Abstract
Genome editing using site-specific nucleases, such as transcription activator-like effector nucleases (TALENs) and clustered regularly interspaced short palindromic repeat–CRISPR-associated protein 9 (CRISPR-Cas9), is a powerful technology for crop breeding. For plant genome editing, the genome-editing reagents are usually expressed in plant cells from stably integrated transgenes within the genome. This requires crossing processes to remove foreign nucleotides from the genome to generate null segregants. However, in highly heterozygous plants such as potato, the progeny lines have different agronomic traits from the parent cultivar and do not necessarily become elite lines. Agrobacteria can transfer exogenous genes on T-DNA into plant cells. This has been used both to transform plants stably and to express the genes transiently in plant cells. Here, we infected potato, with Agrobacterium tumefaciens harboring TALEN-expression vector targeting sterol side chain reductase 2 (SSR2) gene and regenerated shoots without selection. We obtained regenerated lines with disrupted-SSR2 gene and without transgene of the TALEN gene, revealing that their disruption should be caused by transient gene expression. The strategy using transient gene expression by Agrobacterium that we call Agrobacterial mutagenesis, developed here should accelerate the use of genome-editing technology to modify heterozygous plant genomes.
Keywords: genome editing, potato, steroidal glycoalkaloids, TALEN, transient gene expression
Introduction
Site-specific nucleases, such as zinc finger nucleases, transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeat (CRISPR)–CRISPR-associated protein 9 (CRISPR-Cas9) systems, recognize specific sequences in the genomic sequence and introduce DNA double-strand breaks (DSBs). DSBs are incorrectly repaired by the endogenous non-homologous end-joining pathway, and targeted mutagenesis is introduced into the target sequence (reviewed in Voytas 2013; Voytas and Gao 2014). Before the development of genome-editing technology using site-specific nucleases, there was no way to disrupt a target gene in the plant genome. The generation of mutant lines for scientific research or agriculture depended completely on random mutations caused by radiation or mutagenic chemicals. It is time-consuming and laborious to select objective mutants (McCallum et al. 2000). By using genome-editing reagents, we can easily generate plants that have objective mutations in the genome. Genome editing in plants began with the transformation of the expression vector for site-specific nucleases. After introducing targeted mutagenesis in the target gene, the transgenes are removed by genetic segregation to generate null segregants (harboring the targeted mutation without the transgenes) (Wolt et al. 2016). When highly homozygous plants are the subject materials, no problems arise. Vegetatively propagated crops have a highly heterozygous plant genome, such as a tetraploid potato genome. Their genomic information and agricultural traits must be changed completely through segregation and crossing-over. One possible strategy to overcome this problem is transient expression of site-specific nucleases in plant cells without integration of a transgene in the genome. Polyethylene-glycol-mediated protoplast transfection and particle bombardment are usually employed to generate transgenic plants with antibiotic selection. Nicolia et al. (2015), Clasen et al. (2016), and Zhang et al. (2016) used these techniques to deliver nucleotide molecules encoding site-specific nucleases, incubated the transfected plant cells under no selection, and succeeded in generating genome-edited plants without transgene integration in the genome. Agrobacterium-mediated transformation is the method used most widely for generating transgenic plants (Hwang et al. 2017). During the transformation process, T-DNA on a binary vector is processed and transferred into plant cells. Cells that have stable integrated T-DNA can be selected for under different conditions, in the presence of antibiotics or herbicides, for example. Agrobacteria is also used for transient expression in plants. Using the Agrobacterium-mediated transient expression method for plants, strong gene expression can be observed after cultivation for a few days transiently (Fischer et al. 1999; Kapila et al. 1997). Since there is strong transient gene expression in the infected region, we hypothesized that Agrobacterium-mediated transient site-specific nuclease expression in plant cells under non-selection conditions is suitable for plant genome editing. In this study, we infected potato stems with Agrobacterium tumefaciens harboring the TALEN-expression vector. All regenerated shoots on medium containing plant hormones without any selection reagents, were checked for gene modification in the target sequence, and evaluated the transgene integration in the genome. We found the shoots contained genome-edited line by Agrobacterium-transient expression.
Materials and methods
Plant material
Murashige and Skoog (MS) medium containing 3% sucrose solidified with 0.8% agar was used for in vitro cultivation of Solanum tuberosum cv. Sassy. Shoots were subcultured every 3 to 4 weeks.
