Abstract
Background
Placental mosaicism is one of the major reasons for noninvasive prenatal testing (NIPT) discrepancy. Herein, we discovered a rare case of placenta with complex karyotypes that caused false‐positive and false‐negative results in noninvasive prenatal testing.
Methods
Next‐generation sequencing (NGS) and Quantitative fluorescent polymerase chain reaction (QF‐PCR) were performed on the cord blood sample, fetal tissues, and eight placental biopsies. Fluorescent In Situ Hybridization (FISH) and karyotyping were also carried to confirm the fetal genome status.
Results
The results suggested that the fetal chromosome was 47,XXX and the placenta had three karyotypes of 48,XXX,+21, 47,XX,+21, and 47,XXX. QF‐PCR indicated that the extra chromosome 21 and chromosome X were all from the father. It is speculated that the zygote may have 48,XXX,+21 karyotype and trisomy rescue could be the main mechanism for the development of the homogeneous fetus and complex mosaic placenta.
Conclusion
Overall, the complicated nature of our case underlines the importance of discussing with parents the possibility of both atypical and discordant results during preconfirmatory amniocentesis counseling and consent.
Keywords: chromosomal abnormalities, confined placental mosaicism, NIPT, prenatal diagnosis, trisomy
The composition of the unusual placental mosaicsim that caused NIPT discrepancy was complex. The karyotype for the paternal sperm may be an unusual diploid gamete (25,XX,+21) and zygote is likely to be a double trisomy (48,XXX,+21). After the fifth cleavage, the zygote grows into a morula and then into a blastocyst. The blastocyst contains two major part of compositions. One is the inner cell mass (yellow), which develops into the embryo (the homogeneous 47,XXX). The other is trophoblast cells (blue), which develops with some inner cell mass into the placenta (the mosaic 47,XXX, 47,XX,+21 or 48,XXX,+21).
1. CLINICAL REPORT
The pregnant woman is a 34‐year‐old mother who had one previous pregnancy that resulted in the delivery of a normal, healthy boy. Results of her second trimester combined test (AFP: 0.80MoM and Free‐hCGβ: 2.52MoM) indicated that the pregnancy (gravida 3, para 1) was at high risk of Down syndrome (1/20). The noninvasive prenatal testing (NIPT) was then performed at the 20 + 3 week as the second screening test and the result also revealed high risk of trisomy 21 with a Z‐score of 18.51 for chromosome 21. After consulting, the patient agreed to take a further amniocentesis to confirm the screening results. Fluorescence in situ hybridization (FISH), copy number variation using next‐generation sequencing (CNV‐seq) and G‐banded karyotype were used to analyze the amniocytes cultivation. All of the results showed 47,XXX in fetal chromosome, which implied that NIPT gave a false positive result of T21 and a false negative result of trisomy X. To further analyze the inconsistent results, a second NIPT test was carried out at the 23 + 4 week of gestation and an additional CNV‐seq test for the pregnant woman was also performed. The result of the second NIPT still showed high risk of trisomy 21 (Z‐score 18.75) and the CNV‐seq analysis of maternal peripheral blood was normal, which eliminated the possibility of maternal T21 mosaicism or large chromosome fragment duplication on chromosome 21. Additionally, ultrasonography at the 27 + 3 week did not show any structural abnormalities nor intrauterine growth retardation (IUCR). To our knowledge, this inconsistency may occur when the placental have a status of mosaic T21 and trisomy X. After posttest consultation, the couple decided to terminate pregnancy and agreed to carry out additional research.
2. MATERIALS AND METHODS
2.1. Karyotype
Amniotic cells were conducted on G‐banded metaphase chromosomes at a resolution of approximately 450 bands. Twenty metaphases were analyzed for nonmosaic cells and 60 metaphases were analyzed for mosaic cells or suspected mosaic cases. Nomenclatures were assigned for each karyotype according to the international system for human cytogenetic nomenclature (ISCN 2013).
