Abstract
An infusion prepared from the aerial parts of Salvia amarissima Ortega inhibited the enzyme protein tyrosine phosphatase 1B (PTP-1B) (IC50~88 and 33 μg/mL, respectively). Phytochemical analysis of the infusion yielded amarisolide (1), 5,6,4′-trihydroxy-7,3′-dimethoxyflavone (2), 6-hydroxyluteolin (3), rutin (4), rosmarinic acid (5), isoquercitrin (6), pedalitin (7) and a new neo-clerodane type diterpenoid glucoside, named amarisolide G (8a,b). Compound 8a,b is a new natural product, and 2–6 are reported for the first time for the species. All compounds were tested for their inhibitory activity against PTP-1B; their IC50 values ranged from 62.0 to 514.2 μM. The activity was compared to that of ursolic acid (IC50 = 29.14 μM). The most active compound was pedalitin (7). Docking analysis predicted that compound 7 has higher affinity for the allosteric site of the enzyme. Gas chromatography coupled to mass spectrometry analyses of the essential oils prepared from dried and fresh materials revealed that germacrene D (15) and β-selinene (16), followed by β-caryophyllene (13) and spathulenol (17) were their major components. An ultra-high performance liquid chromatography coupled to mass spectrometry method was developed and validated to quantify amarisolide (1) in the ethyl acetate soluble fraction of the infusion of S. amarissima.
Keywords: Salvia amarissima, PTP-1B activity, amarisolide G, diabetes
1. Introduction
Type 2 diabetes mellitus is a metabolic disease characterized by chronic hyperglycemia due to insulin resistance, or the relative absence of the hormone. The prevalence of the disease is continuously increasing, with approximately 463 million people living with diabetes nowadays. Mexico is one of the countries more affected by type 2 diabetes mellitus, with more than 12 million cases. The population sector more affected is the indigenous people owing to variations in its traditional way of life and the effects of industrial developments [1]. Mexican population employs more than 300 plant species to treat the symptoms of diabetes; in some cases, the patients combine allopathic therapies with the botanical remedies [2]. These plants are an essential part of the country’s alternative medical care, and the best testimony of their efficacy is their persistence in Mexican markets and other places for crude or fresh drug selling. Therefore, it is crucial to analyze these plants to establish their composition, security, efficacy, and to develop a suitable methodology for quality control of the crude drug following good practice guidelines.
Quality control of herbal drugs is the base for their efficacy and safety. Quality control of herbal drugs aims to define their identity, purity and content of active principles or marker compounds. The chemical composition of plants, and hence of their therapeutic preparations, is variable, so standardization is necessary to guarantee comparable therapeutic effects. To prove the constant composition of herbal preparations, there are appropriate standard analytical methods to establish relevant criteria for uniformity. Standard analytical techniques include, among others, high-performance liquid chromatography. For many years, the World Health Organization (WHO) has encouraged all its country members to elaborate pharmacopeic monographs providing comprehensive scientific information on the quality of their most commonly used medicinal plants. Following WHO guidelines, Mexico has developed the Mexican Herbal Pharmacopeia, which contained monographs with definitions, analytical techniques for identity and composition, as well as storage regulations of the most widely used Mexican Herbal drugs [2,3].
Like other pharmaceutical products, herbal drugs should fulfill the basic requirements of being efficacious and safe. To establish herbal drugs’ efficacy and safety is necessary to perform preclinical and clinical assays, including those of the healer or medical doctor in rural communities who apply locally produced herbal preparations. When assessing the efficacy of the plants, it is essential to study both the traditional preparations and their components; this is because sometimes the efficacy is attained with the combinations of compounds in the preparations, which may be acting through synergy, network pharmacology or by targeting several nonrelated proteins involved in the pathology of a disease. Altogether, these studies can lead to the development of standardized phytomedicines of good quality and discover good drug candidates or molecules useful for lead optimization or even fragment-based drug discovery [3].
Among the species highly valued in Mexico for treating diabetes is Salvia amarissima Ortega (syn. Salvia circinata Cav.) belonging to the mint family. It is a perennial aromatic shrub native to Mexico, listed as medicinal in the catalog of plants from the Royal Botanical Expeditions to New Spain. Like many other New World Salvia species, S. amarissima is melittophilous (bee-pollinated). A tea brewed from dried aerial parts of the plant is useful in Mexican folk medicine for treating diabetes, ulcers and helminthiases [4,5]. The species is commonly regarded as “insulina” (insulin), referring to its efficacy to improve the diabetic condition [5]. Previous phytochemical studies allowed the isolation of some neo-clerodane diterpenoids, including amarisolide (1) [6], a few seco-clerodane diterpenoids [6,7,8,9,10] and some flavonoids [6,8]. The traditional preparation of the plant collected in Puebla, Mexico, as well as amarisolide (1), and some of the flavonoids showed inhibitory activity against mammal α-glucosidases in vitro and in vivo. The preparation and compounds were hypoglycemic and reduced the postprandial peak significantly during an oral sucrose tolerance test in healthy mice [8]. Some of the seco-clerodane diterpenoids were cytotoxic against a few human cancer cell lines, had modulatory activity in a breast cancer cell line resistant to vinblastine and exhibited antiprotozoal action [7,9,10,11]. Furthermore, the traditional preparation lack toxicity when tested according to the Lorke criteria. [8]. More recently, the antinociceptive properties of an aqueous extract of the plant, 1 and 7 were demonstrated [12].
Based on the above considerations, this investigation aimed: (i) to determine the effect of the traditional preparation (infusion) of the plant collected in Oaxaca and their components on the activity of the protein tyrosine phosphatase (PTP-1B) in order to assess a new molecular target, and get a better insight in the in vivo hypoglycemic effect previously demonstrated [8]. This target was chosen, considering that this enzyme acts as a negative regulator of insulin and leptin dependent signal cascades holding therapeutic utility in type 2 diabetes mellitus and obesity [13]. (ii) To analyze the chemical composition of the essential oil. (iii) To set up an appropriate procedure using Ultra-High-Performance Liquid Chromatography (UHPLC) to quantify one active or marker compound of the plant’s infusion. The chemical composition of the plant’s essential oil and the UHPLC procedure will allow developing a pharmacopeic monograph of S. amarissima, as they represent valuable identity and composition tests, respectively.
2. Results and Discussion
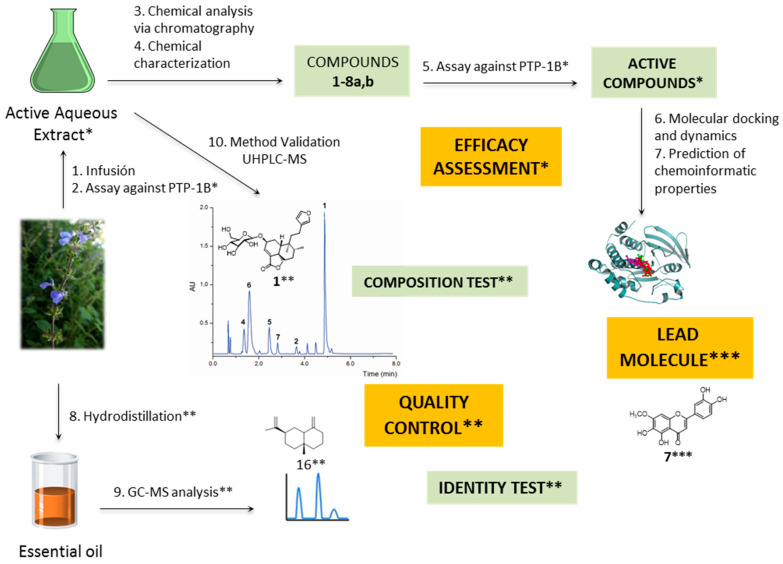
Scheme 1 summarizes the whole work.
Scheme 1.
Workflow diagram of this study. * Efficacy studies. ** Quality control analyses. *** Discovery of a lead molecule.
