Abstract
Paper‐based sensors, microfluidic platforms and electronics have attracted attention in the past couple of decades because they are flexible, can be recycled easily, environmentally friendly and inexpensive. Here, we report a paper‐based potentiometric sensor to detect the whole Zika virus with a minimum sensitivity of 0.26 nV/Zika and a minimum detectable signal (MDS) of 2.4x107 Zika. Our paper sensor works very similar to a P‐N junction where a junction is formed between two different regions with different electrochemical potentials on the paper. These two regions with slightly different ionic contents, ionic species and concentrations, produce a potential difference given by the Nernst equation. Our paper sensor consists of 2‐3 × 10 mm segments of paper with conducting silver paint contact patches on two ends. The paper is dipped in a buffer solution containing aptamers designed to bind to the capsid proteins on Zika. We then added the Zika (in its own buffer) to the region close to one of the silver paint contacts. The Zika virus (40 nm diameter with 43 kDa or 7.1 × 10−20 gm weight) became immobilized in the paper's pores and bonded with the resident aptamers creating a concentration gradient. Atomic force microscopy and Raman spectroscopy were carried out to verify that both the aptamer and Zika become immobilized in the paper. The potential measured between the two silver paint contacts reproducibly became more negative upon adding the Zika. We also showed that a liquid crystalline display (LCD) powered by the sensor can be used to read the sensor output.
1. INTRODUCTION
Sensors built with paper as a substrate have short response time, are low‐cost (Määttänen, 2013) and are flexible (Siegel et al., 2010; Tobjörk & Österbacka, 2011). Moreover, they are biodegradable and suitable for mass deployment in resource‐limited areas and can be easily used by unskilled operators. Paper is also a great medium for immobilization and trapping and, in some cases, for binding with biomolecules. Its porous structure with large connected pores composed of cellulose fibres allows transporting liquid by means of capillary forces that result in short response time. The porous structure of paper also allows any nano‐ and microparticles to remain immobilized in the paper structure. Paper can be functionalized with certain materials such as nitrocellulose paper used for immobilizing nucleic acids for selective sensing (Novell. Guinovart, Blondeau, Rius, & Andrade, 2014). Paper‐based potentiometric sensors are reported for detecting many ions and proteins (Cui, Lisak, Strzalkowska, & Bobacka, 2014; HuJun, Stein, & Bühlmann, 2016; Novell et al., 2014; Szucs & Gyurcsányi, 2012). Potentiometric paper‐based sensors (Määttänen, 2013), (Ding, He, Lisak, Qin, & Bobacka, 2017; Jin et al., 2018; Kawahara, Sahatiya, Badhulika, & Uno, 2018; Kofler, Nau, & List‐Kratochvil, 2015; Lisak, Cui, & Bobacka, 2015; Nery & Kubota, Apr., 2016; Parrilla, Cánovas, & Andrade, 2017; Ping et al., 2013; Sjöberg et al., 2016) utilize gradients of ion distributions that generate open‐circuit voltages (Voc) that are measured to transduce analytes. Paper‐based pathogen and virus sensors are also easy to incinerate. They can be used as one‐time‐use front end in sensor systems that can be peeled off and replaced.
There are many Zika sensors and detection methods reported in the literature. These include serum analysis using the antibody detection assays (Al‐Qahtani, Nazir, Al‐Anazi, Rubino, & Al‐Ahdal, 2016; Haug, Kieny, & Murgue, 2016) and detection of viral RNA using molecular‐based techniques like conventional polymerase chain reaction (PCR). Real‐time reverse transcription–polymerase chain reaction (RT‐PCR) (Calvet et al., 2016; Faye et al., 2008; Gourinat, O’Connor, Calvez, Goarant, & Dupont‐Rouzeyrol, 2015; Klungthong, 2007; Murray et al., 2017; Musso et al., 2015; Shin, Cherstvy, & Metzler, 2014) for Zika has also been reported. These techniques involve using viral antibody or DNA/RNA extraction, followed by labelled detection using fluorescent probes. They provide high sensitivity and specificity but require specialized equipment and expensive procedures, and are time‐consuming. The paper‐based sensors are fast, inexpensive and can be used in regions with limited resources. We show that a simple liquid crystalline display (LCD) can be used to measure the sensor output with the power generated from the sensor itself. The paper sensor reported here can be modified by replacing its aptamer with other virus aptamers, such as COVID‐19.
Our work shows for the first time that a potentiometric paper sensor can reliably detect a whole virus using standard printer papers functionalized with antigens or aptamers. Aptamers are single‐strand DNA structures that are artificially synthesized using a combinatorial selection process called systematic evolution of ligands by exponential enrichment (SELEX) (Tuerk & Gold, 1990) to detect biomolecules and pathogens with high specificity (Lee & Zeng, 2017; Jenison, Gill, Pardi, & Polisky, 1994). In our sensors, when Zika was added to one side of the paper, it resulted in a concentration gradient with the associated electrochemical potential difference. Electrical contacts to the paper sensor can be made with graphene (Kawahara et al., 2018; Ping et al., 2013) conducting glues and epoxies and silver paint. We also developed a proof‐of‐concept device (Figure 2a) that can be printed with silver paint contacts and show that the paper‐based Zika sensor can be connected to an LCD for electronic readout.
Figure 2.