2.2. Fish
FISH was performed on cytogenetic preparations fixed in 3:1 (v:v) methanol/glacial acetic acid. Slide preparations were hybridized with the locus‐specific probe using co‐denaturation and 2 hr hybridization. FISH was performed according to the manufacturers’ instructions. FISH probe was used LSI PML/RARA dual color translocation probe (FastProbe, Healthcare biotechnology Inc, Wuhan, China).
2.3. NIPT
NIPT screening was performed using the shallow‐depth whole genome sequencing‐based noninvasive prenatal subchromosomal copy number variation detection (NIPSCCD) method as previously reported (Yu et al., 2019). The first NIPT was performed at the 20 + 3 week of gestation and the second one was carried out at the 23 + 4 week, samples were sequenced on the Nextseq550AR platform (Annoroad Gene Technology, Beijing, China).
2.4. CNV‐seq
CNV‐seq was carried out by following the protocol mentioned in Qi et al.(2018) using the Nextseq550AR platform (Annoroad Gene Technology, Beijing, China). Genomic DNA extracted from peripheral blood leukocytes of the pregnant woman and tissues of the fetus and placenta was used. The resolution of chromosome structure variation detection is 100 kb.
2.5. QF‐PCR
Multiplex quantitative fluorescent polymerase chain reaction (QF‐PCR) was using a Devyser compact v3 kit (Devyser AB, Sweden). Selected polymorphic STR markers were used to analyze chromosomes 13, 18, 21, X, and Y. Each STR marker was labeled with either 5’‐6‐carboxy fluorescein (FAM) or 5’‐hexachloro fluorescein (HEX). Labeled PCR products were resolved on a 3,130 Genetic Analyzer (Applied Biosystems) and allelic profiles plotted by GeneScan software. Data analysis was performed by following the ACC QF‐PCR for diagnosis of aneuploidy best practice guidelines (2012) v3.01 (Association for Clinical Cytogenetics, 2012).
2.6. Ethical compliance
The study was approved by the Office of Research Ethics, Zhongnan Hospital (approval number (2019111)). Informed consent was obtained from the participant in this study.
3. RESULTS
A cord blood sample, three fetal tissues (skin, muscle, and umbilical cord), and eight placental biopsies (four from the fetal side and four from the maternal side) were taken for the CNV‐seq analysis. QF‐PCR was also performed to evaluate maternal DNA pollution and identify the origin of the extra chromosome 21 and chromosome X. Most samples were successfully analyzed, except the cord blood sample was polluted by the maternal DNA as indicated by the QF‐PCR.
Overall, CNV‐seq analysis revealed the existence of heterogeneity between fetal and placental tissues (Table 1). Results of the eight placental biopsies showed complex combinations. Among the placental samples, trisomy 21 was observed in 5 of 8 positions, trisomy X was observed only once (Sample ID: ED135‐10), and double trisomy of 21 and X was observed in two positions (Sample ID: ED135‐8, and ED135‐9). However, because the trophoblast of the placenta was not investigated separately, the karyotype of possible cell lines existed in the layer could not be determined.
Table 1.
The result of CNV‐seq analysis in different samples
| Sample ID | Sample | Karyotype |
|---|---|---|
| ED133‐1 | Maternal peripheral blood | 46,XX |
| ED135‐7 |
Centre of the placenta (fetal side) |
47,XX,+21 |
| ED135‐1 |
12 o'clock of the placenta (fetal side) |
47,XX,+21 |
| ED135‐2 |
4 o'clock of the placenta (fetal side) |
47,XX,+21 |
| ED135‐3 |
8 o'clock of the placenta (fetal side) |
47,XX,+21 |
| ED135‐8 |
Centre of the placenta (maternal side) |
48,XXX,+21 |
| ED135−9 |
12 o'clock of the placenta (maternal side) |
48,XXX,+21 |
| ED135‐10 |
4 o'clock of the placenta (maternal side) |
47,XXX |
| ED135‐11 |
8 o'clock of the placenta (maternal side) |
47,XX,+21 |
| ED135‐12 | Fetal skin | 47,XXX |
| ED135‐14 | Fetal muscle | 47,XXX |
| ED135‐5 | Umbilical cord | 47,XXX |
In comparison, all fetal tissues (skin, muscle, and umbilical cord) demonstrated a homogeneous trisomy X (Sample ID: ED135‐12, ED135‐14 and ED135‐5 respectively), indicating that the fetal tissues were different from the placental tissues. FISH analysis confirmed the result of 47,XXX in three fetal tissues. The extra CNVs detected in the placenta was also presented in the fetus and showed a normal inheritance pattern. Moreover, QF‐PCR analysis revealed that the extra chromosome X in fetal tissues and the extra chromosome 21 and X in placenta tissues were all from the father (Table 2).
