Abstract
Coronavirus epidemic 2019 (COVID‐19), instigated by SARS‐CoV‐2 virus, is recently raising worldwide and inspiring global health worries. The main 3‐chymotrypsin‐like cysteine protease (3CLPro) enzyme of SARS‐CoV‐2, which operates its replication, could be used as a medication discovery point. We therefore theoretically studied and docked the effects of 19 hydrolysable tannins on SARS‐CoV‐2 by assembling with the catalytic dyad residues of its 3CLpro using molecular operating environment (MOE 09). Results discovered that pedunculagin, tercatain, and castalin intensely interacted with the receptor binding site and catalytic dyad (Cys145 and His41) of SARS‐CoV‐2. Our analyses estimated that the top three hits might serve as potential inhibitor of SARS‐CoV‐2 leading molecules for additional optimization and drug development process to combat COVID‐19. This study unleashed that tannins with specific structure could be utilized as natural inhibitors against COVID‐19.
Practical applications
The 3CLPro controls SARS‐CoV‐2 copying and manages its life series, which was targeted in case of SARS‐CoV and MERS‐CoV coronavirus. About 19 hydrolysable tannins were computed against 3CLpro of SARS‐CoV‐2. Pedunculagin, tercatain, and castalin interacted with Cys145 and His41 of SARS‐CoV‐2‐3CLpro. Pedunculagin‐SARS‐CoV‐2‐3CLpro remain stable, with no obvious fluctuations. We predicted that the understandings gained in the current research may evidence valued for discovering and unindustrialized innovative natural inhibitors for COVID‐19 in the nearby future.
Keywords: COVID‐19, hydrolysable tannins, main 3‐chymotrypsin‐like cysteine protease, molecular docking, structural‐relationship activity
About 19 hydrolyzable tannins were computed against 3CLpro enzyme of 2019‐nCoV. It was found that pedunculagin, tercatain, and castalin interacted with Cys145 and His41 of 2019‐nCoV‐3CLpro. Likewise, pedunculagin‐2019‐nCoV‐3CLpro remain stable, with no obvious fluctuations. We predicted that the understandings obtained in the current study may evidence valued for discovering and unindustrialized novel natural anti‐COVID‐19 therapeutic agents in the near future.

Abbreviations
- 3CLpro
papain‐like protease
- COVID‐19
coronavirus epidemic 2019
- MD
molecular dynamic simulations
- MOE 09
molecular operating environment
- SARS‐CoV‐2
novel coronavirus
1. INTRODUCTION
Freshly, the new coronavirus (SARS‐CoV‐2) was discovered on the late of 2019 (Xu et al., 2020). COVID‐19 suddenly appeared, leading not only the health authorities, but also the scientific community to swift actions toward it. As a result, the entire‐genome series of SARS‐CoV‐2 was issued and subsequently it was scientifically spotlighted (Jiang et al., 2020; Stebbing et al., 2020). The SARS‐CoV‐2 belongs to the β‐coronavirus group, sharing ancestry with bat coronavirus HKU9‐1, alike to SARS, and regardless of arrangement variety its Spike protein intensely networks with human ACE2‐receptor (Ul Qamar, Alqahtani, Alamri, & Chen, 2020). By July 13, the global death toll exceeded 574,000, with 13,200.000 confirmed cases, and 7,600.000 thousand recovered cases in 213 countries (https://www.worldometers.info/coronavirus).
Recent studies emphasized that SARS‐CoV‐2 genes stake <80% nucleotide uniqueness and 89.1% nucleotide likeness with SARS‐genes (Kim, Kim, & Lee, 2020; Moorthy, Restrepo, Preziosi, & Swaminathan, 2020). Usually, β‐coronaviruses yield a ~800 kDa polypeptide upon dictation of the genome. This polypeptide is proteolytically slashed to create numerous proteins, and the proteolytic diversion is facilitated by papain‐like protease, and slicing the polyprotein at 16 different spots to create several nonoperational proteins that are vital for the viral reproduction (Macchiagodena, Pagliai, & Procacci, 2020). Thus, this protease portrays a key task in the virus duplication, and distinct structural/fitment protein‐encoding genes situated at the 3′ end that exhibit extreme inconsistency (Rut et al., 2020). Therefore, 3CLPro enzyme may assist as a possible target for natural inhibitors against COVID‐19.
Structure‐based effect analyses and high output findings have recognized possible inhibitors of SARS‐ and MERS‐3CLpro (Pillaiyar, Meenakshisundaram, & Manickam, 2020). Bioactive substances, especially hydrolysable tannins, have attracted significant attention which could be valorized to develop medications without side‐effects (Liu et al., 2016). Hydrolysable tannins are also measured to be valuable in eliminating the opposing effects of several chemotherapeutic stuffs as well as in extending permanency and achieving positive overall health with anticancer possible and can be utilized as a substitute cancer‐drug supply (Buzzini et al., 2008; Lin et al., 2011; Aires, 2020; Jia et al., 2019; Zhu, Khalifa, Peng, & Lic, 2018). Hydrolysable tannins, including punicalagin, punicalin, and geraniin, exerted antiviral effects toward B virus by averting the creation of cccDNA and indorsing cccDNA decline, which may aid as chief components for the expansion of novel agents to remedy HBV‐infection (Liu et al., 2016). Likewise, tannin‐type compounds, such as epiacutissimins A and B, castalin, vescalin, chebulagic acid, and punicalagin showed anti‐herpesvirus activity via targeting viral glycoprotein‐glycosaminoglycan binding to inhibit access and cell‐to‐cell feast (Lin et al., 2011; Aires, 2020). Most importantly, it was hypothesized that the biological activities, including antiviral effects, of tannins are structural‐dependent. For example, additional reactions, such as hydroxylation, methylation, glycosylation, galloylation, and polymerization are the main factors affecting the biological activity of tannins. Therefore, we selected 19 different structure tannins in term of representing the structure‐relationship activity of tannins.
