Abstract
Host-associated microbial communities have profound impacts on animal physiological function, especially nutrition and metabolism. The hypothesis of ‘symmorphosis’, which posits that the physiological systems of animals are regulated precisely to meet, but not exceed, their imposed functional demands, has been used to understand the integration of physiological systems across levels of biological organization. Although this idea has been criticized, it is recognized as having important heuristic value, even as a null hypothesis, and may, therefore, be a useful tool in understanding how hosts evolve in response to the function of their microbiota. Here, through a hologenomic lens, we discuss how the idea of symmorphosis may be applied to host-microbe interactions. Specifically, we consider scenarios in which host physiology may have evolved to collaborate with the microbiota to perform important functions, and, on the other hand, situations in which services have been completely outsourced to the microbiota, resulting in relaxed selection on host pathways. Following this theoretical discussion, we finally suggest strategies by which these currently speculative ideas may be explicitly tested to further our understanding of host evolution in response to their associated microbial communities.
This article is part of the theme issue ‘The role of the microbiome in host evolution’.
Keywords: host–microbe interactions, symmorphosis, adaptive modulation, hologenome theory, physiology
1. Introduction
Host-associated microbial communities provide numerous services and benefits to their hosts, especially in regard to nutrition and metabolism. Owing to these many services, the gut microbiome was often referred to as a ‘forgotten organ’ [1,2]. We now recognize the complex ecological and evolutionary factors that make the microbiome more than just an organ, and the microbiome has been thought of as a community, an ecosystem, a ‘second genome’, etc. [3]. While none of these terms may perfectly describe the microbiome, there may still be heuristic value in considering some aspects of the microbiome under the lens of organ function and evolutionary physiology. For example, we have previously discussed how the microbiome integrates well into the ‘Grand Challenges of Comparative Physiology’ [4,5]. Here, we focus on whether the actions of the microbiome and the host might be coordinated and tuned to one another. Specifically, do hosts evolve in response to the capabilities or services provided by their microbiome? Our discussion below is largely focused on the bacterial members, owing to their overall dominance in the gut community [6]; however, it should be noted that other microbial groups can provide substantial physiological services to hosts [7].
Understanding the integration of physiological processes across organ systems, tissues, and levels of biological organization is recognized as one of the ‘Grand Challenges’ in animal physiology [5]. In the 1980s and 1990s, there was heavy debate around the idea of optimization within physiological systems. The hypothesis of ‘symmorphosis’ posits that animals' structural or physiological elements are regulated to satisfy but not exceed the requirements of a functional system [8,9]. The idea of symmorphosis has been well studied in respiratory physiology, where an animal's heart rate, capillary volume and mitochondrial density can be coordinated to determine an animal's peak metabolic rate [9]. Similarly, the field of digestive physiology has heavily researched the ‘adaptive modulation hypothesis', which states that intestinal enzymes and transporters should match the relative levels of their substrates in an animal's diet [10]. Such matching promotes optimal digestion, while not wasting available membrane space and synthetic energy on enzymes/transporters in excess of need [10]. Indeed, on both a plastic and evolutionary time scale this matching between dietary substrates and enzymes/transporters has been observed in numerous systems [10–12]. Such symmorphosis has even been observed at the molecular level, where total activities of enzymes in the Krebs cycle vary across tissues, yet are found in consistent proportions [13].
However, the idea of symmorphosis or optimal matching has received substantial criticism from evolutionary biologists [14]. First, resounding support for symmorphosis has not been demonstrated. For example, while there is substantial coordinated activity between the heart, capillaries, and mitochondria, the diffusion capacity in the lungs does not match this trend [8]. Second, genetic and evolutionary constraints or trade-offs may preclude perfect coordination across many enzymes, tissues, and organs [14,15]. Third, many physiological elements perform multiple functions, and so perfect coordination may not be likely for every downstream function [14,15]. Last, the hypothesis of symmorphosis relies heavily on natural selection as the force driving optimization. However, genetic drift, sexual selection or variable selective forces may hinder optimization [15,16].
Regardless of the ubiquity of symmorphosis, both originators and criticizers of this hypothesis argue that it has powerful heuristic value, even if as a null hypothesis, and is most useful when studying the integration of functional systems [8,14,16,17]. Here, we argue that host–microbe interactions could be viewed under the lens of these integrated physiological systems, given the considerable connections between the gut microbiome and host physiology [4,18]. Specifically, these questions could be applied to host–microbe interactions to understand whether hosts evolve and adapt to the capacities or functions of their microbiome.
2. Hologenome theory and symmorphosis
The recognition that microbial communities vary within and across host species, and impact whole-animal performance, has given rise to the concept of ‘hologenomic evolution’, which considers the collection of host and microbial genomes—known as ‘the holobiont’—a biological unit of organization upon which natural selection can act [19]. In this framework, microbial symbionts may be gained, lost, or change in frequency and abundance within a population, which occurs in parallel with random mutations and changes in allele frequencies in nuclear genes. These events provide the raw genetic variation, at multiple levels, necessary for natural selection to act. Over time, this strict definition has been adapted and expanded to accommodate a growing body of work on this topic, such as the importance of horizontal gene transfer between microbes, and community heritability [20–22]. Despite similarities between hologenome theory and traditional ideas of evolution, the concept has been met with considerable criticism. For example, questions regarding patterns of transmission in the microbiome, and fidelity among partners remain to be resolved [23].
Through the lens of hologenome theory, microbial symbionts and their functions represent another physiological system than can be optimized as predicted under the hypothesis of symmorphosis. A main criticism of symmorphosis is that evolutionary constraints may prevent the optimal matching between a host's physiology and their functional demands [14]. However, when the microbiome is considered as a component of the holobiont, these constraints may be lessened. For example, unique characteristics of microbes, such as horizontal gene transfer and rapid evolution [24], may allow the holobiont to respond and adapt much more quickly to changing functional demands than could be accommodated by the host's physiological systems alone. Indeed, certain gut microbes of many Japanese people have acquired seaweed-degrading porphyranases via horizontal gene transfer from marine bacteria, while these genes are absent in populations that do not commonly ingest seaweed [25]. Despite the potential contribution of the microbiome to optimal matching, it is important to remember that microbes, like other host physiological systems, perform many functions. For example, the common human gut bacterium Bacteroides thetaiotamicron is important for both carbohydrate and lipid metabolism [26], and also has immunomodulatory effects on the host [27,28]. Therefore, optimization of any one particular function may reduce the capacity of another [14]. However, a unique property of the microbiome as compared to host physiological systems is a high degree of functional redundancy [29]. Therefore, the loss or reduction of a particular function by one microbe may have little consequence to performance of the overall system.
