Abstract
Molecular similarity is an elusive but core “unsupervised” cheminformatics concept, yet different “fingerprint” encodings of molecular structures return very different similarity values, even when using the same similarity metric. Each encoding may be of value when applied to other problems with objective or target functions, implying that a priori none are “better” than the others, nor than encoding-free metrics such as maximum common substructure (MCSS). We here introduce a novel approach to molecular similarity, in the form of a variational autoencoder (VAE). This learns the joint distribution p(z|x) where z is a latent vector and x are the (same) input/output data. It takes the form of a “bowtie”-shaped artificial neural network. In the middle is a “bottleneck layer” or latent vector in which inputs are transformed into, and represented as, a vector of numbers (encoding), with a reverse process (decoding) seeking to return the SMILES string that was the input. We train a VAE on over six million druglike molecules and natural products (including over one million in the final holdout set). The VAE vector distances provide a rapid and novel metric for molecular similarity that is both easily and rapidly calculated. We describe the method and its application to a typical similarity problem in cheminformatics.
Keywords: cheminformatics, molecular similarity, deep learning, variational autoencoder, SMILES
1. Introduction
The concept of molecular similarity lies at the core of cheminformatics [1,2,3]. It implies that molecules of “similar” structure tend to have similar properties. Thus, a typical question can be formulated as follows: “given a molecule of interest M, possibly showing some kind of chemical activity, find me the nearest 50 molecules from a potentially huge online collection to purchase that are most similar to M so I can assess their behaviour in a relevant quantitative-structure-activity (QSAR) analysis”.
The most common strategies for assessing molecular similarity involve encoding the molecule as a vector of numbers, such that the vectors encoding two molecules may be compared according to their Euclidean or other distance. In the case of binary strings, the Jaccard or Tanimoto similarity (TS) is commonly used [4] as a metric (between zero and one). One means for obtaining such a vector for a molecule is to calculate from the structure (or measure) various properties of the molecule (“descriptors” [5,6,7]), such as clogP or total polar surface area, and then to concatenate them. However, a more common strategy for obtaining the encoding vector of numbers is simply to use structural features directly and to encode them as so-called molecular fingerprints [8,9,10,11,12,13,14,15,16,17]. Well-known examples include MACCS [18], atom pairs [19], torsion [20], extended connectivity [21], functional class [22], circular [23], and so on. The similarities so encoded can also then be compared as their Jaccard or Tanimoto similarities. Sometimes a “difference” or “distance” is discussed and formulated as 1-TS (a true metric). An excellent and widely used framework for performing all of this is RDKit (Pathon v3.6.8) (www.rdkit.org/) [24], that presently contains nine methods for producing molecular fingerprints.
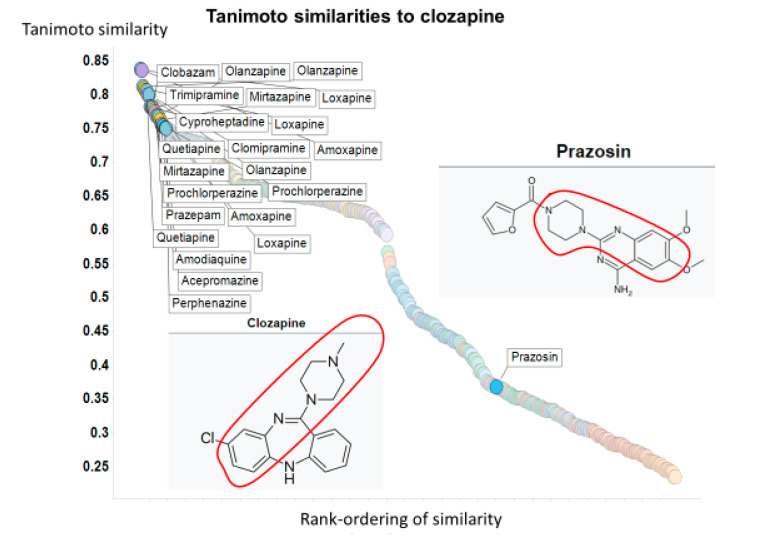
The problem comes from the fact that the “most similar” molecules to a target molecule often differ wildly both as judged by their structures observable by eye and quantitatively in terms of the value of the TS of the different fingerprints [25]. As a very small and simple dataset, we take the set of molecules observed by Dickens and colleagues [26] to inhibit the transporter-mediated uptake of the second-generation atypical antipsychotic drug clozapine. These are olanzapine, chlorpromazine, quetiapine, prazosin, lamotrigine, indatraline, verapamil and rhein. Of the FDA-approved drugs, we assessed the top 50 drugs in terms of their structural similarity to clozapine using the nine RDKit encodings, with the results shown in Table 1 and Figure 1 (see also Supplementary Figure S1). Only the first four of these are even within the top 50 for any encoding, and only olanzapine appears for each of them. By contrast, the most potent inhibitor is prazosin (which is not even wholly of the same drug class, being both a treatment for anxiety and a high-blood-pressure-lowering agent); however, it appears in the top 50 in only one encoding (torsion) and then with a Tanimoto similarity of just 0.37. That said, visual inspection of their “Kekularised” structures does show a substantial common substructure between prazosin and clozapine (marked in Figure 1). It is clear that the similarities, as judged by standard fingerprint encodings, are highly variable, and are prone to both false negatives and false positives when it comes to attacking the question as set down above. What we need is a different kind of strategy.