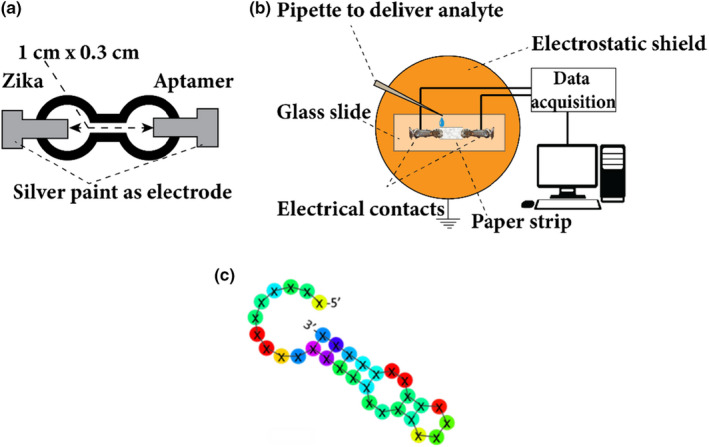
Schematic of the (a) proposed printed paper device (distance between the electrode is 1.5 cm and width of the narrow region between the electrode is 0.3 cm). (b) Schematic of the experimental set‐up. (c) The structure of the aptamer provided by Base Pair Biotechnologies Inc. [Colour figure can be viewed at wileyonlinelibrary.com]
2. THEORETICAL BACKGROUND
The charge distribution and ionic transport of differently charged species in the paper device and the resulting open‐circuit voltage can be explained using the phase‐boundary model formed by two different ionic species (aptamer in the background and Zika added on the anode side). The total electrochemical potential difference (Voc) (Figure 1a) is the sum of the phase‐boundary potentials (Bakker, Bühlmann, & Pretsch, 2004) between the aptamer and electrode (Voc1), the liquid junction potential (Hickman, 1970), (Harper, 1985) between the Zika/aptamer and aptamer (Voc2) and between the Zika/aptamer and electrode (Voc3): Voc=Voc1+ Voc2 +Voc3. Figure 1.a shows the schematic of the phase boundary between electrodes and the paper and liquid junction potential at the centre. The phase‐boundary potential between the aptamer–electrode (Voc1) and Zika/aptamer–electrode (Voc3) is obtained from the Nernst equation:
| (1) |
| (2) |
where is the standard potential, R is the gas constant, T is temperature, F is the Faraday constant (Coul./mol), z1 =1 is the valence of the aptamer, and z3=1 is the valence of the Zika. Zika is a complex extended pathogen with surface proteins, DNA, RNA, etc., and has many oxidation/reduction potentials corresponding to more than one charge transfer that we have indicated in Equation 1. However, under the small voltages and chemically ‘mild’ conditions of our paper sensors, it is reasonable to assume the single valency indicated in Equation 1.
Figure 1.
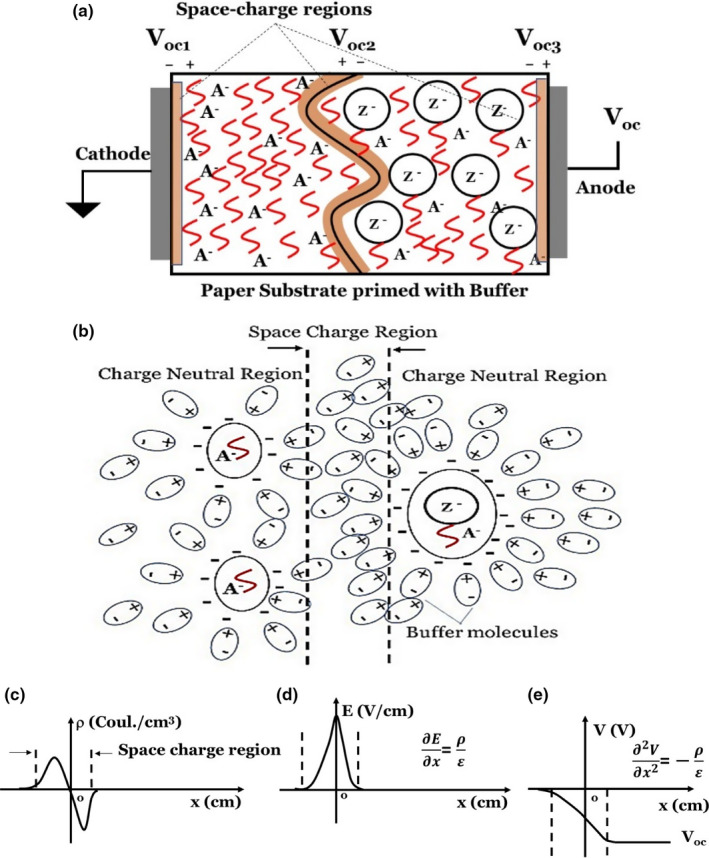
(a) and (b), Schematic of the phase‐boundary potential model with the electrodes and liquid wherein the potential is given by the uneven distribution of the sample ions/particles. Here, A‐ refers to the aptamer and Z‐ refers to that of the Zika virus. (c), (d) and (e), Space charge, resulting in the electric field and potential at the boundary of Zika in the centre of the paper device. [Colour figure can be viewed at wileyonlinelibrary.com]
The liquid junction potential between Zika–aptamer and aptamer in the middle of the device (Figure 1) can be written as (Base Pair Biotechnologies ‐ Aptamer Discovery Company,; Harper, 1985):
| (3) |
where is the valence of the particular sample species (in our case, z's for aptamers and the Zika are equal to 1), denotes the activity of the ions/sample species, and is the transference number and signifies the fractional conductance of the i th ion/sample species. The transference number is defined by , where u is the mobility of the i th ion, and Mj is the molar concentration, with j ranging over all the ions. We note that there are 3 space‐charge regions that form (Figure 1a) between the anode/cathode and the paper and at the boundary of the Zika/aptamer–aptamer regions in the middle.
