As a result of the clinical similarity of coronavirus disease 2019 (COVID-19) with other flulike syndromes, patients were tested for other pathogens of respiratory tract infections as well, especially in the first weeks of the epidemic. In those weeks, we used the multiplex PCR BioFire FilmArray Pneumonia panel (BioFire Diagnostics; bioMérieux, Marcy l’Étoile, France), allowing the detection of 33 respiratory pathogens with a run time of 1 hour. Here we report the aetiologic pattern of respiratory tract infections diagnosed at the Amedeo di Savoia Hospital, the regional reference centre for infectious diseases of Piedmont, north-western Italy, during the first weeks of the COVID-19 epidemic.
We retrospectively reviewed clinical and microbiologic records generated via routine clinical practice (ethical approval not required) from 162 consecutive patients (93 male; median age, 64 years; range, 12–93 years) from 12 February to 31 March 2020 with respiratory symptoms who were screened with FilmArray and real-time reverse transcriptase PCR for COVID-19 diagnosis after multiple sample collection in Copan UTM transport medium (Copan, Brescia, Italy). For the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) specific RNA, a multiplex Real Time PCR Kit (Liferiver, San Diego, CA, USA) was used (detection limit, 1 × 10³ copies/mL). The kit measures three virus genes simultaneously: SARS-CoV-2 gene E, N and ORF1ab. The ABI Prism 7500 thermal cycler was used for PCR amplification (Thermo Fisher Scientific, Waltham, MA, USA) [1]. The results of the two tests were compared by the chi-square test.
We included nasopharyngeal swab (123/162, 75.9%), bronchoalveolar lavage (17/162, 10.5%), bronchoaspirate (12/162, 7.4%) and sputum (10/162, 6.2%) samples. SARS-CoV-2 RNA was detected in 56 (34.6%) of 162 samples, while FilmArray showed the presence of viruses and/or bacteria in 63 (38.9%) of 162. FilmArray results (28 samples with multiple microorganisms) identified bacteria in 53 (84.1%) of 63 patients and viruses in 22 (34.9%) of 63, as follows: Staphylococcus aureus (25/63, 39.7%), Haemophilus influenzae (15/63, 23.8%), group B streptococci (6/63, 9.5%), influenza A virus (5/63, 7.9%) and influenza B, coronaviruses and rhinoviruses/enteroviruses (each 4/63, 6.3%).
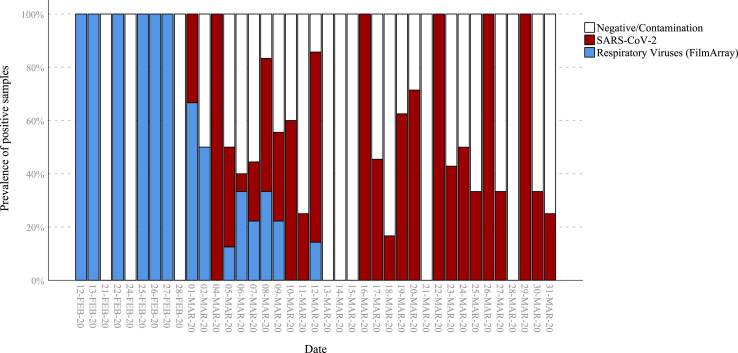
FilmArray results were positive in 10 of 56 SARS-CoV-2–infected patients (17.8% vs. 50% in SARS-CoV-2–negative samples, χ2 = 15.9, p <0.001); interestingly, no other virus was found in SARS-CoV-2–positive patients (0 vs. 20.7%, χ2 = 13.4, p <0.001), but only Staphylococcus aureus (7/10), Haemophilus influenzae (2/10), Escherichia coli (2/10), Moraxella catarrhalis, group B streptococci, Klebsiella pneumoniae and Enterobacter cloacae (1/10 each), most probably as a result of colonization or bacterial superinfection in two COVID-19 patients whose bronchoaspirate tested positive for Haemophilus influenzae plus Escherichia coli and Staphylococcus aureus, respectively (Table 1 ). Soon after the onset of the global SARS-CoV-2 epidemic, the presence of other respiratory viruses declined and even disappeared (Fig. 1 ).
Table 1.