Table 2.
The result of QF‐PCR analysis in different samples
| Sample ID | Sample types | Markers of chromosome 21 | Markers of chromosome X | |||||
|---|---|---|---|---|---|---|---|---|
| 21B | 21C | 21D | 21I | 21H | X1 | X3 | ||
| ED133‐1 | Maternal peripheral blood | 239,265 | 304,308 | 464,466 | 120,131 | 380,396 | 143,155 | 294,294 |
| ED135‐7 |
Centre of the placenta (fetal side) |
257,257,265 | 300,300,304 | 464,470,470 | 120,131,131 | 392,392,396 | 143,147 | 285,294 |
| ED135‐1 |
12 o'clock of the placenta (fetal side) |
257,257,265 | 300,300,304 | 464,470,470 | 120,131,131 | 392,392,396 | 143,147 | 285,294 |
| ED135‐2 |
4 o'clock of the placenta (fetal side) |
257,257,265 | 300,300,304 | 464,470,470 | 120,131,131 | 392,392,396 | 143,147 | 285,294 |
| ED135‐3 |
8 o'clock of the placenta (fetal side) |
257,257,265 | 300,300,304 | 464,470,470 | 120,131,131 | 392,392,396 | 143,147 | 285,294 |
| ED135‐8 |
Centre of the placenta (maternal side) |
257,257,265 | 300,300,304 | 464,470,470 | 120,131,131 | 392,392,396 | 143,147,147 | 285,285,294 |
| ED135‐9 |
12 o'clock of the placenta (maternal side) |
257,257,265 | 300,300,304 | 464,470,470 | 120,131,131 | 392,392,396 | 143,147,147 | 285,285,294 |
| ED135‐10 |
4 o'clock of the placenta (maternal side) |
257,265 | 300,304 | 464,470 | 120,131 | 392,396 | 143,147,147 | 285,285,294 |
| ED135‐11 |
8 o'clock of the placenta (maternal side) |
257,257,265 | 300,300,304 | 464,470,470 | 120,131,131 | 392,392,396 | 143,147 | 285,294 |
| ED135‐12 | Fetal skin | 257,265 | 300,304 | 464,470 | 120,131 | 392,396 | 143,147,147 | 285,285,294 |
| ED135‐14 | Fetal muscle | 257,265 | 300,304 | 464,470 | 120,131 | 392,396 | 143,147,147 | 285,285,294 |
| ED135‐5 | Umbilical cord | 257,265 | 300,304 | 464,470 | 120,132 | 392,396 | 143,147,147 | 285,285,294 |
4. DISCUSSION
In this case, we reported discordant results between NIPT and fetal karyotyping. The placenta has the mosaicism of 48,XXX,+21, 47,XXX, and 47,XX,+21, but we could not find any evidence showing the fetus contained trisomy 21 nor double trisomy 21 and X. Therefore, according to the results of CNV‐seq and FISH on the fetal skin and muscle tissues, the karyotype of the fetus was homogeneous 47,XXX. We believe this confined placental mosaicism (CPM) lead to the false 47,XX,+21 NIPT result (Choi et al., 2013). However, since the trophoblast and mesenchymal core were not separately examined and the exact karyotype of cells in the trophoblast could not be identified, we therefore can only speculate that the missing trisomy X in fetal cfDNA might also be caused by the mosaic placenta due to the insufficient concentration for NIPT detection as demonstrated in the previous report (Mao et al., 2014; Wang et al., 2019).