Table 1.
Tanimoto similarity to clozapine using nine different RDKit encodings and their ability to inhibit clozapine transport (data extracted from [26]). A shaded cell means that the molecule was not judged to be in the “top 50” using that encoding.
| Drug | % Inhiclozapine Uptake | TS Atom Pair | TS Avalon | TS Feat Morgan | TS Layered | TS MACCS | TS Morgan | TS Pattern | TS RDKit | TS Torsion |
|---|---|---|---|---|---|---|---|---|---|---|
| Olanzapine | 41 | 0.68 | 0.47 | 0.55 | 0.77 | 0.8 | 0.53 | 0.81 | 0.74 | 0.66 |
| Chlorpromazine | 75 | 0.53 | - | 0.35 | - | 0.66 | 0.3 | 0.74 | - | 0.33 |
| Quetiapine | 65 | 0.51 | 0.57 | 0.42 | 0.78 | - | 0.35 | 0.8 | - | 0.48 |
| Prazosin | 94 | - | - | - | - | - | - | - | - | 0.37 |
| Lamotrigine | 26 | - | - | - | - | - | - | - | - | - |
| Indatraline | 35 | - | - | - | - | - | - | - | - | - |
| Veraapamil | 83 | - | - | - | - | - | - | - | - | - |
| Rhein | 39 | - | - | - | - | - | - | - | - | - |
Figure 1.
Tanimoto similarities of various molecules to clozapine using the Torsion encoding from RDKit.
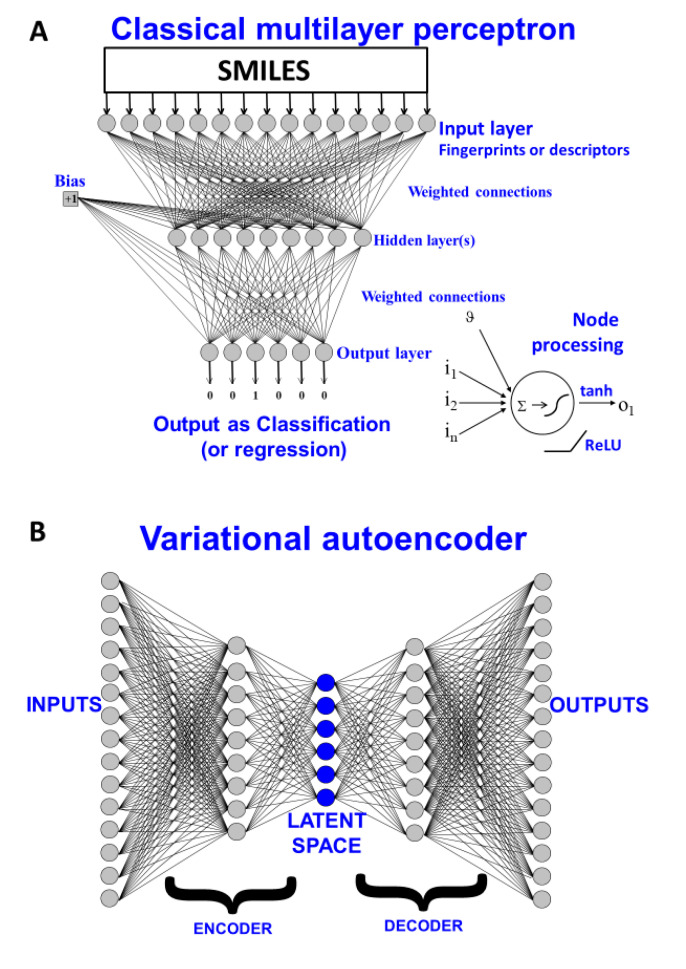
The typical structure of a QSAR type of problem is provided in Figure 2A, where a series of molecules represented as SMILES strings [27] are encoded as molecular fingerprints and used to learn a nonlinear mapping to produce an output in the form of a classification or regression estimation. The architecture of this is implicitly in the form of a multilayer perceptron (a classical neural network [28,29,30,31]), in which weights are modified (“trained”) to provide a mapping from input SMILES to a numerical output. Our fundamental problem stems from the fact that these types of encoding are one-way: the SMILES string can generate the molecular fingerprint but the molecular fingerprint cannot generate the SMILES. Put another way, it is the transition from a world of discrete objects (here molecules) with categorical representations (here SMILES strings) to one of a continuous representation (vectors of weights) that is seemingly irreversible in this representation. One key element is the means by which we can go from a non-numerical representation (such as SMILES or similar [32]) to a numerical representation or “embedding” (this, as we shall see, is typically constituted by vectors of numbers in the nodes and weights of multilayer neural networks) [33,34,35,36,37,38]. Deep learning has also been used for the encoding step of 2D chemical structures [39,40].