Agrobacterium
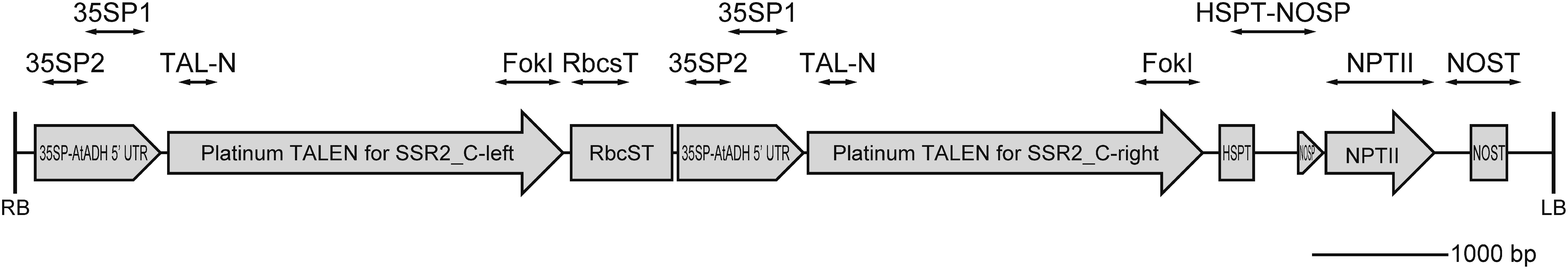
The Agrobacterium tumefaciens EHA105 strain was transformed with the Platinum Gate TALEN expression vector pYS_026-SSR2 (Yasumoto et al. 2019). The T-DNA region in this expression vector contained the coding sequence for the TALENs targeting the sterol side chain reductase 2 (SSR2) gene under the 35S promoter (Figure 1). Transformed Agrobacterium clones were selected on YEB medium (1 g/l yeast extract, 5 g/l beef extract, 5 g/l peptone, 5 g/l sucrose, 0.5 g/l MgSO4 7H2O, pH 7.0) containing 50 mg/l kanamycin and stored in 25% glycerol at −80°C.
Figure 1. Schematic illustration of the T-DNA region of the Platinum transcription activator-like effector nuclease (TALEN) expression construct. RB and LB, T-DNA right and left borders, respectively; 35SP-AtADH-5'UTR, Cauliflower mosaic virus 35S promoter 5'-untranslated region of A. thaliana alcohol dehydrogenase; RbcsT, Rubisco small subunit terminator; HSPT, heat shock protein terminator from Arabidopsis thaliana; NOSP, nopaline synthase promoter; NPTII, neomycin phosphotransferase II; NOST, nopaline synthase terminator. The double-headed arrows indicate the amplified regions in Figure 2.
Agrobacterium infection of potato stems
The glycerol stock was inoculated into 2–3 ml YEB containing kanamycin and cultured at 28°C overnight at 180–200 rpm shaking. Agrobacterium cells were collected by centrifugation and suspended in the appropriate amount of MS medium containing 3% sucrose. Potato stem segments were prepared by cutting an in vitro culture and incubated with Agrobacterium solution. After removing the excess solution with sterilized filter paper, the segments were co-cultivated on 3C5ZR plates (Sheerman and Bevan 1988) for 3 to 7 days. To eliminate bacteria cells, the segments were transferred to 3C5ZR plates containing 250 mg/l carbenicillin every 2 weeks. The shoots that regenerated from the segments were isolated and cultured in plant boxes containing solidified MS medium with 3% sucrose and cefotaxime.
Heteroduplex mobility assay (HMA)
The leaves were removed from the regenerated shoots and incubated in buffer solution (100 mM Tris-HCl, pH 9.5, 1 M KCl, 10 mM EDTA) at 95°C for 10 min. The supernatant was used as a polymerase chain reaction (PCR) template. The regions surrounding the TALEN target sites were amplified using the primer set pY365/pY366 and KOD FX Neo DNA polymerase (Toyobo, Japan). The PCR products were analyzed on a micro-tip electrophoresis machine (MultiNA; Shimadzu, Japan) or 5% polyacrylamide gels.
PCR analysis for transgene integration
Shoots that showed extra bands in the HMA assay were subcultured at least three times on MS medium containing 3% sucrose and 250 ppm cefotaxime or carbenicillin, and maintained on MS medium containing 3% sucrose without antibiotics. The genomic DNA was extracted and purified from each regenerated line using a NucleoSpin® Plant II kit (Takara Bio, Japan) or DNAzol reagent (Invitrogen, USA). Genomic PCR reactions were performed as follows. Each 20 µl PCR reaction solution contained 10 µl of Hot Start Taq 2× Master Mix (New England Biolabs, USA), 0.4 µl of 10 µM forward and reverse primers, and approximately 10 ng of purified genomic DNA. The genomic DNA from non-transgenic Sassy and plasmid DNA were used for control reactions. Eight primer pairs were used to amplify the corresponding regions [35S promoter_1 (35SP_1), 35S promoter_2 (35SP_2), TALEN-N-terminal (TAL-N), TALEN-C-terminal (FokI), ribulose-1,5-biphosphate carboxylase small subunit terminator (RbcsT), heat shock protein terminator-nopaline synthase promoter (HSPT-NOSP), neomycin phosphotransferase II (NPTII), and nopaline synthase terminator (NOST)] in T-DNA on the TALEN expression binary vector, and one primer pair was used to amplify the potential off-target site in SSR1, a homologue of SSR2 (Figure 1, Table 1). The PCR reaction mixture was incubated at 95°C for 30 s, followed by 34 cycles of denaturing at 95°C for 15 s, annealing at annealing temperature for 15s, and extension at 68°C for 30 to 60 s. The PCR reaction mixtures were run on the MultiNA instrument. Table 1 lists the primer sequences used in this study.