2.1. Chemical Constituents of the Aqueous Extract
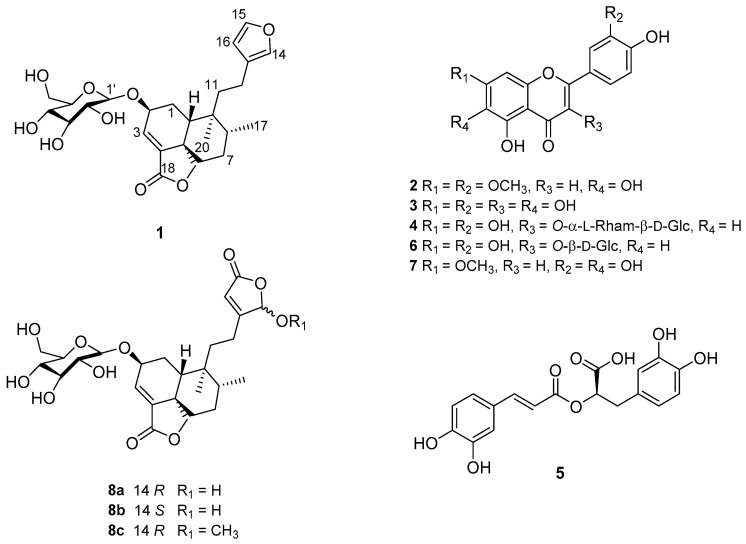
An aqueous extract (AE) from S. amarissima inhibited the PTP-1B activity significantly, with an IC50 value of 88.6 ± 5.4 μg/mL. Workup of AE by solvent partitioning and repeated chromatography afforded eight compounds (Figure 1), namely amarisolide (1), 5,6,4′-trihydroxy-7,3′-dimethoxyflavone (2), 6-hydroxyluteolin (3), rutin (4), rosmarinic acid (5), isoquercitrin (6), pedalitin (7) and a neo-clerodane type diterpenoid glycoside, named amarisolide G (8a,b). Compound 8a,b, is a new natural product characterized by conventional spectroscopic and spectrometric techniques. The known compounds were identified by comparing their spectroscopic data with those previously described (Figures S1–S11, Supplementary Material) [8,14,15]. Compounds 2–6 are reported for the first time for this species. In addition, thin layer chromatographic (TLC) analysis revealed that these compounds were present in the AE prepared from the fresh material.
Figure 1.
Structures of compounds (1–8a,b) from S. amarissima.
Product 8a,b was obtained as a white solid optically active. DART-HRMS (Direct Analysis In Real Time-High Resolution Mass Spectrometry) established its molecular formula as C26H36O11. The IR spectrum included bands for hydroxyl (3365 cm−1) and α,β-unsaturated-γ-lactone (1749 cm−1) functionalities (Figure S12, Supplementary Material) [8]. The NMR spectra of 8a,b (Table 1; Figures S13–S18, Supplementary Material) were closely similar to those of amarisolide D (8c), a neo-clerodane type of diterpenoid with an α,β-unsaturated-γ-lactone at C-4/C-5, a five-membered keto-γ-lactol methyl ether and a β-D-glucopyranosyloxy moiety at C-2 [8]. Thus, in compound 8a,b the signals for the five-membered keto-γ-lactol methyl ether were replaced by those of a keto-γ-lactol moiety. Accordingly, the NMR spectra showed signals at δH 5.90, 6.05 (brs, H-14)/δC 99.4, 99.9 (C-14), δH 5.96 (brs, H-16)/δC 116.1 (C-16), δC 171.4 (C-13) and δC 171.9 (C-15) (Table 1). Since the resonances for H-14 appeared as two separate signals, the intensity of each corresponding to one-half proton, compound 8a,b is a 1:1 mixture of C-14 epimers. The doubling of C-14 signal in the 13C-NMR spectra provided further evidenced (Table 1). The 13C-NMR chemical shifts (Table 1) of C-11-C-16 and the key HMBC (Heteronuclear Multiple Bond Correlation) correlations of H-12 and H-14 established the point of attachment of the ethyl fragment (C-11-C-12) to the keto−γ-lactol ring. The most relevant HMBC correlations were H-12a (δH 1.72) with C-14 (δC 99.4 and 99.9); H-12b (δH 1.56) with C-14 (δC 99.4 and 99.9); and H-14 (δH 5.90 and 6.05) with C-12 (δC 33.8). The NOESY (Nuclear Overhauser Effect Spectroscopy) interactions revealed that the relative configuration at the stereogenic centers of 8a,b was identical to that of amarisolides A–D [8]. The electronic circular dichroism spectrum of 8a,b showed negative Cotton effects at ~212 and ~250 nm due to the electronic transitions π→π* and n→π*, respectively, of the α,β-unsaturated-γ-lactone. The latter data indicated that the absolute configuration at the stereogenic centers C-2, C-5, C-8, C-9 and C-10 of compound 8a,b was S, S, R, R and R, respectively. The D configuration of the β-glucopyranosyloxy moiety was established as previously described [8]. On the basis of these evidences, compound 8a,b was characterized as (2S,5S,8R,9R,10R,14R,S)-2-(O-β-d-glucopyranosyl)-neo-clerodan-14-hydroxy-3, 13-diene-14,15;18,19-diolide (8a,b) and was designated with the trivial name of amarisolide G.
Table 1.
1H and 13C NMR spectroscopic data for compound 8a,b in DMSO-d6.
| Position | δH a (J/Hz) | δC b |
|---|---|---|
| 1 | α: 1.35 dd (13.6, 3.2) | 26.3 |
| β: 1.81 brd (13.7) | ||
| 2 | 4.44–4.49 m | 70.0 |
| 3 | 6.66 d (6.4) | 131.1 |
| 4 | 143.0 | |
| 5 | 45.4 | |
| 6 | α: 1.72 td (12.7,3.3) | 33.8 |
| β: 1.22–1.26 m | ||
| 7 | α: 1.53–1.58 m β: 1.65−1.70 m |
27.6 |
| 8 | 1.65–1.70 m | 36.2 |
| 9 | 37.8 | |
| 10 | 2.21 d (15.1) | 40.5 |
| 11 | 1.59–1.64 m | 27.6 |
| 12 | α: 1.71–1.74 m | 33.8 |
| β: 1.53–1.64 m | ||
| 13 | 171.4 | |
| 14 | α: 5.90 brs β: 6.05 brs |
99.4 99.9 |
| 15 | 171.9 | |
| 16 | 5.96 brs | 116.1 |
| 17 | 0.80 d (6.5) | 15.9 |
| 18 | 169.2 | |
| 19a | 4.39 d (8.2) | 70.9 |
| 19b | 4.02 d (8.2) | |
| 20 | 0.57 s | 17.7 |
| 1′ | 4.31 d (7.7) | 102.8 |
| 2′ | 2.96–3.05 m | 74.1 |
| 3′ | 3.13–3.16 m | 77.2 |
| 4′ | 2.96–3.05 m | 70.7 |
| 5′ | 3.13–3.16 m | 77.4 |
| 6′ | α: 3.43–3.47 m β: 3.63–3.69 m |
61.6 |
| 14-OH | 7.75 brs | |
| 2′-OH | 4.92 brs | |
| 3′-OH | 4.92 brs | |
| 4′-OH | 4.92 brs | |
| 6′-OH | 3.33 s |
Assignments based on the analysis of the HSQC (Heteronuclear single quantum coherence spectroscopy) and HMBC experiments; J values (Hz) in parentheses. a 700 MHz. b 175 MHz.
2.2. Chemical Constituents of the Essential Oil
Dried and fresh plant materials were analyzed to assess any change during the drying process. Since it is an aromatic plant, the chemical profile of the essential oil is valuable as an identity test. The essential oil content of distilled aerial parts was 0.02% and 0.03% for fresh and dried material, respectively. In each case, eight major compounds were identified, representing 99.96 and 99.97% of the composition of the essential oil, respectively. As observed in Table 2 and Figures S19 and S20 (Supplementary Material), the major components in both samples were germacrene D (15) and β-selinene (16), followed by β-caryophyllene (13) and spathulenol (17). The only mutually exclusive constituents were 3-methoxy-p-cymene (9) present in the dried material, and δ-elemene (11) found only in the fresh plant. These differences are not due to seasonal changes because the plant material was from the same batch.
Table 2.
GC-MS identified compounds from the essential oils of S. amarissima.
| Compound | CAS Number | Peak Area (%) | ||
|---|---|---|---|---|
| RI [a] | Sa-Batch 1 | Sa-Batch 2 | ||
| 3-Methoxy-p-cymene (9) | 1076-56-8 | 1219 | 4.40 | |
| (E)-Pinocarvyl acetate (10) | 1686-15-3 | 1313 | 5.98 | 2.10 |
| δ-Elemene (11) | 20307-84-0 | 1329 | 1.99 | |
| α-Bourbonene (12) | 5208-58-2 | 1378 | 4.24 | 4.11 |
| β-Caryophyllene (13) | 87-44-5 | 1413 | 15.05 | 21.27 |
| α-Caryophyllene (14) | 1139-30-6 | 1447 | 7.68 | 4.21 |
| Germacrene D (15) | 37839-63-7 | 1476 | 25.09 | 24.26 |
| β-Selinene (16) | 17066-67-0 | 1491 | 28.35 | 30.16 |
| Spathulenol (17) | 6750-60-3 | 1576 | 11.59 | 9.45 |
| Total peak area (%) | 99.97 | 99.96 | ||
[a] RI retention indices relative to series of n-alkanes (C8-C20) on a DB-5 column.
