Abstract
Vernalization accelerates flowering after prolonged winter cold. Transcriptional and epigenetic changes are known to be involved in the regulation of the vernalization response. Despite intensive applications of next-generation sequencing in diverse aspects of plant research, genome-wide transcriptome and epigenome profiling during vernalization response has not been conducted. In this work, we present the first comprehensive analyses of transcriptomic and epigenomic dynamics during the vernalization process in Arabidopsis thaliana. Six major clusters of genes exhibiting distinctive features were identified. Temporary changes in histone H3K4me3 levels were observed that likely coordinate photosynthesis and prevent oxidative damage during cold. In addition, vernalization induced a stable accumulation of H3K27me3 over genes encoding many development-related transcription factors, resulting in either inhibition of transcription or a bivalent status of the genes. Lastly, FLC-like and VIN3-like genes were identified that appear to be novel components of the vernalization pathway.
Keywords: Vernalization, Transcriptome, Histone modification, RNA-seq, ChIP-seq, Arabidopsis
Introduction
Temperature is an important environmental cue that, coupled with day length, cues plants to initiate flowering. For most winter-annual and biennial plants, prevention of flowering before winter and induction of flowering after winter is required for successful reproduction. Cold itself is not sufficient since temperature fluctuations in fall might be falsely taken as the passing of winter. A timing mechanism is needed to distinguish long-term winter cold from short-term chilling stress. Therefore, the vernalization process evolved which accelerates flowering only after prolonged cold exposure. In winter-annual Arabidopsis thaliana, vernalization is regulated by two major loci: FLOWERING LOCUS C (FLC) and FRIGIDA (FRI) (Shindo et al., 2005, Werner et al., 2005, Coustham et al., 2012). FLC encodes a MADS-box transcription factor that represses the expression of downstream targets (Michaels and Amasino, 1999, Hepworth et al., 2002, Lee and Lee, 2010). FRI acts with other proteins in a complex to upregulate FLC expression (Johanson et al., 2000, Choi et al., 2005, Schmitz et al., 2005, Kim et al., 2006, Geraldo et al., 2009, Jiang et al., 2009, Hu et al., 2014, Li et al., 2018). High level of FLC and its clade members prevent flowering by repressing floral integrator genes such as FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) (Sheldon et al., 2000, Hepworth et al., 2002, Moon et al., 2003, Michaels et al., 2005, Helliwell et al., 2006, Searle et al., 2006, Gu et al., 2013) and also feedback regulations operate between FLC and floral integrators (Chen and Penfield, 2018, Luo et al., 2019), forming intricate regulatory networks that control flowering. FLC is stably repressed by prolonged winter cold, thereby enables rapid induction of flowering under favorable day length in spring. The vernalization-triggered FLC repression is mitotically stable and it is reset only during meiosis to ensure the requirement of vernalization in the next generation (Sheldon et al., 2008). This “memory” of winter indicates the involvement of epigenetic regulation. Indeed, studies performed during the past decade have begun to elucidate the role of histone modification and chromatin structural dynamics in FLC repression (Sung and Amasino, 2004, Mylne et al., 2006, Sung and Amasino, 2006, Sung et al., 2006, De Lucia et al., 2008, Kim et al., 2010, Heo and Sung, 2011, Crevillen et al., 2013, Rosa et al., 2013, Questa et al., 2016, Kim and Sung, 2017).
Before vernalization, FLC chromatin is enriched with active histone marks, including histone acetylation, H3K4me3, H3K36me3, and etc., which are likely deposited by FRI complexes (Kim et al., 2005, Schmitz et al., 2005, Jiang et al., 2009, Tamada et al., 2009, Whittaker and Dean, 2017). Early in vernalization, the expression of antisense noncoding RNAs are induced at FLC locus. Expression of these RNAs, termed COOLAIR (cold induced long antisense intragenic RNA) correlates with the reduction in expression of the FLC sense transcript, and COOLAIR physically associates with FLC chromatin resulting in depletion of H3K36me3 (Swiezewski et al., 2009, Csorba et al., 2014). Recently, expression of VP1/ABI3-LIKE1 (VAL1) was shown to be necessary for vernalization-mediated reduction of histone acetylation at FLC. VAL1 is a B3 domain protein recruited to FLC through its direct binding to RY motifs within the nucleation region. VAL1 recruits histone deacetylase HDA19 to FLC chromatin (Questa et al., 2016, Yuan et al., 2016).
In late stage of vernalization, prolonged cold induces sufficient amount of VERNALIZATION INSENSITIVE 3 (VIN3), a PHD-finger domain protein, which forms heterodimer with VIN3-LIKE 1 (VIL1) and together recruit POLYCOMB REPRESSIVE COMPLEX 2 (PRC2) to the nucleation region in the first intron of FLC. This PHD-PRC2 complex catalyzes the tri-methylation of histone H3K27, a well-characterized repressive mark (De Lucia et al., 2008). At this stage, H3K27me3 modifications are confined within the nucleation region. Meanwhile, expression of another noncoding RNA, termed COLDAIR (cold assisted intronic noncoding RNA) is induced from the sense direction of the first intron of FLC. Loss of COLDAIR results in a vernalization-insensitive phenotype (Heo and Sung, 2011). COLDAIR interacts with CURLY LEAF (CLF), the enzymatic core of PRC2, to facilitate its sequence-specific binding at the FLC locus (Heo and Sung, 2011, Kim and Sung, 2017). When temperatures warm, VIN3 levels decline rapidly, but VIL1-PRC2 remains bound to the FLC locus. H3K27me3 spreads until it covers the entire genomic region of FLC. It is not clear how or why the spreading of repressive marks occurs only when the temperature warms. The accelerated enzymatic activity of histone modifying complexes at higher temperatures might explain this phenomenon. LIKE HETEROCHROMATIN PROTEIN 1 (LHP1) proteins are enriched at FLC following PRC2 action, and these proteins are necessary for stable maintenance of the epigenetically repressed state of FLC in warm conditions. VAL1 recruits LHP1 to FLC through direct protein-protein interactions (Yuan et al., 2016). The repressive state of FLC is stably inherited through many cycles of cell division during subsequent growth and development.
In addition to changes in histone modifications, chromatin structural changes also occur at FLC locus during vernalization. Vernalization induces physical clustering of FLC alleles in the nucleus, which requires Polycomb complex components VERNALIZATION INSENSITIVE 2 (VRN2) and VIL1, but not LHP1 (Rosa et al., 2013). An interaction between the 5’ and 3’ regions of the FLC chromatin is formed before cold and is disrupted during the early stage of vernalization. The mechanism of formation this loop is not clear. It is known that the transcriptional status of FLC is not relevant to this process and that the components of PRC2 complex are not necessary (Crevillen et al., 2013). An intragenic chromatin loop is also induced by vernalization, which could be responsible for the vernalization-induced spreading of H3K27me3 marks along FLC chromatin (Kim and Sung, 2017). A non-coding RNA derived from the FLC promoter called COLDWRAP is involved in the formation of the intragenic chromatin loop.
Given the quantitative nature of vernalization response, it would be helpful to have a comprehensive picture of the transcriptome and epigenome changes that occur during the vernalization process. To date, few vernalization-related next-generation sequencing datasets have been generated, and most come from food crops such as pak choi (Brassica rapa subsp. chinensis) and radish (Raphanus sativus L.) (Sun et al., 2015, Liu et al., 2016). The RNA-seq and ChIP-seq analyses collected at multiple time points during vernalization described in this work represent the first comprehensive profiling of the transcriptome and epigenome dynamics of vernalization in Arabidopsis thaliana.
Results
Transcriptional dynamics of vernalization in Arabidopsis thaliana
To capture genome-wide transcriptional dynamics during the vernalization process, seven samples were collected, termed NV (without cold exposure), V1h (1-hour cold), V1d (1-day cold), V10d (10-day cold), V20d (20-day cold), V40d (40-day cold), and T10 (40-day cold followed by 10-day normal growth temperature). The well-known patterns of FLC repression and VIN3 induction were successfully captured by the RNA-seq (Fig. 1A, 1B and Table S1). FLC belongs to a small gene family, including FLC and the MADS AFFECTING FLOWERING genes MAF1 (also known as FLOWERING LOCUS M), MAF2, MAF3, MAF4, and MAF5. The RNA-seq data showed relatively similar dynamics of MAF1 and FLC that differed from the patterns of expression of MAF2 and MAF3 (Fig. 1A). MAF4 and MAF5 were of too low abundance for a pattern of expression to be confidently differentiated by RNA-seq. Of the VIN family members, VIL2 showed the highest expression, whereas VIL3 was barely detected. Levels of VIL1 and VIL2 were largely stable across vernalization (Fig. 1B).
Figure 1. Changes in the Arabidopsis transcriptome during the course of vernalization.