Table 1. PCR primers and their applications.
| Primer name | Primer sequence (5′→3′) | Application | Annealing temp. (°C) | Extention time (s) |
|---|---|---|---|---|
| pY365/1682/check-for | TGTTCTCTGACACTGTTGTAGCACT | Amplifying the SSR2 target site | 55 | 30 |
| pY366/1683/check-rev | TCGAAGCATACATACCGGTCATCAT | |||
| 1748/StDWF1_C-Forward | TGTTCTCAGACACTGTTGTGTCATA | Amplifying the SSR1potential off target site | 52 | 30 |
| 1749/StDWF1_C-Reverse | TTGAAGCATATCTACCAGTCATGCA | |||
| pY515/35SP-C-1F | AAGACTGGCGAACAGTTCATACAGAG | Detectting TALEN construct (35SP_1) | 56 | 30 |
| pY516/35SP-C-1R | GTCTTGCGAAGGATAGTGGGATTGTG | |||
| pY517/35SP-C-2F | CCCACAGATGGTTAGAGAGGCTTACG | Detecting TALEN construct (35SP_2) | 55 | 30 |
| pY518/35SP-C-2R | CTCTGTATGAACTGTTCGCCAGTC | |||
| pY519/TALN-C-1F | GAAGGACGCAAGTGGTTGGTCTAG | Detecting TALEN construct (TAL-N) | 57 | 30 |
| pY520/TALN-C_1R | CTCTGGCAACGCCGTGATTATGTG | |||
| pY27/FokI-for | CGCAAGAAATTCAACTCAGGATAGA | Detecting TALEN construct (FokI) | 51 | 30 |
| pY28/FokI-rev | CCGTTATTAAATTTCCTTCTCACTTCCT | |||
| 1658/RbcsT-1-seq | TCCCCTTTCTGGAATATTCAGC | Detecting TALEN construct (RbcsT) | 51 | 30 |
| 1659/RbcsT-400-rev-seq | CCAAATCTCCAATGGCTATGTC | |||
| pY521/HSPT-C_1F | GGCTTGTTGTGTTATGAATTTGTGG | Detecting TALEN construct (HSPT-NOSP) | 52 | 40 |
| pY522/NOSP-C_1R | GGAATTTATGGAACGTCAGTGGAGC | |||
| pY465/NPTII-F2 | ATGATTGAACAAGATGGATTGCACGC | Detecting TALEN construct (NPTII) | 55 | 60 |
| pY466/NPTII-R2 | TCAGAAGAACTCGTCAAGAAGGCG | |||
| pY523/NOSTU-C_1F | TATGAAAGGTTGGGCTTCGGAATCG | Detecting TALEN construct (NOST) | 56 | 40 |
| pY524/NOSTD-C_1R | CGAGATAGGGTTGAGTGTTGTTCCA |
Sequence analysis
The regions surrounding the TALEN target sites were amplified using the primer set pY365/pY366, and Q5 Hot Start High-Fidelity 2× Master Mix (New England Biolabs) from genomic DNA extracted from genome-edited plants without detectable transgenes. The amplicons were cloned into pJET1.2/blunt Cloning Vector (Thermo Fisher Science, USA), and randomly selected colonies were used for sequencing analysis.
Steroidal glycoalkaloid (SGA) measurements of SSR2-edited potato plants
The leaves of in vitro cultured SSR2-edited potato plants were harvested and lyophilized. α-Solanine and α-chaconine were extracted and quantified as described previously (Nakayasu et al. 2017).