The current study was designed to find out a potent inhibitor against COVID‐19 from 19 structural different hydrolysable tannins which could target the main protease of SARS‐CoV‐2 using in silico approaches (molecular docking and drug‐likeness scan). The findings of our study will enable the researchers to rally the status of natural therapeutics against COVID‐19.
2. MATERIALS AND METHODS
Structure‐based simulated inspection tactic was performed using high‐performing computing work‐station with the subsequent stipulations (Intel(R) Core‐(TM) i7‐3210M CPU @ 2.50 GHz, 5 Core(s) CPU of 4.00 GB RAM and 64‐bit Windows‐10 Functioning System). Structure‐based medication inspection was done using Molecular Operating Environment (MOE 09).
2.1. Ligand database arrangement
The structures of 19 hydrolysable tannins including, castalin (CID‐99973), bicornin (CID‐71308161), grandinin (CID‐492392), tercatain (CID‐14411424), granatin A (CID‐131752596), tellimagradin I (PubChem CID‐73179), geraniin (CID‐3001497), casuarinin (CID‐13834145), strictinin (CID‐73330), pedunculagin (CID‐442688), punicalin (CID‐5388496), chebulagic acid (CID‐250397), casuarictin (CID‐73644), ß‐pedunculagin (CID‐5320441), potentillin (CID‐5315734), isoterchebin (CID‐442685), roxbin B (CID‐176131), repandusinic acid A (CID‐147900), and terchebin (CID‐3084341) were retrieved from PubChem database. The tannins structure was optimized for docking by adding partial charges and energy minimization via Protonate‐3D and MMFF94X strength field, separately. Adjusted ligands archives for the 19 tannins were kept in ligand catalog which was lastly utilized as an involvement file for docking investigations.
2.2. Refinement of 3CLpro of 2019‐nCoV structure
Likewise, three‐dimensional structure of 3CLpro of SARS‐CoV‐2 (PDB: 6y84) was regained from Protein‐Data‐Bank (http://www.rcsb.org) with 2.16 Å resolution. To enhance the 3CLpro‐structure, previously interacted ligands and H2O substances were detached from 3CLpro‐structure, 3D‐protonation, and energy minimalization were dedicated in MOE‐09. The minimalized structure was utilized for further docking operation in the following steps.
2.3. Molecular docking and drug likeness analyses
MOE docking tool was used to dock the 19 structural different tannins toward allosteric ligand interacting spot of SARS‐CoV‐2‐3CLpro. The possible binding compact was identified using site locater tool of MOE‐09 and was then occupied through docking procedures. The 10 finest docked postures were created applying a scoring job London dG. Enhancement of docking process was ended by operating forcefield algorithm which saves the receptor firm. From them, finest relating ligands and components were selected based on Root‐Mean‐Square Deviation (RMSD) which is often computed in Angstrom (Å), and docking score. Ligand receptor interacting analysis was performed using LigX tool in MOE‐09. It is intended to display possible residues binding with each ligand realistically. It creates 2D‐images signifying the forces steadying ligands within binding pouches of receptors (Khan et al., 2017). To investigate the medication likeliness of 19 tannins, all were further clarified based on behavior suitable molecular characters to be candidate inhibitor against COVID‐19.
2.4. Molecular dynamics simulations and pharmacophore analysis
Molecular dynamics simulations (MD) were done to refine our docking results and to analyze the assembling performance and constancy of top three hydrolysable tannins using SARS‐CoV‐2 3CLpro crystal model. GROMOS software was used to engage 80 ns MD‐simulations succeeding the similar procedure as explained before (Ul Qamar et al., 2020). The ligand topology files were built using CHARMM force field through CGenFF server64,65,66. Ligand‐protein complexes were solvated in octahedron box with TIP3P water model. The predicted pharmacophore of pedunculagin was also created (Khaerunnisa, Kurniawan, Awaluddin, Suhartati, & Soetjipto, 2020).