Figure 2.
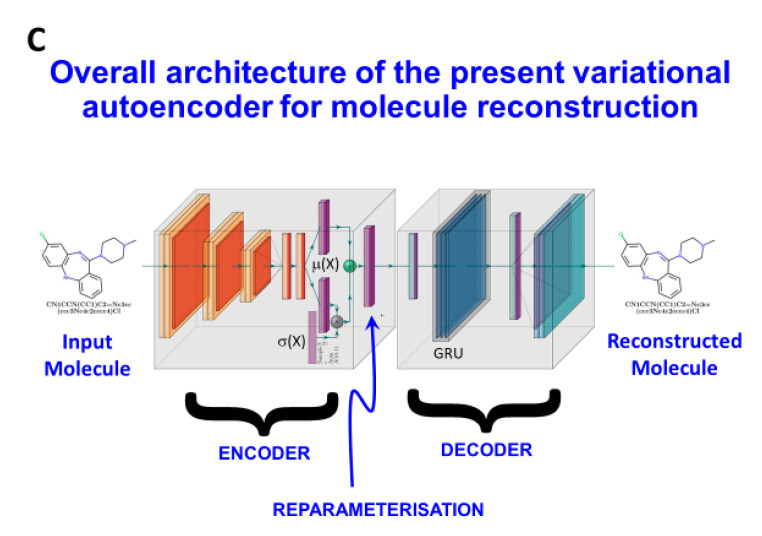
Two kinds of neural architecture. (A) A classical multilayer perceptron representing a supervised learning system in which molecules encoded as SMILES strings can be used as paired inputs with outputs of interest (whether a classification or a regression). The trained model may then be interrogated with further molecules and the output ascertained. (B) A variational autoencoder, is a supervised means of fitting distributions of discrete models in a way that reconstructs them via a vector in a latent space. (C) The variational autoencoder (VAE) architecture used in the present work.
More recently, it was recognised that various kinds of architectures could, in fact, permit the reversal of this numerical encoding so as to return a molecule (or its SMILES string encoding a unique structure). These are known as generative methods [41,42,43,44,45,46,47,48,49,50,51,52,53,54,55], and at heart their aim to generate a suitable and computationally useful representation [56] of the input data. It is common (but cf. [57,58]) to contrast two main flavours: generative adversarial networks [59,60,61,62,63,64,65,66] and (especially variational) autoencoders (VAEs) [41,42,67,68,69,70,71,72,73,74,75,76]. We focus here on the latter, illustrated in Figure 2B.
VAEs are latent-variable generative models that define a joint density pθ(x,z) between some observed data x ∈ Rdx and unobserved or latent variables z ∈ Rdz [77], given some model parameters θ. They use a variational posterior (also referred to as an encoder), qϕ(z|x), to construct the latent variables with variational parameters ϕ, and a combination of p(z) and p(x|z) to create a decoder that has the opposite effect. Learning the posterior directly is computationally intractable, so the generic deep learning strategy is to train a neural network to approximate it. The original “error” backpropagated was based on the Kullback–Leibler (KL) divergence between the desired (log likelihood reconstruction error) and the predicted output distributions [67]. A very great many variants of both architectures and divergence metrics have been proposed since then (not all discernibly better [78]), and it is a very active field (e.g., [63,64,79,80,81,82,83]). Since tuning is necessarily domain-specific [84], and most work is in the processing of images and natural languages rather than in molecules, we merely mention a couple, such as transformers (e.g., [85,86]) and others (e.g., [87,88]). Crucial to such autoencoders (that can also be used for data visualisation [89]) is the concept of a bottleneck layer, that as a series of nodes of lower dimensionality than its predecessors or successors, serves to extract or represent [56] the crucial features of the input molecules that are nonetheless sufficient to admit their reconstruction. Indeed, such strategies are sometimes referred to as representational learning.
A higher-level version of the above might state that a good variational autoencoder will project a set of discrete molecules into a continuous latent space represented for any given molecule by the vector representing the values of the outputs of the nodes in the bottleneck layer when it (or its SMILES representation) is applied to the encoder as an input. As with the commonest neural net training system (but cf. [90,91,92,93,94]), we use backpropagation to update the network so as to minimise the difference between the predicted and the desired output, subject to any other constraints that we may apply. We also recognise the importance of various forms of regularisation, that are all designed to prevent overfitting [49,95,96,97,98].