It is worth mentioning that the oils from other Salvia species analyzed also contains sesquiterpenes. In this context, Salvia ceratophylla, S. aethiopis L., S. palaestina Bentham and S. xanthocheila Boiss. ex Benth., are rich sources of germacrene D (15) [16]; β-caryophyllene (13) is the major component of S. nemorosa L., S. verticillata L., S. virgata Ortega and S. hydrangea DC. ex Benth. Finally, germacrene B was the major compound of S. syriaca L. [17,18].
2.3. Evaluation of Compounds 1–8a,b on the PTP-1B Inhibitory Activity
One of the major causes of type 2 diabetes mellitus is insulin resistance, which occurs when the hormone cannot activate signaling pathways in major metabolic tissues (muscles, fat and liver). Insulin resistance involves several inhibitory molecules that interfere with tyrosine phosphorylation of the insulin receptor. Among those, PTP-1B is a promising therapeutic target since it inactivates crucial signaling effectors in the insulin- and leptin-signaling cascades by dephosphorylating their tyrosine residues [13]. Therefore, natural products 1–8a,b were tested against PTP-1B (Table 3). Among the flavonoids tested, the most active component was pedalitin (7) with an IC50 of 62.0 ± 4.1 μM (19.0 μg/mL), whereas of the diterpenoids was amarisolide (1) (279.9 ± 26.0 μM; 137.9 μg/mL). In both cases, the inhibitory effect was concentration-dependent. The inhibitory activity against PTP-1B reported for compounds 4 and 6 correlated well with that found in this work [19,20]. On the other hand, rosmarinic acid (5) is an ingredient of an active extract of Rosmarinus officinalis against PTP-1B [21]. However, in this study we report for the first time the effect of the pure 5. The activity of the isolates 1–8a,b was compared to that of ursolic acid (UA, IC50 = 28.1 ± 1.2 μM), which in other studies displayed lower IC50 values (~3.1 μM) [22].
Table 3.
Protein tyrosine phosphatase 1B (PTP-1B) inhibitory activity of compounds (1–8a,b) from S. amarissima expressed as IC50.
| μg/mL ± SD | μM ± SD | |
|---|---|---|
| 1 | 137.9 ± 12.8 | 279.9 ± 26.0 |
| 2 | 62.5 ± 2.3 | 189.4 ± 7.0 |
| 3 | 24.2 ± 0.7 | 80.1 ± 2.2 |
| 4 | 197.3 ± 10.1 | 323.4 ± 16.6 |
| 5 | 49.4 ± 6.7 | 137.1 ± 18.7 |
| 6 | 120.7 ± 14.7 | 259.9 ± 31.7 |
| 7 | 19.6 ± 1.3 | 62.0 ± 4.1 |
| 8a,b | 269.7 ± 3.6 | 514.2 ± 6.8 |
| UA [a] | - | 28.1 ± 1.2 |
[a] Positive control. Values are representative of three independent determinations.
The results of the PTP-1B are significant. They suggest that the traditional preparation of S. amarissima, with hypoglycemic and antihyperglycemic properties demonstrated in vivo [8] (i.e., the overall action), contains compounds such as 1–8a,b that might weakly target different proteins (i.e., PTP-1B and others) within the same signaling network thus shutting insulin signaling cascade process by network pharmacology. It is also possible that compounds 1–6 and 8a,b, with weaker activity than compound 7, altogether put forth a biochemical effect by synergism (i.e., a synergy between weakly active compounds against PTP-1B). Finally, molecules like compounds 1, 4, 5 and 7 can exert their action binding different targets such as PTP-1B and α-glycosidases, among others (polypharmacology). The fact that rutin (4) [23] and rosmarinic acid (5) [21] are multitarget antidiabetic compounds, and compounds 1 and 7 inhibited α-glycosidases in vivo [8] strengthen any of these possibilities.
2.4. Docking Study
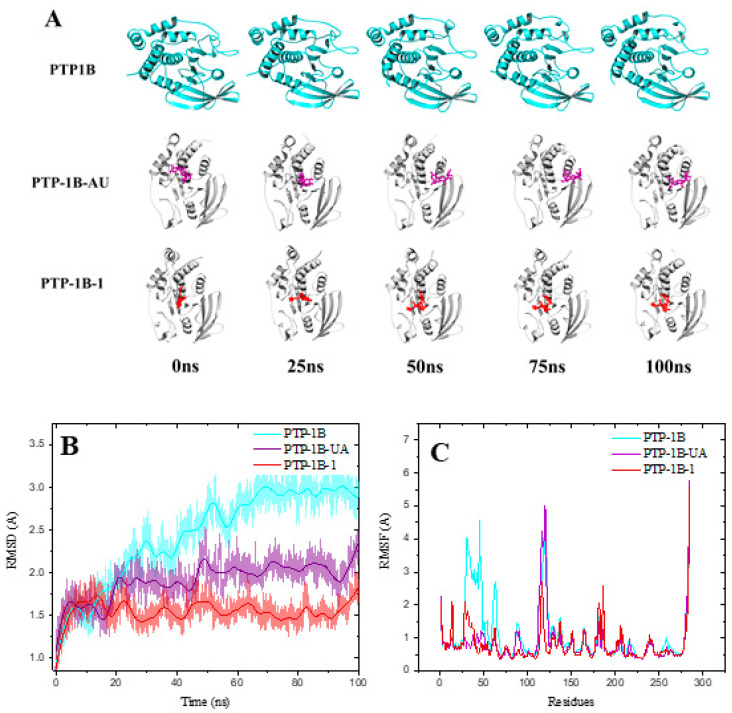
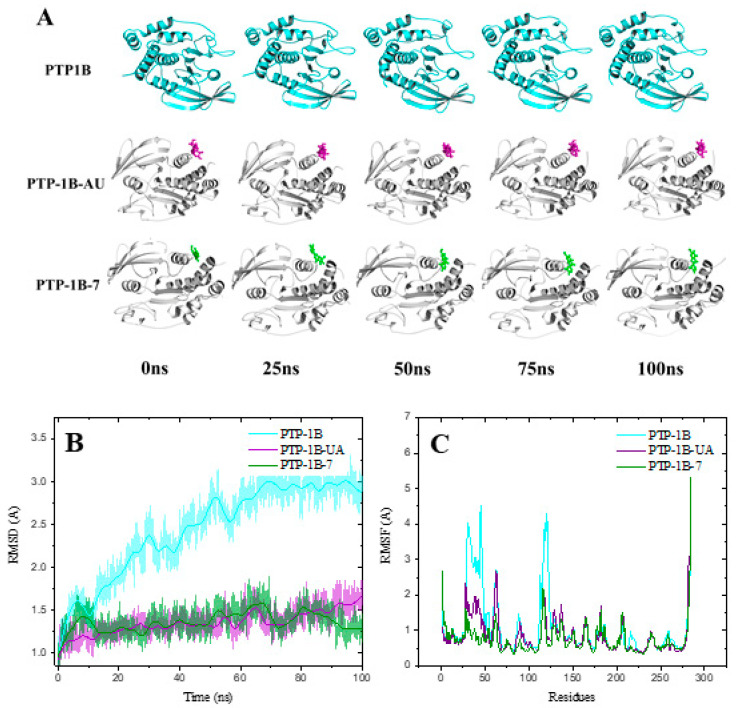
To predict the preferred binding orientation of compounds 1, UA and 7 into PTP-1B, we performed a docking analysis. Compound 1 was not among the most active compounds but included for comparative purposes. These substances 1, 7 and UA were docked with the co-crystallized structure of PTP-1B at the catalytic (PDB ID: 1G7F) and allosteric (PDB ID: 1T49) sites. The results in Figure 2 and Table 4 indicate that all ligands bind at the same site as the co-crystallized ligands 892 (3-(3,5-dibromo-4-hydroxy-benzoyl)-2-ethyl-benzofuran-6-sulfonic acid (4-sulfamoyl-phenyl)-amide) and INZ (2-{4-[(2s)-2-[({[(1s)-1-carboxy-2-phenylethyl]amino}carbonyl)amino]-3-oxo-3-(pentylamino)propyl]phenoxy}malonic acid) [24,25]. The estimated energy binding is different for each compound. Compound 1 has a higher affinity for the catalytic site, which is more and conserved site, while compound 7 and UA targeted its more hydrophobic and less conserved allosteric site. The RMSD values for ligands 1 and 7 are in the range of 2 to 3 Å, which indicates that the ligands do not precisely overlap but maintain the correct orientation [26,27]. The amino acids interacting with compounds 1 and 7, as well as AU are similar to those previously reported for other inhibitors (Table 4; Figures S30 and S31 of the Supplementary Material) [24,25]. At the allosteric site, compound 7 has hydrophobic interactions with Ala189, Leu192, Phe280 and Phe196 while AU with Leu192, Phe280 and Phe196. Therefore, pedalitin (7) behaves as UA regarding its higher affinity for the allosteric site. For UA, the key structural feature is the pentacyclic core displaying a nonpolar characteristic, which interacts with nonpolar residues in the allosteric site [28]. For compound 7, apparently beside the tricyclic structure, the lack of oxygen at C-3 of the flavonoid core seems to be essential. It will be necessary to pursue a kinetic analysis to determine if experimentally compound 7 is an allosteric inhibitor of PTP-1B. It is essential to mention, that the less-conserved PTP-1B allosteric site is an ideal target for a compound to inhibit PTP-1B activity because the problems associated with inhibition at catalytic site will disappear. Thus, this study may provide an important lead among flavones for the development of novel PTP-1B allosteric inhibitors.