3. Symmorphosis between the microbiome and host physiology
Here, we will focus on two ways in which host physiology may evolve to coordinate with the functions of the gut microbiome: collaboration and relaxation. We define collaboration as processes where the gut microbiome and host physiology interact with one another to yield a physiological function (such as fibre degradation). Relaxation will be discussed as scenarios when physiological functions are completely provided by the gut microbiome. In these contexts, hosts may experience relaxed selection, and may lose enzymatic pathways as a means to conserve biosynthetic energy [30]. For each of these ideas, we start by briefly discussing support for animal evolution in the field of ecological physiology (often independent of the microbiome) as a proof of concept that physiology can evolve in response to varying nutrients. Next, we will discuss any relevant studies on complex microbiomes. However, given that the microbiome field is still developing, especially in the context of natural systems [31], some of these directions may be highly speculative at this point.
(a). Collaboration
In the field of animal physiology, the ‘adaptive modulation hypothesis' posits that animals can adjust their digestive physiology in response to diet composition, typically by regulating the expression of hydrolases and transporters when there is an increase or decrease in the intake of relevant substrates [10]. For example, feeding on a diet rich in carbohydrates may result in increased activity of disaccharidases, such as maltase and sucrase, while a protein-rich diet may increase activity of protein-degrading enzymes, like aminopeptidase-N [32]. This phenomenon occurs on ecological time scales in response to diet switching [33,34], as well as over evolutionary time scales when species lack plasticity in digestive enzyme expression [12]. In response to diet switching, the ability to maintain plasticity in the activity of all or some digestive enzymes may be an evolved trait. Indeed, dietary generalists are typically able to modulate digestive enzymes to a greater extent than specialists [35], and carnivores can adjust the activity of amino acid, but not sugar, transporters [36]. Together, these results suggest that animals have evolved to match their enzyme production with the intake of their expected dietary substrates, consistent with the predictions of symmorphosis and the adaptive modulation hypothesis.
Thus, when considering host–microbe interactions, we may predict that hosts will evolve to optimally interact with microbial products. Indeed, extensive collaboration and coordinated evolution has been demonstrated between insect hosts and endosymbiotic microbes [37]. For example, in pea aphids (Acyrthosiphon pisum), both nucleotide metabolism and biosynthesis of essential amino acids are conducted by coordinated activity of host and microbial enzymes [38–40]. Furthermore, coordination between gene expression of hosts and microbes may be important in maintaining these symbioses. For example, aphids that maintain high loads of amino acid producing microbes invest less in their own amino acid producing enzymes, and the inverse is also true (i.e. aphids that maintain low titers of amino acid producing microbes invest more in their own amino acid biosynthesis) [40]. However, insects and their endosymbionts are obligate symbioses.
Evidence for similar evolutionary coordination in interactions between hosts and complex microbial communities may, at first, appear more diffuse than those between insects and their endosymbionts owing to the high degree of inter-individual variation in the composition of these communities [41]. However, functional redundancy in the gut microbiome results in more homogeneous communities across individuals at the functional level, as opposed to the taxonomic level [29]. For example, human twins may share only approximately 10% of bacterial operational taxonomic units in their gut based on 16S rRNA gene sequences, but exhibit approximately 70% overlap in sequences when the entire metagenome is considered [42]. Furthermore, mammalian hosts with similar dietary strategies also share similar dominant microbial functions, such as carbohydrate or protein digestion [43]. We would expect evolutionary matching of host physiology to the functional capabilities of the microbiome, rather than their taxonomic composition, especially in relation to microbially derived energy sources.
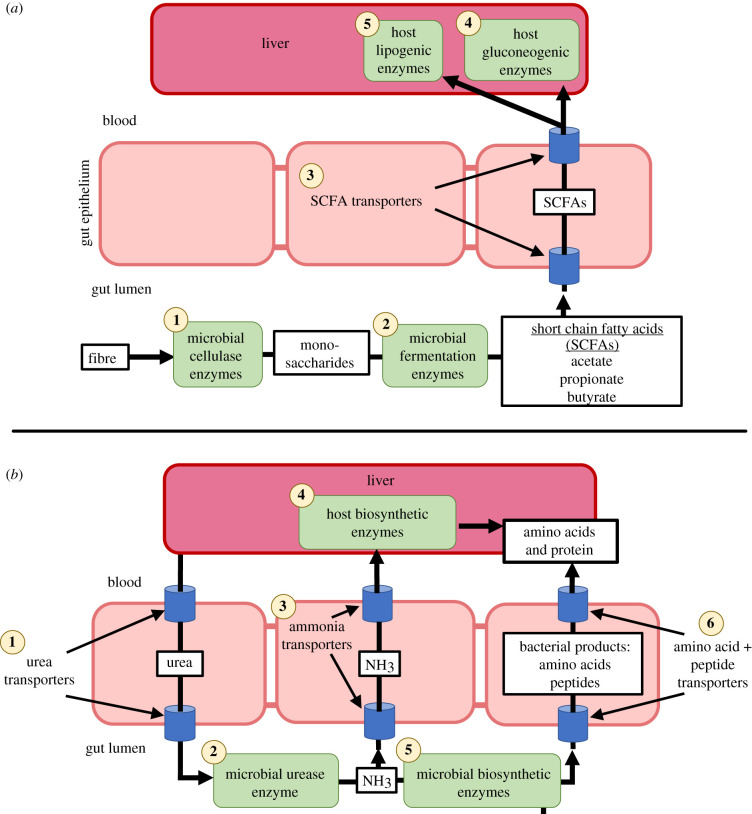
Short-chain fatty acids (SCFAs), such as acetate, butyrate and propionate, are metabolites produced as by-products of microbial fermentation of plant material in the gastrointestinal tract of vertebrates. These molecules can be transported throughout the body and are used as energy sources in a variety of tissues (figure 1a). In a similar fashion to their response to dietary substrates, animals may modulate the activity of SCFA transporters in response to an increase in their production. For example, monocarboxylate transporter 1 (MCT1), an important transporter of SCFAs across colonic luminal membranes, is upregulated in human colon cells by the direct addition of butyrate [44]. Furthermore, MCT1 expression is elevated in mice upon increased intake of dietary fibre, which stimulates the production of SCFAs by gut microbiota [45]. Over evolutionary time periods, some hosts have become more dependent on microbially derived energy than others. For example, ruminants obtain greater than 80% of energy requirements from microbial fermentation, while this value ranges from only 5 to 10% in omnivorous species, including humans [46]. Ruminants are also capable of modulating rates of SCFA absorption in response to dietary starch content, and preferentially incorporate SCFA-derived carbon into adipose tissue, while omnivorous animals use glucose more readily [46].