Because the outputs of the nodes in the bottleneck layer both (i) encode the molecule of interest and (ii) effectively represent where molecules are in the chemical space on which they have been trained, a simple metric of similarity between two molecules is clearly the Euclidean or other comparable distance (e.g., cosine distance) between these vectors. This thus provides for a novel type of similarity encoding, that in a sense relates the whole chemical space on which the system has been trained and that we suspect may be of general utility. We might refer to this encoding as the “essence of molecules” (EM) encoding, but here, we refer to it as VAE-Sim.
Thus, the purpose of the present article is to describe our own implementation of a simple VAE and its use in molecular similarity measurements as applied, in particular, to the set of drugs, metabolites and natural products that we have been using previously [25,99,100,101,102,103,104] as our benchmark for similarity metrics. A preprint was deposited at bioRxiv [105].
2. Results
Autoencoders that use SMILES as inputs can return three kinds of outputs: (i) the correct SMILES output mirroring the input and/or translating into the input molecular structure (referred to as “perfect”), (ii) an incorrect output of a molecule different from the input but that is still legal SMILES (hence will return a valid molecule), referred to as “good”, and (iii) a molecule that is simply not legal SMILES. In practice, our VAE after training returned more than 95% valid SMILES in the test (holdout) set, so those that were invalid could simply be filtered out without significant loss of performance.
Specifically, we partitioned the dataset into 50% for training (3,101,207 samples), 20% for validation (1,240,483 samples) and 30% for testing (1,860,725 samples). The results for our VAE model for ZINC molecule reconstruction are as follows (Table 2):
Table 2.
Data partitioning of training, validation and test sets, and their generalization.
| Data Partition | Total Samples | Valid Reconstructed Samples | Accuracy |
|---|---|---|---|
| Train | 3,101,207 | 2,964,749 | 95.60 |
| Validation | 1,240,483 | 1,170,827 | 94.38 |
| Test | 1,860,725 | 1,757,079 | 94.42 |
Following training, each molecule (SMILES) could be associated with a normalised vector of 100 dimensions, and the Euclidean distance between them could be calculated.
| (1) |
| (2) |
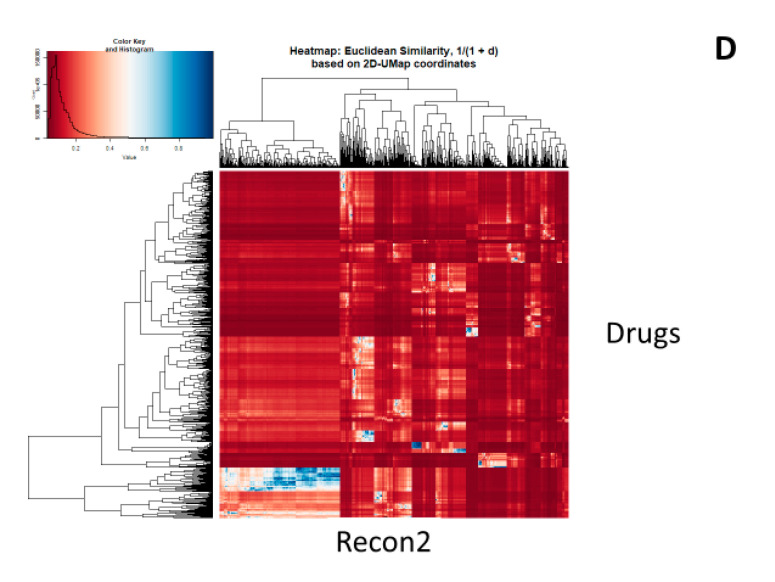
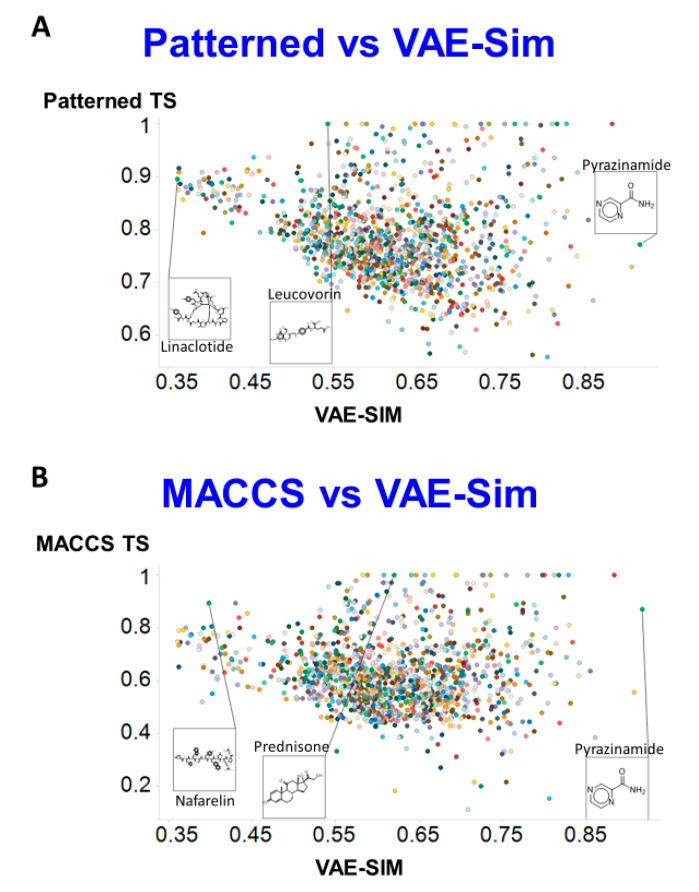