3. RESULTS AND DISCUSSION
Analysis of allosteric binding of 19 structural different tannins with COVID‐19 protease initially revealed many poses and we used the best pouches with small S‐score. Reiterative docking of the finest docked molecules within receptor compact formerly employed by co‐crystallized inhibitor. The chemical structure of our 19 structural different tannins are shown in Figure 1. By analyzing the physicochemical parameters of SARS‐CoV‐2‐3CLpro, it was found that the enzyme consisted of 306 residues with chain type of polypeptide(L) with a molecular weight of 33.796 kDa, cataloging the protein as a steady, hydrophilic molecule able to establish H‐bonds with other ligands (Figure 2). Recently, it was found that comparing the sequence of SARS‐CoV‐2‐3CLpro protein gathered with bat SARS‐like, disclosing 99.02% of sequence individuality (Ul Qamar et al., 2020). Furthermore, it was found that SARS‐CoV‐2 is much comparable to SARS than MERS, and imparts a mutual forebear with bat‐based coronaviruses (Ji, Wang, Zhao, Zai, & Li, 2020; Xu et al., 2020). The findings also discovered that SARS‐CoV‐2 had a Cys‐His catalytic dyad (Cys145 and His41), reliable with SARS‐3CLpro (Cys145 and His41), TGEV‐3CLpro (Cys144 and His41), and HCoV‐3CLpro (Cys144 and His41) (Yang et al., 2003). It was disclosed that SARS‐CoV‐2‐3CLpro receptor‐binding concise configuration and be similar with that of SARS‐3CLpro binding compact and increases the opportunity that inhibitors projected for SARS‐3CLpro may also mitigate the motion of SARS‐CoV‐2‐3CLpro.
FIGURE 1.
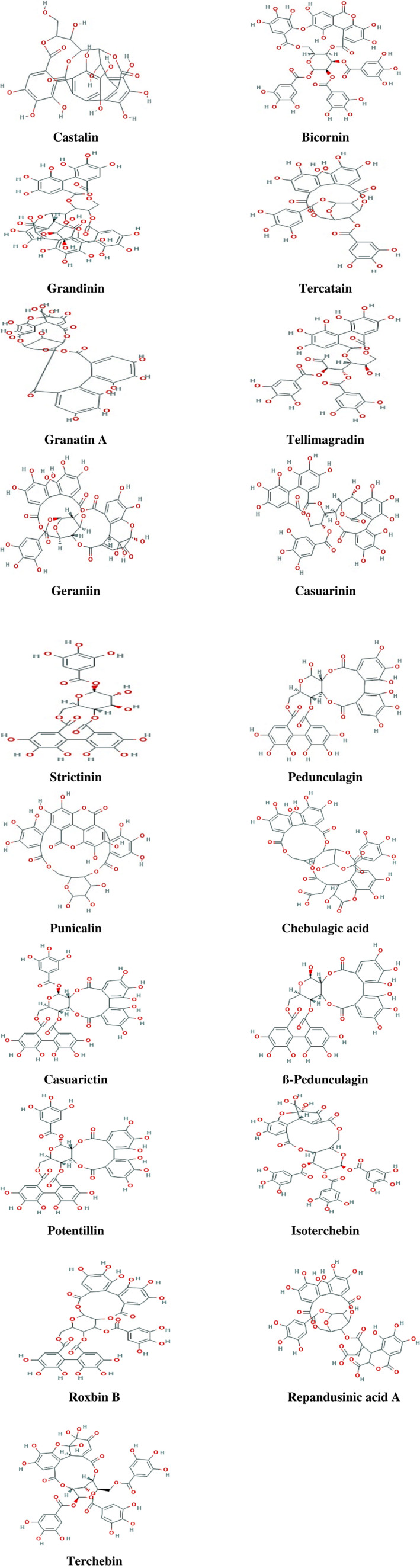
2D‐chemical structures of 19 selected hydrolysable tannins
FIGURE 2.
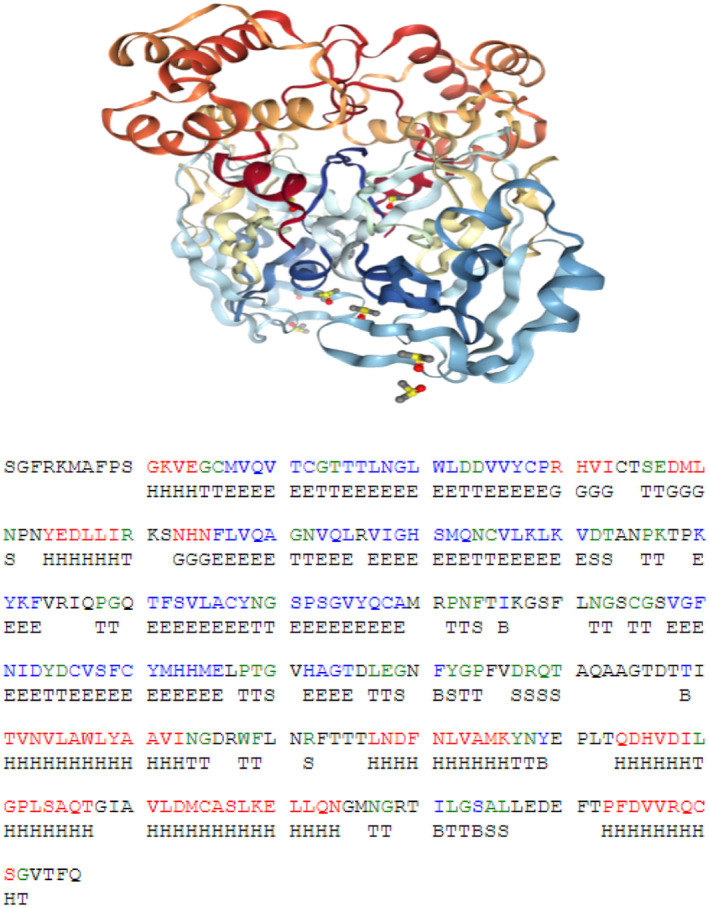
The 3D‐chemical structures of 3CLpro enzyme of SARS‐CoV‐2 and its sequence
To manage with the continuous essential of new and operative small molecule as natural therapeutics against COVID‐19 with negligible side‐effects, studies are now directing more on computational drug finding (Desai et al., 2008). Several reports testified the antiviral possible of tannins via computer‐assisted medication strategy (Buzzini et al., 2008; Lin et al., 2011). To speed the drug agreement process and to realize more effectual inhibitors with novel frameworks that can progress the antiviral‐based therapeutics status, computational drug discovery methods are extremely steadfast. From the innovative aspect of structural variety among hydrolysable tannins, simulated selection was ended to realize new allosteric compounds as inhibitors against COVID‐19. Allosteric directive has been described as an operative tactic to conquer irretrievable inhibition. Spatial alignment and dock score of present stated four top‐ranked chiefs discloses effectual interacting of functional residues with greatest binding affinity. These tannins fit with the drug prospect norms tabulated in Table. 1 and S1 and may attest exceptional perceptive to use as a preparatory point.