Figure 1.
Interconnected processes of the host and microbiome involved in (a) fibre fermentation and (b) nitrogen recycling. These examples are specific to vertebrate physiology, though similar processes may occur in other animal groups. (Online version in colour.)
SCFAs are transported throughout the body, and therefore, may result in modulation of enzymes in a variety of systemic tissues. For example, in humans, up to 70% of microbially produced acetate is used by the liver, and this value ranges from 30 to 60% for propionate, depending on the host species, where these SCFAs act as substrates for hepatic gluconeogenesis and lipogenesis [47,48]. Colonization of germ-free mice with gut microbiota stimulates glycogenesis and triglyceride synthesis in the liver, while altering the expression of enzymes that regulate lipid absorption [49]. Furthermore, SCFA production stimulates AMP-activated protein kinase activity in the liver, which proceeds to regulate transcriptional control of enzymes involved in glucose, lipid and cholesterol metabolism [47]. Again, flexibility of hepatic enzymes in response to differing concentrations of SCFAs may vary by animal group, depending on how necessary SCFAs are to overall energy metabolism. For example, hepatic metabolism of propionate can provide 50–75% of glucose requirements in ruminants, while this value is as low as 7% in non-ruminant herbivores [46]. Propionate metabolizing enzymes of ruminants are sensitive to food intake, and an increase in high-grain diets increases their activity [46]. Thus, over evolutionary time, the expression of transporters and enzymes may be matched to the levels normally produced by the microbiome, consistent with the predictions of symmorphosis.
In addition to provisioning SCFAs, the gut microbiota also contributes to host energy metabolism through the degradation of protein into amino acids and peptides [50]. Although animal hosts are also capable of metabolizing proteins, it appears they outsource some of these processes to bacteria. In both mice and zebrafish, germ-free animals upregulate amino acid and peptide transporters in comparison to conventional individuals, potentially to compensate for a lack of microbial processing of these substrates [51]. Across species, host diet also influences the role of the gut microbiota in protein metabolism. For example, carnivorous mammals exhibit an enrichment of bacterial genes involved in amino acid degradation, while in herbivores, bacterial genes that synthesize amino acids are enriched [43]. Thus, dominant microbial functions may match the unique needs of the host. Understanding the downstream adaptations that hosts may have to coordinate with these microbial functions requires further study.
Another important function conducted by collaboration between the microbiome and host physiology is the recycling of nitrogen. For example, mammals produce urea as a metabolic waste product, which is primarily excreted through the kidney into the urine to avoid toxic buildup. However, this process results in the loss of important nitrogen. A number of animals, such as herbivores that feed on low-nitrogen foods, have salvaging mechanisms to recover this nitrogen, such as transporting it into the gut for microbial recycling (figure 1b) [52]. Additionally, some amphibians catabolize muscle tissue and use urea as protective antifreeze during winter hibernation [53]. In spring, these amphibians may rely on microbial urea recycling to use this nitrogen and rebuild tissues [53]. Given that vertebrates do not produce enzymes capable of carrying out this urea recycling, nitrogen salvaging is only able to take place through host–microbe interactions.
Studies in ruminants show that both host and microbial steps in these processes are responsive to dietary protein levels [54,55]. For example, in sheep, microbial urease activity is higher when animals are fed low nitrogen diets, presumably to assist with nitrogen recycling [55]. Adaptations and differences in urea transporter expression, affinity, etc. have been well studied in kidneys, given that herbivores, omnivores, and carnivores maintain distinct levels of plasma urea owing to different levels of protein in their diets [56]. Specific adaptations within the gut microbiota, and physiological interactions have been less studied. While examples of nitrogen hydrolyzing bacteria are widespread across animals, robust comparative approaches investigating multiples steps in the process of urea recycling have not, to our knowledge, been conducted [52]. The hypothesis of symmorphosis might predict that host intestinal urea transporters, microbial urease enzymes, and pathways for the microbial synthesis of amino acids would be more abundant and coordinated in herbivores as compared to carnivores.