As previously described [25], we compared the similarities between all drugs and all metabolites using the datasets made available in [99]. We here focus on just the MACCS and Patterned encodings of RDKit, and compare them with the normalised Euclidean distances according to the latent vector obtained from the VAE. As before, we rank ordered each drug in terms of its closest similarity to any metabolite. First, Figure 3A (reading from right to left) shows the Tanimoto similarities for the Patterned and MACCS fingerprints, as well as the VAE-Sim values as judged by two metrics. The first, labelled E-Sim Equation (1), is the Euclidean similarity, based on the raw 100-dimensional hidden vectors, while the second, EU-Sim Equation (2), used the Uniform Manifold Approximation and Projection (UMAP) dimension reduction algorithm [106,107] based on the first two UMAP dimensions was used for purposes of visualisation; clearly, as with other encodings, they do not at all follow the same course, and one that may be modified according to the similarity measure used. Figure 3B,C show the “all-vs.-all” heatmaps for two of the encodings, indicating again that the VAE-Sim encoding falls away considerably more quickly, i.e., that similarities are judged in a certain sense more “locally”.
Figure 3.
Top similarities between drugs and metabolites as judged by a fingerprint encoding (RDKit patterned) and our new VAE-Sim metric. (A) Rank ordering. (B) Heatmap for Tanimoto similarities using RDKit patterned encoding. (C) Heatmap of Euclidean similarities E-Sim (Equation (1)) for VAE-Sim in the 100-dimensional latent vector). (D) Heatmap of Euclidean similarities EU-Sim (Equation (2)) for VAE-Sim in 2-dimensional uniform manifold approximation and projection (UMAP) space.
Figure 4A shows the Patterned similarity for the “most similar” metabolite for each drug (using TS) compared to that for VAE-Sim (using Euclidean distance), while Figure 4B shows the same for the MACCS encoding. These, again, illustrate how the new encoding provides a quite different readout from the standard fingerprint encodings.
Figure 4.
Comparison of similarities between two RDKit fingerprint methods and VAE-Sim Using Tanimoto similarity for fingerprints and Euclidean d100 similarity for VAE-Sim. (A) Patterned encoding. (B) MACCS encoding.
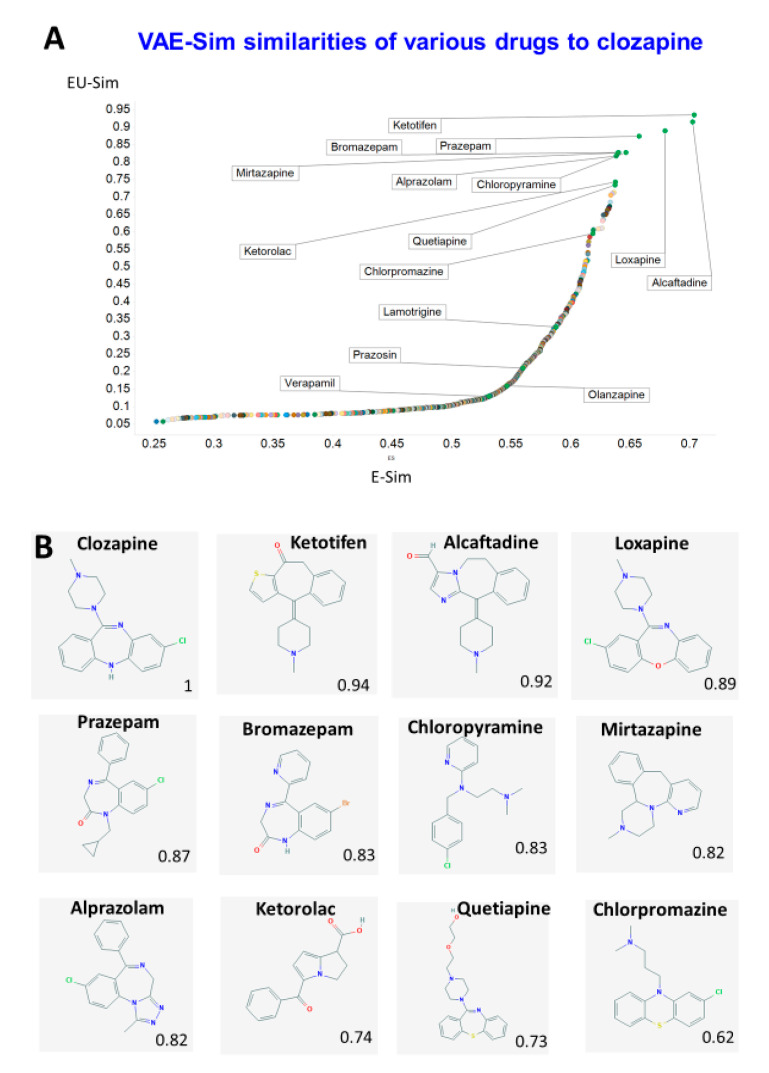
Finally, we used our new metrics to determine the similarity to clozapine of other drugs. Figure 5 shows the two similarity scores based on VAE-Sim, calculated as in Figure 3. Gratifyingly, and while structural similarity is, in part, in the eye of the beholder, a variety of structurally and functionally related antipsychotic drugs such as loxapine, mirtazapine and quetiapine were indeed among the most similar to clozapine, while others not previous considered (such as the antihistamines ketotifen and alcaftadine, and the anti-inflammatory COX inhibitor ketorolac) were also suggested as being similar, providing support for the orthogonal utility of the new VAE-Sim metric. However, the rather promiscuous nature of clozapine binding (e.g., [108,109]), along with that of many of the other drugs (e.g., [110,111,112,113,114,115,116]), means that this is not the place to delve deeper.