Figure 2.
Structural models of the binding sites of the PTP-1B ligand complexes. In cyan cartoons (PTP-1B protein), red sticks (1), green sticks (7) purple sticks (UA) at the catalytic (A) and allosteric sites (B). Images created with Pymol.
Table 4.
Results of the dockings analyses.
| Catalytic Site | Allosteric Site | |||||||
|---|---|---|---|---|---|---|---|---|
|
Ki (μM) 1 |
BE (kcal/mol) |
RMSD (Å) 3 | Residues 2 |
Ki (μM) 1 |
BE (kcal/mol) | RMSD (Å) 3 | Residues 2 | |
| 1 | 0.55 | −8.5 | 2.89 | Tyr46, Asp48, Val49, Phe182, Gly183, Cys215, Ser216, Ala217, Gly220, Arg221, Gln262, Thr263 and Gln266 | 8.39 | −6.8 | 2.15 | Phe196, Lys197, Arg199, Glu200, Gly277, Phe280 and Ile281 |
| 7 | 9.94 | −6.8 | 2.73 | Tyr46, Asp48, Val49, Trp179, Asp181, Phe182, Gly183, Ala217, Ile219, Arg221, Gln262 and Gln 266 | 2.16 | −7.7 | 2.27 | Ala189, Leu192, Glu276, Gly277, Phe280, Phe196, Lys197 and Glu200 |
| UA | 2.56 | −7.6 | 1.35 | Phe182, Gly183, Cys215, Ala217, Gly218, Ile219, Gly220, Arg221, Gln262, Thr263 and Gln266 | 0.78 | −8.3 | 1.73 | Asn193, Phe196, Asp263, Phe280, Leu192 and Ile281 |
1Ki values were calculated using the equation ΔG = RT lnKi [26] 2 Residues interacting 4 Å of the compound; 3 RMSD were calculated from the co-crystallized ligands for UA; and for 1 and 7 from UA. BE, binding energy.
2.5. Molecular Dynamics of PTP-1B-Compounds 1, 7 and UA Complexes
Molecular dynamics (MD) studies of the complexes PTP-1B-compounds 1, 7 and UA were carried out to evaluate the stability of the docked complexes illustrate in Figure 3. Table 5 shows the theoretical parameters calculated from 100 ns of MD. All PTP-1B-compound complexes show negative ∆G (affinity parameter) consistent with their stability. Compound 1 has a ∆G similar to UA at the catalytic site, while compound 7 has a lower ∆G than AU at the same site. Figure 3 and Figure 4 show the structural models, RMSD and Root Mean Square Fluctuations (RMSF) of the molecular trajectories for compounds 1 and 7, respectively. The RMSD of the complexes is lower with respect to PTP-1B in both the catalytic and allosteric MDs, which indicates the conformational stability of the complexes. In the RMSF analysis, it is observed an area between residues 27–50 (corresponding to a loop), which is stabilized with the ligands. The data obtained with the MDs are complementary and conscious with the docking data and experimental results.
Figure 3.
Molecular dynamics at the catalytic site of PTP-1B for compound 1. (A) Structural models, (B) RMSD and (C) Root Mean Square Fluctuations (RMSF) from molecular trajectories. AU = UA.
Table 5.
Calculations of the theoretical energy components from the trajectories of molecular dynamics.
| Complex. | ∆Evdw | ∆Eele | ∆GGB | ∆GNP | ∆Gsolv | ∆Gbind |
|---|---|---|---|---|---|---|
| PTP-1B-UA 1 | −21.88 ± 0.44 | −20.02 ± 1.08 | 27.60 ± 1.63 | −41.91 ± 1.26 | 25.39 ± 1.04 | −16.51 ± 0.36 |
| PTP-1B-UA 2 | −28.67 ± 0.23 | −1.21 ± 0.46 | 10.89 ± 0.42 | −27.78 ± 0.48 | 7.73 ± 4.21 | −20.05 ± 0.21 |
| PTP-1B-1 | −36.22 ± 0.47 | −18.53 ± 0.77 | 41.62 ± 0.55 | −54.76 ± 1.00 | 36.84 ± 0.53 | −17.92 ± 0.61 |
| PTP-1B-7 | −35.95 ± 0.45 | −4.27 ± 0.31 | 12.90 ± 0.33 | −40.22 ± 0.55 | 9.06 ± 0.33 | −31.16 ± 0.46 |
1 Catalytic site; 2 Allosteric site; ∆Evdw, contributions by van der Waals interactions; ∆Eele, electrostatic energy; ∆GGB, polar solvation energy; ∆Gnp, nonpolar solvation energy; ∆Gsolv, desolvation free energy (∆Gsolv = ∆GGB + ∆Gnonpol); ∆Gbind, binding affinity.
Figure 4.
Molecular dynamics at the allosteric site of PTP-1B for compound 7. (A) Structural models, (B) RMSD and (C) RMSF from molecular trajectories.
2.6. Drug Likeness for Compounds 1, 7 and UA
According to the SwissTargetPrediction (http://www.swisstargetprediction.ch/index.php), and Molinspiration (http://www.molinspiration.com/cgi-bin/properties) databases which predict the most probable targets of small bioactive molecules, compounds 1, 7 and UA 1, 7 could target any protein with percentages of 26.7, 40 and 40 %, respectively. SwissTargetPrediction predicted PTP-1B inhibition for these compounds with probabilities of 0, 0.1266 and 0.95, respectively. According to these predictions, UA should have been 7.5 times more active than compound 7; however, experimentally, UA was only 2.2 times more than compound 7 (Figures S31–S34).
Next, using the Osiris Property Explorer server (http://cheminformatics.ch/propertyExplorer), relevant properties for compounds 1, 7 and UA were calculated and summarized in Table 6. These properties indicate whether a molecule is a potential drug. The logP value is a measure of a compound’s hydrophilicity. Low hydrophilicity and, therefore, high logP values cause poor absorption. For a compound being well absorbed, its logP value must not be greater than 5.0; logP values between 1.35 and 1.8 indicate perfect oral and intestinal absorption. Thus, compounds 1 and 7 could have proper absorption, but not UA.
Table 6.
Physicochemical properties of compounds 1, 7 and UA.
| 1 | 7 | UA | |
|---|---|---|---|
| LogP | 1.01 | 1.92 | 6 |
| Solubility (LogS) | −3.48 | −2.58 | −6.11 |
| Molecular weight | 492.56 | 316.26 | 456.71 |
| Druglikeness | −2.96 | 1.8 | −3.66 |
| H bond acceptor | 9 | 7 | 3 |
| H bond donor | 4 | 4 | 2 |
| Nb stereocenters | 10 | 0 | 10 |
| Nb rotable bonds | 6 | 2 | 1 |
| Drug score | 0.40 | 0.52 | 0.26 |
The data was calculated using the OSIRIS Property Explorer server (http://www.cheminfo.org/Chemistry/Cheminformatics/Property_explorer/index.html).
The aqueous solubility of a compound influences its absorption and distribution characteristics; a low solubility goes along with inadequate absorption. More than 80% of the drugs on the market have a logS value higher than −4. Compounds 1 and UA present −3.48 and −6.11, respectively. The drug-likeness parameter is a complex balance of various molecular properties and structural features that determine whether a molecule is similar to the known drugs. These properties, mainly hydrophobicity, electronic distribution, hydrogen bonding characteristics, molecule size and flexibility, and of course presence of various pharmacophoric features influence the behavior of molecule in a living organism, including bioavailability, transport properties, affinity to proteins, reactivity, toxicity, metabolic stability and many others. A positive value indicates that a molecule contains predominantly fragments, which are frequently present in commercial drugs [29]; compound 7 has a drug-likeness value of 1.8. The H bond acceptor and H bond donor’s parameters indicate a molecule’s ability to interact to a greater or lesser degree with a protein; compounds 1 and 7 present a higher number of possible hydrogen bridges than UA. Finally, the drug-score is an indicator that qualifies the potential of a compound for being a drug based on all the calculated parameters; compound 7 has the best drug-score (0.52), which is in harmony with the experimental and theoretical data.