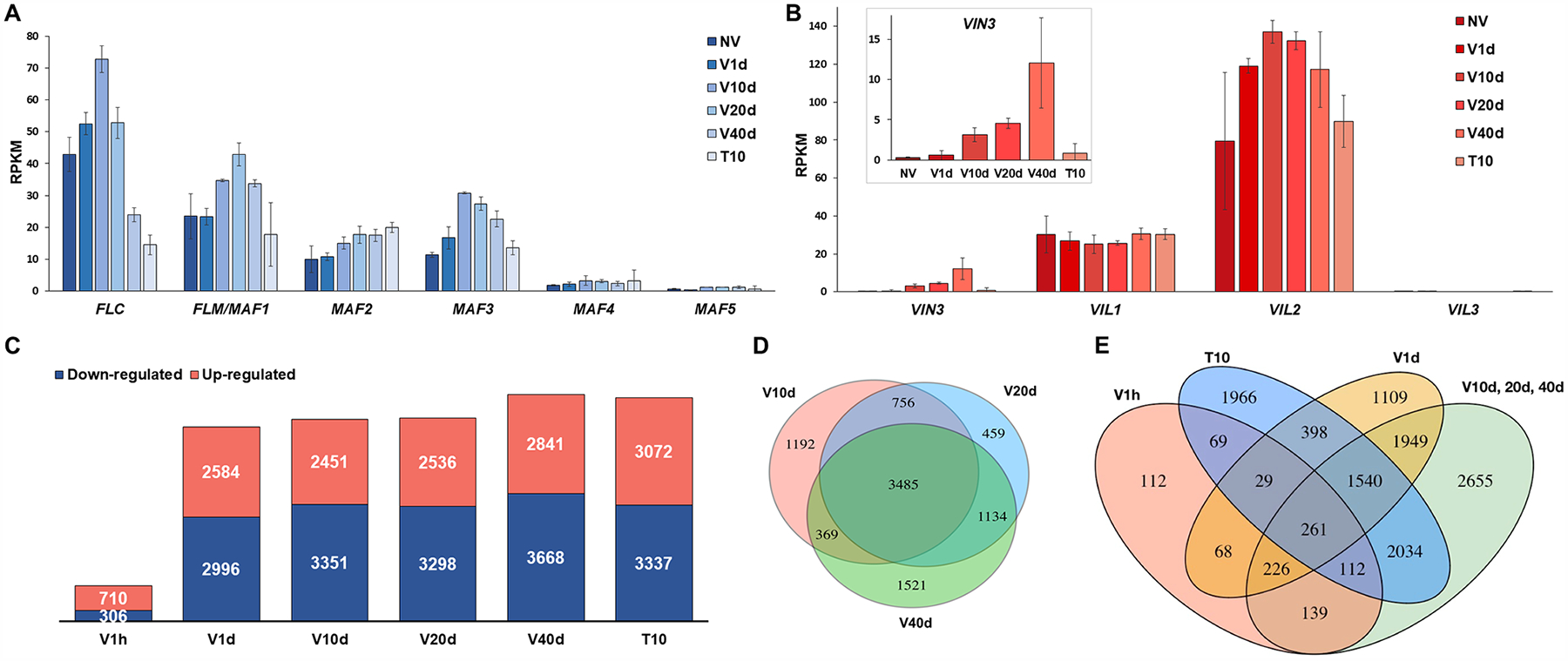
(A) Quantitative measurement of expression levels of FLC family genes over a time course during vernalization as in Reads Per Kilobase of transcript, per Million mapped reads (RPKM). Error bars were generated based on normalized read counts within each locus from 2 biological replicates. (B) Quantitative measurement of expression levels of VIN3 family genes over a time course during vernalization as in RPKM. Error bars were generated based on normalized read counts within each locus from 2 biological replicates. (C) Bar graph showing total numbers of differentially up-regulated (red) and down-regulated (blue) genes at each time point relative to NV. (D) Venn diagram showing the overlapping and uniquely differentially regulated genes at V10d, V20d, and V40d. (E) Venn diagram showing the overlapping and uniquely differentially regulated genes at V1h, V1d, V10d/20d/40d, and T10.
Differentially expressed genes (DEGs) were by comparison of vernalized samples to NV samples. All the time points, except V1h, showed similar numbers of up- and down-regulated genes (Fig. 1C). Only 710 up-regulated and 306 down-regulated genes were identified in V1h samples, indicating that the downstream cascades of cold-regulated genes were initiated by a limited number of early responsive genes. V10d, V20d, and V40d shared 3,485 differentially regulated genes in common (Fig. 1D), suggesting that expression of many cold-regulated genes was stably maintained regardless of the duration of cold. That 3,976 of the 5,580 genes expressed during V1d were also expressed at one or more of the V10d, V20d, and V40d time points indicate that long-term responses built up within just one day of cold exposure are maintained (Fig. 1E).
To fully explore the time-course dynamics, differentially expressed genes from all time points were clustered based on expression patterns. Six major clusters with distinct transcriptional dynamics were identified (Fig. 2). Cluster 1 consisted of a small number of early responsive genes (545) that were up- or down-regulation within just 1 hour of cold treatment (Fig. 2A). Gene Ontology (GO) analysis revealed that this cluster was enriched in hormone-related genes, including ethylene, abscisic acid, cytokinin, and salicylic acid (Fig. 2A), which is consistent with the fact that plant hormones are usually among the “first responders” upon environmental changes and stresses. Members of cluster 2 (2,272 genes) and cluster 3 (1,744 genes) exhibited relatively constant up- and down-regulation, respectively, at time points V1d to V40d (Fig. 2B, 2C), indicating that these genes are regulated during cold. GO analysis showed that up-regulated genes in cluster 2 were enriched in translation-related terms (Figure 2B), such as ribosome biogenesis, translation initiation, RNA secondary structure unwinding, and rRNA processing, suggesting that protein synthesis is boosted during prolonged cold, probably in order to make up for the reduced enzymatic activity at low temperature.
Figure 2. Clustering analysis of transcriptome data collected during vernalization.
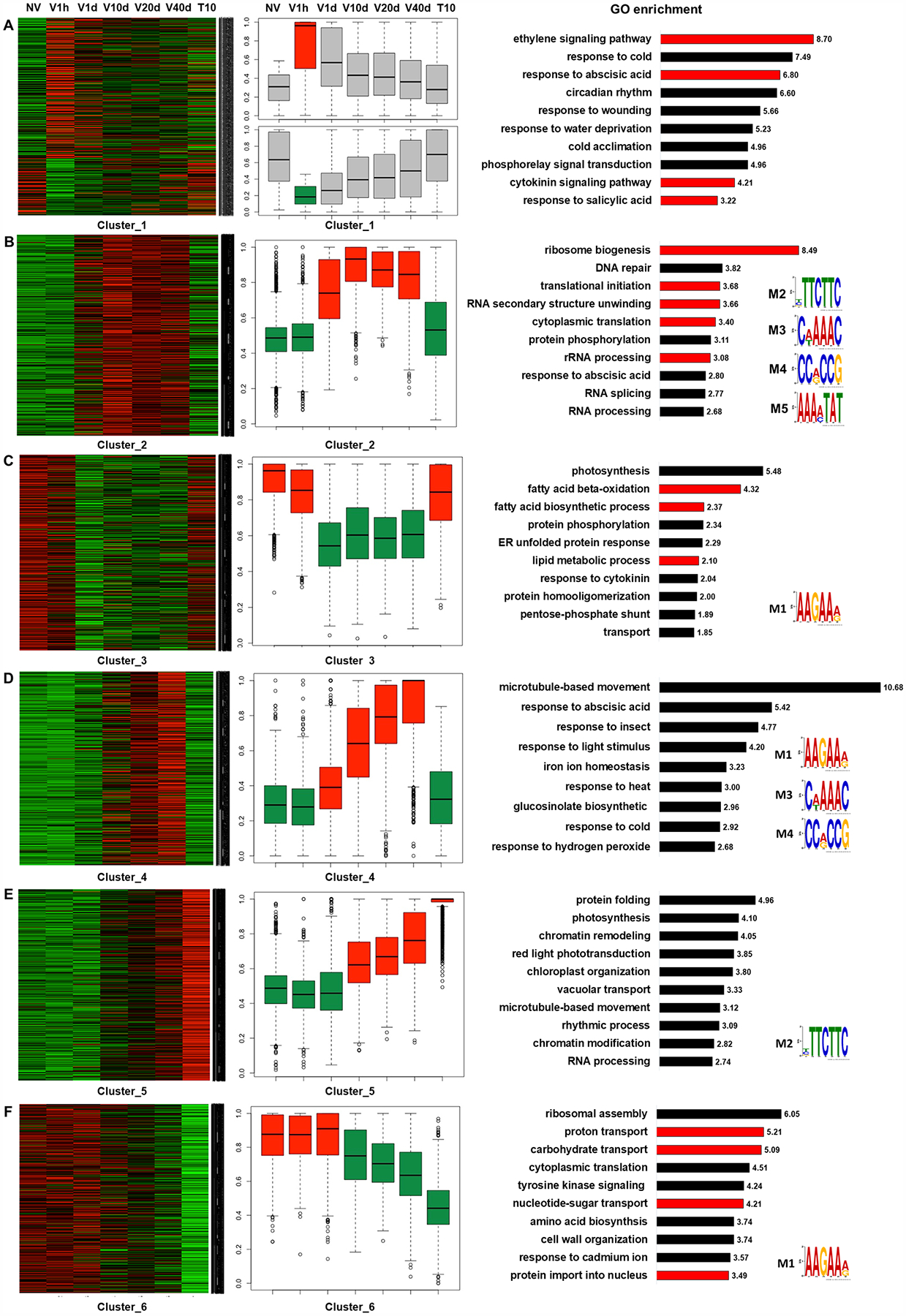
(A-F) Clusters 1 to 6, respectively, were generated from k-means clustering of transcription profiles obtained over the time course of vernalization. Shown from left to right for genes in the indicated cluster are heatmaps of gene expression at each time point, normalized box plots of genes expression at each time point, enriched GO terms, and motifs enriched within clustered genes if detected. Grey boxes indicate no-significant difference, red boxes indicate significant up-regulation, and green boxes indicate significant down-regulation in normalized box plots of genes expression.