Results
Targeted genome editing in regenerated shoots
SSR2 is the first genome-edited gene in potatoes and the key gene in the steroidal glycoalkaloid (SGA) biosynthetic pathway (Sawai et al. 2014). We infected potato stems with Agrobacterium harboring the TALEN expression vector targeting to the gene (Figure 1) and induced the regenerated shoots without selection for transformed cells. The targeted mutagenesis was first screened by HMA with crude template DNA. Of the 371 regenerated shoots produced in four independent experiments, 24 samples showed extra bands that were not detected in the controls, suggesting the presence of mutations in the TALEN target site (Table 2). To confirm the stability of the mutations, HMA was conducted using purified genomic DNA extracted from the samples after at least three times subculture. As a result, 19 lines still showed the extra-bands, indicating stable mutations of SSR2 in the regenerated shoots (Figure 2).
Table 2. Genome editing efficiency in regenerated shoots from Agrobacterium infected stem explants without selection.
| Experiment | No. of infected stem explants | Co-cultivation time with Agrobacterium | No. of stem without Agrobacterium overgrowth | No. of isolated regenerated shoots | No. of HMA positive samples (crude genomic DNA, 1st assay) | No. of HMA positive samples (purified genomic DNA, 2nd assay) | No. of genome edited samples without transgene integration |
|---|---|---|---|---|---|---|---|
| 1 | 100 | 3 days | 100 | 67 | 5 | 3 | 1 (1.5%) |
| 2 | 100 | 7 days | 52 | 28 | 4 | 3 | 1 (3.6%) |
| 3 | 200 | 6 days | 200 | 132 | 11 | 7 | 2 (1.5%) |
| 4 | 190 | 3 days | 187 | 144 | 4 | 6 | 3 (2.1%) |
| Total | 590 | — | 539 | 371 | 24 (6.5%) | 19 (5.1%) | 7 (1.9%) |
Figure 2. Polymerase chain reaction (PCR) verification of the non-transgenic sterol side chain reductase 2 (SSR2) mutant lines. Purified genomic DNA extracted from regenerated shoots was used as a PCR template. SSR2 and SSR1 are the TALEN target and potential off-target, respectively. Eight primer sets were used to amplify the partial T-DNA regions between the left and right border sequences. The amplified regions are shown in Figure 1. The plasmid pYS_026-SSR2_C was used as a positive control for the transgene amplifications. NT, non-transgenic Sassy. The seven lines did not show amplification of any transgene tested were marked by asterisks.
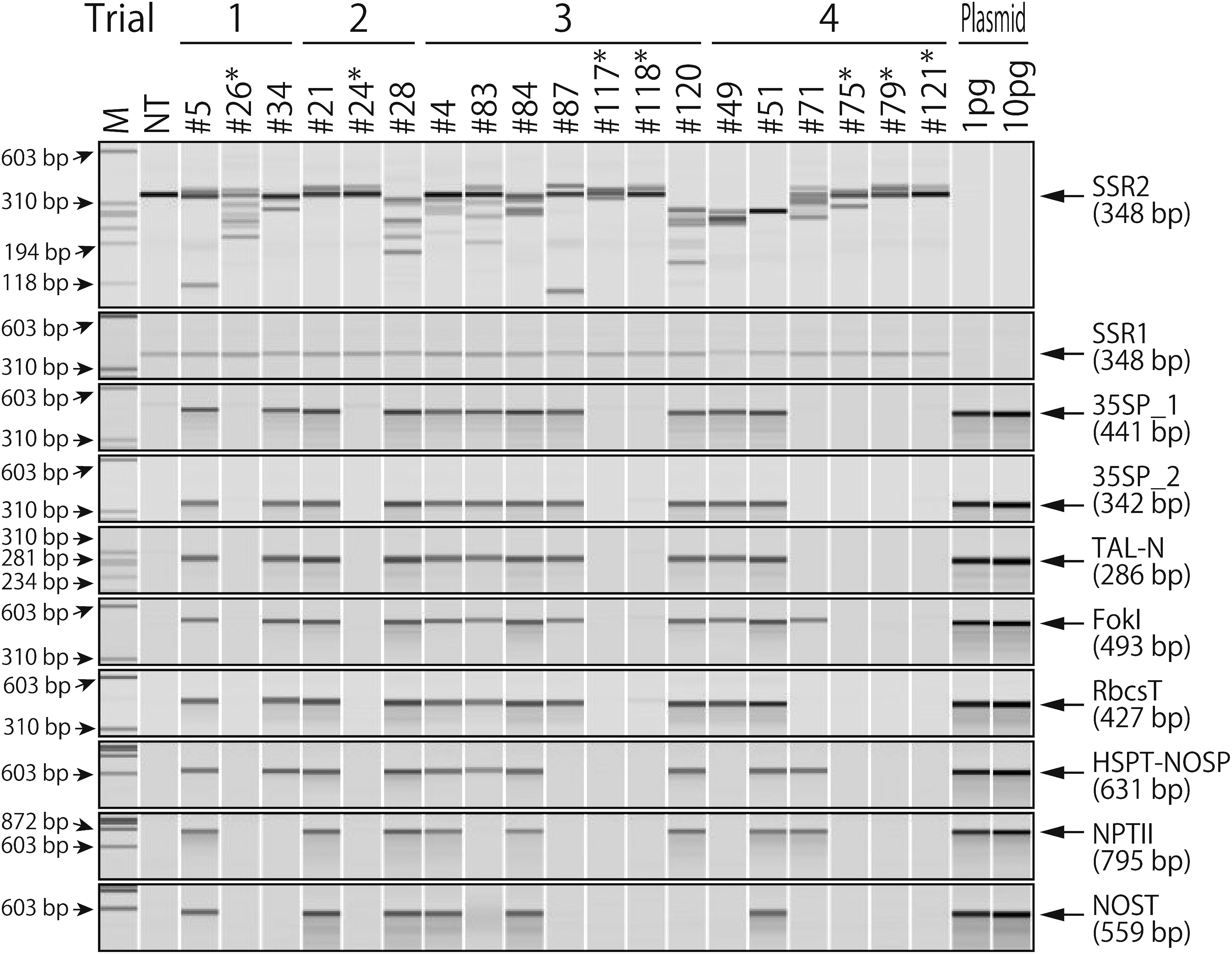
No T-DNA insert into the regenerated lines