2.7. Development and Validation of an UHPLC-MS Method for Quantifying 1
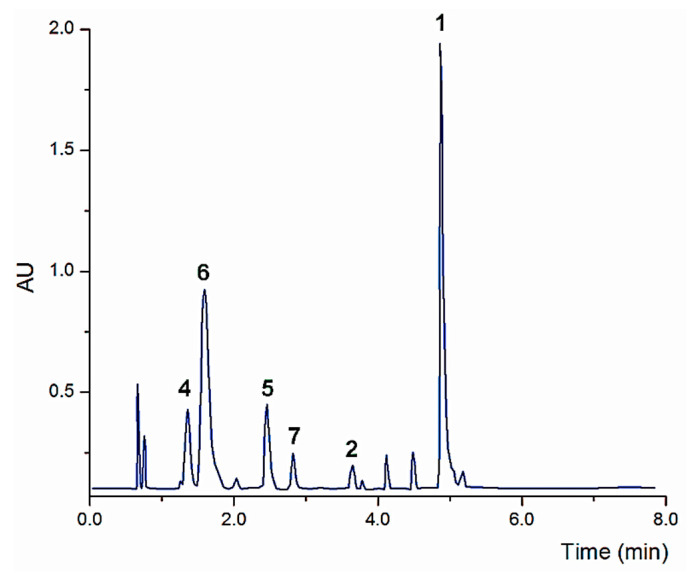
Initial assessments about the complexity of samples of AE were based on visual comparisons of their chromatographic profiles. The ethyl acetate soluble fraction of AE yielded the best profile. Chromatographic separation was performed on Acquity UHPLC® BEH Shield C18 column (2.1 × 100 mm, 1.7 μm) applying a binary gradient elution of water (0.1% formic acid) and MeCN. The total run time was 10 min. As illustrated in Figure 5 compounds 1, 2 and 4–7 are present in the chromatogram. All compounds were identified by their retention times and m/z values corresponding to [M − H]− ions. Compounds 1, 2 and 4–7 showed an effective baseline resolution. The pseudo molecular ions of these compounds appeared at m/z 609.54 [M − H]− (4; RT 1.36 min), 463.46 [M − H]− (6; RT 1.60 min), 359.23 [M − H]− (5; RT 2.46 min), 315.48 [M − H]− (7; RT 2.82 min), 329.70 [M − H]− (2; RT 3.65 min) and 491.23 [M − H]− (1; RT 4.89 min). The main component of the ethyl acetate fraction was amarisolide (1), then selected as a marker for validation. So far amarisolide (1) has been only isolated from this species, which makes it an excellent marker compound for quality control. It is worth mentioning that the neo-clerodanes type compounds detected in the infusion were 1 and 8a,b, but not the minor diterpenoids we previously isolated from the organic extract of the plant [8].
Figure 5.
Representative LC chromatogram of the ethyl acetate soluble fraction prepared from the infusion of S. amarissima (Detection: 270 nm). Peak identification: 4: RT 1.36 min; 6: RT 1.60 min; 5: RT 2.46 min; 7: RT 2.82 min; 2: RT 3.65 min; 1: RT 4.89 min.
The analytical method was validated in terms of precision, accuracy, linearity and recovery according to the Q2 (R1) guideline published by the International Conference on Harmonisation (ICH) [30]. The linearity of the system was tested using a concentration range of 1 between 5 to 100 μg/mL and was found to be linear (R2 = 0.9994 (UV) and 0.9921 (Electrospray Ionization Mass Spectrometry, ESI-MS) in the concentration range used. The CV was less than 0.13% at each concentration level analyzed. The limit of identification (LOD) and quantification (LOQ) values were 1.22 and 3.70 μg/mL, respectively for UV detection; and 0.60 and 1.82 μg/mL, respectively for ESI-MS detection.
The linearity of the method was tested by recovery assay; the linear regression equation were found to be y = 5636.35x − 4337.55 (UV detection) and y = 5252.80x + 64,101.76 (ESI-MS detection). The recovery ranges for the standard were expressed as the concentration detected as a percentage of the expected concentration and were found in the ranges of 100.7–101.7 % for UV detection and 83.7–95.7% for ESI-MS detection. The reproducibility and repeatability of the analytical method were evaluated in terms of the intermediate precision by analyzing 6 replicates of the stock solution (50 μg/mL) in two different days. The relative SD (RSD; n = 6) was calculated for each sample evaluated. The results indicated that their chromatographic patterns were similar showing the presence of amarisolide (1). The CV values for accuracy were less than 0.11%. Subsequently, compound 1 was quantified, and the mean concentration calculated was 116 mg/g in dry matter.
3. Materials and Methods
3.1. General Procedures
IR spectra were recorded using a Bruker Tensor 27 FT-IR spectrophotometer (Bruker Corp., Billerica, MA, USA). Optical rotations were recorded at the sodium d-line wavelength using a Perkin Elmer model 343 polarimeter at 20 °C (Perkin Elmer, MA, USA). NMR spectra were registered on a Bruker AVANCE III HD with TCI CryoProbe 700 H-C spectrometer at 700 MHz (1H) or 175 MHz (13C), using TMS as an internal standard (Bruker Corp., Billerica, MA, USA). DARTHRMS were acquired with a JEOL AccuTOF-DART JMS-T100LC (JEOL Ltd., Tokyo, Japan) spectrometer in positive mode. For GC-MS analyses, an Agilent 6890N series gas chromatograph coupled to a LECO (Laboratory Equipment Corporation) time-of-flight mass spectrometer detector (MS-TOF; Agilent Technology, Santa Clara, CA, USA) was used. UHPLC-MS analyses were performed on a Waters Acquity UHPLC-H® Class system (Waters, Darmstadt, Germany) equipped with a quaternary pump, sample manager, column oven and photodiode array detector (PDA) interfaced with an SQD2 single mass spectrometer detector with an electrospray ion source. Column chromatography (CC) was carried out on Sephadex LH-20 (GE Healthcare, IL, USA). Thin layer chromatographic (TLC) analyses were performed on silica gel 60 F254 plates (Merck, Darmstadt, Germany), or C18-silica gel matrix plates Analtech plates (Merck, Darmstadt, Germany), visualization of the plates was carried out using an (NH4)4Ce(SO4)4 (10%) solution in H2SO4. Reagent-grade EtOAc, CHCl3, CH2Cl2 and MeOH were purchased from J.T. Baker (Avantor Performance Materials, PA, USA). MeCN, MeOH and water LC-MS or HPLC grades were purchased from Honeywell Burdick & Jackson (Morristown, NJ, USA). All other analytical grade solvents and reagents were obtained from various commercial sources. Amarisolide (1) was isolated from the species S. amarissima in the present study. The purity was determined to be more than 98% by UHPLC-MS.
3.2. Plant Material
Salvia amarissima was collected in Capulálpam de Méndez, Ixtlán de Juárez, Oaxaca, in January 2019 (Sa-Batch 1 (fresh) and Sa-Batch 2 (air-dried)). A voucher specimen (Number 1502277) was deposited at the National Herbarium of Mexico (MEXU), Instituto de Biología, UNAM. R. Torres-Colin achieved the botanical identification of the plant. The plant was air-dried and ground to a fine powder (2 mm) in a Thomas Wiley Model 4 Mill.
3.3. Extracts and Essential Oils Preparation
AE from S. amarissima (dried aerial parts) was prepared with 250 mL of boiling water and 12.5 g of the crude drug for 30 min. After filtration, the aqueous extract was concentrated in vacuo to obtain 0.1 g of a green residue. This process was repeated as necessary to prepare 10 g of AE. The ethyl acetate soluble fraction was prepared via partitioning with EtOAc (3 × 250 mL) from the aqueous extract. The resulting organic phase was dried over anhydrous sodium sulfate and concentrated in vacuo to yield 130 mg of a brown residue (yield 1.0%).
EOs were prepared from fresh (Sa-batch 1) and air-dried (Sa-batch 2) plant material (200 g in 1.5 L of distilled water) by hydrodistillation in a modified Clevenger type apparatus for 3 h. In both cases, the hydrodistilled was extracted with CH2Cl2 (3 × 2 L). The resulting organic phases were dried over Na2SO4 and concentrated in vacuo to yield an oily yellow residue (0.040 g, yield 0.02% in the case of the fresh material, and 0.063 g, yield 0.03% for the dried plant). All samples were stored at −4 °C until chemical analysis.
