW, Thy1 (+) and (−) lung fibrosis subpopulations in LEW and F344 rats, Eur Respir J 7(12) (1994) 2131–8. [DOI] [PubMed] [Google Scholar]
- [82].Paxson JA, Gruntman AM, Davis AM, Parkin CM, Ingenito EP, Hoffman AM, Age Dependence of Lung Mesenchymal Stromal Cell Dynamics Following Pneumonectomy, Stem Cells Dev (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Cohen PY, Breuer R, Zisman P, Wallach-Dayan SB, Bleomycin-Treated Chimeric Thy1-Deficient Mice with Thy1-Deficient Myofibroblasts and Thy-Positive Lymphocytes Resolve Inflammation without Affecting the Fibrotic Response, Mediators Inflamm 2015 (2015) 942179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Kumar A, Bhanja A, Bhattacharyya J, Jaganathan BG, Multiple roles of CD90 in cancer, Tumour Biol 37(9) (2016) 11611–11622. [DOI] [PubMed] [Google Scholar]
- [85].Tan C, Jiang M, Wong SS, Espinoza CR, Kim C, Li X, Connors E, Hagood JS, Soluble Thy-1 reverses lung fibrosis via its integrin-binding motif, JCI Insight 4(21) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Nicola T, Hagood JS, James ML, Macewen MW, Williams TA, Hewitt MM, Schwiebert L, Bulger A, Oparil S, Chen YF, Ambalavanan N, Loss of Thy-1 inhibits alveolar development in the newborn mouse lung, Am J Physiol Lung Cell Mol Physiol 296(5) (2009) L738–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Sanders YY, Kumbla P, Hagood JS, Enhanced myofibroblastic differentiation and survival in Thy-1(−) lung fibroblasts, Am J Respir Cell Mol Biol 36(2) (2007) 226–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Fiore VF, Strane PW, Bryksin AV, White ES, Hagood JS, Barker TH, Conformational coupling of integrin and Thy-1 regulates Fyn priming and fibroblast mechanotransduction, J Cell Biol 211(1) (2015) 173–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Fiore VF, Wong SS, Tran C, Tan C, Xu W, Sulchek T, White ES, Hagood JS, Barker TH, alphavbeta3 Integrin drives fibroblast contraction and strain stiffening of soft provisional matrix during progressive fibrosis, JCI Insight 3(20) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Sakiyama S, Kondo K, Matsuoka H, Yoshida M, Miyoshi T, Yoshida S, Monden Y, Fatal air embolism during computed tomography-guided pulmonary marking with a hook-type marker, J Thorac Cardiovasc Surg 126(4) (2003) 1207–9. [DOI] [PubMed] [Google Scholar]
- [91].Yuan T, Volckaert T, Chanda D, Thannickal VJ, De Langhe SP, Fgf10 Signaling in Lung Development, Homeostasis, Disease, and Repair After Injury, Front Genet 9 (2018) 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Arman E, Haffner-Krausz R, Gorivodsky M, Lonai P, Fgfr2 is required for limb outgrowth and lung-branching morphogenesis, Proc Natl Acad Sci U S A 96(21) (1999) 11895–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].De Moerlooze L, Spencer-Dene B, Revest J, Hajihosseini M, Rosewell I, Dickson C, An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis, Development 127(3) (2000) 483–92.10631169 [Google Scholar]
- [94].Ramasamy SK, Mailleux AA, Gupte VV, Mata F, Sala FG, Veltmaat JM, Del Moral PM, De Langhe S, Parsa S, Kelly LK, Kelly R, Shia W, Keshet E, Minoo P, Warburton D, Bellusci S, Fgf10 dosage is critical for the amplification of epithelial cell progenitors and for the formation of multiple mesenchymal lineages during lung development, Dev Biol 307(2) (2007) 237–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Volckaert T, Campbell A, Dill E, Li C, Minoo P, De Langhe S, Localized Fgf10 expression is not required for lung branching morphogenesis but prevents differentiation of epithelial progenitors, Development 140(18) (2013) 3731–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Rawlins EL, Hogan BL, Intercellular growth factor signaling and the development of mouse tracheal submucosal glands, Dev Dyn 233(4) (2005) 1378–85. [DOI] [PubMed] [Google Scholar]