TABLE 1.
The top five poses of the interaction between 19 hydrolysable tannins with the catalytic dyad residues of 3CLpro of SARS‐CoV‐2 and their docking properties
| Hydrolysable tannins | Mol | S | E‐conf | E‐place | E‐score |
|---|---|---|---|---|---|
| Castalin |
|
−14.04 | 156.57 | −98.32 | −15.69 |
| Bicornin |
|
−24.36 | 122.35 | −82.19 | −14.60 |
| Grandinin |
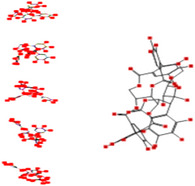
|
−21.86 | 220.85 | −127.95 | −16.66 |
| Tercatain |
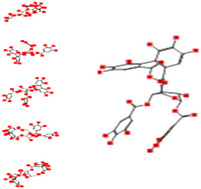
|
−23.11 | 85.43 | −133.94 | −16.82 |
| Granatin A |
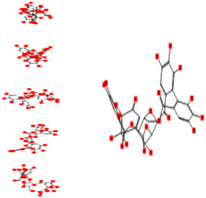
|
−15.38 | 187.54 | −98.06 | −13.28 |
| Tellimagradin I |
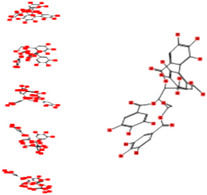
|
−29.53 | 81.89 | −107.41 | −13.83 |
| Geraniin |
|
−22.17 | 152.74 | −93.78 | −12.09 |
| Casuarinin |
|
−24.15 | 122.36 | −61.90 | −15.24 |
| Strictinin |
|
−23.96 | 98.31 | −96.30 | −16.82 |
| Pedunculagin |
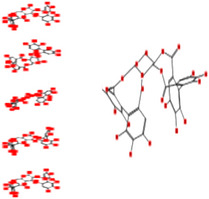
|
−18.58 | 110.04 | −70.23 | −12.96 |
| Punicalin |
|
−18.01 | 81.58 | −61.70 | −14.78 |
| Chebulagic acid |
|
−22.35 | 132.94 | −124.68 | −13.74 |
| Casuarictin |
|
−24.88 | 112.87 | −112.35 | −16.09 |
| ß‐Pedunculagin |
|
−19.05 | 99.45 | −116.44 | −15.71 |
| Potentillin |
|
−17.40 | 124.16 | −44.06 | −17.23 |
| Isoterchebin |
|
−25.38 | 120.42 | −139.58 | −16.83 |
| Roxbin B |
|
−23.82 | 100.86 | −97.63 | −19.67 |
| Repandusinic acid A |
|
−24.15 | −41.28 | −167.41 | −13.87 |
| Terchebin |
|
−29.06 | 119.45 | −60.55 | −13.21 |
Mol: An output pose, S: The final score, which is the score of the last stage that was not set to None, E‐conf: The energy of the conformer, E‐place: Score from the placement stage, E‐score: Score from the rescoring stage.