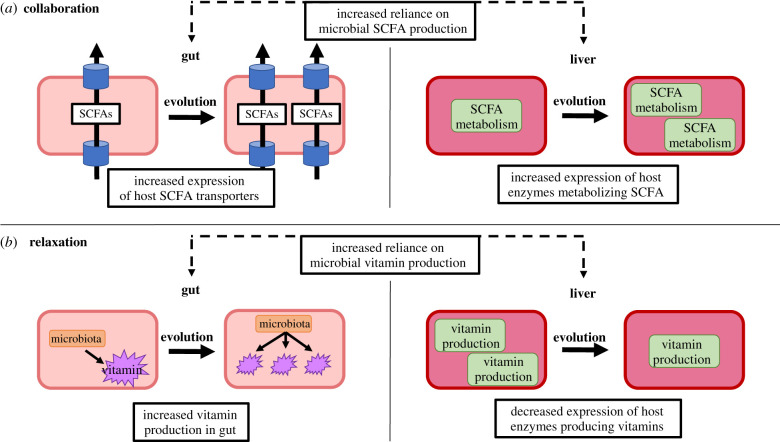
While speculative at this point, it is possible that over evolutionary time, the consistent presence of microbial metabolites has resulted in constitutive increases in the physiological machinery necessary to process them. For example, as occurs in response to dietary substrates [12], sustained increases and reliance on microbially produced SCFAs may eventually lead to evolved increases in expression of their transporters in the gut, and enzymes needed for their metabolism in the liver (figure 2a). This process could occur through genetic assimilation [57], whereby an initially plastic increase in transporters or enzymatic activity becomes fixed under constant selection pressure (i.e. sustained inputs of SCFAs). From a mechanistic standpoint, single nucleotide polymorphism (SNPs) and gene duplication events have previously explained evolutionary changes in digestive capacity [58]. For example, multiple human populations have convergently evolved mutations in the enhancer region of the lactase gene, permitting lactose tolerance in these individuals [59]. The human salivary amylase gene, AMY1, originated from duplication of the pancreatic amylase gene, AMY2, and its copy number is positively correlated with dietary starch levels, the primary substrate of its encoded enzyme [60]. Furthermore, gene duplication has also resulted in the evolution of lysozymes that function not as antimicrobials, but as digestive enzymes, in several foregut-fermenting animals (including monkeys, ruminants and birds), aiding in their dietary consumption of bacteria [61]. These findings suggest that animals would have the capacity to evolve digestive capacities that meet the demands imposed by bacteria. However, to our knowledge, no studies have explored whether these events have occurred in response to relationships with commensal bacteria (e.g. gene duplication events or SNPs in enhancer regions of genes encoding SCFA transporters). Using symmorphosis as a conceptual hypothesis (even if as a null hypothesis), will be informative to guide future research in this area.
Figure 2.
Potential evolutionary consequences of increased host reliance on microbial metabolite production in the gut and liver. These examples are specific to vertebrate physiology, though similar processes may occur in other animal groups. (a) When hosts become more reliant on microbially produced SCFAs as an energy source, they may evolve increased expression of both SCFA transporters in the gut and SCFA metabolizing enzymes in the liver, an example of a collaborative process. (b) When hosts become more reliant on microbially produced vitamins, there may be relaxed selection on host processes that produce these compounds. This would result in an increased concentration of microbially produced vitamins in the gut and a decreased expression of host vitamin producing enzymes in the liver. (Online version in colour.)
(b). Relaxation
Again, the hypothesis of ‘symmorphosis’ posits that animals' structural or physiological elements are regulated to satisfy but not exceed the requirements of a functional system [8,9]. Thus, to avoid exceeding functional requirements, there may be downregulation or loss of physiological systems. For example, when nutritional enzymatic pathways are no longer needed, they can be downregulated or lost entirely to conserve biosynthetic energy or preserve membrane space [10,11]. Numerous animal groups that acquire sufficient vitamin C from their diet, such as some fishes, bats and birds, have lost the enzymatic machinery to synthesize this necessary cofactor in the liver [62–64]. Additionally, cats are hypercarnivores that consistently consume adequate levels of many nutrients, resulting in the loss of enzymes that synthesize vitamin A, and extremely low hepatic expression of enzymes that produce the amino acid taurine [65]. When cats are fed diets that lack these nutrients, they suffer extreme health consequences [65].
Given that microbes are known to provide hosts with many essential nutrients, it is possible that hosts have evolved to lose enzymatic pathways that have been outsourced to microbes. For example, pea aphids house bacterial endosymbionts that conduct arginine synthesis, and as a result the pea aphid genome has lost enzymes required for these processes [39]. Additionally, herbivorous ruminants express different levels of glucose transporters on their gut lining depending on their reliance on microbial communities. For example, early-evolved ruminants such as deer, giraffes and some antelope species have relatively simpler stomachs, and do not rely heavily on microbial fermentation [66]. Thus, these animals digest simple sugars from the cell contents of plant material and absorb monosaccharides through high expression of sodium-glucose co-transporters in the small intestine [67]. Later-evolving ruminants such as sheep and bovids exhibit extensive morphological adaptations and rely heavily on microbial fermentation in the rumen. Here, microbial communities ferment plant fibres but also use the majority of sugars from plant cell contents, leaving few simple sugars to enter the small intestine. Resultantly, ruminant species that rely more on ruminal fermentation have lower or absent expression of the sodium-glucose co-transporters in the small intestine [67]. These patterns of host gene expression support the notion that hosts can downregulate gene expression to coordinate with microbial activities.
While demonstrated examples of relaxation by complex microbial communities are generally lacking, one potential area of research might be in the synthesis of vitamin A. Plant tissues lack vitamin A, but often contain the important precursors (carotenoids) that can be converted into vitamin A [68]. These conversion steps can be conducted by the host (intestinal tissue or liver), or by the gut microbiome [69]. As written above, carnivorous felines regularly consume sufficient levels vitamin A, and so have lost the enzymatic machinery for these conversions [65]. In herbivores, it could be that some species rely more heavily on microbial vitamin A synthesis, and so similarly they have lower expression of these enzymes in the liver (figure 2b) as a means to conserve biosynthetic energy, or complete loss of these conversion pathways through relaxed selection [30]. It would be interesting to test whether herbivores might develop vitamin deficiencies when their microbiome is altered through antibiotics or other techniques. Such data would reveal whether herbivores have optimized their own physiology to match the nutritional processes conducted by their gut microbiome.
4. Testing for symmorphosis between hosts and their microbiota
Although we suspect hosts have evolved expression of physiological systems to match processing demands imposed by incoming microbial metabolites, this idea remains to be explicitly tested. We argue that comparative approaches will capitalize on the diversity and variation of host-microbe interactions and will be powerful for investigating coordination between microbial functions and host physiology. For example, metagenomic studies have shown differences in microbial capabilities across animals with different feeding strategies [43]. Additionally, reliance on microbial communities may vary across hosts. The roughly 180 species of herbivorous ruminants vary in their stomach morphology, such that some species exhibit extensive morphological adaptations and can consume highly fibrous materials, while others have stomach morphologies that are less adapted for microbial fermentation and actively avoid consuming high-fibre plants [66,70]. Furthermore, in ants, the absolute abundance or density of microbes varies by diet and habitat [71]. There are even examples where hosts may not rely on microbial communities at all [72,73]. Similarly, physiological studies demonstrate that some herbivorous mammals obtain greater than 80% of energy requirements from microbial fermentation, while this value ranges from only 5 to 10% in omnivorous mammals [46]. Using diverse study systems that vary in their reliance on microbes will be crucial for testing for optimized coordination between microbial communities and host physiology. We discuss a number of modern approaches that may provide potential avenues for this investigation.