To assess T-DNA integration in the genome-edited regenerated plants, we performed genomic PCR using eight primer pairs for transgenes (Figure 1). The endogenous SSR1 gene was also amplified from all samples to check the quality and quantity of the template genomic DNA and off-target activity. Of the 19 lines, only six (#5, #21, #28, #4, #84, and #51) showed amplification of all the transgenes tested. Another six lines (#34, #83, #120, #49, and #71) showed partial T-DNA insertion (Figure 2). The remaining seven lines (#26, #24, #117, #118, #75, #79, and #121) did not show amplification of any transgene tested (Figure 2, asterisks). Therefore, these seven lines did not contain transgene and had been genome-edited by transient expression. We did not observe clear changes in genome-editing efficiency among the different times of co-cultivation with Agrobacterium (Table 2).
Targeted mutations and SGA accumulation in regenerated plants
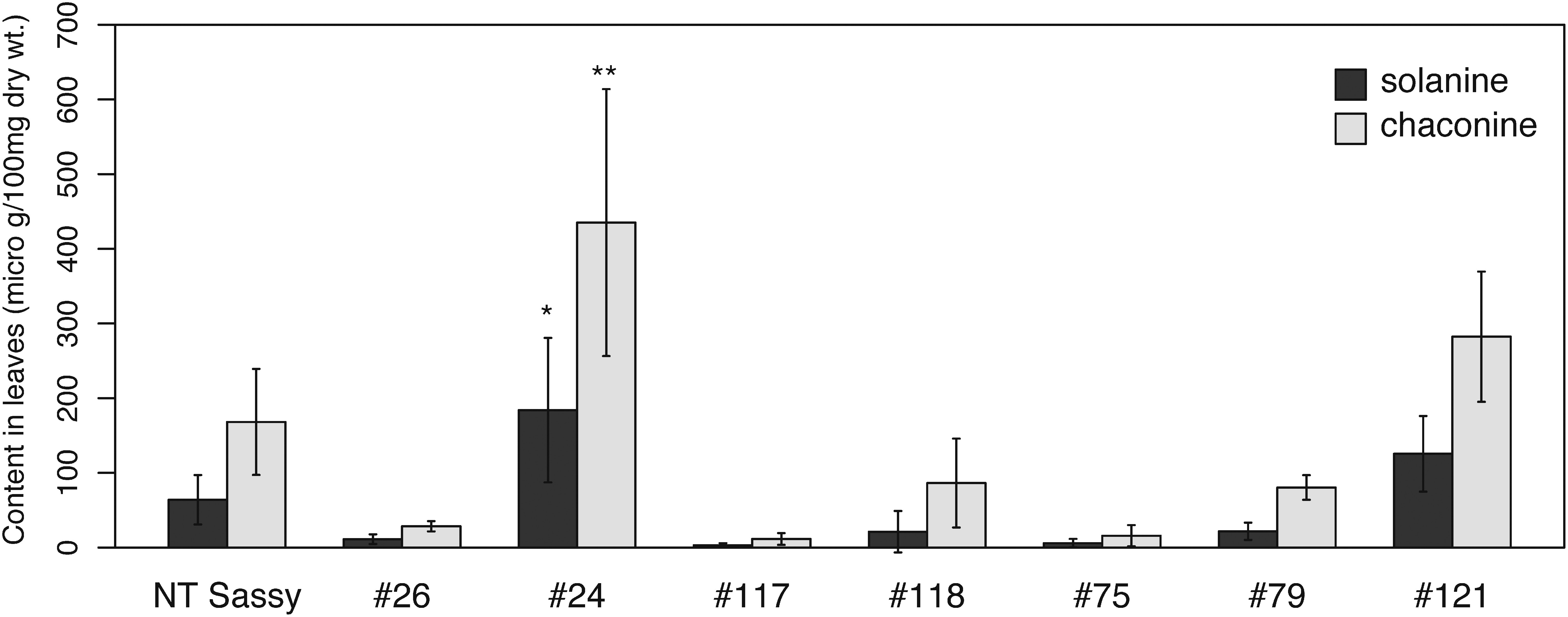
To evaluate the nucleotide sequence of the genome-edited SSR2 allele, sequences from fifteen or sixteen independent E. coli clones were determined. The results indicated that five lines (#26, #24, #118, #79, and #121) had both mutated and intact SSR2 alleles in their genomes (Figure 3). In lines #75 and #117, three nucleotide deletions (−5, −8, and −51 bp) and three mutations (+1, −2, and −21 bp) were detected respectively (Figure 3). No intact sequence was found among both sequences. As previously reported (Sawai et al. 2014; Yasumoto et al. 2019), we could not observe any difference in growth among the SSR2-knockout lines and the control plants (data not shown). We performed liquid chromatography-mass spectrometry (LC-MS) analysis to evaluate the SGA levels in the regenerated plants. The levels of the predominant SGAs, α-solanine, and α-chaconine in the regenerated lines that had no intact SSR2 sequences (#75 and #117) were much lower than in the non-transgenic sample (Figure 4). The other regenerated lines tested, which had intact SSR2 without transgene integration, showed varying levels of SGA accumulation. The results suggest the lines #75 and #117 should have complete disrupted-SSR2 allele.
Figure 3. The target SSR2 sequence in the regenerated lines with no detectable transgenes. Multiple alignment of the SSR2 target sequence and PCR products amplified from regenerated lines #26, #24, #117, #118, #75, #79, and #121. Gray uppercase letters indicate TALEN-recognition sites. Dashes and black uppercase letters in the sequence alignments indicate nucleotide deletions and insertions, respectively. Deletion sizes are indicated to the right of the alignment.

Figure 4. The steroidal glycoalkaloid (SGA) contents of regenerated lines with mutated SSR2 sequences. The α-solanine and α-chaconine levels in non-transformed Sassy and genome-edited lines without detectable transgenes (#26, #24, #117, #118, #75, #79, and #121). The leaves from in vitro culture were analyzed for each line. Error bars indicate the standard deviation of three technical replicates. Dunnett’s test indicate statistical significance (* p<0.05, ** p<0.01).
Discussion