- [97].Volckaert T, Yuan T, Chao CM, Bell H, Sitaula A, Szimmtenings L, El Agha E, Chanda D, Majka S, Bellusci S, Thannickal VJ, Fassler R, De Langhe SP, Fgf10-Hippo Epithelial-Mesenchymal Crosstalk Maintains and Recruits Lung Basal Stem Cells, Dev Cell 43(1) (2017) 48–59 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Yuan T, Volckaert T, Redente EF, Hopkins S, Klinkhammer K, Wasnick R, Chao CM, Yuan J, Zhang JS, Yao C, Majka S, Stripp BR, Gunther A, Riches DWH, Bellusci S, Thannickal VJ, De Langhe SP, FGF10-FGFR2B Signaling Generates Basal Cells and Drives Alveolar Epithelial Regeneration by Bronchial Epithelial Stem Cells after Lung Injury, Stem Cell Reports 12(5) (2019) 1041–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Mailleux AA, Kelly R, Veltmaat JM, De Langhe SP, Zaffran S, Thiery JP, Bellusci S, Fgf10 expression identifies parabronchial smooth muscle cell progenitors and is required for their entry into the smooth muscle cell lineage, Development 132(9) (2005) 2157–66. [DOI] [PubMed] [Google Scholar]
- [100].Al Alam D, El Agha E, Sakurai R, Kheirollahi V, Moiseenko A, Danopoulos S, Shrestha A, Schmoldt C, Quantius J, Herold S, Chao CM, Tiozzo C, De Langhe S, Plikus MV, Thornton M, Grubbs B, Minoo P, Rehan VK, Bellusci S, Evidence for the involvement of fibroblast growth factor 10 in lipofibroblast formation during embryonic lung development, Development 142(23) (2015) 4139–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].El Agha E, Moiseenko A, Kheirollahi V, De Langhe S, Crnkovic S, Kwapiszewska G, Szibor M, Kosanovic D, Schwind F, Schermuly RT, Henneke I, MacKenzie B, Quantius J, Herold S, Ntokou A, Ahlbrecht K, Braun T, Morty RE, Gunther A, Seeger W, Bellusci S, Two-Way Conversion between Lipogenic and Myogenic Fibroblastic Phenotypes Marks the Progression and Resolution of Lung Fibrosis, Cell Stem Cell 20(4) (2017) 571. [DOI] [PubMed] [Google Scholar]
- [102].Benjamin JT, Smith RJ, Halloran BA, Day TJ, Kelly DR, Prince LS, FGF-10 is decreased in bronchopulmonary dysplasia and suppressed by Toll-like receptor activation, Am J Physiol Lung Cell Mol Physiol 292(2) (2007) L550–8. [DOI] [PubMed] [Google Scholar]
- [103].Hokuto I, Perl AK, Whitsett JA, Prenatal, but not postnatal, inhibition of fibroblast growth factor receptor signaling causes emphysema, J Biol Chem 278(1) (2003) 415–21. [DOI] [PubMed] [Google Scholar]
- [104].Chao CM, Yahya F, Moiseenko A, Tiozzo C, Shrestha A, Ahmadvand N, El Agha E, Quantius J, Dilai S, Kheirollahi V, Jones M, Wilhem J, Carraro G, Ehrhardt H, Zimmer KP, Barreto G, Ahlbrecht K, Morty RE, Herold S, Abellar RG, Seeger W, Schermuly R, Zhang JS, Minoo P, Bellusci S, Fgf10 deficiency is causative for lethality in a mouse model of bronchopulmonary dysplasia, J Pathol 241(1) (2017) 91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Srisuma S, Bhattacharya S, Simon DM, Solleti SK, Tyagi S, Starcher B, Mariani TJ, Fibroblast growth factor receptors control epithelial-mesenchymal interactions necessary for alveolar elastogenesis, Am J Respir Crit Care Med 181(8) (2010) 838–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Li R, Herriges JC, Chen L, Mecham RP, Sun X, FGF receptors control alveolar elastogenesis, Development 144(24) (2017) 4563–4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Weinstein M, Xu X, Ohyama K, Deng CX, FGFR-3 and FGFR-4 function cooperatively to direct alveogenesis in the murine lung, Development 125(18) (1998) 3615–23. [DOI] [PubMed] [Google Scholar]
- [108].Boucherat O, Franco-Montoya ML, Thibault C, Incitti R, Chailley-Heu B, Delacourt C, Bourbon JR, Gene expression profiling in lung fibroblasts reveals new players in alveolarization, Physiol Genomics 32(1) (2007) 128–41. [DOI] [PubMed] [Google Scholar]