Interacting affinity analysis of 19 tannins through LigX revealed in Figure 3. It was revealed that 19 tannins differentially interacted with different residues of SARS‐CoV‐2‐3CLpro. In details, pedunculagin are the best hydrolysable tannins, where it could bind with 12 amino acid residues of SARS‐CoV‐2‐3CLpro and there are 5 H‐bonding forces have been occurred. Most importantly, pedunculagin directly interacted with His41 and Cys145 through H‐bonding forces, showing its ability to interact with the catalytic dyad residues of 3CLpro. Likewise, castalin and tercatain directly networked with His41 and Cys145 via arene–arene interactions and H‐bonding forces. Bicornin, Repandusinic acid A, grandinin, granatin A, roxbin B, and terchebin also precisely interacted with His41 through H‐bonding or arene–arene interactions, showing their influence on the catalytic dyad residues of 3CLpro. Results also showed that there are some of the hydrolysable tannins, including tellimagradin I, strictinin, punicalin, chebulagic acid, casuarictin, ß‐pedunculagin, potentillin, and isoterchebin have been secondarily interacted with both of His41 or Cys145, showing their partial influence on the catalytic dyad residues of 3CLpro. On the contrary, both of geraniin and casuarinin neither interact with His41 nor Cys145; however, both of them have been networked with eight and seven amino acid residues of 3CLpro via arene–arene and H‐bonding. The vital catalytic residues of compact spatial site of pedunculagin is stabilized within the pocket with the deepest S‐score (−18.58) and binding with C145, H41, N142, S46, T45, T24, T25, Q189, E166, H164, M165, and M43. Castalin interacted with C145, H41, L27, M49, G143, Q189, N142, S46, T45, T25, and C44. The difference in the binding ability of each tannin with SARS‐CoV‐2‐3CLpro may be due to their structure‐based relationship activity.
FIGURE 3.
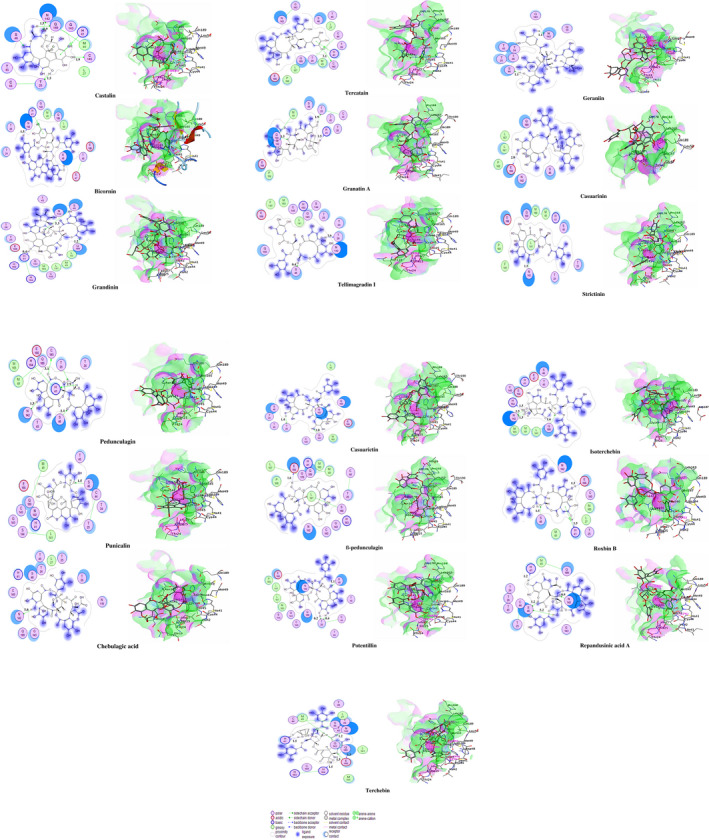
LigX interaction diagram representing binding pattern of 19 hydrolysable tannins with binding pocket residues of 3CLpro enzyme of SARS‐CoV‐2
These results identified at least three novels harmless, druggable tannins that are envisaged to interact with the receptor binding spot and catalytic dyad (Cys‐145 and His‐41) of SARS‐CoV‐2‐3CLpro. Among these hydrolysable tannins, pedunculagin, strongly interacted with the catalytic dyad residues (Cys‐145 and His‐41) of SARS‐CoV‐2‐3CLpro, with sense binding affinity, docking score, and ADMET properties. As supplemented in Table S2, pedunculagin is an excellent compound regarding solubility, and having high intestinal absorption, and does not shows any mutation. This molecule is not inhibitor of HERG l and HERG ll that means it is not harmful to cardiac muscles (heart muscles). Previously, pedunculagin, tercatain, and castalin which could be extracted from pomegranates, walnut, Indian gooseberry, oak wood, leaves of Melaleuca quinquenervia, Terminalia catappa, and Combretum glutinosum showed antiviral activity and other biological activities (Chang et al., 1995; Silva et al., 2016; Zuo, LI, Chen, & Xu, 2005). In a related study, Park et al. (2017) isolated 10 polyphenols from Broussonetia papyrifera inhibitors versus 3‐chymotrypsin‐like and papain‐like coronavirus cysteine proteases. Jo, Kim, Kim, Shin, and Kim (2019) also examined four types of flavonoids against MERS‐CoV and he reported that quercetin 3‐β‐d‐glucoside, herbacetin, isobavachalcone, and helichrysetin blocked the enzymatic activity of MERS‐3CLpro.