In our experiments, the paper was uniformly primed with the buffer solution and aptamers. We assume that the anode paper and cathode paper Nernst potentials (Voc1 and Voc3 in Figure 1a) are approximately equal and are determined by the electrochemical potential differences between these electrodes and the primed paper. Thus, Voc1=Voc3 and the total Voc between the cathode and the anode is approximately equal to Voc2 (Figure 1a). The space‐charge region at the Zika boundary (Figure 1a, b) is composed of charges that are induced by the Zika–aptamer–buffer on one side of this junction and aptamer–buffer on the other side (Figure 1b). The boundary is controlled by the amount of Zika added and the flow of the buffer that accompanies the Zika. We note that aptamers and Zika viruses become immobilized in the pores of the paper. According to our experiments, the aptamer and Zika had negative residual charges. Each of these analytes is charge‐neutral in their respective medium, but when they are added together, because of their different polarizability, they exhibit different residual charges. In the case of Zika +aptamer, there is an additional charge re‐distribution because of the hydrogen binding. The binding process does not happen instantly and proceeds as a function of time. In our experiments, the Zika–aptamer complex provided higher Voc change than Zika alone. The selectivity of the sensor to detect Zika is primarily due to its preferential binding with the aptamer. When we added the Zika to the anode region, Voc became negative, suggesting that the space‐charge region has a charge, electric field and potential distributions schematically shown in Figure 1c‐e.
3. MATERIALS AND METHODS
The devices used in our work (Figure 2) consisted of a sample holder with two clips and electrodes mounted on a glass slide. The devices were connected to a data acquisition system (NI‐USB 6341 DAQ) and a computer with a custom LabVIEW program (Figure 2b). We measured the open‐circuit voltage (Voc) of the paper device as a function of time as buffer, aptamer and Zika were added to the sensor. The glass slide was located inside a grounded copper enclosure to shield and improve the signal‐to‐noise ratio. Paper strips (0.2‐0.3 cm x 1 cm) used in our devices were manually cut from a standard printer paper and were contacted with silver paint (from Ted Pella). These paper devices were then coated (impregnated) with appropriate analytes such as de‐ionized (DI) water/buffer/aptamer, and the buffer/Zika solutions were added from the anode side.
We used a 1x concentration of phosphate‐buffered solution (PBS) with 1 mM magnesium chloride as a buffer solution (pH ~7.3). The 1 mM magnesium chloride was used for keeping the aptamer stable (manufacturer recommended). The Zika aptamers were reconstituted as per Base Pair Biotechnologies Inc by mixing the dried aptamers in resuspension buffer also provided by the manufacturer. The aptamer solution was then diluted to a working concentration of 100 µM using an aptamer folding buffer followed by heating to 90°C −95°C for 5 minutes. The resulting aptamer solution was then diluted to 1 µM solution using the buffer. 1 µM aptamer had ~1.2 × 1012 aptamers in 2 µL volume (calculated from the Avogadro number present in 1 M concentration in 1 L). The aptamer used in our experiments had a thiol end group and consisted of 32 nucleotide chain that folded to bind with the Zika SF9 envelope protein (Zika Envelope Recombinant Antigen). Figure 2c shows the schematic of the structure of the aptamer. The inactivated Zika virus (0810521CFHI) was obtained from the ZeptoMetrix Corporation. 40 µl of TCID_50 Zika stock solution was diluted with 160 µl buffer. The number of Zika viruses in the diluted 2 µl volume was ~7 × 107, estimated in the same manner as in Ref. (Dolai & Tabib‐Azar, 2020).
4. RESULTS AND DISCUSSIONS
The open‐circuit voltage (Voc) of the sensor containing DI water with the buffer solution introduced on the anode is shown in Figure 3a. The Voc, in this case, was measured immediately after the buffer was added to the anode region. Voc diminishes as a function of time (Figure 3b) as the buffer diffuses in the paper, and its concentration gradient diminishes.
Figure 3.
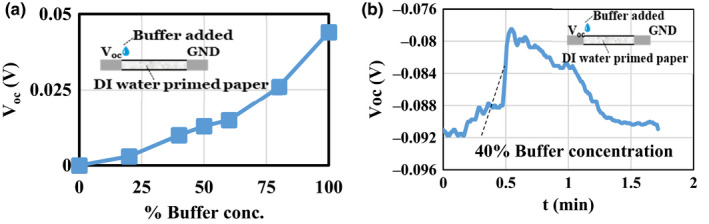
(a) Voc vs percentage volume concentration of the buffer with one side of the paper dipped in DI water and the other side with the diluted buffer solution. (b) Voc vs t of the response of the paper sensor primed with DI water with buffer (40% by volume concentration) introduced at t = 0.5 min. [Colour figure can be viewed at wileyonlinelibrary.com]
The Zika virus has a small residual negative charge (Figure 4a) that reduces the Voc as soon as it is added to the sensor. Figure 4 shows the response of the sensor when Zika was added. The initial rise in Figure 4a is due to the pipette tip approaching the sensor to deliver the Zika. Figure 4b shows another run with the Zika added in the beginning and later at 3.1 min. In both cases, the sensor output reached a steady‐state value. This indicates that the concentration gradient introduced by the addition of Zika did not change as a function of time, and it appears that the Zika became immobilized in the paper. In the second case (Figure 4b), the paper was not primed with the aptamer, and the change in Voc was due to the residual charge of the Zika and buffer ions that accompany the Zika as part of its solution. It is also possible that the printer paper we are using has its own impurities, which in the presence of Zika +buffer became ionized. Zika is spherical with 40 nm diameter, and it is expected to get entangled in the paper pores. If Zika was mobile, the resulting Voc would diminish as a function of time as the Zika diffuses and reduces its concentration gradient. In separate experiments with buffer solutions alone (Figure 3b), the sensor response diminished after the initial introduction of the buffer at the anode, indicating that the buffer ions could diffuse in the paper, decreasing their initial concentration gradients.
