Abstract
Abstract
Infectious bronchitis virus (IBV) is a member of genus gamma-coronavirus in the family Coronaviridae, causing serious economic losses to the poultry industry. Reverse genetics is a common technique to study the biological characteristics of viruses. So far, there is no BAC reverse genetic system available for rescue of IBV infectious clone. In the present study, a new strategy for the construction of IBV infectious cDNA clone was established. The full-length genomic cDNA of IBV vaccine strain H120 was constructed in pBAC vector from four IBV fragment subcloning vectors by homologous recombination, which contained the CMV promoter at the 5′ end and the hepatitis D virus ribozyme (HDVR) sequence and bovine growth hormone polyadenylation (BGH) sequence after the polyA tail at the 3′ end of the full-length cDNA. Subsequently, using the same technique, another plasmid pBAC-H120/SCS1 was also constructed, in which S1 gene from IBV H120 strain was replaced with that of a virulent SC021202 strain. Recombinant virus rH120 and rH120/SCS1 were rescued by transfecting the plasmids into BHK cells and passaged in embryonated chicken eggs. Finally, the pathogenicity of both the recombinant virus strains rH120 and rH120/SCS1 was evaluated in SPF chickens. The results showed that the chimeric rH120/SCS1 strain was not pathogenic compared with the wild-type IBV SC021202 strain and the chickens inoculated with rH120/SCS1 could resist challenge infection by IBV SC021202. Taken together, our results indicate that BAC reverse genetic system could be used to rescue IBV in vitro and IBV S1 protein alone might not be the key factor for IBV pathogenicity.
Key points
• BAC vector was used to construct IBV full-length cDNA by homologous recombination.
• Based on four subcloning vectors, a recombinant chimeric IBV H120/SCS1 was constructed and rescued.
• Pathogenicity of H120/SCS1 was similar to that of H120, but different to that of SC021202.
Keywords: Infectious bronchitis virus, Reverse genetics, Chimeric S1 gene, Pathogenicity
Introduction
Infectious bronchitis virus (IBV) is a member of the Coronaviridae family in the order Nidovirales. It is a highly pathogenic virus that causes serious economic losses to the world poultry industry (Cavanagh 2005; Cook et al. 2012). IBV is an enveloped virus having a single-stranded, positive-sense RNA genome of 27.6 kb in size (Boursnell et al. 1987). Because of the high degree of genome sequence variation, IBV has many genotypes circulating at any time in poultry farms worldwide (Jackwood 2012), making it difficult to control or prevent its spread through vaccination.
Reverse genetics is a common in vitro method to study virus biological characteristics. The development of this technique allowed researchers to manipulate the IBV genome and rescue of virus in vitro (Shan et al. 2018). Previously, the most common technique to obtain the full-length cDNA of IBV was the cDNA ligation (Youn et al. 2005; Zhou et al. 2013). By using this method, small pieces of IBV genomic cDNAs were amplified, digested with restriction enzymes (RE), and then ligated. The full-length cDNA with T7 promoter attached to 5′ end of the IBV genome was transcribed and transfected into BHK cells by electroporation to rescue the virus (Casais et al. 2001).
Another reverse genetic system for IBV was based on targeted RNA recombination. This method enabled the modification of the IBV genome at its 3′ terminal by targeted RNA recombination (van Beurden et al. 2017). By using this method, a recombinant H52 strain carrying the spike glycoprotein ectodomain of MHV was constructed, which had the capability to grow in a mammalian cell lines (van Beurden et al. 2017). However, targeted RNA recombination is only useful for the modification of 3′-end of the IBV genome and is difficult to modify the 5′-end polymerase gene, because the construction of donor RNA vectors entering this region is hindered (van Beurden et al. 2017).
Bacterial artificial chromosomes (BAC) vector reverse genetic system is also a common method to rescue coronaviruses in vitro. The first coronavirus full-length genomic cDNA in BAC was constructed for TGEV (Almazan et al. 2000). Later on, several coronavirus, such as SARS-CoV (Almazan et al. 2006), FIPV (Balint et al. 2012), and MERS-CoV (Scobey et al. 2013), were rescued successfully using that technique. However, there is no BAC reverse genetic system currently available for the rescue of IBV.
The spike protein of IBV is posttranslationally cleaved into two subunits, S1 and S2, where S2 is anchored into the viral envelop and is important for the membrane fusion. S1 comprises the head domain of spike and is responsible for host receptor binding (Wickramasinghe et al. 2011). The S1 subunit is the important immunogenic component that contains epitopes for neutralizing antibody (Ignjatovic and Sapats 2005).
In this study, a new strategy for construction of IBV infectious clone was established. A nephropathogenic strain SC021202 (close to IBV GI-22 genotype), which causes severe kidney damage and mortality to infected chickens (Zhou et al. 2004) was selected as donor of IBV S1 gene. First, infectious clone of IBV vaccine strain H120 (rH120) was rescued using the BAC reverse genetic system. Then, a chimeric IBV virus rH120/SCS1 with the H120 backbone replacing its S1 subunit with that of virulent SC021202 isolate was constructed. Lastly, the rH120/SCS1 was used to inoculate specific pathogen free (SPF) chickens for the preliminary study of the function of S1 of IBV field isolate on virus pathogenicity and the potential of recombinant virus as a vaccine candidate was also evaluated.