Comparative genomics has been successful in identifying convergence in patterns of gene evolution among animals that inhabit environments with similar selective pressures [74,75]. This approach could be applied to groups of animal hosts that depend on microbial products for nutrition. For example, are there consistent signatures of rapid evolution in genes coding for SCFA transporters across herbivore lineages? Further, comparative genomics can be combined with metagenomic studies to determine how evolution in both host and microbial genes have influenced present phenotypes. In a study of blood-feeding bats, Mendoza et al. [76] used comparative genomics and metagenomics to uncover adaptation in both host and microbial genomes which, combined, allow the holobiont to deal with a suite of physiological challenges associated with this diet, such as a lack of essential nutrients and exposure to blood-borne pathogens. These approaches could be applied to more host-microbe associations to uncover examples of host evolution in relation to the microbiome.
Experimental evolution is another powerful approach that has been used to address ecological and evolutionary questions across a variety of model systems, and may be quite useful in the context of host–microbe interactions [77]. For example, bank-voles that have been selected for the ability to maintain body mass on high fibre diets for 15 generations have distinct microbial communities from randomly bred control lines [78]. It would be interesting to compare the microbial functions and potential host evolution in these lines. Additionally, germ-free hosts can be inoculated with known microbiota and the resulting effects on host evolution observed. This approach has been used in studies of Drosophila melanogaster, where experimental evolution in a field setting has revealed that populations diverge genetically and phenotypically depending on the composition of their original microbiota [79]. Researchers could experimentally evolve hosts with microbial communities with varying nutritional capabilities (fibre fermentation, urea recycling), and search for coordinated evolution in host metabolic pathways.
Last, comparative germ-free models (i.e. closely related species that are all able to be reared in sterile conditions) may prove powerful for explicitly testing aspects of host evolution. Across distantly related hosts (mice and flies), inoculation of germ-free hosts has demonstrated some conserved aspects of host gene expression, such as increases in genes associated with epithelial proliferation and innate immunity [51]. Additionally, germ-free flies and germ-free mice both express higher levels of amino acid transporters compared to their conventional counterparts, suggesting that germ-free animals might need to compensate for the inability to use consumed dietary proteins [51]. Such studies could be conducted in more closely related species, as has been done with various species of germ-free Drosophila flies [80]. Interestingly, when hosts are colonized with heterologous microbial communities, they often suffer decreases in performance or survival, suggestive of some incompatibilities between microbiota and host [80–82]. Suboptimal ratios of nutrients (also known as nutritional imbalance) can also have detrimental effects on host physiology [83,84], and so it would be interesting to test whether a mismatch between microbial products and host physiology underlie the importance of hosts maintaining a specific microbial community.
5. Conclusion
Although currently speculative, we have provided evidence to support the idea that host animals may evolve in response to their associated microbiota, in accordance with the hypothesis of symmorphosis. Specifically, owing to their reliance on microbial products for important physiological functions, hosts are likely to regulate the expression of genes needed to interact with these substrates, and these expression patterns may become constitutively fixed over evolutionary time. Additionally, hosts may experience losses of function in pathways that have been outsourced to microbes completely. Furthermore, these patterns may vary across animal groups that differ in their degree of reliance on microbial metabolites. Experimental methods, like those discussed above, will need to be employed to test these ideas and elucidate how the physiological systems of hosts and microbes have integrated with one another to potentially optimize performance.
Acknowledgements
We thank Dr William Karasov for reviewing the manuscript and providing feedback.
Data accessibility
This article has no additional data.
Authors' contributions
S.S.F. and K.D.K. contributed equally to all aspects of this manuscript.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by the National Science Foundation (grant no. IOS-1942587 to K.D.K. and a Graduate Research Fellowship to S.S.F.).