Genome editing is a powerful tool not only for generating mutants for basic plant science research, but also for crop breeding for agriculture (e.g., Chandrasekaran et al. 2016; Sawai et al. 2014; Yasumoto et al. 2019; Wang et al. 2014). Since it is difficult to deliver genome-editing reagents as RNA or protein into plant cells, genome-editing research in plant science has had to depend almost completely on the generation of transgenic plants expressing site-specific nucleases using stable transformation methods. However, the genome-edited plants produced by stable transformants must be treated as genetically modified organisms (GMOs). The generation of null segregants (harboring the targeted mutation without transgenes) by crossing mutants is one way to overcome this problem (Pacher and Puchta 2017; Wolt et al. 2016). In highly homozygous plants such as rice and soybean, genome-edited plants are usually crossed to generate null segregants (Haun et al. 2014; Li et al. 2012). However, the progenies of highly heterozygous crops, such as potato, have completely different agricultural traits from their parent lines, such as different yields. In this study, we infected potato stem explants with Agrobacterium to transiently express TALEN proteins in potato cells. Unlike conventional plant genome editing research, no antibiotics or herbicides were added to the regeneration medium for selection of transgenic cells. The shoots regenerated from the sample without selection were analyzed by HMA to confirm targeted mutations in the SSR2 SGA biosynthetic gene; 5.1% (19 lines from 371 regenerated shoots) had stable SSR2 mutations. We confirmed the presence of T-DNA insertions in the genomes of these lines. Since we could not amplify any of the transgenes tested in seven regenerated lines, this suggested that the introduced T-DNA harboring the TALEN expression cassette was transiently transcribed and translated, and introduced mutations in SSR2 that were then degraded by endogenous nucleases before the T-DNA was inserted into the genome. The contribution of each SSR2 allele in SGA biosynthesis is unknown. Therefore, it is possible that the varying levels of SGAs in genome-edited plants with intact SSR2 sequences is caused by their mutation pattern. It is unknown what caused the # 24 SGA content to be higher than that of control. It is possible that SSR2 was activated by the mutation. Generally, SGA is rarely low, but occasionally SGA content is increased by the environmental stimuli (such as light and wounding). Since at least the disruption of all SSR2 alleles is enough for reducing SGA levels in potato, two genome-edited plants (#75 and #117) with no intact SSR2 sequence showed much lower accumulation levels than that of control. We generated SSR2 mutants with the desired reduced SGA phenotype and no detectable transgenes.