Materials and methods
Vector, cells, and viruses
The strains of IBV SC021202 (GenBank No.: EU714029.1) and H120 (GenBank No.: FJ888351.1) were stored in our laboratory. Viruses were propagated and titrated in the allantoic cavity of 10-day-old embryonated SPF chicken eggs. Baby hamster kidney (BHK-21) cell line was stored in our laboratory and cultured at 37 °C in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS, Biological Industries, Israel), penicillin (100 U/mL), and streptomycin (100 μg/mL). Primary chicken embryonated egg kidney (CEK) cells were prepared from 17–19-day-old SPF chicken embryonated eggs and maintained in DMEM supplemented with 10% FBS at 37 °C in 5% CO2 atmosphere. Competent cells were prepared according to the instructions provided by Ultra-Competent Cell Preps Kit (Sang Biotech, China). pBeloBAC11 (called pBAC in the subsequent texts) vector was a gift from Professor Yaowei Huang of Zhejiang University.
Construction scheme of a full-length cDNA clone of IBV H120
To construct the full-length cDNA of IBV H120 strain, total RNA was extracted from the allantoic fluid of SPF chicken embryonated eggs infected with H120 and transcribed to cDNA by reverse transcriptase using the Thermo Scientific RevertAid First-Strand cDNA Synthesis Kit (Thermo Fisher, USA). Four cDNA fragments (f1, F2, F3, and f4) covering the whole H120 genome sequence were amplified separately from the cDNA template with the primer sets (f1-F and f1-R, F2-F and F2-R, F3-F and F3-R, f4-F and f4-R) shown in Table 1, which also contained sequences for homologous recombination.
Table 1.
Primers used for amplification and construction of IBV genomic cDNA
| Primers | Primer sequence (5′–3′) | Position in genome (length) | Purpose |
|---|---|---|---|
| f1-F | agctcggtacccggggatccCTTGTTTTGCCGTGTCTC | 192–6282 bp (6091 bp) | To amplify F1, F2, F3, and F4 fragments carrying homology arm sequences for ligation into BAC vector, respectively. |
| f1-R | tcaagcttgcatgcctcgagATGCCTGTTACTGCGTGC | ||
| F2-F | agctcggtacccggggatccGGTTTCTATTTCTGGCTCT | 6172–13,192 bp (7021 bp) | |
| F2-R | tcaagcttgcatgcctcgagTCACTACAGTCACGGCAG | ||
| F3-F | agctcggtacccggggatccTTATGATCTCCTCAAGTATG | 13,085–20,019 bp (6935 bp) | |
| F3-R | tcaagcttgcatgcctcgagAATCCTGTGCAATGTCATA | ||
| f4-F | agctcggtacccggggatccGATTATGTTTCTGACGCAC | 19,752–27,397 bp (7646 bp) | |
| f4-R | tcaagcttgcatgcctcgagAGACAGATTAGACATTTCCC | ||
| f1A5472C-F | TATTGTTGGCTCCAGTGTTGTTACTACA | / | To introduce a silent mutation site 5472 A → C. |
| f1A5472C-R | AACACTGGAGCCAACAATAGCTTTCTTA | ||
| 5’UTR-F | agctcggtacccggggatccACTTAAGATAGATATTAATATA | 1–293 bp (293 bp) | To fuse the 5′UTR to the F1 subcloning vector. |
| 5’UTR-R | ACCAGAGGTCCTTCGCACT | ||
| 3’UTR-F | GAATCAAGGAAGATAGGC | 27,328–27,652 bp (325 bp) | To fuse the 3′UTR to the F4 subcloning vector. |
| 3’UTR-R | tcaagcttgcatgcctcgagTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTGCTCTAACTCT | ||
| CMV-F | agctcggtacccggggatcc TTAATTAACTTGACATTG | 610 bp | To add the CMV promoter to the 5′ end of F1 fragment |
| CMV-R | tatttatatcacacttaagt ACGGTTCACTAAACGAGC | ||
| HB-F | tagagcaaaaaaaaaaaaaaaaaaaaaaaaaaaaaa GGCCGGCATGGTCCCAGC | 304 bp | To add the sequence of BGH and HDVR to the 3′ end of F4 fragment |
| HB-R | tcaagcttgcatgcctcgag CCACCGCATCCCCAGCAT | ||
| (F1-F2)-F | GGTTTCTATTTCTGGCTCT | 6172–13,192 bp (7021 bp) | To fuse F2 to the (CMV) F1 subcloning vector via the F1 homology arm. |
| (F1-F2)-R | tcaagcttgcatgcctcgagTCACTACAGTCACGGCAG | ||
| (F3-F4)-F | GATTATGTTTCTGACGCAC | 7901 bp | To fuse F4 (HB) to the F3 subcloning vector via the F3 homology arm. |
| (F3-F4)-R | tcaagcttgcatgcctcgagTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTGCTCTAACTCT | ||
| (F34-F12)-F | GATTATGTTTCTGACGCAC | (14,872 bp) | To fuse F34 (HB) to the (CMV) F12 subcloning vector via the F2 homology arm. |
| (F34-F12)-R | tcaagcttgcatgcctcgag CCACCGCATCCCCAGCAT | ||
| H120-F4-△S1-F | GTTTTAGACGTTCTATTA | 13,518 bp | To prepare the linearized vector by deleting H120 S1 from pBeloBAC11-H120F4 (HB) |
| H120-F4-△S1-R | CTCTTACCAGTAACTTAC | ||
| SCS1-F | TGGTAAGTTACTGGTAAGAGATGTTGGGGAAGTCACTGTT | 20,296–21,931 bp (1635 bp) | To amplify the sequence of SCS1 |
| SCS1-R | AGTAATAGAACGTCTAAAACGACGTGTCTCTTTAGTGAGT | ||
| RM-F | AAAAGCGCCAGTCTACTACCC | 621 bp | To detect the mutant oligonucleotide sites (rescue marker) |
| RM-R | GGACCACATAAAGAACCCTCA | ||
| H120/SC-S-F | ACTGGTAAGAGATGTTGGTAA | 3937 bp | To amplify the S gene sequence of rH120/SCS1 |