Figure 4.
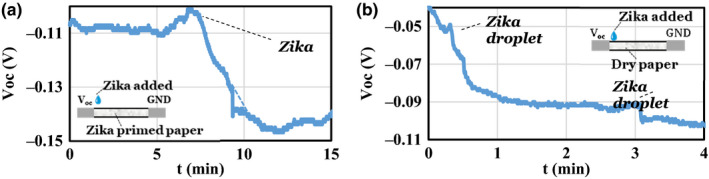
Zika introduced (a) at t = 7.45 min to paper device presoaked in Zika to demonstrate net negative charge of the virus and (b) at t = 0.5 min and t = 3.1 min to a dry paper device to demonstrate that as the Zika is applied on the positive side, it stays immobilized in the applied region as the buffer spreads maintaining the voltage level. [Colour figure can be viewed at wileyonlinelibrary.com]
Figure 5 shows the Voc change ∆Voc(t)~ Voc(t = 0) – Voc as a function of Zika concentration using a paper device primed with aptamers and buffer. Once all the resident aptamers bind with Zika, the only changes in the device charge content and its gradient are introduced by the additional Zika alone that has negative residual charges. Successive additions of Zika (used to increase its concentration) introduced larger changes in Voc followed by smaller changes once all the resident aptamers were saturated (Figure 5b), and ∆Voc became smaller at the end. The average ‘point’ sensitivity in these devices was 0.26 nV/Zika calculated at ∆Voc of ~18 mV for 2 µl Zika (~7x107 number of Zika per 2 µl). The sensitivity was nearly the same for 4 μl Zika in device P1 but not in device P2 (Figure 5). The paper‐based sensor's output response had a rms noise voltage of 2 mV, and the minimum detectable voltage was 2 mV that resulted in the MDS of 2.4 × 107 Zika.
Figure 5.
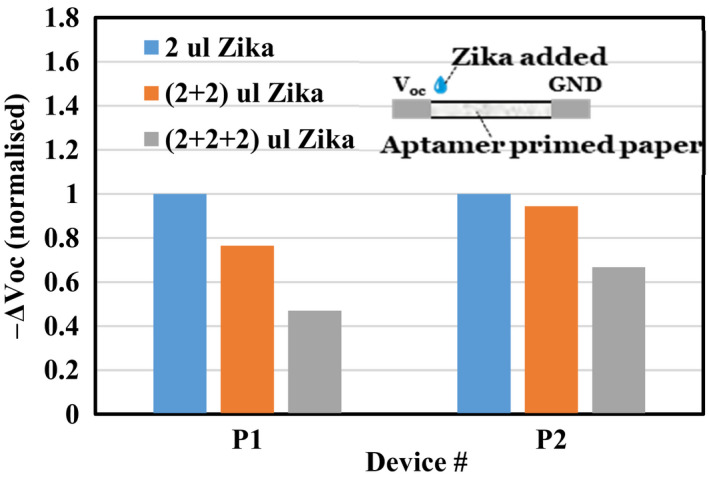
2 µl of diluted Zika added to aptamer‐soaked paper at each instance (total 3 times) repeated on 2 separate paper devices (P1 and P2). The ΔVoc is normalized against the ΔVoc(max) obtained for 2 µl Zika of each device. [Colour figure can be viewed at wileyonlinelibrary.com]
To verify the presence of aptamers and Zika in the paper‐based devices, we carried out two additional experiments. The first series of experiments used atomic force microscopy (AFM) to image Zika‐coated regions of the paper‐based device, as shown in Figure 6a. Although one can see large Zika complexes, individual Zika viruses could not be resolved. The paper has large surface roughness in excess of 2 µm rms that prevented performing high spatial resolution scans. We then performed AFM scans on aptamers and Zika deposited on gold‐coated glass samples (Dolai & Tabib‐Azar, 2020). As can be seen in Figure 6b, one could resolve individual Zika viruses on gold.
Figure 6.