Positive FilmArray results for respiratory tract microbiologic characterization in patients according to SARS-CoV-2 infection test result
| Parameter | FilmArray POSITIVE SARS-COV-2 NEGATIVE | FilmArray POSITIVE SARS-COV-2 POSITIVE |
|---|---|---|
| No. of patients | 53 | 10 |
| Age (years) (±SD) | 62.4 ± 18 | 63.3 ± 18 |
| Sex (F/M) | 25/28 | 4/6 |
| No. of nasopharyngeal swabs | 40 | 8 |
| No. of other type of specimens | 13a | 2b |
| No. of pathogens detected | ||
| Staphylococcus aureus | 18 | 7 |
| Moraxella catarrhalis | 7 | 1 |
| Haemophilus influenzae | 13 | 2 |
| Streptococcus agalactiae | 5 | 1 |
| Mycoplasma pneumoniae | 4 | — |
| Streptococcus pneumoniae | 3 | — |
| Streptococcus pyogenes | 3 | — |
| Pseudomonas aeruginosa | 2 | — |
| Enterobacter cloacae | 3 | 1 |
| Klebsiella oxytoca | 1 | — |
| Klebsiella pneumoniae | 4 | 1 |
| Escherichia coli | 3 | 1 |
| Adenovirus | 1 | — |
| Influenza virus B | 4 | — |
| Influenza virus A | 5 | — |
| RSV | 2 | — |
| Rhinovirus/enterovirus | 4 | — |
| Coronavirus (OC43, NL63, 229E) | 4 | — |
| Metapneumovirus | 3 | — |
| Total pathogens | 89 | 14 |
SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Sputum (n = 6), bronchoalveolar lavage (n = 4) and brochoaspirates (n = 3).
Bronchoaspirates (n = 2).
Fig. 1.
Prevalence of positive samples by date. SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Our data are consistent with the absence of other respiratory virus coinfection in SARS-CoV-2 patients and a low bacterial coinfection rate mostly reflecting a carriage status.
Recently several articles have reported the possible concomitant presence of respiratory pathogens in subjects with COVID-19. Viral and bacterial pathogens have been described in case reports, case series and four cohorts: a low prevalence was reported (1.6–6.5%), with the exception of a single recent report (24/116, 20.7%) [2]. Phenomena like viral interference, common receptor usage, different inoculum size or simply resource competition might explain why dual or multiple concurrent viral respiratory infections are rare [3,4]. Few articles have addressed the clinical features of coinfected patients, with the relevant exception of a child with SARS-CoV-2, metapneumovirus and respiratory syncytial virus admitted to an intensive care unit, with no increased risk of severe disease reported [5]. In line with this, a 9-year-old child included in the French Alps cluster was coinfected with SARS-CoV-2, rhinovirus/enterovirus and influenza H1N1 and asymptomatic, but no secondary cases were found among his contacts [6].
In conclusion, a multiplex PCR system for the rapid diagnosis of respiratory infections revealed that there is virtually no concomitant infection in patients positive for SARS-CoV-2. In addition, interestingly, with the spread of SARS-CoV-2, other respiratory pathogens have seemed to vanish.
Transparency declaration
AC has received honoraria from AbbVie, BMS, Gilead, Janssen-Cilag, MSD and Viiv and he is currently receiving research grants from BMS, Gilead and Viiv. GDP and SB have received honoraria from AbbVie, BMS, Gilead, Janssen-Cilag, MSD and Viiv. The remaining authors report no conflicts of interest relevant to this letter.
Editor: L. Leibovici
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2020.08.012.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W. Detection of 2019 novel coronavirus (2019-nCoV) by realtime RT-PCR. Euro Surveill Bull Eur Sur Mal Transm Eur Commun Dis Bull. 2020;25:23–30. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim D., Quinn J., Pinsky B., Shah N.H., Brown I. Rates of coinfection between SARS-CoV-2 and other respiratory pathogens. JAMA. 2020:E1–E2. doi: 10.1001/jama.2020.6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pinky L., Dobrovolny H.M. Coinfections of the respiratory tract: viral competition for resources. PLoS One. 2016;11:1–19. doi: 10.1371/journal.pone.0155589. e0155589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goka E.A., Vallely P.J., Mutton K.J., Klapper P.E. Single, dual and multiple respiratory virus infections and risk of hospitalization and mortality. Epidemiol Infect. 2015;143:37–47. doi: 10.1017/S0950268814000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Danis K., Epaulard O., Bénet T., Gaymard A., Campoy S., Bothelo-Nevers E. Cluster of coronavirus disease 2019 (COVID-19) in the French Alps, 2020. Clin Infect Dis. 2020;71:825–832. doi: 10.1093/cid/ciaa424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang S., Liu P., Xiong G., Yang Z., Wang M., Li Y. Coinfection of SARS-CoV-2 and multiple respiratory pathogens in children. Clin Chem Lab Med. 2020:1–2. doi: 10.1515/cclm-2020-0434. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.