| H120/SC-S-R | GAAAACTTCCATTCTCCTCTA |
Sequences in 5′ end with lowercase letters are the sequences of homology arms
The procedure for the construction of full-length cDNA of IBV H120 has been shown in Fig. 1a. The amplified 4 fragments (f1, F2, F3, and f4) carrying the same homologous recombination sequences as the pBAC vector were cloned into modified linearized pBAC vector. Homologous recombination reaction was carried out according to the instructions provided with ClonExpress one-step cloning kit (Vazyme Biotechnology, China). The resulting plasmids were named as pBAC-f1, pBAC-F2, pBAC-F3, and pBAC-f4. BamHI enzyme site in the first fragment was mutated as a marker (A5472C) for identification of rescued virion through amplification of the F1 fragment with primers f1A5472C-F and f1A5472C-R containing mutation site (Table 1).
Fig. 1.
Strategy for the construction of full-length cDNA of IBV. (a) Construction of full-length cDNA of H120. (b) Construction of full-length cDNA of chimeric H120/SCS1
For the 5′UTR and 3′UTR fragments, the primers (5′UTR-F and 5′UTR-R, 3′UTR-F and 3′UTR-R) containing the homology arms were designed (Table 1), and the nucleotide sequences were separately amplified. The first and fourth subcloning vectors pBAC-f1 and pBAC-f4 were digested with BamHI and XhoI restriction enzymes (Takara, Japan), respectively, resulting in a linearization of the vectors (Fig. 1a). The 5′UTR was fused to the first subcloning vector construct, and the 3′UTR was fused to the fourth construct by homologous recombination to obtain the first subcloning pBAC-F1 and fourth subcloning pBAC-F4. The first subcloning vector construct was modified by adding the CMV (cytomegalovirus) promoter sequence in the 5′ terminal of the first fragment, and then HDVR (hepatitis delta virus ribozyme) sequence and BGH (bovine growth hormone polyadenylation signal) sequence were added to the 3′ terminal of the fourth fragment after the poly(A) by homologous recombination with the primers (CMV-F and CMV-R, HB-F and HB-R) listed in Table 1.
Subsequently, the F2 and F4 fragments were amplified with the primer sets (F2-F1)-F and (F2-F1)-R and primer sets (F4-F3)-F and (F4-F3)-R (Table 1), respectively. They were fused to the linearized plasmids pBAC-F1 and pBAC-F3 digested by XhoI to obtain two semi-subclones pBAC-F12 and pBAC-F34, respectively (Fig. 1a). Finally, two semi-subclones were fused using the same method with the primer sets (F34-F12)-F and (F34-F12)-R (Table 1) to complete the full-genome cDNA construction of IBV H120 (Fig. 1a).
Replacement strategy for chimeric recombinant H120/SCS1 and generation of full-length cDNA
Procedure used to construct chimeric H120 recombinant strain has been shown in Fig. 1b. The linearized plasmid DNA pBAC-F4(HB)-ΔS1 lacking the S1 gene was reverse amplified from the plasmid pBAC-F4 with primer set H120-F4-△S1-F and H120-F4-△S1-R (Table 1) as a linearized vector. The SC021202 S1 gene containing same homology arms to those of the H120F4 was amplified from total RNA of allantoic fluid of SPF chicken embryonated eggs infected with IBV SC021202 by RT-PCR with the primers SCS1-F and SCS1-R listed in Table 1 and was then inserted into pBAC-F4-△S1 by the same homologous recombination process using the ClonExpress one-step cloning kit (Vazyme Biotechnology, China). Then, the third segment of IBV genome and the pBAC-F4/SCS1 was fused to obtain two semi-subclones. Subsequently, both of these semi-subclones were fused to complete the construction of full-genome cDNA of IBV H120/SCS1.
Transfection and recovery of infectious virus
BHK-21 cells were cultured in 6-well plates. When the cells grew to 60% confluency, 4 μg (200 μL/well) of pBAC-H120 or pBAC-H120/SCS1 was transfected to the cells using jetPRIME transfection reagent (Polyplus, France) according to the manufacturer’s instructions. Transfected cells were incubated for 6 h at 37 °C. After the incubation, the culture medium was replaced with DMEM with 2% FBS and cells were incubated at 37 °C for 48 h. After completion of the incubation period, culture medium and cells were harvested by repeated freeze-thaw (three times) and named as rH120 and rH120/SCS1 P0 generation of the rescued virus. This P0 generation was inoculated into 10-day-old SPF chicken embryos. After 48 h, the allantoic fluid of chicken embryo was collected and blindly passaged for 3 generations. EID50 of passaged virus was determined by the method of Reed and Muench.