Photosynthesis and lipid processing genes were enriched in cluster 3 (Figure 2C), indicating that in Arabidopsis photosynthesis is repressed during cold. In evergreen plants, winter cold inhibits the efficiency of photosynthetic CO2 assimilation, which could lead to over-excitation and increased photo-oxidative damage if plants continue to absorb light energy. Therefore, down-regulation of light absorption balances the supply and utilization of energy during cold and protect plants from photo-oxidative damage (Oquist and Huner, 2003). Indeed, the photosynthesis-related genes in cluster 3 mostly encode components of light harvesting complexes, suggesting that Arabidopsis utilizes a similar strategy as evergreens during winter cold.
Genes in cluster 4 (911 genes) had expression that was gradually induced during cold instead as opposed to the constant high levels observed for genes in cluster 2 (Figure 2D). This pattern resembles that of VIN3 during vernalization. Genes related to microtubule movement were present in this cluster. Genes in cluster 5 (1,828 genes) and cluster 6 (1,650 genes) had gradually increased or decreased expression during cold, respectively, and levels of these genes were maintained after the return to warm temperature (Figure 2E, 2F). The pattern of expression of genes in cluster 6 resembled that of FLC during vernalization. No functional terms showed obvious enrichment in these two clusters.
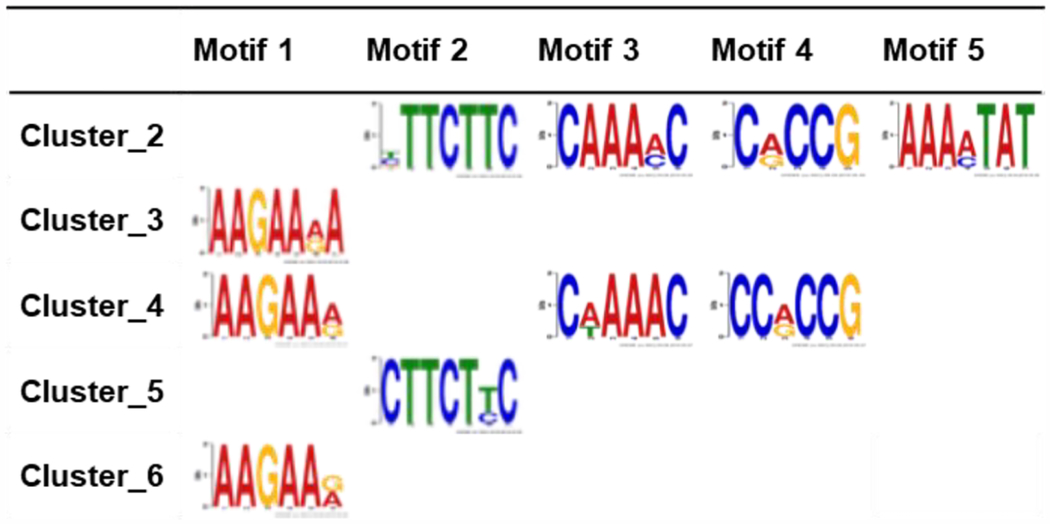
To identify potential protein binding motifs enriched in the six major clusters, 3 kilobases of promoter sequence for each gene were extracted and analyzed using the MEME program for motif discovery and analysis. Five major motifs were discovered with distinct and overlapping enrichment among the clusters (Fig. 2, far right; Table 1). Motif 1 (M1) was enriched in clusters 3, 4, and 6 and motif 2 (M2) in clusters 2 and 5. Motif 3 (M3) and motif 4 (M4) were both enriched in clusters 2 and 4, and motif 5 (M5) was only enriched in cluster 2. Overall, gene clusters up-regulated during vernalization showed higher motif enrichment, suggesting that induction of genes was regulated by the combination of transcription factors, whereas repression might require distinct mechanisms. The transcription factors with binding motifs that match those enriched in genes differentially expressed during vernalization are listed in Table 2. Many of these transcription factors are involved in salt stress, hormone signaling, and flowering regulation. Motif 4 was of great interest since it is the binding motif for the ERF/AP2 transcription factors involved in hypoxia signaling (Yang et al., 2011, Gasch et al., 2016), and VIN3 was reported to be induced by hypoxia (Bond et al., 2009). It is also noteworthy that a recent finding showed that hypoxia also stabilizes the VRN2-containing PRC2 complex to mediate the repression of FLC during vernalization (Gibbs et al., 2018), implicating biological relevance between hypoxia and vernalization.
Table 1.
Motifs enriched in each cluster of genes differentially regulated during vernalization.
|
Table 2.
Transcription factors (TFs) with binding motifs similar to those identified in genes differentially regulated during vernalization.
| Motif 1 (M1) | Motif 2 (M2) | Motif 3 (M3) | Motif 4 (M4) | Motif 5 (M5) | |
|---|---|---|---|---|---|
| Matched TFs | NTL4, NAC2 | NTL8 | CRC | ERF17, ERF38, ERF74, WIND3, WIMD4 | RVE1, RVE4, RVE5, RVE8 LHY, EPR1 |
| Family | NAC domain family | NAC domain family | Plant-specific YABBY family | DREB subfamily of ERF/AP2 family | Homeodomain-like family |
| Reported functions | drought response, JA biosynthesis, salt stress, embryogenesis, stamen development | salt stress, flowering trichome formation | fatty acid biosynthesis, carpel development | hypoxia response, osmotic stress, ethylene signaling, redox sensing | circadian clock, photoperiodic flowering, auxin signaling, chlorophyll synthesis |
Histone modification changes during vernalization
Three well-studied histone modifications, H3K27me3, H3K4me3, and H3K36me3, were analyzed by ChIP-seq at NV, V40d, and T10 (Fig. 3 and Table S2). We first analyzed the distribution of histone marks on FLC chromatin at these time points (Fig. 3A). An enrichment of H3K27me3 was observed around the FLC transcription start site at V40d compared to NV. The gene body of FLC exhibited a minor increase of H3K27me3 during cold, whereas the major spreading and coverage of repressive marks occurred only after plants were moved back to warm temperature at T10 (Fig. 3A). Consistent with the increase of H3K27me3, a decrease of H3K36me3 along the gene body of FLC was observed as a function of time, although the overall enrichment of H3K36me3 was much lower than that of H3K27me3 at all stages. The H3K4me3 marks showed little change during and after vernalization (Fig. 3A). Besides the transcription start site, a minor H3K4me3 peak was observed around the 3’-end of FLC. This may be involved in the formation of chromatin loop or the expression of antisense transcripts (Swiezewski et al., 2009, Csorba et al., 2014). Indeed, a minor peak of H3K4me3 in 3’ FLC, as well as the localization of the H3K4 methyltransferase COMPASS-like in this region, have been reported (Li et al., 2018). In addition, the 3’ localized COMPASS-like appears to be involved in 5’ to 3’ gene looping (Li et al., 2018).
Figure 3. Genome-wide analysis of histone modifications during the course of vernalization.