Previously, we used the same TALEN expression vector to generate genome-edited potato lines by stable transformation. As a result, all the three transgenic potato lines had the targeted mutations in SSR2 (Yasumoto et al. 2019). The frequencies of genome editing in the present study was lower than that of conventional stable transformation approach. However, we obtained genome edited lines without transgene by transient TALEN expression. The efficiency of transgene-free genome editing in the present study (7 transgene free mutated shoots/371 regenerated shoots, 1.9%) was comparable with that of previous studies using particle bombardment in wheat or protoplast transfection in potato (Clasen et al. 2016; Zhang et al. 2016). We used highly active Platinum TALEN (Sakuma et al. 2013; Yasumoto et al. 2019). It may be difficult to achieve high genome-editing efficiency using other site-specific nucleases. Chen et al. (2018) performed genome editing using Agrobacterium-mediated transient CRISPR-Cas9 expression in tobacco, and reported about several percent mutation rate for all regenerated shoots. Their efficiency of transgene-free genome editing was lower than that of our study. They used the phytoene desaturase gene as the target gene; this makes it easy to screen the regenerated shoots with targeted mutations based on their albino phenotype. Because the albino phenotype should be due to knockout of all four PDS alleles, the genome editing efficiency might be underestimated in that report. In our study, we clearly showed that mutants with invisible phenotypes (e.g., metabolite changes) from Agrobacterium-infected samples can be screened simply by PCR with comparable genome-editing efficiency. Many factors would be optimized, including bacterial strain, cell concentration, and co-culture time, to ensure higher efficiency of transgene-free genome editing using Agrobacterium.
We used PCR to detect insert T-DNA segments in genome-edited potatoes. Although PCR is highly sensitive, it is difficult to test complete T-DNA region because it depends on the primers used. Recently, a paper on detection of foreign DNA segments in genome-edited rice using high-throughput DNA sequencing has been published (Itoh et al. 2020). We expect that a system that can easily assess transgenes in the genome edited crops will be authorized.
The new varieties generated in this study should have almost the same genome as the parent cultivar, Sassy, except for the SSR2 sequences and SGA content, since we did not cross the genome-edited plants to remove the integrated transgenes. The strategy used here will make a substantial contribution to the fine-tuning breeding of vegetative crops, including potato. The strategy is like mutagenesis, so we named Agrobacterial mutagenesis to genome editing using transient gene expression by Agrobacterium. Since it is difficult to assess SGA levels in many progenies, the levels are usually evaluated late in the breeding cycle. Lenape, a potato variety released in 1968, was withdrawn in 1970 since it had unsafe solanine levels (Akeley et al. 1968; Anonymous 1970). By applying our strategy, a potato line with much SGA level could be improved by disrupting the SGA biosynthesis gene to breed novel cultivars with both unchanged agricultural traits and lower SGA levels. Our strategy could also be applied to modify other important traits of potato breeding, such as cold-induced sweetening by vacuolar invertase, enzymatic browning by polyphenol oxidase, etc., by knocking out single genes.
Acknowledgments
This work was supported by the Cross-Ministerial Strategic Innovation Promotion Program (SIP; Council for Science, Technology and Innovation, Cabinet Office, Japan) and the administration of individual commissioned project study (Development of new varieties and breeding materials in crops by genome editing, the Ministry of Agriculture, Forestry and Fisheries, Japan). We thank Chika Shimadzu for technical support.