RT-PCR and sequencing analysis of rescued viruses
The P0 generation rescue virus and allantoic fluid of infected chicken embryo were used for RNA extraction. Reverse transcription reaction was carried out using a reverse transcription kit (Thermo Fisher, USA) to obtain cDNA. IBV H120 strain was used as template for designing primers (RM-F and RM-R, Table 1) before and after the introduced rescue marker (RM) to amplify a 621 bp fragment spanning between 5211 and 5831 bp of the viral genome. The PCR products were purified and ligated into pMD-18T vector (Takara, Japan) and then transformed into competent cells, and positive clones were picked for sequencing to identify rescue marker sites. The rescue virus was further inoculated into the SPF chicken embryonated eggs and observed for development of dwarf embryo lesion. To examine the stability of rescued viruses, S1 fragment of the virus of different passages was amplified from total RNA extracted from allantoic fluid of infected embryonated eggs by RT-PCR using the primers H120SC-S-F and H120SC-S-R (Table 1). The amplified fragments were then sequenced and analyzed.
Immunofluorescence microscopy
CEK cells prepared from 18-day-old chicken embryos were infected with H120, rH120, and rH120/SCS1. The medium was removed 48 hpi, and infected cells were fixed with methanol:acetone (1:1) at − 20 °C. Considering that membrane protein (M protein) is one of the abundant structural proteins in coronavirus (Neuman et al. 2011), in-house mouse monoclonal antibody 2B3 to IBV M protein (1:1000) was used as primary antibody in immunofluorescence assay, which were produced by our research group by the method described previously (Hu et al. 2007). Bound primary antibody was detected with fluorescein-labeled antibody to mouse lgG (H + L) (1:500) (Sigma-Aldrich, USA) and stained cells were observed under fluorescence microscope.
Western blotting
The 10-day-old SPF chicken embryonated eggs were inoculated with IBV H120, rH120, and rH120/SCS1. After 48 h, allantoic fluids were collected and subjected to SDS-PAGE analysis. The separated proteins were then transferred to a nitrocellulose membrane at 400 mA for 30 min using 25 mM Tris–192 mM glycine buffer (pH 8.3) containing 20% methanol. The membrane was blocked with 5% skim milk in PBS (20 mM Tris-HCl, pH 7.5, 150 mM NaCl) for 1 h and then washed three times with wash buffer (PBS-Tween 20). After that, the membrane was incubated with monoclonal antibody 2B3 against IBV M protein (dilution 1:1000) for 1 h. The membrane was again washed three times with wash buffer at room temperature. After that, bound primary antibody was detected with horseradish peroxidase-conjugated goat anti-mouse secondary antibody (dilution 1:5000) (Sigma-Aldrich, USA). Signals were analyzed by enhanced chemiluminescence using the AMI600 system (GE Healthcare, USA).
Pathogenicity analysis of rescued viruses
A total of fifty 7-day-old SPF chickens were divided into five groups and housed in different negative pressure isolators. Ten chickens in each group were inoculated with 0.3 mL of 104.5 EID50 of IBV H120, rH120, rH120/SCS1, and SC021202 via intranasal route. Birds in the control group were inoculated with the same volume of sterilized PBS. After every 2 days, oropharyngeal and cloacal swabs were collected and subjected to RT-PCR to check the virus shedding status. The dead chickens were examined by necropsy, and the tissues displaying gross lesions were collected. Meanwhile, one chicken in the groups without mortality was randomly selected for dissection and observation of gross lesions in tissues. The tissues of trachea and kidney were sent to Wuhan Servicebio Technology Co., Ltd. (Hangzhou, China) for processing hematoxylin and eosin (H&E) staining, and pathogenic lesion was observed under the microscope.
Immunoprotection experiment
A total of forty 7-day-old SPF chicks were divided into four groups and 10 chickens in every group were immunized with 0.3 mL of 104.5 EID50 of IBV H120, rH120, and rH120/SCS1 by intranasal inoculation, and chickens in the control group were inoculated with the same volume of sterilized PBS. After 7 and 14 days post-immunization, serum was collected and the antibody against IBV was detected by ELISA. Then, the chickens were challenged with 0.3 mL of 105.85 EID50 of IBV SC021202 strain. Clinical signs and mortality of chickens after the challenge were recorded every day.
ELISA
IBV N recombinant protein (1 μg/well) in 0.01 M PBS (pH 7.4) was coated on a 96-well microtiter plates (Canada JET Biochemicals Int’l. Inc.) at 4 °C overnight, followed by blocking with 200 μL blocking buffer for 2 h at 37 °C. The plate was washed three times with PBST and then incubated with 100 μL chicken serum samples diluted in blocking buffer (1:500) for 1 h at 37 °C. After washing three times with PBST, plates were incubated with 100 μL HRP-conjugated goat anti-chicken IgG (KPL, USA) in blocking buffer for 1 h at 37 °C. After washing three times with PBST, the colorimetric reaction was developed after incubating the plates with 100 μL chromogenic substrate for 10 min at 37 °C. Color development was stopped with 50 μL 2 M H2SO4, and an optical density at 450 nm (OD 450 nm) was recorded using ELx800 universal microplate reader (Bio-Tek Instruments, Inc., USA).