References
- 1.Bäckhed F, Ley RE, Sonnenburg JS, Peterson DA, Gordon JI. 2005. Host-bacterial mutualism in the human intestine. Science 307, 1915–1920. ( 10.1126/science.1104816) [DOI] [PubMed] [Google Scholar]
- 2.O'Hara AM, Shanahan F. 2006. The gut flora as a forgotten organ. EMBO Rep. 7, 688–693. ( 10.1038/sj.embor.7400731) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morar N, Bohannan BJM. 2019. The conceptual ecology of the human microbiome. Q. Rev. Biol. 94, 149–175. ( 10.1086/703582) [DOI] [Google Scholar]
- 4.Kohl KD. 2018. A microbial perspective on the grand challenges in comparative animal physiology. mSystems 3, e00146-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mykles DL, Ghalambor CK, Stillman JH, Tomanek L. 2010. Grand challenges in comparative physiology: integration across disciplines and across levels of biological organization. Integr. Comp. Biol. 50, 6–16. ( 10.1093/icb/icq015) [DOI] [PubMed] [Google Scholar]
- 6.Qin J, et al. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Comtet-Marre S, et al. 2017. Metatranscriptomics reveals the active bacterial and eukaryotic fibrolytic communities in the rumen of dairy cow fed a mixed diet. Front. Microbiol. 8, 67 ( 10.3389/fmicb.2017.00067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weibel ER, Taylor CR, Hoppeler H. 1991. The concept of symmorphosis: a testable hypothesis of structure-function relationship. Proc. Natl Acad. Sci. USA 88, 10 357–10 361. ( 10.1073/pnas.88.22.10357) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor CR, Weibel ER. 1981. Design of the mammalian respiratory system. Respir Physiol. 44, 1–164. ( 10.1016/0034-5687(81)90073-6) [DOI] [PubMed] [Google Scholar]
- 10.Karasov WH. 1992. Test of the adaptive modulation hypothesis for dietary control of intestinal transport. Am. J. Physiol. 267, R496–R502. [DOI] [PubMed] [Google Scholar]
- 11.Hume ID. 1998. Optimization in design of the digestive system. In Principles of animal design: the optimization and symmorphosis debate (eds Weibel ER, Taylor CR, Bolis L), pp. 212–219. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 12.Kohl KD, Brzęk P, Caviedes-Vidal E, Karasov WH. 2011. Pancreatic and intestinal carbohydrases are matched to dietary starch levels in wild passerine birds. Physiol. Biochem. Zool. 84, 195–203. ( 10.1086/658146) [DOI] [PubMed] [Google Scholar]
- 13.Pette DG, Klingenberg G, Bucher T. 1962. Comparable and specific proportions in the mitochondrial enzyme activity pattern. Biochem. Biophys. Res. Commun. 7, 425–429. ( 10.1016/0006-291X(62)90328-5) [DOI] [PubMed] [Google Scholar]
- 14.Dudley R, Gans C. 1991. A critique of symmorphosis and optimality models in physiology. Physiol. Zool. 64, 627–637. ( 10.1086/physzool.64.3.30158197) [DOI] [Google Scholar]
- 15.Garland T. 1991. Conceptual and methodological issues in testing the predictions of symmorphosis. In Principles of animal design: the optimization and symmorphosis debate (eds Weibel ER, Taylor CR, Bolis L), pp. 40–47. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 16.Gordon MS. 1991. Evolution of optimal systems: overview. In Principles of animal design: the optimization and symmorphosis debate (eds Weibel ER, Taylor CR, Bolis L), pp. 37–39. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 17.Weibel ER. 1998. Symmorphosis and optimization of biological design: introduction and questions. In Principles of animal design: the optimization and symmorphosis debate (eds Weibel ER, Taylor CR, Bolis L), pp. 1–10. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 18.Kohl KD, Carey HV. 2016. A place for host–microbe symbiosis in the comparative physiologist's toolbox. J. Exp. Biol. 219, 3496–3504. ( 10.1242/jeb.136325) [DOI] [PubMed] [Google Scholar]
- 19.Zilber-Rosenberg I, Rosenberg E. 2008. Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol. Rev. 32, 723–735. ( 10.1111/j.1574-6976.2008.00123.x) [DOI] [PubMed] [Google Scholar]
- 20.Bordenstein SR, Theis KR. 2015. Host biology in light of the microbiome: ten principles of holobionts and hologenomes. PLoS Biol. 13, e1002226 ( 10.1371/journal.pbio.1002226) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenberg E, Zilber-Rosenberg I. 2018. The hologenome concept of evolution after 10 years. Microbiome 6, 78 ( 10.1186/s40168-018-0457-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Theis KR, et al. 2016. Getting the hologenome concept right: an eco-evolutionary framework for hosts and their microbiomes. mSystems 1, e00028-16 ( 10.1128/mSystems.00028-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Douglas AE, Werren JH. 2016. Holes in the hologenome: why host-microbe symbioses are not holobionts. MBio 7, e02099–15 ( 10.1128/mBio.02099-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koskella B, Hall LJ, Metcalf CJE. 2017. The microbiome beyond the horizon of ecological and evolutionary theory. Nat. Ecol. Evol. 1, 1606 ( 10.1038/s41559-017-0340-2) [DOI] [PubMed] [Google Scholar]
- 25.Hehemann J-H, Correc G, Barbeyron T, Helbert W, Czjzek M, Michel G. 2010. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature 464, 908–912. ( 10.1038/nature08937) [DOI] [PubMed] [Google Scholar]
- 26.Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, Reddy DN. 2015. Role of the normal gut microbiota. World J. Gastroenterology: WJG. 21, 8787 ( 10.3748/wjg.v21.i29.8787) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelly D, Campbell JI, King TP, Grant G, Jansson EA, Coutts AGP, Pettersson S, Conway S. 2004. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-γ and RelA. Nat. Immunol. 5, 104–112. ( 10.1038/ni1018) [DOI] [PubMed] [Google Scholar]
- 28.Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. 2003. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat. Immunol. 4, 269–273. ( 10.1038/ni888) [DOI] [PubMed] [Google Scholar]
- 29.Moya A, Ferrer M. 2016. Functional redundancy-induced stability of gut microbiota subjected to disturbance. Trends Microbiol. 24, 402–413. ( 10.1016/j.tim.2016.02.002) [DOI] [PubMed] [Google Scholar]