3.4. Separation of Active Compounds from the Ethyl Acetate Soluble Fraction
The ethyl acetate soluble fraction (100 mg) was subjected to CC on Sephadex LH-20 (400 g) using MeOH as eluent; fractions were pooled into 20 secondary fractions (F1–F20) according to their TLC profiles. From fraction F16 (73 mg) crystallized 60 mg of amarisolide (1). Preparative RP-TLC of fraction F20 (10 mg) yielded 1.2 mg of 5,6,4′-trihydroxy-7,3′-dimethoxyflavone (2) and 4.2 mg of 6-hydroxyluteolin (3).
3.5. Separation of Active Compounds from AE
AE (4.3 g) was fractionated via CC on Sephadex LH-20 using a gradient system of methanol–water (water 40–100%); this process gave 12 secondary fractions (AE1–AE12). From fraction AE9 (35 mg) crystallized 30 mg of rutin (4; m.p. 241–242 °C). From fraction AE11 (15 mg) crystallized 5 mg of isoquercitrin (6). Fraction AE6 (220 mg) was further purified on a Sephadex CC, eluting with MeOH, to yield 16 mg of rosmarinic acid (5). Preparative TLC on silica gel [ethyl acetate-methanol (85:15)] of fraction AE5 (11 mg) afforded 1 mg of pedalitin (7). Finally, preparative RP-TLC of AE6 (10 mg, MeOH) afforded 4 mg of an epimeric mixture of amarisolide G (8a,b).
Amarisolide G (8a,b): White solid; m.p. 133–135 °C. = –149 (c = 1 mg/mL, MOH). UV (MeOH): λmax (log ε) 206 (0.612) nm. IR (KBr): νmax 3365, 1749 cm−1. ECD (c 0.2 mM, MeOH): λmax (Δε) 212 (–5.21), 250 (–4.65) nm. 1H and 13C-NMR: see Table 1. HRESIMS: m/z 525.2317 [M + H]+ (calc. 525.2330 for C26H37O11).
3.6. Enzymatic Hydrolysis of 8a,b
Compound 8a,b (2 mg) was mixed with β-glucosidase (2 mg, Sigma-Aldrich, MO, USA) in phosphate buffer solution (2 mL, 100 mM at pH 7); and kept at 40 °C for 15 days. Subsequently, the reaction mixture was extracted with CHCl3, and the aqueous phase was concentrated to dryness and subjected to TLC analysis. d-Glucose was identified by comparison of the retention factor and optical rotation value with those of the authentic sample.
3.7. UHPLC-MS Analysis and Method Validation
The analytical method (Figure 5) was developed using an Acquity UHPLC® BEH Shield C18 column (2.1 × 100 mm, 1.7 μm) at 40 °C. The mobile phase consisted of (A) water (0.1% formic acid) and (B) acetonitrile with a linear gradient elution program: 0–10 min, 20–100% (B); 10–10.5 min, 20% (B); 10.5–13 min, 20% (B). The flow rate was set to 0.3 μL/min, and the sample injection volume was 3.0 μL; detection was achieved with a PDA detector at 270 nm. For the identification of compounds, each sample was analyzed with the electrospray ion source operating in both positive (ESI+) and negative (ESI−) ionization modes. The ESI-MS conditions consisted of capillary voltage at 3.0 or 2.5 kV in positive and negative ion modes, respectively; dry heater temperature 150 °C; and nitrogen as the sheath gas flow. MS spectra were obtained within a mass range of m/z 100–1000 using nitrogen as the collision gas. The MassLynk software (version 4.1) was used to control of the UHPLC-MS system and for data acquisition and processing.
The method was validated according to the ICH guidelines [30]. For linearity, amarisolide (1) was accurately weighed and dissolved in dioxane-methanol (v/v, 1:1) to prepare stock solution at a final concentration of 1 mg/mL. Six working solutions in the range of 5–100 μg/mL for the standard were prepared from serial dilutions from the stock solution. Each concentration was prepared in sextuplicate. The linearity was assessed estimating the slope, y-intercept and coefficient of determination (R2) using the least squares method. Limits of detection (LOD) and quantification (LOQ) for the standard were determined at signal-to-noise (S/N) ratios of 3 and 10, respectively. Recovery experiments were carried out to evaluate the accuracy, assaying independently three amounts equivalent to 50 (ca. 10 μg/mL), 100 (ca. 50 μg/mL) or 125% (ca. 75 μg/mL). At each level, compound 1 was added simultaneously to the ethyl acetate soluble fraction (50 μg/mL). Each sample was injected twice and analyzed according to the method previously described. The mean percentage recovery for the standard was found to be between 98 and 102% by means of Fisher’s F test [30]. Finally, the repeatability and inter-day precision was evaluated by testing six identical samples according to the above described method on two consecutive days and by two different analysts by triplicate. The relative standard deviation (RSD) was calculated for each determination as a measure of precision and repeatability.
3.8. GC-MS Analysis of the Essential Oils
For GC-MS analyses, compounds were separated on a DB-5 capillary column (Supelco, Bellefonte, PA, USA) with the following temperature program: oven temperature was programmed from 40 to 260 °C at 4 °C/min during 20 min, and finally up to 340 °C for 20 min isothermally; injector and MS transfer line temperatures were set at 200 and 300 °C, respectively; Helium was used as the carrier gas at a constant flow rate of 1 mL/min; split ratio, 1:20. A mixture of the homologous series of n-alkanes (C8–C20) in CH2Cl2 was directly injected into the GC under the above temperature program, in order to calculate the linear retention indices (RI). All mass spectra were acquired in EI mode (scan range m/z 40–400, ionization energy 70 eV). The components were identified using retention index (RI) of peaks in the chromatogram [31,32] and by comparison of their mass spectra with those of standard library data (NIST) of the GC-MS system and literature data or with those of authentic samples available commercially. All determinations were performed in triplicate.
3.9. Protein Tyrosine Phosphatase 1B Inhibition Assay
The expression and purification of hPTP-1B was performed as previously described [33]. Aqueous extract (AE), ethyl acetate soluble fraction, pure compounds and positive control were dissolved in DMSO or MeOH or Tris buffer solution (Tris-HCl, 20 mM, pH 7). Aliquots of 0–10 μL of testing materials (triplicated) were incubated for 5 min with 20 μL of enzyme stock solution in Tris-HCl (22 nM). After incubation, 10 μL of p-nitrophenylphosphate (pNPP; 5 mM) was added and further incubated for 15 min at 25 °C; then, the absorbance was determined (λmax 415 nm). For all samples, the inhibitory activity was determined as a percentage in comparison to the blank (Tris-HCl) according to the following equation:
| (1) |
where % PTP-1B is the percentage of inhibition, A415t is the corrected absorbance of the extracts, fraction, or compounds under testing (A415end − A415initial) and A415C is the absorbance of the blank (A415end blank − A415initial blank). The IC50 was calculated by regression analysis, using the following equation:
| (2) |
where A100 is the maximum inhibition, I is the inhibitor concentration, IC50 is the concentration required to inhibit the activity of the enzyme by 50% and S is the cooperative degree.
3.10. Docking Studies
To perform the docking at the catalytic site, the PTP1B-INZ complex corresponding to the PDB 1G7F was used, which has a resolution of 1.8 A. For the allosteric site, the PDB 1T49 (resolution of 1.9) was used. The two PDBs used were selected considering the resolution and that they had co-crystallized ligands at the sites of interest. All compounds were built using the HyperChem 8.0 release program and optimized geometrically using the Gaussian 09 program, revision A.02 (Gaussian Inc., Wallingford, CT, USA) at DFT B3LYP/3-21G level of theory. The protein and ligands were further prepared using the utilities implemented by AutoDockTools 1.5.4 (http://mgltools.scripps.edu/). The protein was adding polar hydrogen atoms, Kollman united-atom partial charges, and to the ligands computing Gasteiger–Marsilli formalism charges, rotatable groups which were assigned automatically as were the active torsions. Blind docking was carried out using AutoDock Vina version 2.0 [34]. The root mean square deviation (RMSD) values were obtained by comparing the best pose generated in AutoDock. The initial parameters used for the active site were a grid box size was 42 Å × 40 Å × 40 Å in the x, y and z dimensions and grid center 9.73, 18.00, 971 to 1G7F.PDB. For the allosteric site docking were a grid box size was 42 Å × 40 Å × 40 Å in the x, y and z dimensions and grid center 9.73, 18.00, 971 used 1T49.pdb. For both sites, the exhaustiveness was 25, and the ten best poses were obtained. The analysis of the docking was made with PyMol (Maestro, Schrödinger, LLC, New York, NY) [35].