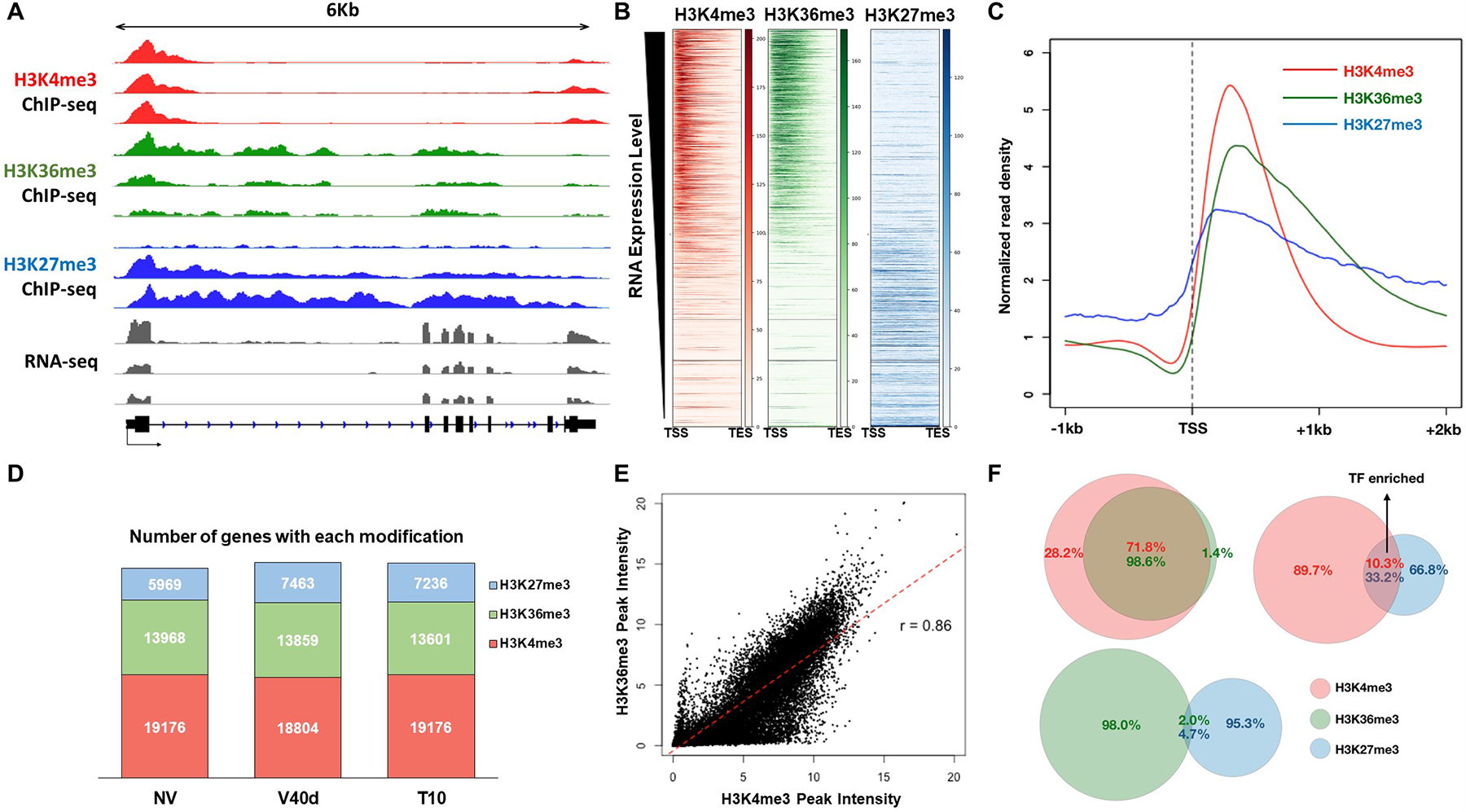
(A) Genome browser illustration of normalized ChIP-seq and RNA-seq results at FLC locus. H3K4me3 tracks are shown in red, H3K36me3 in green, and H3K27me3 in blue. RNA-seq results are shown in grey colors. (B) Heatmaps of H3K4me3 (red), H3K36me3 (green), and H3K27me3 (blue) over all coding genes in Arabidopsis genome. Each row represents the normalized read density from transcription start site (TSS) to transcription end site (TES) of each gene, ranked by transcription level from the highest (top) to the lowest (bottom). (C) Averaged profiles of H3K4me3 (red), H3K36me3 (green), and H3K27me3 (blue) distributions around TSS regions over all coding genes in Arabidopsis genome. (D) Bar graph showing total number of peaks called by MACS2 within each sample. (E) Correlation plot of genome-wide H3K4me3 and H3K36me3 densities. (F) Venn diagrams showing overlapped among different histone marks from all three time points.
At the genome-wide level, H3K4me3 and H3K36me3 were enriched on actively transcribed genes, whereas H3K27me3 was observed over genes expressed at low levels and over silenced genes (Fig. 3B). H3K4me3 peaks were confined around transcription start sites with an average span of about 2 kilobases, whereas H3K36me3 and H3K27me3 were diffused into gene bodies (Fig. 3C). Most of the H3K4me3 peaks did not change much in terms of location or intensity during vernalization (Fig. 3D). H3K36me3 largely followed the pattern of H3K4me3 distribution (Fig. 3E, 3F) as expected since both are active histone marks. In total, 19,176, 18,804, and 19,176 peaks were called for H3K4me3 in NV, V40d, and T10 samples, respectively, and 13,968, 13,859, and 13,601 peaks were called for H3K36me3 at these time points (Fig. 3D). These numbers represent two-thirds of coding genes in Arabidopsis genome, which roughly matches the number of actively transcribed genes. Thus, nearly every actively transcribed gene has an H3K4me3 peak located at their transcription start site. The lower numbers of genes marked by H3K36me3 compared to H3K4me3 are probably due to the overall lower enrichment levels for H3K36me3 compared to H3K4me3 detected in our ChIP-seq analysis. As expected due to the synergistic function of these modifications in transcriptional regulation, 98.6% of H3K36me3 peaks overlapped with an H3K4me3 peak (Fig. 3F). H3K36me3 mark is known to prevent cryptic transcription and facilitate RNA polymerase elongation through gene bodies (Wagner and Carpenter, 2012).
A much smaller number of peaks were called for H3K27me3 than for the active histone marks, with 5,969, 7,463, and 7,236 peaks in NV, V40d, and T10 samples, respectively (Fig. 3D). Only 2.0% to 4.7% of peaks, depending on the time point, overlapped between H3K36me3 and H3K27me3 marks. Surprisingly, a large portion of H3K27me3 peaks (33.2%) overlapped with H3K4me3 marks (Fig. 3F), resulting in the so-called “bivalent” status for the underlying genes (Vastenhouw and Schier, 2012, Voigt et al., 2013, Harikumar and Meshorer, 2015, Zaidi et al., 2017). GO analysis indicated that transcription factors were highly enriched in the group of genes with bivalent histone marks, suggesting that the combination of H3K27me3 and H3K4me3 could be required for flexible regulation of transcription factors in Arabidopsis. The transcription factors with bivalent marks are listed in Supplemental Table S3.
Vernalization causes an overall increase of H3K27me3 in Arabidopsis genome
Vernalization had minimal effect on H3K36me3 distribution, as peaks from V40d and T10 correlated almost perfectly with NV samples (Fig. 4A). A temporary effect of vernalization on H3K4me3 was enhanced diffusion at V40d; patterns of H3K4me3 at T10 were similar to those at NV. In contrast, vernalization-induced H3K27me3 changes observed at V40d were maintained after plants were moved back to warm temperature at T10 (Fig. 4A). To quantify the differential peaks among samples, reads within each peak were extracted and converted to digital counts for statistical analysis. Consistent with the correlation analysis, only 9.9% of H3K36me3 peaks were differentially regulated; a slightly higher percentage of H3K4me3 peaks (15.6%) were differentially regulated. In contrast, over one-third of H3K27me3 peaks (36.6%) were differentially regulated by vernalization (Fig. 4B). Surprisingly, the direction of change of these differentially regulated peaks was not evenly distributed: Cold induced an overall decrease of H3K4me3 and increase of H3K27me3 at V40d (Fig. 4C). The absence of down-regulated H3K27me3 peaks indicated a potential unidirectional action of H3K27me3 for switching off genes and suggests that, once added, the H3K27me3 mark is difficult to remove. To confirm the ChIP-seq results, several genes were randomly chosen for validation. Quantitative real-time PCR (qRT-PCR) showed validated the ChIP-seq analysis (Fig. S1).
Figure 4. Characteristics of histone modification-enriched loci during the course of vernalization.
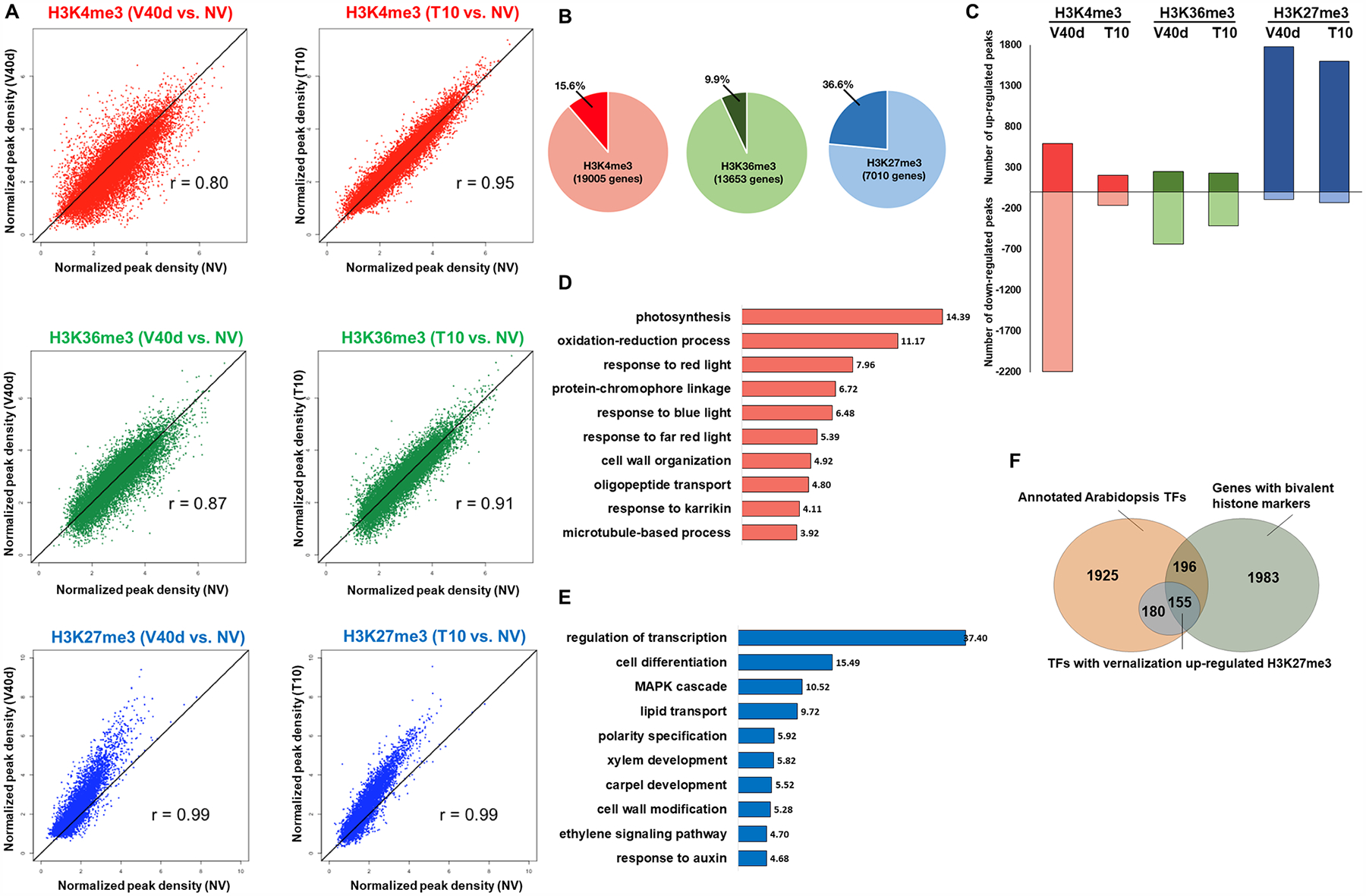
(A) Correlation plots of densities of H3K4me3 in red, H3K36me3 in green, and H3K27me3 in blue in V40d vs. NV (left) and T10 vs. NV (right) samples. (B) Pie graph showing the percentages of histone modification peaks differentially regulated during vernalization. (C) Bar graph showing the number of vernalization up-regulated (darker hues) and down-regulated (lighter hues) H3K4me3 (red), H3K36me3 (green), and H3K27me3 (blue) peaks. (D) Bar graph showing top GO terms ranked by enrichment score from H3K4me3 temporarily down-regulated loci with p-value. (E) Bar graph showing top GO terms ranked by enrichment score from H3K27me3 up-regulated loci with p-value. (F) Venn diagrams showing overlaps among all annotated transcription factors (TFs), genes with bivalent histone marks, and TFs with increased H3K27me3 by vernalization.