- 30.Lahti DC, Johnson NA, Ajie BC, Otto SP, Hendry AP, Blumstein DT, Coss RG, Donohue K, Foster SA. 2009. Relaxed selection in the wild. Trends Ecol. Evol. 24, 487–496. ( 10.1016/j.tree.2009.03.010) [DOI] [PubMed] [Google Scholar]
- 31.Pascoe EL, Hauffe HC, Marchesi JR, Perkins SE. 2017. Network analysis of gut microbiota literature: an overview of the research landscape in non-human animal studies. ISME J. 11, 2644–2651. ( 10.1038/ismej.2017.133) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sabat P, Lagos JA, Bozinovic F. 1999. Test of the adaptive modulation hypothesis in rodents: dietary flexibility and enzyme plasticity. Comp. Biochem. Physiol. A: Mol. Integr. Physiol. 123, 83–87. ( 10.1016/S1095-6433(99)00042-2) [DOI] [PubMed] [Google Scholar]
- 33.Brzęk P, Kohl KD, Caviedes-Vidal E, Karasov WH. 2009. Developmental adjustments of house sparrow (Passer domesticus) nestlings to diet composition. J. Exp. Biol. 212, 1284–1293. ( 10.1242/jeb.023911) [DOI] [PubMed] [Google Scholar]
- 34.Wang L, Caviedes-Vidal E, Karasov WH. 2019. Diet composition modulates intestinal hydrolytic enzymes in white-footed mice (Peromyscus leucopus). J Mammal. 100, 1512–1521. ( 10.1093/jmammal/gyz110) [DOI] [Google Scholar]
- 35.Diamond J. 1991. Evolutionary design of intestinal nutrient absorption: enough but not too much. Physiology 6, 92–96. ( 10.1152/physiologyonline.1991.6.2.92) [DOI] [Google Scholar]
- 36.Buddington RK, Chen JW, Diamond JM. 1991. Dietary regulation of intestinal brush-border sugar and amino acid transport in carnivores. Am. J. Physiol.-Regulatory, Integr. Comp. Physiol. 261, R793–R801. ( 10.1152/ajpregu.1991.261.4.R793) [DOI] [PubMed] [Google Scholar]
- 37.Zientz E, Dandekar T, Gross R. 2004. Metabolic interdependence of obligate intracellular bacteria and their insect hosts. Microbiol. Mol. Biol. Rev. 68, 745–770. ( 10.1128/MMBR.68.4.745-770.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramsey JS, Macdonald SJ, Jander G, Nakabachi A, Thomas GH, Douglas AE. 2010. Genomic evidence for complementary purine metabolism in the pea aphid, Acyrthosiphon pisum, and its symbiotic bacterium Buchnera aphidicola. Insect. Mol. Biol. 19, 241–248. ( 10.1111/j.1365-2583.2009.00945.x) [DOI] [PubMed] [Google Scholar]
- 39.Wilson ACC, et al. 2010. Genomic insight into the amino acid relations of the pea aphid, Acyrthosiphon pisum, with its symbiotic bacterium Buchnera aphidicola. Insect. Mol. Biol. 19, 249–258. ( 10.1111/j.1365-2583.2009.00942.x) [DOI] [PubMed] [Google Scholar]
- 40.Smith TE, Moran NA. 2020. Coordination of host and symbiont gene expression reveals a metabolic tug-of-war between aphids and Buchnera. Proc. Natl Acad. Sci. USA 117, 201916748 ( 10.1073/pnas.1916748117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benson AK, et al. 2010. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc. Natl Acad. Sci. USA 107, 18 933–18 938. ( 10.1073/pnas.1007028107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferrer M, et al. 2013. Microbiota from the distal guts of lean and obese adolescents exhibit partial functional redundancy besides clear differences in community structure. Environ. Microbiol. 15, 211–226. ( 10.1111/j.1462-2920.2012.02845.x) [DOI] [PubMed] [Google Scholar]
- 43.Muegge BD, Kuczynski J, Knights D, Clemente JC, González A, Fontana L, Henrissat B, Knight R, Gordon JI. 2011. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332, 970–974. ( 10.1126/science.1198719) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cuff MA, Lambert DW, Shirazi-Beechey SP. 2002. Substrate-induced regulation of the human colonic monocarboxylate transporter, MCT1. J. Physiol. 539, 361–371. ( 10.1113/jphysiol.2001.014241) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kirat D, Kondo K, Shimada R, Kato S. 2009. Dietary pectin up-regulates monocaboxylate transporter 1 in the rat gastrointestinal tract. Exp. Physiol. 94, 422–433. ( 10.1113/expphysiol.2009.046797) [DOI] [PubMed] [Google Scholar]
- 46.Bergman E. 1990. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 70, 567–590. ( 10.1152/physrev.1990.70.2.567) [DOI] [PubMed] [Google Scholar]
- 47.den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud D-J, Bakker BM. 2013. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 54, 2325 ( 10.1194/jlr.R036012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tremaroli V, Bäckhed F. 2012. Functional interactions between the gut microbiota and host metabolism. Nature 489, 242–249. ( 10.1038/nature11552) [DOI] [PubMed] [Google Scholar]
- 49.Claus SP, et al. 2011. Colonization-induced host-gut microbial metabolic interaction. MBio 2, e00271–10 ( 10.1128/mBio.00271-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neis E, Dejong C, Rensen S. 2015. The role of microbial amino acid metabolism in host metabolism. Nutrients 7, 2930–2946. ( 10.3390/nu7042930) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rawls JF, Samuel BS, Gordon JI. 2004. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc. Natl Acad. Sci. USA 101, 4596–4601. ( 10.1073/pnas.0400706101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singer MA. 2003. Do mammals, birds, reptiles, and fish have similar nitrogen conserving systems? Comp. Biochem. Physiol. B 134, 543–558. ( 10.1016/S1096-4959(03)00027-7) [DOI] [PubMed] [Google Scholar]
- 53.Wiebler JM, Kohl KD, Lee RE, Costanzo JP. 2018. Urea hydrolysis by gut bacteria in a hibernating frog: evidence for urea-nitrogen recycling in Amphibia. Proc. R. Soc. B 285, 20180241 ( 10.1098/rspb.2018.0241) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marini JC, Van Amburgh ME.. 2003. Nitrogen metabolism and recycling in Holstein heifers. J. Anim. Sci. 81, 545–552. ( 10.2527/2003.812545x) [DOI] [PubMed] [Google Scholar]
- 55.Marini JC, Klein JD, Sands JM, Van Amburgh ME.. 2004. Effect of nitrogen intake on nitrogen recycling and urea transporter abundance in lambs. J. Anim. Sci. 82, 1157–1164. ( 10.2527/2004.8241157x) [DOI] [PubMed] [Google Scholar]
- 56.Yang B, Sands JM. 2014. Urea transporters (ed. Harris R.). Berlin, Germany: Springer. [Google Scholar]