Human embryonic development begins in the early postfertilization stages. The fetus itself is likely to be derived from only a small subset (3 out of 64) of the blastocyst progenitor cells (Bianchi, Wilkins‐Haug, Enders, & Hay, 1993). The remaining cells are believed to give rise to extraembryonic structures. In our case, the results of QF‐PCR analysis revealed that the abnormal chromosome of the oosperm came from the father. Therefore, we speculate that the karyotype for the paternal sperm was an unusual diploid gamete (25,XX,+21) and zygote is likely to be a double trisomy, 48,XXX,+21. There have been a number of cases reported for fetuses with this type of double trisomy and some of which were born and grew up (Balwan, Kumar, Raina, & Gupta, 2008; Shen, Zou, Shang, & Jiang, 2012; Vergara‐Mendez, Talero‐Gutiérrez, & Velez‐Van‐Meerbeke, 2018). Nevertheless, the interesting question in our case is how the double trisomic zygote developed into a homogeneous fetus (47,XXX) and a placenta with three distinct karyotypes (48,XXX,+21, 47,XXX, and 47,XX,+21) rather than a double‐aneuploid abnormal fetus and placenta (48,XXX,+21).
In literature, the formation of chromosomal mosaicism was thought to have two principal mechanisms, trisomy rescue and mitotic nondisjunction. The trisomy rescue could restore a disomic cell line through the loss of one supernumerary chromosome in the cell (Grati et al., 2017). According to the distribution of the different cell lines in our research, we speculate that the trisomy rescue was the main mechanism for the double trisomy zygote to finally differentiate into a homogeneous fetal (47,XXX), and two independent rescue events may exist during the embryonic and placenta development. The first event was thought to be at the beginning of the postfertilization stages, the trisomy rescue precisely progressed on the timing and embryo–fetal localization and diminished the extra chromosome 21 to generate a homogeneous fetal (47,XXX) who was derived from only three ancestral cells at the blastocyst stage (Figure 1). The second event was surmised to exist during the early placenta development, where placenta cells with a karyotype of 47,XX,+21 was generated after rescuing from the extra copy of chromosome X. Since cells with 46,XX in the placenta were not discovered, the two rescues were unlikely to happen in the same cell line, therefore, it was considered to be two independent events.
FIGURE 1.
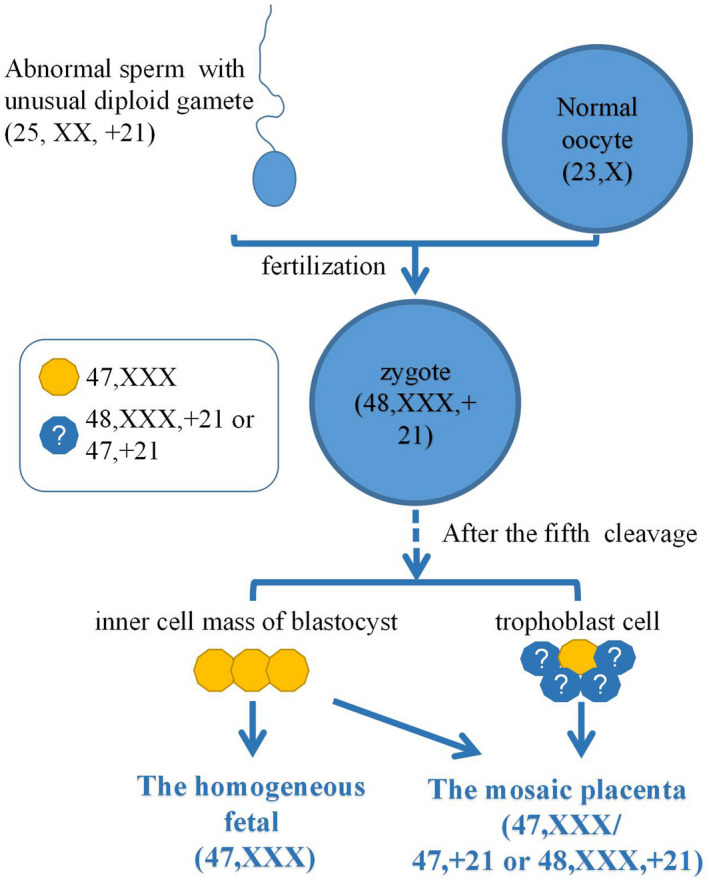
An illustration of speculated embryo and placenta development process of the fetus. The karyotype for the paternal sperm may be an unusual diploid gamete (25,XX,+21) and zygote is likely to be a double‐trisomy (48,XXX,+21). After the fifth cleavage, the zygote grows into a morula and then into a blastocyst. The blastocyst contains two major part of compositions. One is the inner cell mass (yellow), which develops into the embryo (the homogeneous 47,XXX). The other is trophoblast cells (blue), which develops with some inner cell mass into the placenta (the mosaic 47,XXX, 47,XX,+21, or 48,XXX,+21)