Figure 5.
Similarity of drugs to clozapine as judged by the VAE. (A) Rank order of Euclidean similarity in 100 dimensions (E-Sim) or two UMAP dimensions (EU-Sim) as in Figure 3. Some of the “most similar” drugs are labelled, as are some of those in Table 1. (B) Structures of some of the drugs mentioned, together with their Euclidean distances as judged by VAE-Sim.
3. Methods
We considered and tested grammar-based and junction-tree methods such as those used by Kajino [35], that exploited some of the ideas developed by Dai [117], Kusner [118] and by Jin and their colleagues [34]. However, our preferred method as described here used one-hot encoding as set out by Gómez-Bombarelli and colleagues [74]. We varied the number of molecules in the training process from ca 250,000 to over 6 million; the large number of possible hyperparameters would have led to a combinatorial explosion, so exhaustive search was (and is) not possible. The final architecture used here (shown in Figure 2C) required 6 days’ training on a 1-GPU machine. It involved a convolutional neural network (CNN) encoder with the following layers (Figure 2C): convolution (1D): size (in-248 = SMILES string length, 40 possible unique SMILES characters, out-9, kernel_size = 9), ReLU, convolution (1D): size (in-9, out-9, kernel_size = 9) ReLU, convolution (1D): size (in-9, out-10, kernel_size = 11) ReLU, Linear (fully connected): size(140, latent_dims = 100) SeLU, with VAE mean—Linear (fully connected): size(140, latent_dims = 100) and variance—Linear (fully connected): size(140, latent_dims = 100). For the decoder we used a Reparameterization (combined mean and sigma together) such that the output will be the same as the latent dimension (100 in our case), Linear (fully connected): size(latent_dims = 100, latent_dims = 100) SeLU, RNN-GRU (gated neural unit): size (hidden size = 488, num_layers = 3), Linear (fully connected): size(in-488 = hidden_gru_size, out-248 = SMILES length) Softmax. For the loss we used binary cross-entropy + KL-divergence. Neither dropout nor pooling were used. The optimiser was ADAM [119], the fixed learning rate 0.0001, parameters were initialised using the “Xavier uniform” scheme [120], and a batch size of 128. This was implemented in Python using the Pytorch library (Python v3.8.5). (https://pytorch.org/). Most of the pre- and post-processing cheminformatics workflows were written in the KNIME environment (see [121]).
For the UMAP settings, we used the default parameter settings implemented in the RAPIDS toolbox (ArcGIS v10.2.6), as detailed here https://docs.rapids.ai/api/cuml/stable/api.html#umap.