References
- Akeley RV, Mills WR, Cunningham CE, Watts J (1968) Lenape: A new potato variety high in solids and chipping quality. Am Potato J 45: 142–145 [Google Scholar]
- Anonymous (1970) Name of potato variety Lenape withdrawn. Am Potato J 47: 103 [Google Scholar]
- Chandrasekaran J, Brumin M, Wolf D, Leibman D, Klap C, Pearlsman M, Sherman A, Arazi T, Gal-On A (2016) Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol Plant Pathol 17: 1140–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Li W, Katin-Grazzini L, Ding J, Gu X, Li Y, Gu T, Wang R, Lin X, Deng Z, et al. (2018) A method for the production and expedient screening of CRISPR/Cas9-mediated non-transgenic mutant plants. Hortic Res 5: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clasen BM, Stoddard TJ, Luo S, Demorest ZL, Li J, Cedrone F, Tibebu R, Davison S, Ray EE, Daulhac A, et al. (2016) Improving cold storage and processing traits in potato through targeted gene knockout. Plant Biotechnol J 14: 169–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer R, Vaquero-Martin C, Sack M, Drossard J, Emans N, Commandeur U (1999) Towards molecular farming in the future: Transient protein expression in plants. Biotechnol Appl Biochem 116: 113–116 [PubMed] [Google Scholar]
- Haun W, Coffman A, Clasen BM, Demorest ZL, Lowy A, Ray E, Retterath A, Stoddard T, Juillerat A, Cedrone F, et al. (2014) Improved soybean oil quality by targeted mutagenesis of the fatty acid desaturase 2 gene family. Plant Biotechnol J 12: 934–940 [DOI] [PubMed] [Google Scholar]
- Hwang H-H, Yu M, Lai E-M (2017) Agrobacterium-mediated plant transformation: Biology and applications. Arabidopsis Book 15: e0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Onuki R, Tsuda M, Oshima M, Endo M, Sakai H, Tanaka T, Ohsawa R, Tabei Y (2020) Foreign DNA detection by high- throughput sequencing to regulate genome-edited agricultural products. Sci Rep 10: 4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapila J, De Rycke R, Van Montagu M, Angenon G (1997) An Agrobacterium-mediated transient gene expression system for intact leaves. Plant Sci 122: 101–108 [Google Scholar]
- Li T, Liu B, Spalding MH, Weeks DP, Yang B (2012) High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol 30: 390–392 [DOI] [PubMed] [Google Scholar]
- McCallum CM, Comai L, Greene EA, Henikoff S (2000) Targeting induced local lesions IN genomes (TILLING) for plant functional genomics. Plant Physiol 123: 439–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayasu M, Umemoto N, Ohyama K, Fujimoto Y, Lee HJ, Watanabe B, Muranaka T, Saito K, Sugimoto Y, Mizutani M (2017) A dioxygenase catalyzes steroid 16α-hydroxylation in steroidal glycoalkaloid biosynthesis. Plant Physiol 175: 120–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolia A, Proux-Wéra E, Åhman I, Onkokesung N, Andersson M, Andreasson E, Zhu LH (2015) Targeted gene mutation in tetraploid potato through transient TALEN expression in protoplasts. J Biotechnol 204: 17–24 [DOI] [PubMed] [Google Scholar]
- Pacher M, Puchta H (2017) From classical mutagenesis to nuclease-based breeding: Directing natural DNA repair for a natural end-product. Plant J 90: 819–833 [DOI] [PubMed] [Google Scholar]
- Sakuma T, Ochiai H, Kaneko T, Mashimo T, Tokumasu D, Sakane Y, Suzuki K, Miyamoto T, Sakamoto N, Matsuura S, et al. (2013) Repeating pattern of non-RVD variations in DNA-binding modules enhances TALEN activity. Sci Rep 3: 3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai S, Ohyama K, Yasumoto S, Seki H, Sakuma T, Yamamoto T, Takebayashi Y, Kojima M, Sakakibara H, Aoki T, et al. (2014) Sterol side chain reductase 2 is a key enzyme in the biosynthesis of cholesterol, the common precursor of toxic steroidal glycoalkaloids in potato. Plant Cell 26: 3763–3774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheerman S, Bevan MW (1988) A rapid transformation method for Solanum tuberosum using binary Agrobacterium tumefaciens vectors. Plant Cell Rep 7: 13–16 [DOI] [PubMed] [Google Scholar]
- Voytas DF (2013) Plant genome engineering with sequence-specific nucleases. Annu Rev Plant Biol 64: 327–350 [DOI] [PubMed] [Google Scholar]
- Voytas DF, Gao C (2014) Precision genome engineering and agriculture: Opportunities and regulatory challenges. PLoS Biol 12: e1001877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu J-L (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol 32: 947–951 [DOI] [PubMed] [Google Scholar]
- Wolt JD, Wang K, Yang B (2016) The regulatory status of genome-edited crops. Plant Biotechnol J 14: 510–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumoto S, Umemoto N, Lee HJ, Nakayasu M, Sawai S, Sakuma T, Yamamoto T, Mizutani M, Saito K, Muranaka T (2019) Efficient genome engineering using Platinum TALEN in potato. Plant Biotechnol 36: 167–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liang Z, Zong Y, Wang Y, Liu J, Chen K, Qiu J-L, Gao C (2016) Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat Commun 7: 12617. [DOI] [PMC free article] [PubMed] [Google Scholar]