- [109].Franco-Montoya ML, Boucherat O, Thibault C, Chailley-Heu B, Incitti R, Delacourt C, Bourbon JR, Profiling target genes of FGF18 in the postnatal mouse lung: possible relevance for alveolar development, Physiol Genomics (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Chailley-Heu B, Boucherat O, Barlier-Mur AM, Bourbon JR, FGF-18 is upregulated in the postnatal rat lung and enhances elastogenesis in myofibroblasts, Am J Physiol Lung Cell Mol Physiol 288(1) (2005) L43–51. [DOI] [PubMed] [Google Scholar]
- [111].Hagan AS, Zhang B, Ornitz DM, Identification of an FGF18-expressing alveolar myofibroblast that is developmentally cleared during alveologenesis, Development (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Hagan AS, Boylan M, Smith C, Perez-Santamarina E, Kowalska K, Hung IH, Lewis RM, Hajihosseini MK, Lewandoski M, Ornitz DM, Generation and validation of novel conditional flox and inducible Cre alleles targeting fibroblast growth factor 18 (Fgf18), Dev Dyn 248(9) (2019) 882–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Quaggin SE, Schwartz L, Cui S, Igarashi P, Deimling J, Post M, Rossant J, The basic-helix-loop-helix protein pod1 is critically important for kidney and lung organogenesis, Development 126(24) (1999) 5771–83. [DOI] [PubMed] [Google Scholar]
- [114].McGowan SE, McCoy DM, Regulation of fibroblast lipid storage and myofibroblast phenotypes during alveolar septation in mice, Am J Physiol Lung Cell Mol Physiol 307(8) (2014) L618–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Guo M, Bao EL, Wagner M, Whitsett JA, Xu Y, SLICE: determining cell differentiation and lineage based on single cell entropy, Nucleic Acids Res (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Guo M, Du Y, Gokey JJ, Ray S, Bell SM, Adam M, Sudha P, Perl AK, Deshmukh H, Potter SS, Whitsett JA, Xu Y, Single cell RNA analysis identifies cellular heterogeneity and adaptive responses of the lung at birth, Nat Commun 10(1) (2019) 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Adams TS, Schupp JC, Poli S, Ayaub EA, Neumark N, Ahangari F, Chu SG, Raby BA, DeIuliis G, Januszyk M, Duan Q, Arnett HA, Siddiqui A, Washko GR, Homer R, Yan X, Rosas IO, Kaminski N, Single Cell RNA-seq reveals ectopic and aberrant lung resident cell populations in Idiopathic Pulmonary Fibrosis, bioRxiv (2019) 759902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].McGowan SE, McCoy DM, Platelet-derived growth factor-A regulates lung fibroblast S-phase entry through p27(kip1) and FoxO3a, Respir Res 14 (2013) 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].McGowan SE, McCoy DM, Platelet-derived growth factor-A and sonic hedgehog signaling direct lung fibroblast precursors during alveolar septal formation, Am J Physiol Lung Cell Mol Physiol 305(3) (2013) L229–39. [DOI] [PubMed] [Google Scholar]
- [120].Ahlbrecht K, McGowan SE, In search of the elusive lipofibroblast in human lungs, Am J Physiol Lung Cell Mol Physiol 307(8) (2014) L605–8. [DOI] [PubMed] [Google Scholar]
- [121].McGowan SE, McCoy DM, Fibroblast growth factor signaling in myofibroblasts differs from lipofibroblasts during alveolar septation in mice, Am J Physiol Lung Cell Mol Physiol 309(5) (2015) L463–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Gokey J, Snowball J, Green J, Waltamath M, Spinney JJ, Black KE, Hariri LP, Xu Y, Perl AKT, Pretreatment of aged mice with retinoic acid restores alveolar regeneration via upregulation of reciprocal PDGFRA signaling, bioRxiv (2020) 2020.05.06.080994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Kauffman SL, Burri PH, Weibel ER, The postnatal growth of the rat lung. II. Autoradiography, Anat Rec 180(1) (1974) 63–76. [DOI] [PubMed] [Google Scholar]
- [124].Armulik A, Genove G, Betsholtz C, Pericytes: developmental, physiological, and pathological perspectives, problems, and promises, Dev Cell 21(2) (2011) 193–215. [DOI] [PubMed] [Google Scholar]
- [125].Armulik A, Abramsson A, Betsholtz C, Endothelial/pericyte interactions, Circ Res 97(6) (2005) 512–23. [DOI] [PubMed] [Google Scholar]