4. Discussion
Molecular similarity is at the core of much of cheminformatics (e.g., [3,8,122,123,124,125]), but is an elusive concept. Our chief interest typically lies in supervised methods such as QSARs, where we use knowledge of paired structures and activities to form a model that allows us to select new structures with potentially desirable activities. Modern modelling methods such as feedforward artificial neural networks based on multilayer perceptrons are very powerful (and they can in fact fit any nonlinear function—the principle of “universal approximation” [126,127]). Under these circumstances, it is usually possible to learn a QSAR using any of the standard fingerprints. However, what we are focused on here is a purely unsupervised representation of the structures themselves (cf [37] which used substructures), and the question of which of these are the “most similar” to a query molecule of interest. Such unsupervised methods may be taken to include any kinds of unsupervised clustering too (e.g., [128,129,130,131,132]). As with any kind of system of this type, the “closeness” is a function of the weighting of any individual features, and it is perhaps not surprising that the different fingerprint methods give vastly different similarities, even when judged by rank order (e.g., [25] and above). One similarity measure that is independent of any fingerprint encoding is represented by the maximum common substructure (MCSS). However, by definition, the MCSS uses only part of a molecule; it is also computationally demanding [101,102], such that “all-against-all” comparisons such as those presented here are out of the question for large numbers of molecules.
Here, we leveraged a new method that uses only the canonical SMILES encoding of the molecules themselves, leading to its representation as a 100-element vector. Simple Euclidean distances can be used to obtain a metric of similarity that, unlike MCSS, is rapidly calculated for any new molecule, even against the entire set of molecules used in the development of the latent space.
In addition, unlike any of the other methods described, methods such as VAEs are generative: moving around in the latent space and applying the vector so created to the decoder allows for the generation of entirely new molecules (e.g., [41,42,43,44,45,48,50,63,65,73,74,133]). This opens up a considerable area of chemical exploration, even in the absence of any knowledge of bioactivities.
What Determines the Extent to Which VAEs can Generate Novel Examples?
The ability of variational autoencoders to generalise is considered to be based on learning a certain “neighbourhood” around each of the training examples [77,134], seen as a manifold of lower dimensionality than the dimensionality of the input space [56]. Put another way, “the reconstruction obtained from an optimal decoder of a VAE is a convex combination of examples in the training data” [135]. On this basis, an effect of training set size on the improvement of generalisation (here defined simply as being able to return an accurate answer from a molecule not in the training set) is to be expected, and our ability to generalise (as judged by test set error) improved as the number of molecules increased up to a few million. However, although we did not explore this, it is possible that our default architecture was simply too large for the smaller number of molecules, as excessive “capacity” can cause a loss of generalisation ability [135]. This, of course, leaves open the details of the size and “closeness” of that neighbourhood, how it varies with the encoding used (our original problem) and what features are used in practice to determine that neighbourhood. The network described here took nearly a week to train on a well-equipped GPU-based machine, and exhaustive analysis of hyperparameters was not possible. Consequently, because an understanding of the importance of local density will vary as a function of the position and nature of the relevant chemical space, we are not going to pursue them here. What is important is (i) that we could indeed learn to navigate these chemical spaces, and (ii) that the VAE approach permits a straightforward and novel estimation of molecular similarity.
Supplementary Materials
The following are available online at https://www.mdpi.com/1420-3049/25/15/3446/s1, Figure S1: Molecules similar to clozapine as judged by molecular fingerprint encodings.
Author Contributions
Conceptualization, all authors; methodology, all authors; software, S.S., S.O., N.S.; resources, D.B.K.; data curation, S.S., S.O., N.S.; writing—original draft preparation, D.B.K.; writing—review and editing, all authors; funding acquisition, D.B.K., T.J.R. All authors have read and agreed to the published version of the manuscript.
Funding
Present funding includes part of the EPSRC project SuSCoRD (EP/S004963/1), partly sponsored by AkzoNobel. DBK is also funded by the Novo Nordisk Foundation (grant NNF10CC1016517). Tim Roberts is funded by the NIHR UCLH Biomedical Research Centre.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Footnotes
Sample Availability: Samples of the compounds are not available from the authors.