The group of genes with cold-induced reduction of H3K4me3 were enriched with photosynthesis-related terms (Fig. 4D), as was cluster 3 of cold down-regulated genes (Fig. 2C). Therefore, it is likely that in Arabidopsis the temporary removal of H3K4me3 marks at the transcription start site decreases the expression of photosynthesis genes to prevent photo-oxidative damage during cold and quickly restores their activities in warm temperature to ensure normal growth and development. The factors involved in this temperature-induced H3K4me3 change are currently unknown.
Interestingly, transcription factors from almost all families were strongly enriched in the group of genes with H3K27me3 peaks up-regulated during vernalization (Fig. 4E). Of the 335 transcription factor genes that had strongly up-regulated H3K27me3, 155 were marked also with H3K4me3 (Fig. 4F). GO analysis revealed that floral regulator genes were enriched in the group of transcription factors with vernalization-induced H3K27me3 modifications (Table 3), confirming that vernalization promotes the transition from vegetative growth to reproductive growth through epigenetic switching off of regulatory hub genes in Arabidopsis.
Table 3.
Functional annotations of transcription factors with vernalization-induced H3K27me3 up-regulation.
| Functional annotation | Number of genes | Enrichment score | P-value |
|---|---|---|---|
| cell differentiation | 59 | 40.82 | 1.30E-13 |
| ethylene signaling pathway | 30 | 17.4 | 1.00E-19 |
| flower development | 25 | 11.12 | 2.20E-13 |
| carpel development | 10 | 9.4 | 1.30E-11 |
| ovule development | 11 | 6.85 | 5.40E-09 |
| regulation of secondary cell wall biogenesis | 8 | 6.82 | 5.20E-09 |
| vegetative to reproductive phase transition of meristem | 14 | 6.1 | 3.30E-08 |
| gibberellic acid signaling pathway | 11 | 4.82 | 6.50E-07 |
| specification of flora organ identity | 6 | 4.57 | 1.30E-06 |
| trichome differentiation | 7 | 4.32 | 2.40E-06 |
| transmitting tissue development | 4 | 3.42 | 2.20E-05 |
| auxin signaling pathway | 14 | 3.21 | 3.80E-05 |
Identification of FLC-like and VIN3-like transcripts
We hypothesized that any gene with a repression pattern similar to that of FLC or an induction pattern similar to that of VIN3 upon cold treatment could have similar functions during vernalization. Cluster 6 and cluster 4 included genes with patterns of expression similar to those of FLC and VIN3, respectively (Fig. 2D, 2F). A dynamic time warping (DTW) algorithm was used to identify optimal matches within each cluster. DTW was first used in speech recognition for measuring the similarity between soundtracks (Sakoe and Chiba, 1978). The advantage of DTW over simple pairwise comparison is that it allows the stretch and compression of input sequences. In this work, the time-series transcriptional dynamics of two genes were given as inputs, and a distance score was then calculated (Fig. 5A, 5B). The lower the distance score, the higher the similarity of the two expression patterns (Fig. 5B).
Figure 5. Identification of genes with expression similar to that of FLC and VIN3 using the dynamic warping algorithm.
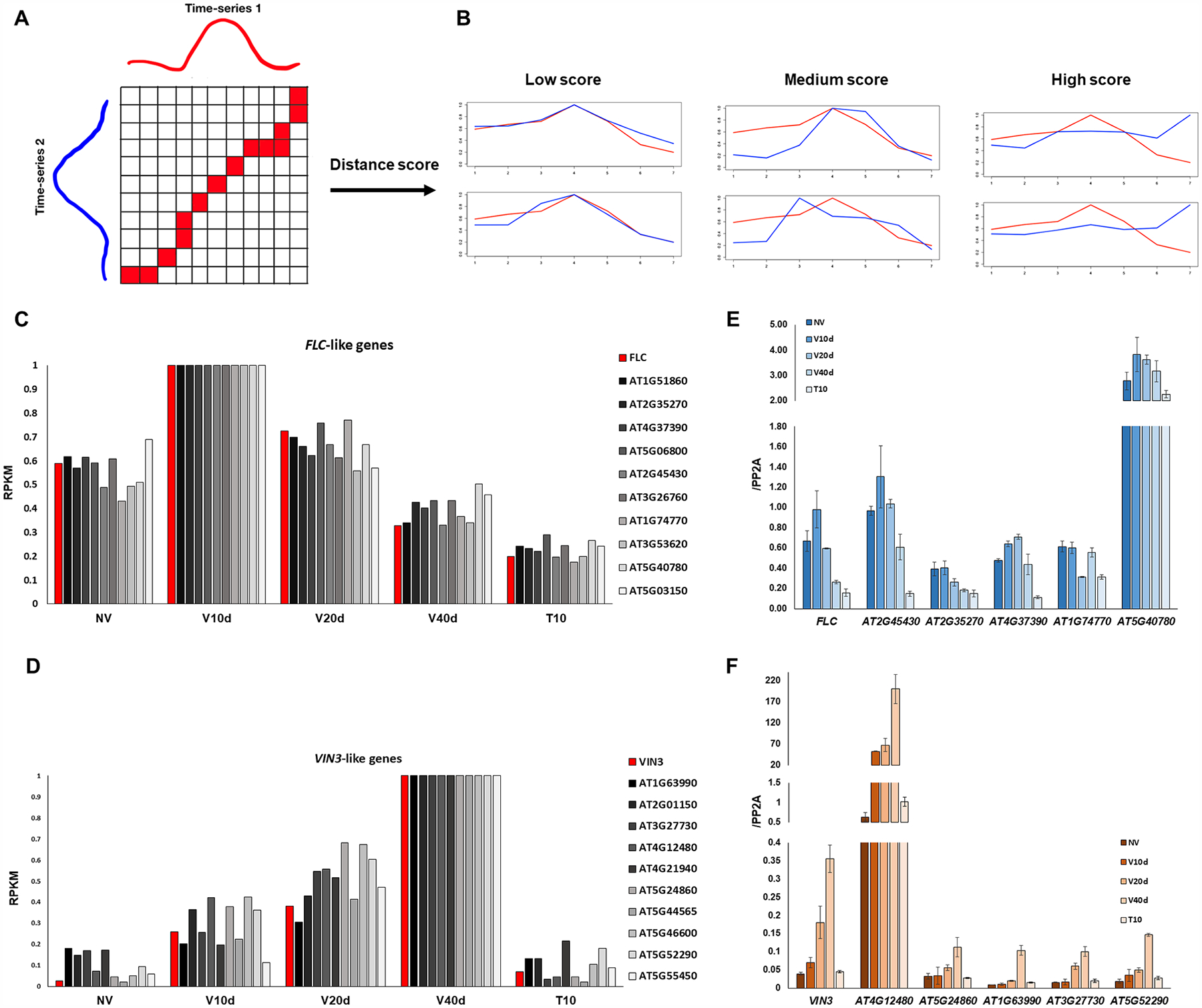
(A) Illustration of Dynamic Time Warping algorithm. (B) Examples of sequences with low (left), medium (middle), and high (right) distance scores. (C) Bar graph showing the RNA-seq results of the 10 genes (in shades of grey) that most closely resemble the vernalization-mediated repression pattern of FLC (red). (D) Bar graph showing the RNA-seq results of the 10 genes (in shades of grey) that most closely resemble the vernalization-mediated induction pattern of VIN3 (red). (E) qRT-PCR validation of expression patterns of FLC-like genes over time. Expression levels are relative to PP2A. (n = 3) (F) qRT-PCR validation of VIN3-like genes. Expression levels are relative to PP2A (n = 3).