3.11. Molecular Dynamics Simulation
All the structural complexes were verified, cleaned and ordered with the pdb4amber scrip before starting the preparation in order to generate suitable topologies from the LEaP module of AMBER 19 [36,37]. Each structure and complex was subjected to the following protocol: hydrogens and other missing atoms were added using the LEaP module with the leaprc.protein.ff19SB parameter set; Cl−or K+ counterions were added to neutralize the system; the complexes were then solvated in an octahedral box of explicit TIP3P model water molecules localizing the box limits at 12 Å from the protein surface. Molecular dynamic simulations were performed at 1 atm and 315 K, maintained with the Berendsen barostat and thermostat, using periodic boundary conditions and particle mesh Ewald sums (grid spacing of 1 Å) for treating long-range electrostatic interactions with a 10 Å cutoff for computing direct interactions. The SHAKE algorithm was used to satisfy bond constraints, allowing the employment of a two fs time step for the integration of Newton’s equations as recommended in the Amber package [36,38]. Amber leaprc.protein.ff19SB force field [39] parameters were used for all residues. All calculations were made using Graphics Processing Units (GPU) accelerated MD engine in AMBER (pmemd.cuda), a program package that runs entirely on CUDA® (Compute Unified Device Architecture)-enabled GPUs [40]. The protocol consisted of performing a minimization of the initial structure, followed by 50ps heating and pressure equilibration at 315 K and 1.0 atm pressure, respectively. Finally, the system is equilibrated with 500ps before starting the production of MD. The production of the MD consisted of 100 ns for each complex. Frames were saved at ten ps intervals for subsequent analysis. All analyses were done using CPPTRAJ [41] part of AMBER19 utilities and OriginPro 9.1. The calculations of RMSD and Root Mean Square Fluctuations (RMSF) were made, considering the C, CA and N. The charts were built with OriginPro 2018 SR1, and the trends were adjusted with the function processing smooth (method lowess span). VMD and PyMOL [35] were used to visualize and create the images from the MD.
3.12. Chemoinformatic Properties of Compounds 1, 7 and AU
The biological and chemoinformatic properties of compounds 1, 7 and AU were explored using the servers Swiss TargetPrediction (http://www.swisstargetprediction.ch/index.php), Molinspiration (http://www.molinspiration.com/cgi-bin/properties) and Osiris Property Explorer server (http://che minformatics.ch/propertyExplorer/) [29,42].
4. Conclusions
The AE of S. amarissima contains rutin (4) and rosmarinic acid (5), which inhibit intestinal glucose absorption, promote glucose uptake in muscle cells and suppress insulin-resistance, among other effects. On the other hand, pedalitin (7), which behave in silico as an allosteric inhibitor of PTP-1B, could contribute to its overall antidiabetic action via α-glucosidase and selective PTP-1B inhibition, and other mechanisms yet to be determined. The overall action of AE could be attained via network pharmacology, synergism and or polypharmacology. Altogether, our studies on S. amarissima tend to support its medicinal use for the treatment of diabetes in Mexican folk medicine. The chromatographic analyses developed and validated in this study will allow the development of a pharmacopeic monograph, and generate standardized preparations of this very Mexican plant. The analytical UHPLC method was suitable for its intended purpose, the quantification of amarisolide (1) according to the Q2 (R1) guideline. Overall, the scientific information generated for this plant will contribute to its rational use in Mexican folk medicine. Like many other New World melittophilous Salvia species, S. amarissima is a rich source of bioactive compounds.
Acknowledgments
The authors recognize the valuable support of M.I. Velásquez-López from Facultad de Medicina; and Georgina Duarte from Facultad de Química. We are indebted to Biol. Itzi Fragoso and Martha Martínez Gordillo (Facultad de Ciencias, UNAM) for their valuable help in the identification of the plant material. We also recognize the support of Dirección General de Cómputo y de Tecnologías de Información y Comunicación (DGTIC, UNAM) for the resources to carry out computational calculations through the Miztli supercomputing system (LANCAD-UNAM-DGTIC-313). E.S.-A. acknowledges the fellowship from CONACyT (289212) to pursue graduate studies.
Supplementary Materials
The following are available online. NMR, IR and MS spectrum of the isolated or identified compounds, TIC chromatograms of the essential oils and Residues of interaction to 4 Å of compounds 1, 7 and UA at the catalytic and allosteric sites of PTP-1B are available online.
Author Contributions
This work is part of the PhD thesis of E.S.-A. who performed the experiments; analysis of spectral data, A.P.-V.; UHPLC-MS method validation, I.R.-C.; docking analysis, M.R.-G.; expression and purification of hPTP-1B, docking, molecular dynamics and parameters calculations, M.G.-A.; botanical characterization of the species, R.T.-C.; conception, funding acquisition, structure elucidation, writing and editing, R.M. All authors have read and agreed to the published version of the manuscript.
Funding
The research was supported by grants from CONACyT CB A1-S-11226 and DGAPA IN 217320 awarded to R.M.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Samples of the compounds are not available from the authors.
References
- 1.International Diabetes Federation (IDF) Diabetes Atlas. [(accessed on 13 March 2020)]; Available online: http://www.diabetesatlas.org.
- 2.Mata R., Figueroa M., Navarrete A., Rivero-Cruz I. Chemistry and Biology of Selected Mexican Medicinal Plants. In: Kinghorn A.D., Falk H., Gibbons S., Kobayashi J., Asakawa Y., Liu J.K., editors. Progress in the Chemistry of Organic Natural Products. Volume 108. Springer Nature Switzerland AG; Cham, Switzerland: 2019. pp. 1–142. [DOI] [PubMed] [Google Scholar]
- 3.Efferth T., Greten H.J. Quality control for medicinal plants. Med. Aromat. Plants. 2012;1:1000e131. [Google Scholar]
- 4.Calderón R.G., Rzedowski J. Flora Fanerogámica del Valle de México. 2nd ed. Instituto de Ecología, A.C. y Comisión Nacional para el Conocimiento y Uso de la Biodiversidad, Pátzcuaro; Mexico City, Mexico: 2010. pp. 1–1406. [Google Scholar]
- 5.López F.C.E., Sánchez D.G., Arrieta B.D., Román G.J.H. Estudio preliminar fitoquímico y de la actividad antimicrobiana de Salvia amarissima Ortega. Cienc. y Tecnol. 2010;9:67–76. [Google Scholar]
- 6.Maldonado E., Cárdenas J., Bojórquez H., Escamilla E.M., Ortega A. Amarisolide, a neo-clerodane diterpene glycoside from Salvia amarissima. Phytochemistry. 1996;42:1105–1108. doi: 10.1016/0031-9422(96)00147-1. [DOI] [Google Scholar]
- 7.Bautista E., Fragoso-Serrano M., Ortiz-Pastrana N., Toscano R.A., Ortega A. Structural elucidation and evaluation of multidrug-resistance modulatory capability of amarissinins A-C, diterpenes derived from Salvia amarissima. Fitoterapia. 2016;114:1–6. doi: 10.1016/j.fitote.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 8.Flores-Bocanegra L., González-Andrade M., Bye R., Linares E., Mata R. α-Glucosidase inhibitors from Salvia circinata. J. Nat. Prod. 2017;80:1584–1593. doi: 10.1021/acs.jnatprod.7b00155. [DOI] [PubMed] [Google Scholar]
- 9.Fragoso-Serrano M., Ortiz-Pastrana N., Luna-Cruz N., Toscano R.A., Alpuche-Solis A.G., Ortega A., Bautista E. Amarisolide F, an acylated diterpenoid glucoside and related terpenoids from Salvia amarissima. J. Nat. Prod. 2019;82:631–635. doi: 10.1021/acs.jnatprod.8b00565. [DOI] [PubMed] [Google Scholar]
- 10.Bautista E., Fragoso-Serrano M., Toscano R.A., García-Peña M.R., Ortega A. Teotihuacanin, a diterpene with an unusual spiro-10/6 system from Salvia amarissima with potent modulatory activity of multidrug resistance in cancer cells. Org. Lett. 2015;17:3280–3282. doi: 10.1021/acs.orglett.5b01320. [DOI] [PubMed] [Google Scholar]