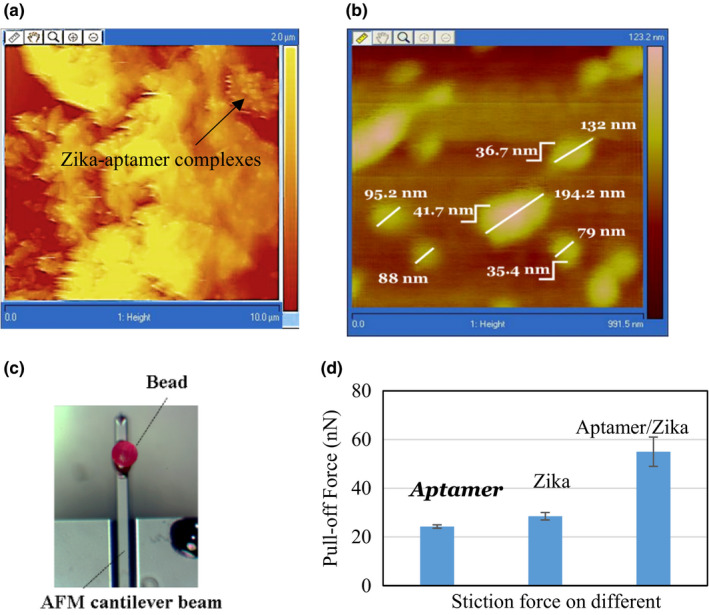
(a) AFM image of the Zika on aptamer‐coated paper. The scale is 10 µm. (b) AFM image of Zika on gold substrate showing the Zika step height of ~40 nm to demonstrate Zika on the aptamer‐coated gold surface. (c) Photograph of the AFM tip with a single microbead (diameter ~150 µm). (d) AFM stiction force measured using the AFM tip (c) with a bead coated with aptamers. [Colour figure can be viewed at wileyonlinelibrary.com]
Next, AFM studies using AFM probes with an aptamer‐functionalized bead (Figure 6c) were carried out. The stiction force (Thoreson, Martin, & Burnham, 2007) was obtained from force–displacement curve using aptamer‐functionalized AFM probe (Figure 6c). The AFM probe has been prepared by gluing a 150‐µm‐diameter polyester microbead to the AFM tip and subsequently coating the microbead with the aptamer (1 µM) for 30 min. It showed larger stiction forces with papers coated with aptamers than Zika papers with aptamers or Zika alone, as shown in Figure 6d. The stiction force of Zika/aptamer/paper was higher due to aptamer‐specific binding with Zika on the aptamer/paper.
The second set of experiments used Raman spectroscopies on paper‐based devices coated with different analytes. The Raman spectra shown in Figure 7 are from different regions of the paper coated with aptamers, Zika and Zika/aptamers. A DeltaNu spectrometer with Examiner 785 Raman unit was used in these experiments. 10 µl, 1 µM aptamer solution was used to prime the paper, and the Zika concentration was 1.7 × 108/µl, with the amount used in each application being 1 µl. The buffer solution was 1 µl of 1× PBS with 1 mM MgCl2. The Raman peak due to Zika was around 1590 cm−1 (Sánchez‐Purrà et al., Oct., 2017).
Figure 7.
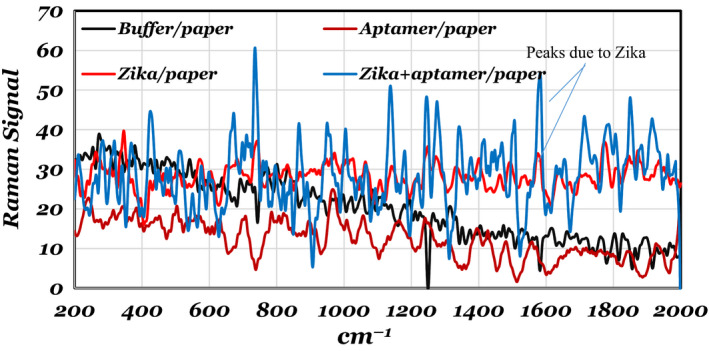
Raman spectra of buffer/paper, aptamer/paper, Zika/paper and Zika/aptamer/paper. The spectra are normalized with dry paper Raman spectrum. These spectra clearly show the difference between aptamers, Zika and buffer deposited on the paper. [Colour figure can be viewed at wileyonlinelibrary.com]
We then used printed patterns on paper, followed by adding silver paint contacts, as shown in Figure 2a and Figure 8 insets. The purpose of the printed device is to clearly show the location of the Zika solution (or fluids that may contain Zika) on the device. Figure 8a shows the Voc–time response for paper‐based devices (D1‐D3). The ∆Voc in these devices is shown in Figure 8b. We verified that the aptamer stayed and was resident in the paper even after rinsing the paper with DI water. Figure 8c shows the response of adding Zika on three different aptamer‐primed sensors (D4‐D6). The Voc levels in Figure 8c are similar and consistent with that of the aptamer‐primed device in Figure 8b.
Figure 8.
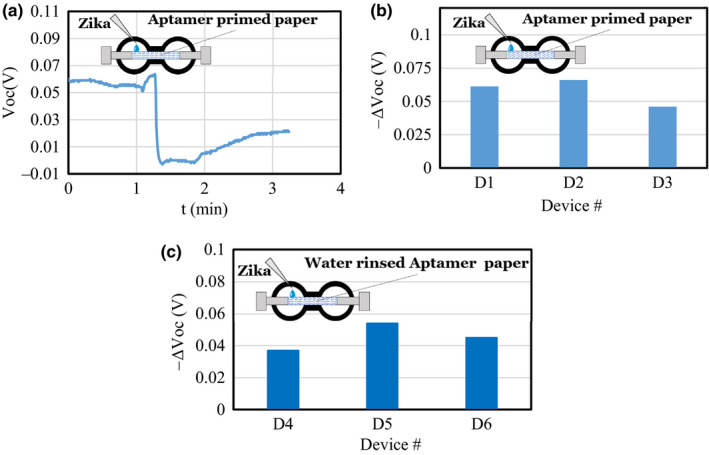
(a) Voc vs time of the printed sensor with Zika added at t = 1.3 min. ΔVoc obtained on adding Zika to (b) aptamer‐coated printed device D1, D2 and D3, and (c) aptamer‐primed and subsequently water‐rinsed devices D4, D5 and D6 showing that the aptamer remained immobilized in the paper after rinsing. [Colour figure can be viewed at wileyonlinelibrary.com]
The actual interactions between different components in the paper matrix are quite complex, and the sensor response depends on the history of the paper, temperature, air current on the sensor and other environmental factors (Kawahara et al., 2018). The results shown here had a standard deviation of ~2 mV from run to run. Uniformity of the electrical contacts can be a concern that can be addressed using 3‐D printing. Another very important source of error was in making sure that the paper sensor was not saturated with the analytes introduced at each step. When the paper was saturated, the addition of Zika resulted in a thin layer of liquid above the paper sensor, and Zika did not incorporate in the paper resulting in unreliable sensor output. The sensor was also very sensitive to spurious environmental electric fields generated by us and other objects nearby that could be addressed by proper grounding.