All genes within cluster 6 were compared to FLC using DTW, and the resulting distance scores were ranked from low to high (Fig. 5C). Genes in cluster 4 genes were ranked for similarity to the VIN3 expression pattern (Fig. 5D). To validate the transcriptional profiles of the FLC- and VIN3-like genes identified from the DTW algorithm, transcripts from five genes from each category were quantified in time-course samples. The results of qRT-PCR were consistent with the RNA-seq profiles (Fig. 5E, 5F). Several of the FLC- and VIN3-like genes are known floral regulators and cold-related genes that could be novel components of the vernalization pathway (Tables 4 and 5). Interestingly, of the top 10 VIN3-like genes, three encode factors involved in meiotic recombination (Table 5), suggesting that VIN3 may have a role in meiotic recombination or may regulate chromatin contact.
Table 4.
Genes with expression patterns similar to FLC during the course of vernalization.
| Locus | Name | Protein domain | Reported function |
|---|---|---|---|
| AT1G51860 | - | LRR, protein kinase | - |
| AT2G45430 | AHL22 | AT-hook DNA-binding | regulation of flowering |
| AT2G35270 | AHL21 | AT-hook DNA-binding | patterning and differentiation of reproductive organs |
| AT4G37390 | AUR3 | GH3 auxin-responsive | negative component in auxin signaling |
| AT5G06800 | - | Myb-like DNA-binding | - |
| AT3G26760 | - | glucose dehydrogenase | - |
| AT1G74770 | BTSL1 | zinc finger | negative regulator of iron deficiency |
| AT3G53620 | PPA4 | pyrophosphatase | regulate pyrophosphate levels |
| AT5G40780 | LHT1 | transmembrane | high-affinity transporter for cellular amino acid uptake |
| AT5G03150 | JKD | zinc finger | epidermal patterning in root meristem |
Table 5.
Genes with expression patterns similar to VIN3 during the course of vernalization.
| Locus | Name | Protein domain | Reported function |
|---|---|---|---|
| AT5G44565 | - | transmembrane | - |
| AT5G55450 | LTP4.4 | - | lipid transport and pathogen resistance |
| AT2G01150 | RHA2B | zinc finger | ABA signaling and drought response |
| AT1G63990 | SPO11–2 | DNA topoisomerase VI | regulate meiotic recombination |
| AT3G27730 | MER3 | DEAD-like helicase | required for meiotic crossover formation |
| AT4G12480 | EARLI1 | plant lipid transfer | resistance to low temperature and fungal infection |
| AT4G21940 | CPK15 | protein kinase | - |
| AT5G52290 | SHOC1 | similar to XPF endonucleases | required for class-I meiotic crossover formation |
| AT5G24860 | FPF1 | - | regulate the competence to flowering |
| AT5G46600 | - | malate transporter | - |
AHL family genes act as floral repressors in vernalization pathway
In the set of 10 most FLC-like genes were two AT-hook family genes, AT-HOOK MOTIF NUCLEAR LOCALIZE PROTEIN 21 (AHL21) and AT-HOOK MOTIF NUCLEAR LOCALIZE PROTEIN 22 (AHL22). qRT-PCR confirmed their expression patterns (Fig. 6B). As found in our analysis of FLC, stable increases of H3K27me3 were observed on both loci during and after vernalization (Fig. 6A). Previous studies have shown that AHL family genes are involved in control of flowering (Ng et al., 2009, Xiao et al., 2009, Yun et al., 2012, Xu et al., 2013). AHL family members exist in nearly all plant species sequenced so far, ranging from moss to higher plants. In Arabidopsis, the AHL family contains 29 members with conserved AT-hook motifs known to bind to AT-rich DNA sequences (Zhao et al., 2013, Zhao et al., 2014). In addition to roles in regulation of flowering AHL family members function in diverse aspects of plant growth and development including hypocotyl elongation, floral development, and light responses (Lim et al., 2007, Street et al., 2008, Ng et al., 2009, Xiao et al., 2009, Yun et al., 2012, Xu et al., 2013).
Figure 6. Phenotypes of AHL22 knockout and overexpression strains.
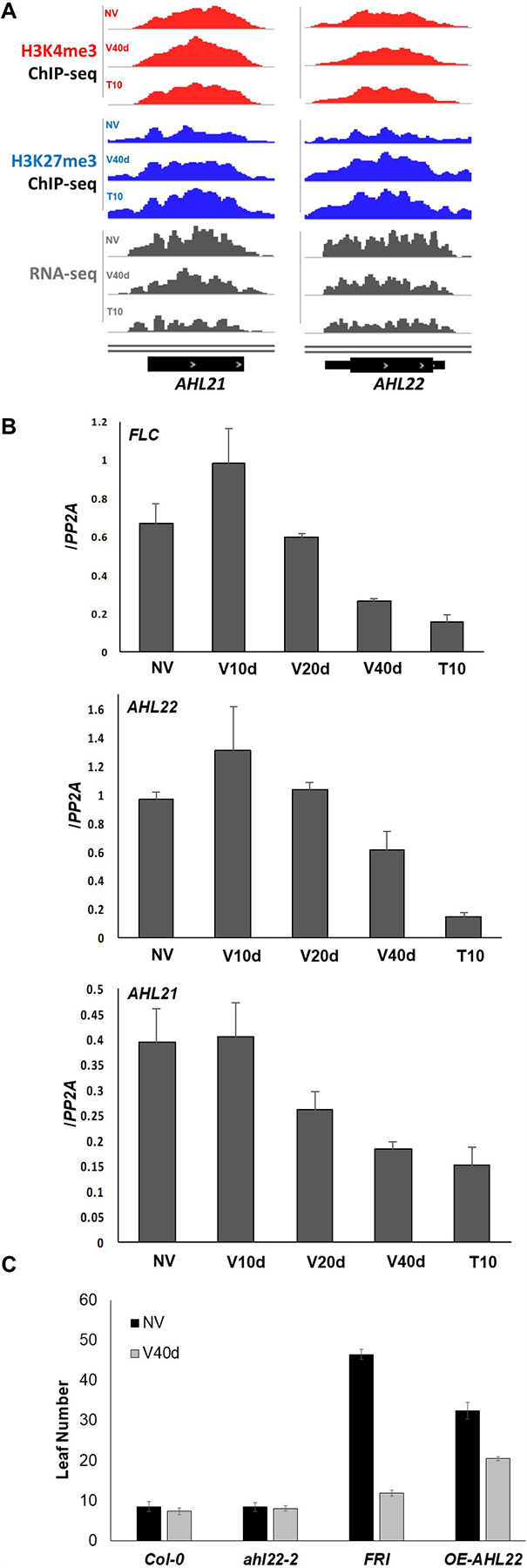
(A) Genome browser tracks showing H3K4me3 (red), H3K27me3 (blue), and RNA-seq (grey) results at AHL21 and AHL22 loci during vernalization. (B) Validation of FLC, AHL21, and AHL22 expression levels using q-RT-PCR. (n = 3) (C) Flowering phenotypes with (40V) or without (NV) vernalization of Col-0, AHL22-null mutant, FRI-null mutant, and AHL22 overexpression lines based on counting rosette leave numbers (n = 12).
AHL genes have evolved into two phylogenetic clades. Clade A are intron-less genes with only one AT-hook motif, whereas clade B are genes containing intron and one or two AT-hook motifs (Fig. S2A) (Zhao et al., 2013). Besides AHL21 and AHL22, several other AHL family members also showed FLC-like transcriptional dynamics during vernalization as well as up-regulated H3K27me3 marks (Fig. S2B), including AHL19, AHL20, AHL23, AHL24, AHL25, AHL27, and AHL29. Interestingly, all of the FLC-like AHLs belong to intron-less clade A, suggesting that clade A of AHL genes could be an ancient family involved in cold response.
To further confirm the biological function of AHL genes in vernalization, we obtained the knockout and overexpression lines of AHL22 to test its flowering phenotype with or without vernalization. The ahl22 mutants were not significantly different from wild-type plants, probably due to the highly redundant functions of AHL family members. However, overexpression of AHL22 in Col-0 rendered the plant late flowering as Col-0 (FRI) without vernalization (Fig. 6C, top). And the flowering was accelerated after 40 days of cold treatment (Fig. 6C, bottom). Quantitative measurement indicated that the overexpression of AHL22 resulted in elevated rosette leaves in Col-0 comparable to but less than that in FRI_Col-0 without vernalization (Fig. 6C, top). Vernalization partially rescued the late-flowering phenotype in AHL22 overexpression line but was less effective than that in FRI_Col-0 (Fig. 6D), suggesting that AHL22 might function in parallel to FLC in regulating downstream floral genes. Altogether, we propose that AHL family genes, especially genes belong to clade A, may be ancient yet novel floral regulators in vernalization pathway which were switched off by prolonged cold-induced H3K27me3 in order to assist the acceleration of flowering in Arabidopsis thaliana.