Results
Construction of full-length cDNA of IBV H120 and its chimeric virus
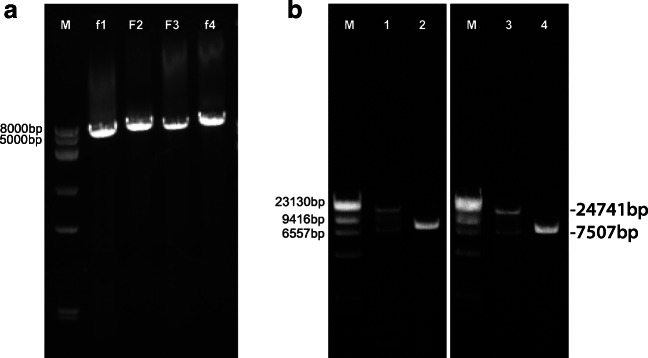
The full-length cDNA of IBV H120 strain was constructed according to the procedure shown in Fig. 1a. Four fragments (f1, F2, F3, and f4) covering the whole genome were amplified by RT-PCR with the total RNA of IBV H120 as template (Fig. 2a). After the addition of CMV promoter to the 5′-terminal of F1 fragment and HB sequence to the 3′-terminal of F4 fragment, all four fragments were individually cloned into pBAC vectors resulting into the generation of four fragment subcloning vectors. Afterwards, those fragments were fused together by homologous recombination. Finally, the full-length cDNA of IBV H120 in pBAC vector (pBAC-IBV-H120FL) was obtained and confirmed by RE digestion, which showed that the full length of pBAC-IBV-H120FL was digested into two major bands with expected molecular size of around 24,741 and 7507 bp by XhoI and BamHI (Fig. 2b). The full-length cDNA was further confirmed by sequencing (data no shown).
Fig. 2.
Amplification of four cDNA fragments of IBV genome and identification of the recombinant plasmids. (a) Amplification of f1, F2, F3, and f4 fragments of H120 cDNA by RT-PCR. (b) Identification of full-length cDNA of H120 and H120/SCS1 by RE digestion with XhoI and BamHI. M, λ-HindIII digest DNA Marker; 1, pBAC-IBV-H120FL; 3, pBAC-IBV-H120/SCS1FL; 2 and 4, pBAC
Additionally, we made another full genome construct in which S1 gene of IBV H120 was replaced with that of strain SC020102 and termed as pBAC-IBV-H120/SCS1. Strategy for the clone construction has been shown in Fig. 1b. The recombinant plasmid pBAC-IBV-H120/SCS1 was also confirmed by RE digestion with similar results to that of pBAC-IBV-H120FL (Fig. 2b). The full-length cDNA was also further confirmed by sequencing (data no shown).
Generation and characterization of rescued virus rH120 and rH120/SCS1
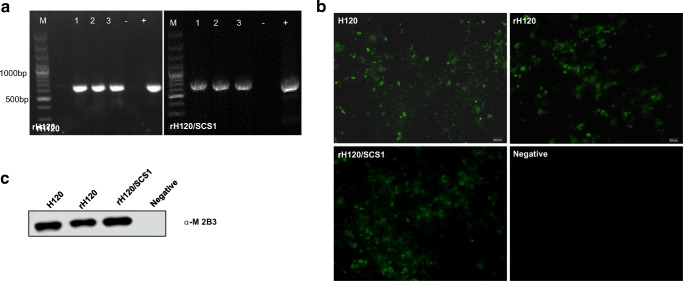
pBAC-IBV-H120 and pBAC-IBV-H120/SCS1 plasmids were first transfected into BHK-21 cells to obtain the P0 generation of rescued virus rH120 and rH120/SCS1. This P0 virus was passaged in SPF chicken embryonated eggs for three times. The P3 generation of the rescued viruses caused typical dwarf embryo lesions (data not shown), and IBV-specific sequences were also amplified using the primers RM-F and RM-R (Table 1) with the cDNA of the P3 generation of the rescued viruses (Fig. 3a). The mutant oligonucleotide site that served as rescue virus marker (GGATCC → GGCTCC) was also confirmed to be in the correct position (data not shown). The S1 gene of rH120/SCS1 was also amplified and sequenced, which was confirmed to be the S1 gene of SC021202 (data not shown), indicating that the S1 gene of H120 was successfully replaced with that of a heterogeneous IBV strain in a chimeric rescued virus. As results shown in Fig. 3b and Fig. 3c, IBV viral protein could be detected in rH120 and rH120/SCS1 infected CEK cells by Western blot analysis and IFA. These results indicated that recombinant virus rH120 and rH120/SCS1 were successfully rescued.
Fig. 3.
Characterization of rescued viruses. (a) Amplification of IBV H120-specific cDNA fragments from different generations of rH120 and rH120/SCS1 passaged in SPF chicken embryonated eggs. 1–3, The first three generations of rH120. 4–6, The first three generations of rH120/SCS1. (b) H120, rH120, and rH120/SCS1 infected CEK cells detected by IFA with mAb against IBV M protein. (c) Western blot analysis of H120, rH120, and rH120/SCS1 infected chicken embryonic allantoic fluid with mAb against IBV M protein
Subsequently, the rescued rH120 and rH120/SCS1 were continuously passaged in chicken embryonated eggs for 8 generations. S1 gene fragment of each passage was sequenced and analyzed. Changes in the S1 gene sequence were not found in the rescued viruses, which indicated that the S1 genes of rH120 and rH120/SCS1 were stable during the repeated passages. The mutant oligonucleotide site (GGATCC → GGCTCC) which served as a rescue virus marker was also present in all generations (data not shown).