On the contrary, it has been reported that hydrolysable tannins are degraded to gallic acid, pyrogallol, phloroglucinol, and finally to acetate and butyrate via sequential actions of different bacterial enzymes. Gallic, hexahydroxydiphenic, and ellagic acids are also the monomer types of hydrolyzed tannins (Serrano, Puupponen‐Pimiä, Dauer, Aura, & Saura‐Calixto, 2009; Smeriglio, Barreca, Bellocco, & Trombetta, 2017). It was informed that ellagitannins release ellagic acid upon hydrolysis in rat’s intestine. Gallic acid is infused via a paracellular route in Caco‐2 cells, with t max of absorption around 1.27 hr in humans. Regarding ellagic acid absorption, some authors have detected ellagic acid in human plasma between 0.5 and 3 hr after oral administration of pomegranate juice. Following absorption, ellagic acid undergoes conjugation, and conjugated forms with methyl, glucuronyl, and sulfate groups have been found in plasma and excreted in urine. Gallotannins are easily degraded by bacteria, fungi, and yeast. Tannase produced by a group of microorganisms is active in galloyl residues of galloyls esters, as well as on hexahydroydi‐phenoyl. Unlike proanthocyanidins, colonic bacteria are capable of metabolizing hydrolysable tannins. The presence of lactobacilli with distinct tannase activity proposes that gallic acid from gallotannins may be available during colonic fermentation. For example, casuarictin incubated with caecal content increase the release of ellagic acid. In rats fed with punicalagin, ellagic acid is transformed by rat microflora to 3,8‐dihydroxy‐6H‐dibenzo (b,d)‐pyran‐6‐one (urolithin B)derivatives, the total urinary excretion accounting for 0.7%–52.7% of the ingested punilagin in rats. When a single dose of ellagitannin‐containing foodstuff was taken by each group of human volunteers, the metabolite excretion ranged from 2.8% to 16.6% of the ingested ellagitannins. Therefore, the health effects of tannins intake are probably a consequence of the biological activity of their metabolites.
Looking at the antiviral drug possible of aforenoted tannins, current study was an endeavor to feat the chemical nature of three hydrolysable tannins as natural inhibitors against COVID‐19. Present molecular docking study has publicized the significant interactions of medicinal tannins with the main protease of COVID‐19, which were successfully block both His41 and Cys145.
To authenticate the drug capacity of selected tannins, ligand characters were considered with LigX tool of MOE. All chosen tannins disclosed constructive results and accomplish the standards of the Lipinski’s regulation of five (Khan et al., 2017). The regulation defines that possible drug‐like components should have about 5 H‐bond donors, maximum 10 H‐bond acceptors, and an octanol water panel coefficient log P not > 5. These results suggest that natural components identified in our study, specially pedunculagin, tercatain, and castalin, may demonstrate more valuable contenders for COVID‐19 drug rehabilitation. To further examine the molecular docking results, pedunculagin was subjected to MD simulation; and RMSD, radius of gyration (RoG), and H‐bond characters were expressed. Pedunculagin‐SARS‐CoV‐2‐3CLpro‐complex did not show any obvious fluctuations, referring to the stability of tannin‐protein complexes with a typical RMSD value of 1.4 ± 0.01 Å (Figure S1a). It was also suggested normal behavior for pedunculagin‐SARS‐CoV‐2‐3CLpro complex; where it was persisted dense and steady during the 80 ns simulations (Figure S1b). Likewise, H‐bonds which are the key stabilizing forces in proteins, proposed that the pedunculagin‐SARS‐CoV‐2‐3CLpro complex remain stable throughout the simulation, with no noticeable fluctuations (Figure S1c). It can be concluded from this study that each of pedunculagin, tercatain, and castalin and their sources such as pomegranates and nuts may assist as possible natural inhibitors against SARS‐CoV‐2. Pedunculagin is a hydrolysable tannin. Secondary metabolite components are regularly found in medicinal plants. We therefore predicted the pharmacophore of pedunculagin based on its active groups. Hydroxy groups (−OH), ketone groups (=O), and phenolic rings in pedunculagin are projected to play parts amino acid residue binding at the energetic spot of SARS‐CoV‐2‐3CLpro (Figure 4). These groups were found to directly interact with the 3CLpro of SARS‐CoV‐2 through H‐bonding and other forces.
FIGURE 4.
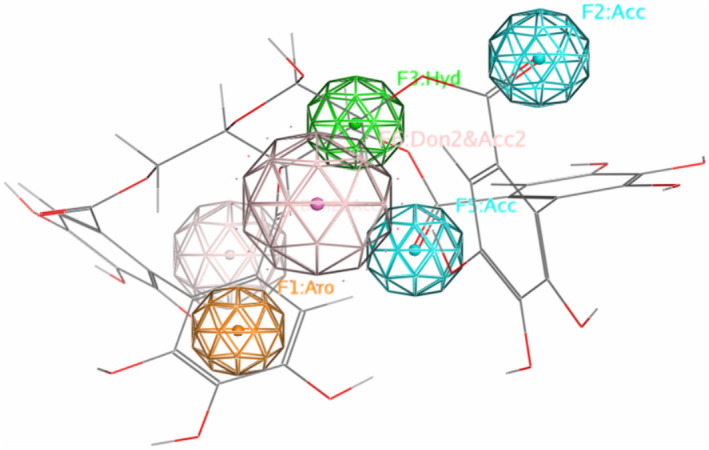
The predicated pharmacophore for pedunculagin
4. CONCLUSION
In conclusion, our study exposed that some medicinal plant rich in hydrolysable tannins, especially pedunculagin, tercatain, and castalin, could be theoretically used to treat the outbreak of COVID‐19. Herein, we screened the structural relationship activity of 19 hydrolysable tannins as potential antiviral components and we chose the top three hits that may inhibit the main protease of SARS‐CoV‐2 and hence virus copying. Further in vitro and in vivo studies are needed to transmute these probable hydrolysable tannins inhibitors into clinical drugs. We predicted that the understandings gained in the current study may evidence valued for discovering and unindustrialized novel natural inhibitors against COVID‐19 in the near future.