Discussion
This work presents the first profile of dynamic transcriptome and epigenome changes during vernalization in Arabidopsis thaliana. RNA-seq data was collected for samples without cold exposure, with 1-hour, 1-day, 10-day, 20-day, and 40-day exposure to cold, and with a 40-day cold followed by 10 days at normal growth temperature. Analyses revealed six major clusters of differentially regulated genes. Plant hormone signaling genes were among those with altered expression immediately after exposure to cold. Throughout the exposure to cold, translation-related genes were up-regulated to enabled efficient protein synthesis when enzymatic activities were limited by low temperature. Also throughout the cold exposure photosynthesis-related genes were down-regulated to prevent photo-oxidative damage caused by excessive energy production. Potential protein-binding motifs within each cluster suggest interesting candidates for further studies.
Genome-wide profiling of histone modifications, including H3K4me3, H3K36me3, and H3K27me3, showed a temporary reduction of H3K4me3 at photosynthesis-related genes after 40 days of exposure to cold and up-regulation of H3K27me3 after 40 days of cold with and without 10 days at optimal growing temperature. About one-third of the H3K27me3 peaks in all loci in the Arabidopsis genome that are marked with H3K27me3 were vernalization regulated; most of these genes encode transcription factors and most harbor bivalent marks of both H3K4me3 and H3K27me3. In mammalian systems, bivalent histone modifications play critical roles in embryonic development and cell lineage commitment (Voigt et al., 2013, Harikumar and Meshorer, 2015, Zaidi et al., 2017). Little is known about the functions of bivalent marks in Arabidopsis, but our finding that thousands of genes, including a large portion of transcription factors, harbor both H3K4me3 and H3K27me3 suggest that “bivalency” may allow rapid switching of transcription status of Arabidopsis genes critical to functions like flowering.
The time-course patterns of transcriptome and epigenome changes allowed us to identify novel components of the vernalization pathway. A number of FLC-like and VIN3-like genes were discovered through classification and pattern recognition. Among them, one AHL family gene was confirmed to be a repressor of flowering that was epigenetically silenced during vernalization. Additional candidates will be interesting targets for further studies.
Methods
Plant materials and growth conditions
The Arabidopsis Col-0 with a functional FRI allele was used as the wild-type strain. Standard growth conditions were 22 ˚C with a 16-h light/8-h dark (long day) photoperiodic cycle under white fluorescent light. Seeds were surface sterilized, placed on agar medium, and grown in the dark at 4 ˚C for 3 days for stratification. For vernalization treatment, seedlings were grown for 7 days at 22 ˚C, and then either harvested as NV or transferred to 4 ˚C under short day (8-h light/16-h dark) for 1 h (V1h), 1 day (V1d), 10 days (V10d), 20 days (V20d), and 40 days (V40d) of treatment. The T10 sample was kept at 4 ˚C for 40 days followed by 10 days at 22 ˚C before harvesting.
RNA extraction and qRT-PCR
Harvested samples were flash-frozen in liquid nitrogen. Total RNA was extracted using the Trizol/chloroform method. Extracted RNA was treated with DNase I to eliminate genomic DNA contamination. Around 2 μg of total RNA was used for cDNA synthesis using M-MLV reverse transcriptase (Promega). qRT-PCR was performed using SYBR green reaction mix (Applied Biosystems) according to the manufacturer’s instructions on a Viia7 Real-Time PCR system (Applied Biosystems).
Chromatin Immunoprecipitation (ChIP)
Seedlings were crosslinked at 4 ˚C with 1% formaldehyde solution under vacuum for 25 min. The reaction was terminated by addition of 0.125 M glycine. Crosslinked seedlings were rinsed in distilled water and then flash frozen in liquid nitrogen. ChIP was performed following the Abcam ChIP protocol (https://www.abcam.com/protocols/chip-using-plant-samples---arabidopsis) with minor adjustments. Aliquots of eluted DNA were used for qRT-PCR and for sequencing.
Library construction and sequencing
Ribosomal RNAs were depleted from the extracted RNA using RiboMinus Plant Kit (Thermo Fisher). The polyA-enrichment procedure was omitted in order to capture the total RNA. Library construction was done using NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB) following the manufacturer’s instructions. Libraries were sequenced on an Illumina Hiseq2500 platform by the Genomic Sequencing and Analysis Facility at the University of Texas at Austin.
Sequence alignment and analysis
The raw reads were trimmed and quality-filtered before aligned to the Arabidopsis thaliana TAIR10 transcriptome by Tophat (RNA-seq) or Bowtie2 (ChIP-seq). Aligned reads were converted to digital counts using Rsubread and were analyzed using edgeR. Differentially expressed genes were identified based on a 0.05 p-value and two-fold difference cut-off. Motif analysis was done by using MEME. Peaks were called by MACS2. GO analysis was done using DAVID. Clustering was done in Python using sikit-learn packages.
Supplementary Material
Table S3. Annotation of transcription factors with bivalent marks
Supplementary Figure S1. Validation of ChIP-Seq by ChIP-qPCR.
Supplementary Figure S2. Characterization of AHL family genes.
Table S1. Summary of RNA-Seq Analysis
Table S2. Summary of ChIP-Seq Analysis
Acknowledgements
We wish to thank Dr. Chung-Mo Park for sharing ahl22-2 and ahl22-OE seeds. The authors acknowledge the Texas Advanced Computing Center (TACC; http://www.tacc.utexas.edu) at The University of Texas at Austin for providing High Performance Computing resources that have contributed to the research results reported within this paper. This work was supported by NIH R01GM100108 and NSF IOS 1656764 to S. S.
Footnotes
Conflict of Interest
The authors declare no conflicts of interest.
Data Availability Statement
The data used in this study found at GSE 130291 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE130291).
References
- Bond DM, Wilson IW, Dennis ES, Pogson BJ and Jean Finnegan E (2009) VERNALIZATION INSENSITIVE 3 (VIN3) is required for the response of Arabidopsis thaliana seedlings exposed to low oxygen conditions. Plant J. [DOI] [PubMed] [Google Scholar]
- Chen M and Penfield S (2018) Feedback regulation of COOLAIR expression controls seed dormancy and flowering time. Science, 360, 1014–1017. [DOI] [PubMed] [Google Scholar]
- Choi K, Kim S, Kim SY, Kim M, Hyun Y, Lee H, Choe S, Kim SG, Michaels S and Lee I (2005) SUPPRESSOR OF FRIGIDA3 encodes a nuclear ACTIN-RELATED PROTEIN6 required for floral repression in Arabidopsis. Plant Cell, 17, 2647–2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustham V, Li P, Strange A, Lister C, Song J and Dean C (2012) Quantitative modulation of polycomb silencing underlies natural variation in vernalization. Science, 337, 584–587. [DOI] [PubMed] [Google Scholar]
- Crevillen P, Sonmez C, Wu Z and Dean C (2013) A gene loop containing the floral repressor FLC is disrupted in the early phase of vernalization. EMBO J, 32, 140–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csorba T, Questa JI, Sun Q and Dean C (2014) Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc Natl Acad Sci U S A, 111, 16160–16165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lucia F, Crevillen P, Jones AM, Greb T and Dean C (2008) A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc Natl Acad Sci U S A, 105, 16831–16836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch P, Fundinger M, Muller JT, Lee T, Bailey-Serres J and Mustroph A (2016) Redundant ERF-VII Transcription Factors Bind to an Evolutionarily Conserved cis-Motif to Regulate Hypoxia-Responsive Gene Expression in Arabidopsis. Plant Cell, 28, 160–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldo N, Baurle I, Kidou S, Hu X and Dean C (2009) FRIGIDA delays flowering in Arabidopsis via a cotranscriptional mechanism involving direct interaction with the nuclear cap-binding complex. Plant Physiol, 150, 1611–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs DJ, Tedds HM, Labandera AM, Bailey M, White MD, Hartman S, Sprigg C, Mogg SL, Osborne R, Dambire C, Boeckx T, Paling Z, Voesenek L, Flashman E and Holdsworth MJ (2018) Oxygen-dependent proteolysis regulates the stability of angiosperm polycomb repressive complex 2 subunit VERNALIZATION 2. Nat Commun, 9, 5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Le C, Wang Y, Li Z, Jiang D and He Y (2013) Arabidopsis FLC clade members form flowering-repressor complexes coordinating responses to endogenous and environmental cues. Nat Commun, 4, 1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harikumar A and Meshorer E (2015) Chromatin remodeling and bivalent histone modifications in embryonic stem cells. EMBO Rep, 16, 1609–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Wood CC, Robertson M, James Peacock W and Dennis ES (2006) The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J, 46, 183–192. [DOI] [PubMed] [Google Scholar]
- Heo JB and Sung S (2011) Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science, 331, 76–79. [DOI] [PubMed] [Google Scholar]