Because the paternal markers for chromosome X and 21, indicated by the QF‐PCR, were all of the same length, the 48,XXX,+21 cell line could also be caused by mitotic nondisjunction when the karyotype of the sperm was normal. If this is the case, the origin of the extra chromosome X and 21 should have several possibilities such as all from the father, all from the mother, or one from each parent, and the chance of becoming our case was small.
In summary, we report a discordant result between fetal karyotyping (47,XXX) and NIPT (47,XX,+21) and subsequently found a rare case of mosaic placenta with trisomy 21, trisomy X, and double trisomy X/+21, as well as a fetus with homogeneous trisomy X examined after pregnancy termination. The complicated nature of our case underlines the importance of discussing with the parents the possibility of both atypical and discordant results during preconfirmatory amniocentesis counseling and consent. In contrast, because NIPT relies on the content of trophoblast cells, it is important to follow‐up the normal or abnormal results. In brief, considering the effect from placenta, the NIPT results should be interpreted as a screening method and combining other clinical tests under comprehensive background information.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTION
J.L. analyzed the patient medical record and wrote the manuscript. M.X. interviewed the patient and collected all the relative medical information. F.W., J.M., J.L., C.C., Z.L., and J.W. performed different experiments and interpreted the data. Y.Z. and Y.L. supervised the study and revised the manuscript. All authors approved the final version of the manuscript.
ACKNOWLEDGMENTS
We thank the parents for their selfless participating in this study. We also thank all the clinicians, nurses, and medical staff for their dedication in this study. Especially, Prof. Li JiaFu, Prof. Ma JianHong, Dr. Ye GuangMing, Liu Juan (Zhongnan Hospital of Wuhan University), Dr. Wang Yun (Suizhou Zendu Hospital of Hubei), and Dr. Wang Qian (The First People's Hospital of Henan) for helps in recruitment of clinical information.
Jin L, Xie M, Wang F, et al. A rare case of NIPT discrepancy caused by the placental mosaicism of three different karyotypes, 47,XXX, 47,XX,+21, and 48,XXX,+21. Mol Genet Genomic Med. 2020;8:e1279 10.1002/mgg3.1279
Funding information
This work was partially supported by the National Natural Science Foundation of China (grant number 81602863) and the Health commission of Hubei Province scientific research project (grant number WJ2019H014).
Contributor Information
Yuanzhen Zhang, Email: zhangyuanzhen@vip.sina.com.
Yirong Li, Email: liyirong838@163.com.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
REFERENCES
- ACC Professional Guidelines for Clinical Cytogenetics . (2012). QF‐PCR for the diagnosis of aneuploidy. Best Practice Guidelines v3.01.