Pathogenicity and immunoprotection of chimeric vaccine strain rH120/SCS1
To examine the pathogenicity of IBV H120 strain carrying S1 gene of virulent isolate, same doses of H120, rH120, rH120/SCS1, and SC0201202 were inoculated into the 7-day-old SPF chickens. Chickens inoculated with sterilized PBS were used as negative control. Results of RT-PCR indicated that the chickens infected with H120, rH120, and rH120/SCS1 showed persistent shedding of virus until 13 dpi (Fig. 4a, Table 2). Clinical signs, such as respiratory distress, were only observed in SC021202 group at 3–7 dpi, and not in H120, rH120, and rH120/SCS1 groups. Fatality occurred only in the SC021202 infection group with mortality rate of 30% (Fig. 4b). Kidneys of dead chickens showed pale discoloration and were swollen with white urate deposits, showing a typical “spotted kidney”-like lesion (Fig. 4c). Mortality and severe gross pathology in kidneys were not observed in H120, rH120, and rH120/SCS1 infection groups as well as the negative control group (Fig. 4b and 4c).
Fig. 4.
Pathogenicity and immunoprotection of rescued viruses. (a) Detection of viral nucleic acid in swabs collected from rH120 inoculated 7 different chickens after 11 days of infection. 1–7, Oropharyngeal swabs. 8–14, Cloacal swabs. (b) Survival rates of chickens inoculated with the rescued viruses. (c) Gross lesions in kidneys of rescued virus infected chickens. (d) Histopathological changes in kidneys and trachea of rescued virus infected chickens. (e) Survival rates of rescued virus immunized chickens challenged with SC021202. (f) Detection of antibodies to IBV N protein of antisera collected one week and two weeks after immunization from IBV H120, rH120, and rH120/SCS1 immunized chicken by ELISA
Table 2.
Detection of virus nucleic acid from swabs collected from virus inoculated chickens
| Group | Sample type | 1 dpi | 3 dpi | 5 dpi | 7 dpi | 9 dpi | 11 dpi | 13 dpi |
|---|---|---|---|---|---|---|---|---|
| H120 | Oropharyngeal | 4/10 | 7/10 | 9/10 | 6/7 | 6/7 | 6/7 | 5/7 |
| Cloacal | 5/10 | 5/10 | 3/10 | 6/7 | 6/7 | 4/7 | 3/7 | |
| rH120 | Oropharyngeal | 10/10 | 6/10 | 6/10 | 4/7 | 4/7 | 7/7 | 6/7 |
| Cloacal | 3/10 | 2/10 | 3/10 | 3/7 | 5/7 | 5/7 | 4/7 | |
| rH120/SCS1 | Oropharyngeal | 8/10 | 9/10 | 10/10 | 4/7 | 4/7 | 7/7 | 5/7 |
| Cloacal | 3/10 | 4/10 | 7/10 | 2/7 | 5/7 | 3/7 | 6/7 | |
| SC021202 | Oropharyngeal | 4/10 | 10/10a | 10/10a | 7/7a | 7/7 | 5/7 | 6/7 |
| Cloacal | 2/10 | 10/10a | 7/10a | 5/8a | 3/7 | 4/7 | 4/7 | |
|
Negative Control |
Oropharyngeal | 0/10 | 0/10 | 0/10 | 0/7 | 0/7 | 0/7 | 0/7 |
| Cloacal | 0/10 | 0/10 | 0/10 | 0/7 | 0/7 | 0/7 | 0/7 |
aObvious breathing distress was observed
Severe histopathological changes were observed only in the kidneys and tracheas of chickens infected with strain SC021202 (Fig. 4d). The kidney section from dead chickens in the SC021202 group showed enlarged renal tubules with epithelial cell exfoliation inside the tubular lumen, necrosis of some epithelial cells, and increased exudate with inflammatory cell infiltration (Fig. 4d). In the tracheal section from dead chicken in the SC021202 group, we observed that the epithelial cells of the mucous layer were exfoliated, degenerated, and necrosed (Fig. 4d). No obvious histopathological changes were observed in H120, rH120, and rH120/SCS1 groups (Fig. 4d).
In order to find out whether the chimeric virus could provide immunoprotection against the virulent IBV isolate, the chickens infected with H120, rH120, and rH120/SCS1 14 dpi were challenged with IBV strain SC021202. The results showed that all the chickens survived except those in the control group (Fig. 4e). The antisera collected from H120, rH120, and rH120/SCS1 immunized chickens at 7 and 14 dpi were analyzed by ELISA. As result showed in Fig. 4f, chickens immunized by H120, rH120, and rH120/SCS1 could produce antibody against IBV at 7 and 14 dpi but not in PBS control group (Fig. 4f).