- Hepworth SR, Valverde F, Ravenscroft D, Mouradov A and Coupland G (2002) Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. Embo J, 21, 4327–4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Kong X, Wang C, Ma L, Zhao J, Wei J, Zhang X, Loake GJ, Zhang T, Huang J and Yang Y (2014) Proteasome-mediated degradation of FRIGIDA modulates flowering time in Arabidopsis during vernalization. Plant Cell, 26, 4763–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Gu X and He Y (2009) Establishment of the winter-annual growth habit via FRIGIDA-mediated histone methylation at FLOWERING LOCUS C in Arabidopsis. Plant Cell, 21, 1733–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R and Dean C (2000) Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science, 290, 344–347. [DOI] [PubMed] [Google Scholar]
- Kim DH and Sung S (2017) Vernalization-Triggered Intragenic Chromatin Loop Formation by Long Noncoding RNAs. Dev Cell, 40, 302–312 e304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Zografos BR and Sung S (2010) Mechanisms underlying vernalization-mediated VIN3 induction in Arabidopsis. Plant Signal Behav, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Choi K, Park C, Hwang HJ and Lee I (2006) SUPPRESSOR OF FRIGIDA4, encoding a C2H2-Type zinc finger protein, represses flowering by transcriptional activation of Arabidopsis FLOWERING LOCUS C. Plant Cell, 18, 2985–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, He Y, Jacob Y, Noh YS, Michaels S and Amasino R (2005) Establishment of the vernalization-responsive, winter-annual habit in Arabidopsis requires a putative histone H3 methyl transferase. Plant Cell, 17, 3301–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J and Lee I (2010) Regulation and function of SOC1, a flowering pathway integrator. J Exp Bot, 61, 2247–2254. [DOI] [PubMed] [Google Scholar]
- Li Z, Jiang D and He Y (2018) FRIGIDA establishes a local chromosomal environment for FLOWERING LOCUS C mRNA production. Nat Plants, 4, 836–846. [DOI] [PubMed] [Google Scholar]
- Lim PO, Kim Y, Breeze E, Koo JC, Woo HR, Ryu JS, Park DH, Beynon J, Tabrett A, Buchanan-Wollaston V and Nam HG (2007) Overexpression of a chromatin architecture-controlling AT-hook protein extends leaf longevity and increases the post-harvest storage life of plants. Plant J, 52, 1140–1153. [DOI] [PubMed] [Google Scholar]
- Liu C, Wang C, Wang G, Becker C, Zaidem M and Weigel D (2016) Genome-wide analysis of chromatin packing in Arabidopsis thaliana at single-gene resolution. Genome Res, 26, 1057–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Chen T, Zeng X, He D and He Y (2019) Feedback Regulation of FLC by FLOWERING LOCUS T (FT) and FD through a 5’ FLC Promoter Region in Arabidopsis. Mol Plant, 12, 285–288. [DOI] [PubMed] [Google Scholar]
- Michaels SD and Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell, 11, 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Himelblau E, Kim SY, Schomburg FM and Amasino RM (2005) Integration of flowering signals in winter-annual Arabidopsis. Plant Physiol, 137, 149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Suh SS, Lee H, Choi KR, Hong CB, Paek NC, Kim SG and Lee I (2003) The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J, 35, 613–623. [DOI] [PubMed] [Google Scholar]
- Mylne JS, Barrett L, Tessadori F, Mesnage S, Johnson L, Bernatavichute YV, Jacobsen SE, Fransz P and Dean C (2006) LHP1, the Arabidopsis homologue of HETEROCHROMATIN PROTEIN1, is required for epigenetic silencing of FLC. Proc Natl Acad Sci U S A, 103, 5012–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng KH, Yu H and Ito T (2009) AGAMOUS controls GIANT KILLER, a multifunctional chromatin modifier in reproductive organ patterning and differentiation. PLoS Biol, 7, e1000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oquist G and Huner NP (2003) Photosynthesis of overwintering evergreen plants. Annu Rev Plant Biol, 54, 329–355. [DOI] [PubMed] [Google Scholar]
- Questa JI, Song J, Geraldo N, An H and Dean C (2016) Arabidopsis transcriptional repressor VAL1 triggers Polycomb silencing at FLC during vernalization. Science, 353, 485–488. [DOI] [PubMed] [Google Scholar]
- Rosa S, De Lucia F, Mylne JS, Zhu D, Ohmido N, Pendle A, Kato N, Shaw P and Dean C (2013) Physical clustering of FLC alleles during Polycomb-mediated epigenetic silencing in vernalization. Genes Dev, 27, 1845–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakoe H and Chiba S (1978) Dynamic-Programming Algorithm Optimization for Spoken Word Recognition. Ieee T Acoust Speech, 26, 43–49. [Google Scholar]
- Schmitz RJ, Hong L, Michaels S and Amasino RM (2005) FRIGIDA-ESSENTIAL 1 interacts genetically with FRIGIDA and FRIGIDA-LIKE 1 to promote the winter-annual habit of Arabidopsis thaliana. Development, 132, 5471–5478. [DOI] [PubMed] [Google Scholar]
- Searle I, He Y, Turck F, Vincent C, Fornara F, Krober S, Amasino RA and Coupland G (2006) The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev, 20, 898–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Hills MJ, Lister C, Dean C, Dennis ES and Peacock WJ (2008) Resetting of FLOWERING LOCUS C expression after epigenetic repression by vernalization. Proc Natl Acad Sci U S A, 105, 2214–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Rouse DT, Finnegan EJ, Peacock WJ and Dennis ES (2000) The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proc Natl Acad Sci U S A, 97, 3753–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo C, Aranzana MJ, Lister C, Baxter C, Nicholls C, Nordborg M and Dean C (2005) Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiol, 138, 1163–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street IH, Shah PK, Smith AM, Avery N and Neff MM (2008) The AT-hook-containing proteins SOB3/AHL29 and ESC/AHL27 are negative modulators of hypocotyl growth in Arabidopsis. Plant J, 54, 1–14. [DOI] [PubMed] [Google Scholar]
- Sun M, Qi X, Hou L, Xu X, Zhu Z and Li M (2015) Gene Expression Analysis of Pak Choi in Response to Vernalization. PLoS One, 10, e0141446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S and Amasino RM (2004) Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature, 427, 159–164. [DOI] [PubMed] [Google Scholar]
- Sung S and Amasino RM (2006) Molecular genetic studies of the memory of winter. J Exp Bot, 57, 3369–3377. [DOI] [PubMed] [Google Scholar]
- Sung S, He Y, Eshoo TW, Tamada Y, Johnson L, Nakahigashi K, Goto K, Jacobsen SE and Amasino RM (2006) Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires LIKE HETEROCHROMATIN PROTEIN 1. Nat Genet, 38, 706–710. [DOI] [PubMed] [Google Scholar]
- Swiezewski S, Liu F, Magusin A and Dean C (2009) Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature, 462, 799–802. [DOI] [PubMed] [Google Scholar]
- Tamada Y, Yun JY, Woo SC and Amasino RM (2009) ARABIDOPSIS TRITHORAX-RELATED7 is required for methylation of lysine 4 of histone H3 and for transcriptional activation of FLOWERING LOCUS C. Plant Cell, 21, 3257–3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw NL and Schier AF (2012) Bivalent histone modifications in early embryogenesis. Curr Opin Cell Biol, 24, 374–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt P, Tee WW and Reinberg D (2013) A double take on bivalent promoters. Genes Dev, 27, 1318–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EJ and Carpenter PB (2012) Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol, 13, 115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner JD, Borevitz JO, Uhlenhaut NH, Ecker JR, Chory J and Weigel D (2005) FRIGIDA-independent variation in flowering time of natural Arabidopsis thaliana accessions. Genetics, 170, 1197–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker C and Dean C (2017) The FLC Locus: A Platform for Discoveries in Epigenetics and Adaptation. Annu Rev Cell Dev Biol, 33, 555–575. [DOI] [PubMed] [Google Scholar]
- Xiao C, Chen F, Yu X, Lin C and Fu YF (2009) Over-expression of an AT-hook gene, AHL22, delays flowering and inhibits the elongation of the hypocotyl in Arabidopsis thaliana. Plant Mol Biol, 71, 39–50. [DOI] [PubMed] [Google Scholar]
- Xu Y, Gan ES, He Y and Ito T (2013) Flowering and genome integrity control by a nuclear matrix protein in Arabidopsis. Nucleus, 4, 274–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CY, Hsu FC, Li JP, Wang NN and Shih MC (2011) The AP2/ERF transcription factor AtERF73/HRE1 modulates ethylene responses during hypoxia in Arabidopsis. Plant Physiol, 156, 202–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Luo X, Li Z, Yang W, Wang Y, Liu R, Du J and He Y (2016) A cis cold memory element and a trans epigenome reader mediate Polycomb silencing of FLC by vernalization in Arabidopsis. Nat Genet, 48, 1527–1534. [DOI] [PubMed] [Google Scholar]
- Yun J, Kim YS, Jung JH, Seo PJ and Park CM (2012) The AT-hook motif-containing protein AHL22 regulates flowering initiation by modifying FLOWERING LOCUS T chromatin in Arabidopsis. J Biol Chem, 287, 15307–15316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Frietze SE, Gordon JA, Heath JL, Messier T, Hong D, Boyd JR, Kang M, Imbalzano AN, Lian JB, Stein JL and Stein GS (2017) Bivalent Epigenetic Control of Oncofetal Gene Expression in Cancer. Mol Cell Biol, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Favero DS, Peng H and Neff MM (2013) Arabidopsis thaliana AHL family modulates hypocotyl growth redundantly by interacting with each other via the PPC/DUF296 domain. Proc Natl Acad Sci U S A, 110, E4688–4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Favero DS, Qiu J, Roalson EH and Neff MM (2014) Insights into the evolution and diversification of the AT-hook Motif Nuclear Localized gene family in land plants. BMC Plant Biol, 14, 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S3. Annotation of transcription factors with bivalent marks
Supplementary Figure S1. Validation of ChIP-Seq by ChIP-qPCR.
Supplementary Figure S2. Characterization of AHL family genes.
Table S1. Summary of RNA-Seq Analysis
Table S2. Summary of ChIP-Seq Analysis