- Balwan, W. K. , Kumar, P. , Raina, T. R. , & Gupta, S. (2008). Double trisomy with 48, XXX+21 karyotype in a Down's syndrome child from Jammu and Kashmir, India. Journal of Genetics, 87(3), 257–259. 10.1007/s12041-008-0039-x [DOI] [PubMed] [Google Scholar]
- Bianchi, D. W. , Wilkins‐Haug, L. E. , Enders, A. C. , & Hay, E. D. (1993). Origin of extraembryonic mesoderm in experimental animals: Relevance to chorionic mosaicism in humans. American Journal of Medical Genetics, 46, 542–550. 10.1002/ajmg.1320460517 [DOI] [PubMed] [Google Scholar]
- Choi, H. , Lau, T. K. , Jiang, F. M. , Chan, M. K. , Zhang, H. Y. , Lo, P. S. S. , … Wang, W. (2013). Fetal aneuploidy screening by maternal plasma DNA sequencing: 'False positive' due to confined placental mosaicism. Prenatal Diagnosis, 33, 198–200. 10.1002/pd.4024 [DOI] [PubMed] [Google Scholar]
- Grati, F. R. , Malvestiti, F. , Branca, L. , Agrati, C. , Maggi, F. , & Simoni, G. (2017). Chromosomal mosaicism in the fetoplacental unit. Best Practice & Research Clinical Obstetrics & Gynaecology, 42, 39–52. 10.1016/j.bpobgyn.2017.02.004 [DOI] [PubMed] [Google Scholar]
- Mao, J. , Wang, T. , Wang, B.‐J. , Liu, Y.‐H. , Li, H. , Zhang, J. , … Chen, Y. (2014). Confined placental origin of the circulating cell free fetal DNA revealed by a discordant non‐invasive prenatal test result in a trisomy 18 pregnancy. Clinica Chimica Acta, 433, 190–193. 10.1016/j.cca.2014.03.011 [DOI] [PubMed] [Google Scholar]
- Qi, H. , Xuan, Z.‐L. , Du, Y. , Cai, L.‐R. , Zhang, H. , Wen, X.‐H. , … Liang, J.‐B. (2018). High resolution global chromosomal aberrations from spontaneous miscarriages revealed by low coverage whole genome sequencing. European Journal of Obstetrics & Gynecology and Reproductive Biology, 224, 21–28. 10.1016/j.ejogrb.2018.03.008 [DOI] [PubMed] [Google Scholar]
- Shen, Z. , Zou, C. C. , Shang, S. Q. , & Jiang, K. W. (2012). Down‐Klinefelter syndrome (48, XXY, +21) in a child with congenital heart disease: Case report and literature review. Internal Medicine, 51(11), 1371–1374. 10.2169/internalmedicine.51.7097 [DOI] [PubMed] [Google Scholar]
- Vergara‐Mendez, L. D. , Talero‐Gutiérrez, C. , & Velez‐Van‐Meerbeke, A. (2018). Double trisomy (XXX+21 karyotype) in a six‐year‐old girl with down phenotype. Journal of Genetics, 97(1), 337–340. 10.1007/s12041-018-0916-x [DOI] [PubMed] [Google Scholar]
- Wang, Z. , Tang, X. , Yang, S. , Yin, T. , Zhao, Y. , Zheng, A. , … Wang, L. (2019). A gradual change of chromosome mosaicism from placenta to fetus leading to T18 false negative result by NIPS. Clinica Chimica Acta, 495, 263–268. 10.1016/j.cca.2019.04.064 [DOI] [PubMed] [Google Scholar]
- Yu, D. , Zhang, K. , Han, M. , Pan, W. , Chen, Y. , Wang, Y. , … Du, Y. (2019). Noninvasive prenatal testing for fetal subchromosomal copy number variations and chromosomal aneuploidy by low‐pass whole‐ genome sequencing. Molecular Genetics & Genomic Medicine, 7(6), e674 10.1002/mgg3.674 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.