CONFLICT OF INTEREST
The authors declared that they have no conflict of interest.
Supporting information
Supplementary Material
ACKNOWLEDGMENTS
This work was self‐supported by the authors. We also acknowledge all laboratories and databases websites mentioned in this study.
Khalifa I, Zhu W, Mohammed HHH, Dutta K, Li C. Tannins inhibit SARS-CoV-2 through binding with catalytic dyad residues of 3CLpro: An in silico approach with 19 structural different hydrolysable tannins. J Food Biochem. 2020;44:e13432. 10.1111/jfbc.13432
Contributor Information
Ibrahim Khalifa, Email: ibrahiem.khalifa@fagr.bu.edu.eg.
Chunmei Li, Email: ibrahiem.khalifa@fagr.bu.edu.eg, Email: lichmyl@mail.hzau.edu.cn.
REFERENCES
- Aires, A. (2020). Tannins as antiviral agents. In Tannins‐structural properties, biological properties and current knowledge (pp. 1‒14). IntechOpen. 10.5772/intechopen.80170 [DOI] [Google Scholar]
- Buzzini, P. , Arapitsas, P. , Goretti, M. , Branda, E. , Turchetti, B. , Pinelli, P. , … Romani, A. (2008). Antimicrobial and antiviral activity of hydrolysable tannins. Mini‐Reviews in Medicinal Chemistry, 8(12), 1179. [DOI] [PubMed] [Google Scholar]
- Chang, J. H. , Cho, J. H. , Kim, H. H. , Lee, K. P. , Lee, M. W. , Han, S. S. , & Lee, D. I. (1995). Antitumor activity of pedunculagin, one of the ellagitannin. Archives of Pharmacal Research, 18(6), 396. 10.1007/BF02976342 [DOI] [Google Scholar]
- Desai, A. G. , Qazi, G. N. , Ganju, R. K. , EL‐Tamer, M. , Singh, J. , Saxena, A. K. , … Bhat, H. K. (2008). Medicinal plants and cancer chemoprevention. Current Drug Metabolism, 9(7), 581–591. 10.2174/138920008785821657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, W. , Wang, W. , Zhao, X. , Zai, J. , & Li, X. (2020). Homologous recombination within the spike glycoprotein of the newly identified coronavirus may boost cross‐species transmission from snake to human. Journal of Medical Virology, 92, 433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, Y. , Khalifa, I. , Hu, L. , Zhu, W. , Li, J. , Li, K. , & Li, C. (2019). Influence of three different drying techniques on persimmon chips’ characteristics: A comparison study among hot‐air, combined hot‐air‐microwave, and vacuum‐freeze drying techniques. Food and Bioproducts Processing, 118, 67–76. 10.1016/j.fbp.2019.08.018 [DOI] [Google Scholar]
- Jiang, F. , Deng, L. , Zhang, L. , Cai, Y. , Cheung, C. W. , & Xia, Z. (2020). Review of the clinical characteristics of coronavirus disease 2019 (COVID‐19). Journal of General Internal Medicine, 35, 1545–1549, 10.1007/s11606-020-05762-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo, S. , Kim, H. , Kim, S. , Shin, D. H. , & Kim, M. S. (2019). Characteristics of flavonoids as potent MERS‐CoV 3C‐like protease inhibitors. Chemical Biology & Drug Design, 94(6), 2023–2030. 10.1111/cbdd.13604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaerunnisa, S. , Kurniawan, H. , Awaluddin, R. , Suhartati, S. , & Soetjipto, S. (2020). Potential inhibitor of COVID‐19 main Protease (Mpro) from several medicinal plant compounds by molecular docking study. Prepr, 1, 1–14. 10.20944/preprints202003.0226. [DOI] [Google Scholar]
- Khan, W. , Ashfaq, U. A. , Aslam, S. , Saif, S. , Aslam, T. , Tusleem, K. , … Ul Qamar, M. T. (2017). Anticancer screening of medicinal plant phytochemicals against Cyclin‐Dependent Kinase‐2 (CDK2): An in‐silico approach. Advancements in Life Sciences, 4(4), 113–119. [Google Scholar]
- Kim, S. , Kim, D.‐M. , & Lee, B. (2020). Insufficient sensitivity of RNA dependent RNA polymerase gene of SARS‐CoV‐2 viral genome as confirmatory test using Korean COVID‐19 cases. Preprint. 10.20944/preprints202002.0424.v1. [DOI] [Google Scholar]