5. ELECTRONIC READOUT
We next explored the possibility of detecting the virus from the paper‐based sensor in the form of direct electronic readout through an LCD instead of measuring the open‐circuit voltage using a digital voltmeter. The LCD was configured to display a letter such as ‘A’ once the paper sensor voltage reached a threshold value generated by the presence of sufficient number of Zika viruses. In order to demonstrate the feasibility of such application, we used an LCD screen from a digital watch and experimentally found the turn‐on voltage for a single display segment to be ~100 mV. From the previous results, we observed that the addition of Zika resulted in ΔVoc ~50 mV. We used 6 paper sensors connected in series to generate ~300 mV (Figure 9a) that was sufficient to power many readout devices, including LCDs. The external LCD load did not draw more than 1 μA current to cause internal sensor resistor to become important. As can be seen in Figure 9b, the LCD displayed ‘A’ upon adding Zika. The LCD can be replaced by an electronic ink display that requires much lower voltages/power eliminating the necessity of using many devices in series.
Figure 9.
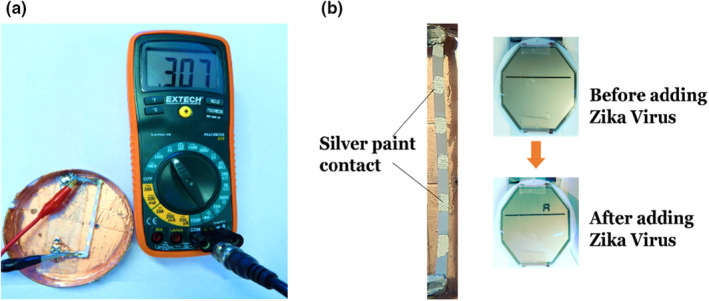
(a) Output from the paper sensor measured using multimeter showing a voltage level of ~300 mV for 6 sensors connected in series. (b) Output from the sensor connected to the LCD readout taken from a watch showing the letter ‘A’ when Zika was added on the sensors. [Colour figure can be viewed at wileyonlinelibrary.com]
The paper‐based sensors we constructed and reported here were very reproducible as long as they were not saturated (with buffer or water) and as long as they were grounded properly. Their ‘point’ sensitivity of 0.26 nV/Zika and their minimum detectable signal of 2.4×107 Zika are quite good given their simple structures and ease of operation. Selectivity of this sensor as well as other aptamer‐based sensors is limited by the selectivity of their respective aptamers that can be modified to achieve better selectivity if needed. In the case of paper‐based sensors discussed here, the selectivity may also be affected by viruses becoming immobilized in the paper even if they do not bind with the resident aptamer. These other viruses can be removed by mildly washing them away that will not affect the Zika–aptamer complexes appreciably. The real‐world application of Zika paper sensors will involve the presence of bodily fluids such as urine or sweat or saliva. These biofluids are very complex, and proper filtering on the sensor will be used to remove their components that may affect the paper sensor operation adversely.
As with any sensor development, the research and development efforts have many different stages. It usually starts with developing a transduction mechanism using modified ‘clean’ samples and progresses towards more realistic samples in the real world. The present work describes the transduction phenomena of ‘sensing pure Zika’ virus using a paper‐based potentiometric device that, to the best of our knowledge, was not done before. The innovation has two parts: a) using paper‐based sensors to detect the whole virus and b) using resident aptamers in the paper to bind with Zika. The sensor's selectivity comes from its resident aptamers. We will measure and report our sensors’ selectivity and verify their operations in the presence of bodily fluid in the near future.
The paper can also be used as an active sensing medium located over electrodes connected to the measurement apparatus. In this approach, the paper can be peeled off and replaced with a fresh paper. Paper itself can be used as a filter preventing larger particles from reaching the sensitive layers in intimate contact with the electrodes.
6. CONCLUSION
This work demonstrates, for the first time, a paper‐based sensor for detecting Zika virus using potentiometry. The sensor had a minimum sensitivity of 0.26 nV/Zika and a minimum detectable signal of 2.4 × 107 Zika. These sensors can be generalized to detect any other viruses, pathogens and bacteria by simply using appropriate aptamers. The most important and unique aspect of the potentiometric paper‐based Zika virus is its structural simplicity and ease of use. This study demonstrates the feasibility of detecting whole Zika viruses and the use of simple electronic readout of paper‐based sensors. Although aptamers are known for their high specificity to capture target pathogens and viruses, more study is required with different viruses to verify and measure the cross‐sensitivity of aptamers and our sensors.