Discussion
Compared with other avian respiratory viruses, such as avian influenza virus and Newcastle disease virus, it is difficult to manipulate IBV genome by reverse genetics due to its large-sized genomic RNA (Lai 2000) and also the availability of very few permissible cell lines for IBV infection (Ferreira et al. 2003). Ligation of restriction enzyme (RE)-digested fragments is a method to obtain the full-length cDNA of IBV genome for rescue of IBV (Casais et al. 2001). However, the procedure for ligation of large DNA fragments to form the full-length genomic cDNA is complicated, time-consuming, and inefficient. On the other hand, the full length of IBV genome is long and selection of the suitable RE for ligation of the full-length cDNA of IBV genome is difficult. Hence, there is a need to establish an efficient reverse genetic system to rescue IBV in vitro. BAC reverse genetic system is a common in vitro method to rescue coronavirus, such as TGEV (Almazan et al. 2000), SARS-CoV (Almazan et al. 2006), FIPV (Balint et al. 2012), and MERS-CoV (Scobey et al. 2013). Ligation of DNA fragments by homologous recombination can be avoided to select many RE enzyme sites. But, so far, no BAC vector combined with homologous recombination has been used for the construction of IBV full-length cDNA.
In this study, four DNA fragments of around 7000–8000 bp covering full-length genome of IBV were separately amplified and cloned into the low-copy vector pBeloBAC11 by homologous recombination. The necessary component sequences of 5′-UTR, 3′-UTR, promoter sequence from CMV as well as HDVR and BGH were added to the 5′ terminal of the first fragment and the 3′ terminal of the last fragment by the same way. Added HDVR at the end of IBV genome will increase the efficiency of rescue of IBV infectious clones by maintaining the integrity of the 3′ terminal of IBV genome. Using the same strategy, two fragments, each were again fused together into the subcloning vector, and finally, both vectors each carrying two fragments were fused into a full-length cloning vector. Construction of subcloning vector by homologous recombination simplifies the operation and improves the efficiency of the ligation. In this reverse genetic system, site mutation, sequence deletion, and insertion can easily be performed based on certain sequence sites in the backbone of subcloning vector.
With the reverse genetics established in this study, recombinant H120 (rH120) strain and its chimeric virus rH120/SCS1 carrying S1 gene of a virulent IBV SC021202 strain were rescued successfully. The pathogenicity of rH120/SCS1 was found to be similar to those of H120 and rH120 and was not able to cause pathological changes as shown in the case of strain SC021202-infected chickens. Previous reports showed that S1 protein of IBV was responsible for pathogenicity, tropism, and antigenicity (Hulswit et al. 2016; Wickramasinghe et al. 2011). However, our result indicates that only the S1 gene of SC021202 isolate is not a determinant of the pathogenicity of IBV since rH120/SCS1 with H120 as the backbone and chimeric SC021202 S1 gene did not cause obvious sickness to the sensitive SPF chickens as caused by the SC021202 strain. The results of this study are in agreement with previous studies showing that the S1 gene has little effect on pathogenicity (Armesto et al. 2011; Hodgson et al. 2004; Jiang et al. 2020). Whether the S1 gene is the determinant of the pathogenicity of IBV might depend on individual isolate, which needs further study to confirm.
Finally, the immunization of H120, rH120, and rH120/SCS1 could protect the chicken against the challenge by IBV strain SC021202, which indicated that the recombinant clones constructed for IBV in this study may be useful for the development of novel IBV vaccine.
In summary, in this study, with homologous recombination technology, an efficient reverse genetic system based on BAC vectors for rescue of IBV infectious clone was established. This will provide a useful tool for further study of the pathogenicity-related genes of IBV and the development of vaccine against the virus.
Author contributions
JYZ, ML, and TTS designed the experiments. CFL, TTS, and PPZ performed the experiments. CFL and TTS analyzed the data. XP, SSC, and YY prepared the reagents. CFL, ML, and NKO drafted the manuscript. All authors read and approved the final manuscript.
Funding information
This work was supported by the National Key Research and Development Program of China (Grant No. 2016YFD0500800), the National Key Technology Research and Development Program of China (Grant No. 2015BAD12B01), and the China Agriculture Research System (Grant No. CARS-40-K13).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Ethical statement
The animal experiment was approved by the Committee on the Ethics of Animal of Zhejiang University (ZJU20170667) and implemented in accordance with the animal care and ethics guidelines. This article does not contain any studies with human participants performed by any of the authors.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Chenfei Lv and Tingting Shi contributed equally to this work.