- [126].Marriott S, Baskir RS, Gaskill C, Menon S, Carrier EJ, Williams J, Talati M, Helm K, Alford CE, Kropski JA, Loyd J, Wheeler L, Johnson J, Austin E, Nozik-Grayck E, Meyrick B, West JD, Klemm DJ, Majka SM, ABCG2pos lung mesenchymal stem cells are a novel pericyte subpopulation that contributes to fibrotic remodeling, Am J Physiol Cell Physiol 307(8) (2014) C684–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Gaskill CF, Carrier EJ, Kropski JA, Bloodworth NC, Menon S, Foronjy RF, Taketo MM, Hong CC, Austin ED, West JD, Means AL, Loyd JE, Merryman WD, Hemnes AR, De Langhe S, Blackwell TS, Klemm DJ, Majka SM, Disruption of lineage specification in adult pulmonary mesenchymal progenitor cells promotes microvascular dysfunction, J Clin Invest 127(6) (2017) 2262–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Spangrude GJ, Aihara Y, Weissman IL, Klein J, The stem cell antigens Sca-1 and Sca-2 subdivide thymic and peripheral T lymphocytes into unique subsets, J Immunol 141(11) (1988) 3697–707. [PubMed] [Google Scholar]
- [129].Kotton DN, Summer RS, Sun X, Ma BY, Fine A, Stem cell antigen-1 expression in the pulmonary vascular endothelium, Am J Physiol Lung Cell Mol Physiol 284(6) (2003) L990–6. [DOI] [PubMed] [Google Scholar]
- [130].Holmes C, Khan TS, Owen C, Ciliberti N, Grynpas MD, Stanford WL, Longitudinal analysis of mesenchymal progenitors and bone quality in the stem cell antigen-1-null osteoporotic mouse, J Bone Miner Res 22(9) (2007) 1373–86. [DOI] [PubMed] [Google Scholar]
- [131].Summer R, Fitzsimmons K, Dwyer D, Murphy J, Fine A, Isolation of an adult mouse lung mesenchymal progenitor cell population, Am J Respir Cell Mol Biol 37(2) (2007) 152–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Martin J, Helm K, Ruegg P, Varella-Garcia M, Burnham E, Majka S, Adult lung side population cells have mesenchymal stem cell potential, Cytotherapy 10(2) (2008) 140–51. [DOI] [PubMed] [Google Scholar]
- [133].Kramann R, Hedgehog Gli signalling in kidney fibrosis, Nephrol Dial Transplant 31(12) (2016) 1989–1995. [DOI] [PubMed] [Google Scholar]
- [134].Fearon ER, Vogelstein B, Feinberg AP, Somatic deletion and duplication of genes on chromosome 11 in Wilms’ tumours, Nature 309(5964) (1984) 176–8. [DOI] [PubMed] [Google Scholar]
- [135].Langerak AW, Williamson KA, Miyagawa K, Hagemeijer A, Versnel MA, Hastie ND, Expression of the Wilms’ tumor gene WT1 in human malignant mesothelioma cell lines and relationship to platelet-derived growth factor A and insulin-like growth factor 2 expression, Genes Chromosomes Cancer 12(2) (1995) 87–96. [DOI] [PubMed] [Google Scholar]
- [136].Colvin JS, White AC, Pratt SJ, Ornitz DM, Lung hypoplasia and neonatal death in Fgf9-null mice identify this gene as an essential regulator of lung mesenchyme, Development 128(11) (2001) 2095–106. [DOI] [PubMed] [Google Scholar]
- [137].Yin Y, Wang F, Ornitz DM, Mesothelial- and epithelial-derived FGF9 have distinct functions in the regulation of lung development, Development 138(15) (2011) 3169–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Sontake V, Kasam RK, Sinner D, Korfhagen TR, Reddy GB, White ES, Jegga AG, Madala SK, Wilms’ tumor 1 drives fibroproliferation and myofibroblast transformation in severe fibrotic lung disease, JCI Insight 3(16) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Sontake V, Shanmukhappa SK, DiPasquale BA, Reddy GB, Medvedovic M, Hardie WD, White ES, Madala SK, Fibrocytes Regulate Wilms Tumor 1-Positive Cell Accumulation in Severe Fibrotic Lung Disease, J Immunol 195(8) (2015) 3978–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Habermann AC, Gutierrez AJ, Bui LT, Yahn SL, Winters NI, Calvi CL, Peter L, Chung M-I, Taylor CJ, Jetter C, Raju L, Roberson J, Ding G, Wood L, Sucre JM, Richmond BW, Serezani AP, McDonnell WJ, Mallal SB, Bacchetta MJ, Loyd JE, Shaver CM, Ware LB, Bremner R, Walia R, Blackwell TS, Banovich NE, Kropski JA, Single-cell RNA-sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis, bioRxiv (2019) 753806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Owens GK, Kumar MS, Wamhoff BR, Molecular regulation of vascular smooth muscle cell differentiation in development and disease, Physiol Rev 84(3) (2004) 767–801. [DOI] [PubMed] [Google Scholar]