ACKNOWLEDGEMENTS
This research was supported by a National Science Foundation Grant under Dr. Leon Esterowitz.
Dolai S, Tabib‐Azar M. Whole virus detection using aptamers and paper‐based sensor potentiometry. Med Devices Sens. 2020;3:e10112. 10.1002/mds3.10112
References
- Al‐Qahtani, A. A. , Nazir, N. , Al‐Anazi, M. R. , Rubino, S. , & Al‐Ahdal, M. N. (2016). Zika virus: A new pandemic threat. The Journal of Infection in Developing Countries, 10(3), 201–207. 10.3855/jidc.8350 [DOI] [PubMed] [Google Scholar]
- Bakker, E. , Bühlmann, P. , & Pretsch, E. (2004). The phase‐boundary potential model. Talanta, 63(1), 3–20. 10.1016/j.talanta.2003.10.006 [DOI] [PubMed] [Google Scholar]
- Base Pair Biotechnologies ‐ Aptamer Discovery Company . Retrieved from https://www.basepairbio.com/ (accessed Sep. 04, 2019)
- Calvet, G. , Aguiar, R. S. , Melo, A. S. O. , Sampaio, S. A. , de Filippis, I. , Fabri, A. , … de Filippis, A. M. B. (2016). Detection and sequencing of Zika virus from amniotic fluid of fetuses with microcephaly in Brazil: a case study. The Lancet Infectious Diseases, 16(6), 653–660. 10.1016/S1473-3099(16)00095-5 [DOI] [PubMed] [Google Scholar]
- Cui, J. , Lisak, G. , Strzalkowska, S. , & Bobacka, J. (2014). Potentiometric sensing utilizing paper‐based microfluidic sampling. The Analyst, 139(9), 2133–2136. [DOI] [PubMed] [Google Scholar]
- Ding, J. , He, N. , Lisak, G. , Qin, W. , & Bobacka, J. (2017). Paper‐based microfluidic sampling and separation of analytes for potentiometric ion sensing. Sensors and Actuators B: Chemical, 243, 346–352. 10.1016/j.snb.2016.11.128 [DOI] [Google Scholar]
- Dolai, S. , & Tabib‐Azar, M. (2020). Microfabricated nano‐gap tunneling current zika virus sensors with single virus detection capabilities. IEEE Sensors Journal, 20(15), 8597–8603. 10.1109/JSEN.2020.2984172 [DOI] [Google Scholar]
- Dolai, S. , & Tabib‐Azar, M. (2020). 433 MHz Lithium Niobate microbalance Aptamer‐coated whole Zika virus sensor with 370 Hz/ng sensitivity. IEEE Sensors Journal, 20(8), 4269–4274. 10.1109/JSEN.2019.2961611 [DOI] [Google Scholar]
- Faye, O. , Faye, O. , Dupressoir, A. , Weidmann, M. , Ndiaye, M. , & Alpha Sall, A. (2008). One‐step RT‐PCR for detection of Zika virus. Journal of Clinical Virology, 43(1), 96–101. 10.1016/j.jcv.2008.05.005 [DOI] [PubMed] [Google Scholar]
- Gourinat, A. C. , O’Connor, O. , Calvez, E. , Goarant, C. , & Dupont‐Rouzeyrol, M. (2015). Detection of zika virus in urine. Emerging Infectious Diseases, 21(1), 84–86. 10.3201/eid2101.140894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, H. W. (1985). Calculation of liquid junction potentials. Journal of Physical Chemistry, 89(9), 1659–1664. 10.1021/j100255a022 [DOI] [Google Scholar]
- Haug, C. J. , Kieny, M. P. , & Murgue, B. (2016). The Zika challenge. New England Journal of Medicine, 374(19), 1801–1803. 10.1056/NEJMp1603734 [DOI] [PubMed] [Google Scholar]
- Hickman, H. J. (1970). The liquid junction potential ‐ the free diffusion junction. Chemical Engineering Science, 25(3), 381–398. 10.1016/0009-2509(70)80037-9 [DOI] [Google Scholar]
- Hu, J. , Stein, A. , & Bühlmann, P. (2016). A disposable planar paper‐based potentiometric ion‐sensing platform. Angewandte Chemie International Edition, 55(26), 7544–7547. 10.1002/anie.201603017 [DOI] [PubMed] [Google Scholar]
- Jenison, R. D. , Gill, S. C. , Pardi, A. , & Polisky, B. (1994). High‐resolution molecular discrimination by RNA. Science, 263(5152), 1425–1429. 10.1126/science.7510417 [DOI] [PubMed] [Google Scholar]
- Jin, J.‐H. , Kim, J. , Lee, S. , Choi, S. , Park, C. , & Min, N. (2018). A Fully Integrated paper‐microfluidic electrochemical device for simultaneous analysis of physiologic blood ions. Sensors, 18(2), 104– 10.3390/s18010104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara, R. , Sahatiya, P. , Badhulika, S. , & Uno, S. (2018). Paper‐based potentiometric pH sensor using carbon electrode drawn by pencil. Japanese Journal of Applied Physics, 57(4S), 4FM08. 10.7567/JJAP.57.04FM08 [DOI] [Google Scholar]
- Klungthong, C. et al (2007). Dengue virus detection using whole blood for reverse transcriptase PCR and virus isolation. Journal of Clinical Microbiology, 45(8), 2480–2485. 10.1128/JCM.00305-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler, J. , Nau, S. , & List‐Kratochvil, E. J. W. (2015). A paper based, all organic, reference‐electrode‐free ion sensing platform. Journal of Materials Chemistry B, 3(25), 5095–5102. 10.1039/c5tb00387c [DOI] [PubMed] [Google Scholar]