- Lin, L.‐T. , Chen, T.‐Y. , Chung, C.‐Y. , Noyce, R. S. , Grindley, T. B. , Mccormick, C. , … Richardson, C. D. (2011). Hydrolyzable tannins (chebulagic acid and punicalagin) target viral glycoprotein‐glycosaminoglycan interactions to inhibit herpes simplex virus 1 entry and cell‐to‐cell spread. Journal of Virology, 85(9), 4386–4398. 10.1128/JVI.01492-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C. , Cai, D. , Zhang, L. , Tang, W. , Yan, R. , Guo, H. , & Chen, X. (2016). Identification of hydrolyzable tannins (punicalagin, punicalin and geraniin) as novel inhibitors of hepatitis B virus covalently closed circular DNA. Antiviral Research, 134, 97–107. 10.1016/j.antiviral.2016.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macchiagodena, M. , Pagliai, M. , & Procacci, P. (2020). Inhibition of the main Protease 3CL‐pro of the coronavirus disease 19 via structure‐based ligand design and molecular modeling. arXiv preprint arXiv:2002.09937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorthy, V. , Restrepo, A. M. H. , Preziosi, M.‐P. , & Swaminathan, S. (2020). Data sharing for novel coronavirus (COVID‐19). Bulletin of the World Health Organization, 98(3), 150. 10.2471/BLT.20.251561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J.‐Y. , Yuk, H. J. , Ryu, H. W. , Lim, S. H. , Kim, K. S. , Park, K. H. , … Lee, W. S. (2017). Evaluation of polyphenols from Broussonetia papyrifera as coronavirus protease inhibitors. Journal of Enzyme Inhibition and Medicinal Chemistry, 32(1), 504–512. 10.1080/14756366.2016.1265519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillaiyar, T. , Meenakshisundaram, S. , & Manickam, M. (2020). Recent discovery and development of inhibitors targeting coronaviruses. Drug Discovery Today, 25(4), 668–688. 10.1016/j.drudis.2020.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rut, W. , Groborz, K. , Zhang, L. , Sun, X. , Zmudzinski, M. , Hilgenfeld, R. , & Drag, M. (2020). Substrate specificity profiling of SARS‐CoV‐2 Mpro protease provides basis for anti‐COVID‐19 drug design. bioRxiv. Preprint. 10.1101/2020.03.07.981928. [DOI] [Google Scholar]
- Serrano, J. , Puupponen‐Pimiä, R. , Dauer, A. , Aura, A. M. , & Saura‐Calixto, F. (2009). Tannins: Current knowledge of food sources, intake, bioavailability and biological effects. Molecular Nutrition & Food Research, 53(S2), S310–S329. 10.1002/mnfr.200900039 [DOI] [PubMed] [Google Scholar]
- Silva, R. M. , Pereira, L. D. , Véras, J. H. , Do Vale, C. R. , Chen‐Chen, L. , & Da Costa Santos, S. (2016). Protective effect and induction of DNA repair by Myrciaria cauliflora seed extract and pedunculagin on cyclophosphamide‐induced genotoxicity. Mutation Research/Genetic Toxicology and Environmental Mutagenesis, 810, 40–47. 10.1016/j.mrgentox.2016.10.001 [DOI] [PubMed] [Google Scholar]
- Smeriglio, A. , Barreca, D. , Bellocco, E. , & Trombetta, D. (2017). Proanthocyanidins and hydrolysable tannins: Occurrence, dietary intake and pharmacological effects. British Journal of Pharmacology, 174(11), 1244–1262. 10.1111/bph.13630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbing, J. , Phelan, A. , Griffin, I. , Tucker, C. , Oechsle, O. , Smith, D. , & Richardson, P. (2020). COVID‐19: Combining antiviral and anti‐inflammatory treatments. The Lancet Infectious Diseases., 20(4), 400–402. 10.1016/S1473-3099(20)30132-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ul Qamar, M. T. , Alqahtani, S. M. , Alamri, M. A. , & Chen, L.‐L. (2020). Structural basis of SARS‐CoV‐2 3CLpro and anti‐COVID‐19 drug discovery from medicinal plants†. Journal of Pharmaceutical Analysis. 10.1016/j.jpha.2020.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X. , Chen, P. , Wang, J. , Feng, J. , Zhou, H. , Li, X. , … Hao, P. (2020). Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Science China Life Sciences, 63(3), 457–460. 10.1007/s11427-020-1637-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H. , Yang, M. , Ding, Y. , Liu, Y. , Lou, Z. , Zhou, Z. , … Rao, Z. (2003). The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proceedings of the National Academy of Sciences, 100(23), 13190–13195. 10.1073/pnas.1835675100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, W. , Khalifa, I. , Peng, J. , & Lic, C. (2018). Position and orientation of gallated proanthocyanidins in lipid bilayer membranes: Influence of polymerization degree and linkage type. Journal of Biomolecular Structure and Dynamics., 36(11), 2862–2875. 10.1080/07391102.2017.1369163 [DOI] [PubMed] [Google Scholar]
- Zuo, G.‐Y. , Li, Z.‐Q. , Chen, L.‐R. , & Xu, X.‐J. (2005). In vitro anti‐HCV activities of Saxifraga melanocentra and its related polyphenolic compounds. Antiviral Chemistry and Chemotherapy, 16(6), 393–398. 10.1177/095632020501600606 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