- [142].Sparrow MP, Lamb JP, Ontogeny of airway smooth muscle: structure, innervation, myogenesis and function in the fetal lung, Respir Physiol Neurobiol 137(2–3) (2003) 361–72. [DOI] [PubMed] [Google Scholar]
- [143].Zhou L, Dey CR, Wert SE, Whitsett JA, Arrested lung morphogenesis in transgenic mice bearing an SP-C-TGF-beta 1 chimeric gene, Dev Biol 175(2) (1996) 227–38. [DOI] [PubMed] [Google Scholar]
- [144].Weaver M, Yingling JM, Dunn NR, Bellusci S, Hogan BL, Bmp signaling regulates proximal-distal differentiation of endoderm in mouse lung development, Development 126(18) (1999) 4005–15. [DOI] [PubMed] [Google Scholar]
- [145].Guo M, Wang H, Potter SS, Whitsett JA, Xu Y, SINCERA: A Pipeline for Single-Cell RNA-Seq Profiling Analysis, PLoS Comput Biol 11(11) (2015) e1004575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Xu Y, Mizuno T, Sridharan A, Du Y, Guo M, Tang J, Wikenheiser-Brokamp KA, Perl AT, Funari VA, Gokey JJ, Stripp BR, Whitsett JA, Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis, JCI Insight 1(20) (2016) e90558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Vieth B, Parekh S, Ziegenhain C, Enard W, Hellmann I, A systematic evaluation of single cell RNA-seq analysis pipelines, Nat Commun 10(1) (2019) 4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Park PW, Introduction to the thematic mini-review series on “Matrix biology in lung health and disease”, Matrix Biol 73 (2018) 1–5. [DOI] [PubMed] [Google Scholar]
- [149].Zhou Y, Horowitz JC, Naba A, Ambalavanan N, Atabai K, Balestrini J, Bitterman PB, Corley RA, Ding BS, Engler AJ, Hansen KC, Hagood JS, Kheradmand F, Lin QS, Neptune E, Niklason L, Ortiz LA, Parks WC, Tschumperlin DJ, White ES, Chapman HA, Thannickal VJ, Extracellular matrix in lung development, homeostasis and disease, Matrix Biol 73 (2018) 77–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Pakshir P, Hinz B, The big five in fibrosis: Macrophages, myofibroblasts, matrix, mechanics, and miscommunication, Matrix Biol 68–69 (2018) 81–93. [DOI] [PubMed] [Google Scholar]
- [151].Schaefer L, Decoding fibrosis: Mechanisms and translational aspects, Matrix Biol 68–69 (2018) 1–7. [DOI] [PubMed] [Google Scholar]
- [152].Karamanos NK, Theocharis AD, Neill T, Iozzo RV, Matrix modeling and remodeling: A biological interplay regulating tissue homeostasis and diseases, Matrix Biol 75–76 (2019) 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Iozzo RV, Gubbiotti MA, Extracellular matrix: The driving force of mammalian diseases, Matrix Biol 71–72 (2018) 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Gokey JJ, Snowball J, Green J, Waltamath M, Spinney JJ, Black KE, Hariri LP, Xu Y, Perl AKT, Pretreatment of aged mice with retinoic acid restores alveolar regeneration via upregulation of reciprocal PDGFRA signaling, BioRXiv (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Mavrogonatou E, Pratsinis H, Papadopoulou A, Karamanos NK, Kletsas D, Extracellular matrix alterations in senescent cells and their significance in tissue homeostasis, Matrix Biol 75–76 (2019) 27–42. [DOI] [PubMed] [Google Scholar]
- [156].Hewlett JC, Kropski JA, Blackwell TS, Idiopathic pulmonary fibrosis: Epithelial-mesenchymal interactions and emerging therapeutic targets, Matrix Biol 71–72 (2018) 112–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Haak AJ, Tan Q, Tschumperlin DJ, Matrix biomechanics and dynamics in pulmonary fibrosis, Matrix Biol 73 (2018) 64–76. [DOI] [PMC free article] [PubMed] [Google Scholar]