References
- Almazan F, Gonzalez JM, Penzes Z, Izeta A, Calvo E, Plana-Duran J, Enjuanes AL. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. PNAS. 2000;97(10):5516–5521. doi: 10.1073/pnas.97.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almazan F, Dediego ML, Galan C, Escors D, Alvarez E, Ortego J, Sola I, Zuniga S, Alonso S, Moreno JL, Nogales A, Capiscol C, Enjuanes L. Construction of a severe acute respiratory syndrome coronavirus infectious cDNA clone and a replicon to study coronavirus RNA synthesis. J Virol. 2006;80(21):10900–10906. doi: 10.1128/JVI.00385-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armesto M, Evans S, Cavanagh D, Abu-Median AB, Keep S, Britton P. A recombinant avian infectious bronchitis virus expressing a heterologous spike gene belonging to the 4/91 serotype. PLoS One. 2011;6(8):e24352. doi: 10.1371/journal.pone.0024352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balint A, Farsang A, Zadori Z, Hornyak A, Dencso L, Almazan F, Enjuanes L, Belak S. Molecular characterization of feline infectious peritonitis virus strain DF-2 and studies of the role of ORF3abc in viral cell tropism. J Virol. 2012;86(11):6258–6267. doi: 10.1128/JVI.00189-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursnell ME, Brown TD, Foulds IJ, Green PF, Tomley FM, Binns MM. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J Gen Virol. 1987;68(Pt 1):57–77. doi: 10.1099/0022-1317-68-1-57. [DOI] [PubMed] [Google Scholar]
- Casais R, Thiel V, Siddell SG, Cavanagh D, Britton P. Reverse genetics system for the avian coronavirus infectious bronchitis virus. J Virol. 2001;75(24):12359–12369. doi: 10.1128/JVI.75.24.12359-12369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Coronaviruses in poultry and other birds. Avian Pathol. 2005;34(6):439–448. doi: 10.1080/03079450500367682. [DOI] [PubMed] [Google Scholar]
- Cook JK, Jackwood M, Jones RC. The long view: 40 years of infectious bronchitis research. Avian Pathol. 2012;41(3):239–250. doi: 10.1080/03079457.2012.680432. [DOI] [PubMed] [Google Scholar]
- Ferreira HL, Pilz D, Mesquita LG, Cardoso T. Infectious bronchitis virus replication in the chicken embryo related cell line. Avian Pathol. 2003;32(4):413–417. doi: 10.1080/0307945031000121167. [DOI] [PubMed] [Google Scholar]
- Hodgson T, Casais R, Dove B, Britton P, Cavanagh D. Recombinant infectious bronchitis coronavirus Beaudette with the spike protein gene of the pathogenic M41 strain remains attenuated but induces protective immunity. J Virol. 2004;78(24):13804–13811. doi: 10.1128/JVI.78.24.13804-13811.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JQ, Li YF, Guo JQ, Shen HG, Zhou JY. Production and characterization of monoclonal antibodies to poly100s1 protein of avian infectious bronchitis virus. Zoonoses Public Health. 2007;54(2):69–77. doi: 10.1111/j.1863-2378.2007.01030.x. [DOI] [PubMed] [Google Scholar]
- Hulswit RJ, de Haan CA, Bosch BJ. Coronavirus spike protein and tropism changes. Adv Virus Res. 2016;96:29–57. doi: 10.1016/bs.aivir.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignjatovic J, Sapats S. Identification of previously unknown antigenic epitopes on the S and N proteins of avian infectious bronchitis virus. Arch Virol. 2005;150(9):1813–1831. doi: 10.1007/s00705-005-0541-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackwood MW. Review of infectious bronchitis virus around the world. Avian Dis. 2012;56(4):634–641. doi: 10.1637/10227-043012-Review. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Cheng X, Zhao X, Yu Y, Gao M, Zhou S. Recombinant infectious bronchitis coronavirus H120 with the spike protein S1 gene of the nephropathogenic IBYZ strain remains attenuated but induces protective immunity. Vaccine. 2020;38:3157–3168. doi: 10.1016/j.vaccine.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MMC. The making of infectious viral RNA—no size limit in sight. PNAS. 2000;97(10):5025–5027. doi: 10.1073/pnas.97.10.5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scobey T, Yount BL, Sims AC, Donaldson EF, Agnihothram SS, Menachery VD, Graham RL, Swanstrom J, Bove PF, Kim JD, Grego S, Randell SH, Baric RS. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc Natl Acad Sci U S A. 2013;110(40):16157–16162. doi: 10.1073/pnas.1311542110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan D, Fang S, Han Z, Ai H, Zhao W, Chen Y, Jiang L, Liu S. Effects of hypervariable regions in spike protein on pathogenicity, tropism, and serotypes of infectious bronchitis virus. Virus Res. 2018;250:104–113. doi: 10.1016/j.virusres.2018.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beurden SJ, Berends AJ, Kramer-Kuhl A, Spekreijse D, Chenard G, Philipp HC, Mundt E, Rottier PJM, Verheije MH. A reverse genetics system for avian coronavirus infectious bronchitis virus based on targeted RNA recombination. Virol J. 2017;14(1):109. doi: 10.1186/s12985-017-0775-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickramasinghe IN, de Vries RP, Grone A, de Haan CA, Verheije MH. Binding of avian coronavirus spike proteins to host factors reflects virus tropism and pathogenicity. J Virol. 2011;85(17):8903–8912. doi: 10.1128/JVI.05112-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn S, Leibowitz JL, Collisson EW. In vitro assembled, recombinant infectious bronchitis viruses demonstrate that the 5a open reading frame is not essential for replication. Virology. 2005;332(1):206–215. doi: 10.1016/j.virol.2004.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JY, Zhang DY, Ye JX, Cheng LQ. Characterization of an avian infectious bronchitis virus isolated in china from chickens with nephritis. J Veterinary Med Ser B. 2004;51(4):147–152. doi: 10.1111/j.1439-0450.2004.00744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YS, Zhang Y, Wang HN, Fan WQ, Yang X, Zhang AY, Zeng FY, Zhang ZK, Cao HP, Zeng C. Establishment of reverse genetics system for infectious bronchitis virus attenuated vaccine strain H120. Vet Microbiol. 2013;162(1):53–61. doi: 10.1016/j.vetmic.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]