- 11.Calzada F., Bautista E. Plants used for the treatment of diarrhoea from Mexican Flora with amoebicidal and giadicidal activity, and their phytochemical constituents. J. Ethnopharmacol. 2020;253:1–49. doi: 10.1016/j.jep.2020.112676. [DOI] [PubMed] [Google Scholar]
- 12.Moreno-Pérez G.F., González-Trujano M.E., Martínez-Gordillo M.J., San Miguel-Chávez R., Basurto-Peña F.A., Dorazco-Gozález A., AguirreHernández E. Amarisolide A and pedalitin as bioactive compounds in the antinociceptive effects of Salvia circinata. Bot. Sci. 2019;97:355–365. doi: 10.17129/botsci.2187. [DOI] [Google Scholar]
- 13.Klaman L.D., Boss O., Peroni O.D., Kim J.K., Martino J.L., Zabolotny J.M., Moghal N., Lubkin M., Kim Y.B., Sharpe A.H. Increased energy expenditure, decreased adiposity, and tissue-specific insulin sensitivity in protein-tyrosine phosphatase 1B-deficient mice. Mol. Cell. Biol. 2000;20:5479–5489. doi: 10.1128/MCB.20.15.5479-5489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voirin B., Viricel M.R., Favre-Bonvin J., Van den Broucke C.O., Lemli J. 5,6,4′-Trihydroxy-7,3′-dimethoxyflavone and other methoxylated flavonoids isolated from Thymus satureiodes. Planta Med. 1985;51:523–525. doi: 10.1055/s-2007-969583. [DOI] [PubMed] [Google Scholar]
- 15.Airoldi C., Sironi E., Dias C., Marcelo F., Martins A., Rauter A.P., Nicotra F., Jimenez-Barbero J. Natural compounds against Alzheimer’s disease: Molecular recognition of Aβ1-42 peptide by Salvia sclareoides extract and its major component, rosmarinic acid, as investigated by NMR. Chem. Asian, J. 2013;8:596–602. doi: 10.1002/asia.201201063. [DOI] [PubMed] [Google Scholar]
- 16.Bagci E., Kocak A. Essential oil composition of the aerial parts of two Salvia, L. (S. multicaulis Vahl. Enum and S. tricochlada Bentham) species from east Anatolian region (Turkey) Int. J. Sci. Tech. 2008;3:13–18. [Google Scholar]
- 17.Salehi P., Tolami L.B., Sefidkon F. Essential oil composition of Salvia xanthocheila Boiss. ex Benth. from Iran. J. Essent. Oil Res. 2005;17:442–443. doi: 10.1080/10412905.2005.9698956. [DOI] [Google Scholar]
- 18.Sonboli A., Babakhani B., Mehrabian A.R. Antimicrobial activity of six constituents of the essential oil from Salvia. Z. Naturforsch. 2006;61:160–164. doi: 10.1515/znc-2006-3-401. [DOI] [PubMed] [Google Scholar]
- 19.Zhao B.T., Le D.D., Nguyen P.H., Ali M.Y., Choi J.S., Min B.S., Shin H.M., Rhee H.I., Woo M.H. PTP1B, α glucosidase, and DPP-IV inhibitory effects for chromene derivatives from the leaves of Smilax china L. Chem. Biol. Interact. 2016;253:27–37. doi: 10.1016/j.cbi.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 20.Xu J., Wang X., Yue J., Sun Y., Zhang X., Zhao Y. Polyphenols from a corn leaves (Quercus liaotungensis) protect pancreatic beta cells and their inhibitory activity against α-glucosidase and protein tyrosine phosphatase 1B. Molecules. 2018;23:2167. doi: 10.3390/molecules23092167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ngoa Y.L., Lau C.H., Chua L.S. Review on rosmarinic acid extraction, fractionation and its anti-diabetic potential. Food Chem. Toxicol. 2018;121:687–700. doi: 10.1016/j.fct.2018.09.064. [DOI] [PubMed] [Google Scholar]
- 22.Oh J., Choe S., Kim D.W., Lee S.H., Na M.K. Rhododendric acid A, a new ursane-type PTP1B inhibitor from the endangered plant Rhododendron brachycarpum G. Don. Bioorg. Med. Chem. Lett. 2012;22:6116–6119. doi: 10.1016/j.bmcl.2012.08.029. [DOI] [PubMed] [Google Scholar]
- 23.Al-Ishaq R.K., Abotaleb M., Kubatka P., Kajo K., Busselberg D. Flavonoids and their anti-diabetic effects: Cellular mechanisms and effects to improve blood sugar levels. Biomolecules. 2019;9:430. doi: 10.3390/biom9090430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiesmann C., Barr J.J., Kung J., Zhu J., Erlanson D.A., Shen W., Fahr B.J., Zhong M., Taylor L., Randal M., et al. Allosteric Inhibition of Protein Tyrosine Phosphatase 1B. Nat. Struct. Mol. Biol. 2004;11:730–737. doi: 10.1038/nsmb803. [DOI] [PubMed] [Google Scholar]
- 25.Shrestha S., Seong S.H., Park S.G., Min B.S., Jung A.H., Chi J.S. Insight into the PTP1B inhibitory activity of arylbenzofurans: An in vitro and in silico study. Molecules. 2019;24:2893. doi: 10.3390/molecules24162893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gohlke H., Klebe G. Approaches to the description and prediction of the binding affinity of small-molecule ligands to macromolecular receptors. Angew. Chem. Int. Ed. Engl. 2002;41:2644–2676. doi: 10.1002/1521-3773(20020802)41:15<2644::AID-ANIE2644>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 27.Ramírez D., Caballero J. Is it reliable to take the molecular docking top scoring position as the best solution without considering available structural data? Molecules. 2018;23:1038. doi: 10.3390/molecules23051038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X.-Y., LI W., Wang J., Li N., Ccheng M.S., Koike K. Protein tyrosine phosphatase 1B inhibitory activities of ursane-type triterpenes from Chinese raspberry, fruits of Rubus chingii. Chin. J. Nat. Med. 2019;17:15–21. doi: 10.1016/S1875-5364(19)30004-4. [DOI] [PubMed] [Google Scholar]
- 29.Khan T., Lawrence A.J., Azad I., Raza S., Joshi S., Khan A.R. Computational drug designing and prediction of important parameters using in silico methods- A review. Curr. Comput. Aided Drug. Des. 2019;15:384–397. doi: 10.2174/1573399815666190326120006. [DOI] [PubMed] [Google Scholar]
- 30.Geneva, Switzerland: Nov 1, 2005. [(accessed on 17 June 2020)]. International Conference on Harmonization (ICH) Q2(R1): Validation of Analytical Procedures: Text and Methodology. In ICH: International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use. Available online: http://academy.gmp-compliance.org/guidemgr/files/Q2(R1).pdf. [Google Scholar]
- 31.Van den Dool H., Kratz P.D. Generalization of the retention index system including linear temperature programmed Gas-Liquid partition Chromatography. J. Chromatogr. A. 1963;11:463–471. doi: 10.1016/S0021-9673(01)80947-X. [DOI] [PubMed] [Google Scholar]
- 32.Adams R.P. Identification of Essential Oil Components by Gas Chromatography/Mass Spectrometry. 4th ed. Allured Publishing Corporation; Carol Stream, IL, USA: 2007. pp. 1–804. [Google Scholar]
- 33.Rangel-Grimaldo M., Macías-Rubalcava M.L., González-Andrade M., Raja H., Figueroa M., Mata R. α-Glucosidase and protein tyrosine phosphatase 1B inhibitors from Malbranchea circinata. J. Nat. Prod. 2020;83:675–683. doi: 10.1021/acs.jnatprod.9b01108. [DOI] [PubMed] [Google Scholar]
- 34.Trott O., Olson A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeLano W.L. Use of PYMOL as a communications tool for molecular science. Abstr. Pap. A.C.S. 2004;228:U313–U314. [Google Scholar]
- 36.Case D.A., Betz R.M., Cerutti D.S., Cheatham T.E., III, Darden T.A., Duke R.E., Giese T.J., Gohlke H., Goetz A.W., Homeyer N., et al. [(accessed on 31 July 2020)];AMBER. 2015 Available online: http://ambermd.org/doc12/Amber16.pdf.
- 37.Case D.A., Cheatham T.E., III, Darden T., Gohlke H., Luo R., Merz K.M., Jr., Onufriev A., Simmerling C., Wang B., Woods R.J. The Amber biomolecular simulation programs. J. Comp. Chem. 2005;26:1668. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walker R.C., Crowley M.F., Case D.A. The implementation of a fast and accurate QM/MM potential method in Amber. J. Comp. Chem. 2008;29:1019. doi: 10.1002/jcc.20857. [DOI] [PubMed] [Google Scholar]
- 39.Tian C., Kasavajhala K., Kellon A., Belfon K., Raguette L., Huang H., Migues A.N., Bickel J., Wang Y., Pincay J., et al. ff19SB: Amino-acid specific protein backbone parameters trained against quantum mechanics energy surfaces in solution. J. Chem. Theory Comput. 2020;16:528–552. doi: 10.1021/acs.jctc.9b00591. [DOI] [PubMed] [Google Scholar]
- 40.Salomon-Ferrer R., Gotz A.W., Poole D., Le Grand S., Walker R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J. Chem. Theory Comput. 2013;9:3878–3888. doi: 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- 41.Roe D.R., Cheatham T.E., III PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. Cheatham. J. Chem. Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 42.Bhal S.K. LogP-making sense of the value. Advanced Chemistry Development, Inc. (ACD/LABS); Toronto, ON, Canada: 2007. pp. 1–4. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.