- 57.Pigliucci M, Murren CJ, Schlichting CD. 2006. Phenotypic plasticity and evolution by genetic assimilation. J. Exp. Biol. 209, 2362–2367. ( 10.1242/jeb.02070) [DOI] [PubMed] [Google Scholar]
- 58.Karasov HW, Douglas AE. 2013. Comparative digestive physiology. Compr. Physiol. 3, 741–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Enattah NS, et al. 2008. Independent introduction of two lactase-persistence alleles into human populations reflects different history of adaptation to milk culture. Am. J. Hum. Genet. 82, 57–72. ( 10.1016/j.ajhg.2007.09.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perry GH, et al. 2007. Diet and the evolution of human amylase gene copy number variation. Nat. Genet. 39, 1256–1260. ( 10.1038/ng2123) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stewart C-B, Schilling JW, Wilson AC. 1987. Adaptive evolution in the stomach lysozymes of foregut fermenters. Nature 330, 401–404. ( 10.1038/330401a0) [DOI] [PubMed] [Google Scholar]
- 62.Cui J, Yuan X, Wang L, Jones G, Zhang S.. 2011. Recent loss of vitamin C biosynthesis ability in bats. PLoS ONE 6, e27114 ( 10.1371/journal.pone.0027114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chatterjee IB. 1973. Evolution and the biosynthesis of ascorbic acid. Science 182, 1271–1272. ( 10.1126/science.182.4118.1271) [DOI] [PubMed] [Google Scholar]
- 64.Birney EC, Jenness R, Ayaz KM. 1976. Inability of bats to synthesize L-ascorbic acid. Nature 260, 626–628. ( 10.1038/260626a0) [DOI] [PubMed] [Google Scholar]
- 65.Morris JG. 2002. Idiosyncratic nutrient requirements of cats appear to be diet-induced evolutionary adaptations. Nutr. Res. Rev. 15, 153–168. ( 10.1079/NRR200238) [DOI] [PubMed] [Google Scholar]
- 66.Hoffman RR. 1989. Evolutionary steps of ecophysiological adaptation and diversification of ruminants: a comparative view of their digestive system. Oecologia 78, 443–457. ( 10.1007/BF00378733) [DOI] [PubMed] [Google Scholar]
- 67.Wood IS, Dyer J, Hofman RR, Shirazi-Beechey SP. 2000. Expression of the Na+/glucose co-transporter (SGLT1) in the intestine of domestic and wild ruminants. Pflügers Archiv. 441, 155–162. ( 10.1007/s004240000404) [DOI] [PubMed] [Google Scholar]
- 68.Robbins CT. 1993. Wildlife feeding and nutrition. San Diego, CA: Academic Press. [Google Scholar]
- 69.Srinivasan K, Buys EM. 2019. Insights into the role of bacteria in vitamin A biosynthesis: future research opportunities. Crit. Rev. Food Sci. Nutr. 59, 3211–3226. ( 10.1080/10408398.2018.1546670) [DOI] [PubMed] [Google Scholar]
- 70.Hoffman RR. 1998. How ruminants adapt and optimize their digestive system ‘blueprint’ in response to resource shifts. In Principles of animal design: the optimization and symmorphosis debate (eds Weibel WR, Taylor CR, Bolis L), pp. 220–229. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 71.Sanders JG, Łukasik P, Frederickson ME, Russell JA, Koga R, Knight R, Pierce NE. 2017. Dramatic differences in gut bacterial densities correlate with diet and habitat in rainforest ants. Integr. Comp. Biol. 57, 705–722. ( 10.1093/icb/icx088) [DOI] [PubMed] [Google Scholar]
- 72.Hammer TJ, Janzen DH, Hallwachs W, Jaffe SP, Fierer N. 2017. Caterpillars lack a resident gut microbiome. Proc. Natl Acad. Sci. USA 114, 9641–9646. ( 10.1073/pnas.1707186114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hammer TJ, Sanders JG, Fierer N. 2019. Not all animals need a microbiome. FEMS Microbiol. Lett. 366, fnz117 ( 10.1093/femsle/fnz117) [DOI] [PubMed] [Google Scholar]
- 74.Partha R, Chauhan BK, Ferreira Z, Robinson JD, Lathrop K, Nischal KK, Chikina M, Clark NL. 2017. Subterranean mammals show convergent regression in ocular genes and enhancers, along with adaptation to tunneling. Elife 6, e25884 ( 10.7554/eLife.25884) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chikina M, Robinson JD, Clark NL. 2016. Hundreds of genes experienced convergent shifts in selective pressure in marine mammals. Mol. Biol. Evol. 33, 2182–2192. ( 10.1093/molbev/msw112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mendoza MLZ, et al. 2018. Hologenomic adaptations underlying the evolution of sanguivory in the common vampire bat. Nat. Ecol. Evol. 2, 659 ( 10.1038/s41559-018-0476-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hoang KL, Morran LT, Gerardo NM. 2016. Experimental evolution as an underutilized tool for studying beneficial animal–microbe interactions. Front. Microbiol. 7, 1444 ( 10.3389/fmicb.2016.01444) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kohl KD, Sadowska ET, Rudolf AM, Dearing MD, Koteja P. 2016. Experimental evolution on a wild mammal species results in modifications of gut microbial communities. Front. Microbiol. 7, 634 ( 10.3389/fmicb.2016.00634) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rudman SM, et al. 2019. Microbiome composition shapes rapid genomic adaptation of Drosophila melanogaster. bioRxiv, 632257.
- 80.Adair KL, Bost A, Bueno E, Kaunisto S, Kortet R, Peters-Schulze G, Martinson VG, Douglas AE. 2020. Host determinants of among-species variation in microbiome composition in drosophilid flies. ISME J. 14, 217–229. ( 10.1038/s41396-019-0532-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brooks AW, Kohl KD, Brucker RM, van Opstal EJ, Bordenstein SR.. 2016. Phylosymbiosis: relationships and functional effects of microbial communities across host evolutionary history. PLoS Biol. 14, e2000225 ( 10.1371/journal.pbio.2000225) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.van Opstal EJ, Bordenstein SR.. 2019. Phylosymbiosis impacts adaptive traits in Nasonia wasps. mBio 10, e00887-19 ( 10.1128/mBio.00887-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Raubenheimer D, Jones SA. 2006. Nutritional imbalance in an extreme generalist omnivore: tolerance and recovery through complementary food selection. Anim. Behav. 71, 1253–1262. ( 10.1016/j.anbehav.2005.07.024) [DOI] [Google Scholar]
- 84.Jensen K, Mayntz D, Toft S, Raubenheimer D, Simpson SJ. 2011. Prey nutrient composition has different effects on Pardosa wolf spiders with dissimilar life histories. Oecologia 165, 577 ( 10.1007/s00442-010-1811-1) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.