- Lee, K. H. , & Zeng, H. (2017). Aptamer‐based ELISA assay for highly specific and sensitive detection of Zika NS1 protein. Analytical Chemistry, 89(23), 12743–12748. 10.1021/acs.analchem.7b02862 [DOI] [PubMed] [Google Scholar]
- Lisak, G. , Cui, J. , & Bobacka, J. (2015). Paper‐based microfluidic sampling for potentiometric determination of ions. Sensors and Actuators B: Chemical, 207, 933–939. 10.1016/j.snb.2014.07.044 [DOI] [Google Scholar]
- Määttänen, A. et al (2013). A low‐cost paper‐based inkjet‐printed platform for electrochemical analyses. Sensors Actuators, B Chem., 177, 153–162. 10.1016/j.snb.2012.10.113 [DOI] [Google Scholar]
- Murray, K. O. , Gorchakov, R. , Carlson, A. R. , Berry, R. , Lai, L. , Natrajan, M. , … Mulligan, M. J. (2017). Prolonged detection of Zika virus in vaginal secretions and whole blood. Emerging Infectious Diseases, 23(1), 99–101. 10.3201/eid2301.161394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musso, D. , Roche, C. , Nhan, T. X. , Robin, E. , Teissier, A. , & Cao‐Lormeau, V. M. (2015). Detection of Zika virus in saliva. Journal of Clinical Virology, 68, 53–55. 10.1016/j.jcv.2015.04.021 [DOI] [PubMed] [Google Scholar]
- Nery, E. W. , & Kubota, L. T. (2016). Integrated, paper‐based potentiometric electronic tongue for the analysis of beer and wine. Analytica Chimica Acta, 918, 60–68. 10.1016/j.aca.2016.03.004 [DOI] [PubMed] [Google Scholar]
- Novell, M. , Guinovart, T. , Blondeau, P. , Rius, F. X. , & Andrade, F. J. (2014). A paper‐based potentiometric cell for decentralized monitoring of Li levels in whole blood. Lab on a Chip, 14(7), 1308–1314. 10.1039/c3lc51098k [DOI] [PubMed] [Google Scholar]
- Parrilla, M. , Cánovas, R. , & Andrade, F. J. (2017). Paper‐based enzymatic electrode with enhanced potentiometric response for monitoring glucose in biological fluids. Biosensors & Bioelectronics, 90, 110–116. 10.1016/j.bios.2016.11.034 [DOI] [PubMed] [Google Scholar]
- Ping, J. , Wang, Y. , Fan, K. , Tang, W. , Wu, J. , & Ying, Y. (2013). High‐performance flexible potentiometric sensing devices using free‐standing graphene paper. Journal of Materials Chemistry B, 1(37), 4781–4791. 10.1039/c3tb20664e [DOI] [PubMed] [Google Scholar]
- Sánchez‐Purrà, M. , Carré‐Camps, M. , De Puig, H. , Bosch, I. , Gehrke, L. , & Hamad‐Schifferli, K. (2017). Surface‐enhanced raman spectroscopy‐based sandwich immunoassays for multiplexed detection of Zika and dengue viral biomarkers. ACS Infectious Diseases, 3(10), 767–776. 10.1021/acsinfecdis.7b00110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, J. , Cherstvy, A. G. , & Metzler, R. (2014). Sensing viruses by mechanical tension of DNA in responsive hydrogels. Physical Review X, 4(2), 10.1103/PhysRevX.4.021002 [DOI] [Google Scholar]
- Siegel, A. C. , Phillips, S. T. , Dickey, M. D. , Lu, N. , Suo, Z. , & Whitesides, G. M. (2010). Foldable printed circuit boards on paper substrates. Advanced Functional Materials, 20(1), 28–35. 10.1002/adfm.200901363 [DOI] [Google Scholar]
- Sjöberg, P. , Määttänen, A. , Vanamo, U. , Novell, M. , Ihalainen, P. , Andrade, F. J. , … Peltonen, J. (2016). Paper‐based potentiometric ion sensors constructed on ink‐jet printed gold electrodes. Sensors Actuators, B Chem., 224, 325–332. 10.1016/j.snb.2015.10.051 [DOI] [Google Scholar]
- Szucs, J. , & Gyurcsányi, R. E. (2012). Towards protein assays on paper platforms with potentiometric detection. Electroanalysis, 24(1), 146–152. 10.1002/elan.201100522 [DOI] [Google Scholar]
- Thoreson, E. J. , Martin, J. , & Burnham, N. A. (2007). Recommendations for the use of an atomic force microscope as an in‐fab stiction monitor. J. Microelectromechanical Syst., 16(3), 694–699. 10.1109/JMEMS.2006.879664 [DOI] [Google Scholar]
- Tobjörk, D. , & Österbacka, R. (2011). Paper electronics. Advanced Materials, 23(17), 1935–1961. 10.1002/adma.201004692 [DOI] [PubMed] [Google Scholar]
- Tuerk, C. , & Gold, L. (1990). Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science, 249(4968), 505–510. 10.1126/science.2200121 [DOI] [PubMed] [Google Scholar]
- Zika Envelope Recombinant Antigen . Zika Virus | ProSpec. Retrieved from https://www.prospecbio.com/zika_envelope_sf9