References
- 1.Gasteiger J. Handbook of Chemoinformatics: From Data to Knowledge. Wiley/VCH; Weinheim, Germany: 2003. [Google Scholar]
- 2.Leach A.R., Gillet V.J. An Introduction to Chemoinformatics. Springer; Dordrecht, The Netherlands: 2007. [Google Scholar]
- 3.Maggiora G., Vogt M., Stumpfe D., Bajorath J. Molecular similarity in medicinal chemistry. J. Med. Chem. 2014;57:3186–3204. doi: 10.1021/jm401411z. [DOI] [PubMed] [Google Scholar]
- 4.Willett P. Similarity-based data mining in files of two-dimensional chemical structures using fingerprint measures of molecular resemblance. Wires Data Min. Knowl. 2011;1:241–251. doi: 10.1002/widm.26. [DOI] [Google Scholar]
- 5.Todeschini R., Consonni V. Molecular Descriptors for Cheminformatics. Wiley-VCH; Weinheim, Germany: 2009. [Google Scholar]
- 6.Ballabio D., Manganaro A., Consonni V., Mauri A., Todeschini R. Introduction to mole db—On-line molecular descriptors database. Math Comput. Chem. 2009;62:199–207. [Google Scholar]
- 7.Dehmer M., Varmuza K., Bonchev D. Statistical Modelling of Molecular Descriptors in QSAR/QSPR. Wiley-VCH; Weinheim, Germany: 2012. [Google Scholar]
- 8.Bender A., Glen R.C. Molecular similarity: A key technique in molecular informatics. Org. Biomol. Chem. 2004;2:3204–3218. doi: 10.1039/b409813g. [DOI] [PubMed] [Google Scholar]
- 9.Nisius B., Bajorath J. Rendering conventional molecular fingerprints for virtual screening independent of molecular complexity and size effects. ChemMedChem. 2010;5:859–868. doi: 10.1002/cmdc.201000089. [DOI] [PubMed] [Google Scholar]
- 10.Owen J.R., Nabney I.T., Medina-Franco J.L., López-Vallejo F. Visualization of molecular fingerprints. J. Chem. Inf. Model. 2011;51:1552–1563. doi: 10.1021/ci1004042. [DOI] [PubMed] [Google Scholar]
- 11.Riniker S., Landrum G.A. Similarity maps—A visualization strategy for molecular fingerprints and machine-learning methods. J. Cheminform. 2013;5:43. doi: 10.1186/1758-2946-5-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vogt M., Bajorath J. Bayesian screening for active compounds in high-dimensional chemical spaces combining property descriptors and molecular fingerprints. Chem. Biol. Drug Des. 2008;71:8–14. doi: 10.1111/j.1747-0285.2007.00602.x. [DOI] [PubMed] [Google Scholar]
- 13.Awale M., Reymond J.L. The polypharmacology browser: A web-based multi-fingerprint target prediction tool using chembl bioactivity data. J. Cheminform. 2017;9:11. doi: 10.1186/s13321-017-0199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geppert H., Bajorath J. Advances in 2d fingerprint similarity searching. Expert Opin. Drug Discov. 2010;5:529–542. doi: 10.1517/17460441.2010.486830. [DOI] [PubMed] [Google Scholar]
- 15.Muegge I., Mukherjee P. An overview of molecular fingerprint similarity search in virtual screening. Expert Opin. Drug. Discov. 2016;11:137–148. doi: 10.1517/17460441.2016.1117070. [DOI] [PubMed] [Google Scholar]
- 16.O’Boyle N.M., Sayle R.A. Comparing structural fingerprints using a literature-based similarity benchmark. J. Cheminform. 2016;8:36. doi: 10.1186/s13321-016-0148-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willett P. Similarity searching using 2d structural fingerprints. Meth. Mol. Biol. 2011;672:133–158. doi: 10.1007/978-1-60761-839-3_5. [DOI] [PubMed] [Google Scholar]
- 18.Durant J.L., Leland B.A., Henry D.R., Nourse J.G. Reoptimization of mdl keys for use in drug discovery. J. Chem. Inf. Comput. Sci. 2002;42:1273–1280. doi: 10.1021/ci010132r. [DOI] [PubMed] [Google Scholar]
- 19.Carhart R.E., Smith D.H., Venkataraghavan R. Atom pairs as molecular-features in structure activity studies—Definition and applications. J. Chem. Inf. Comp. Sci. 1985;25:64–73. doi: 10.1021/ci00046a002. [DOI] [Google Scholar]
- 20.Nilakantan R., Bauman N., Dixon J.S., Venkataraghavan R. Topological torsion—A new molecular descriptor for sar applications—Comparison with other descriptors. J. Chem. Inf. Comp. Sci. 1987;27:82–85. doi: 10.1021/ci00054a008. [DOI] [Google Scholar]
- 21.Rogers D., Hahn M. Extended-connectivity fingerprints. J. Chem. Inf. Model. 2010;50:742–754. doi: 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- 22.Hassan M., Brown R.D., Varma-O’brien S., Rogers D. Cheminformatics analysis and learning in a data pipelining environment. Mol. Divers. 2006;10:283–299. doi: 10.1007/s11030-006-9041-5. [DOI] [PubMed] [Google Scholar]
- 23.Glen R.C., Bender A., Arnby C.H., Carlsson L., Boyer S., Smith J. Circular fingerprints: Flexible molecular descriptors with applications from physical chemistry to adme. IDrugs. 2006;9:199–204. [PubMed] [Google Scholar]
- 24.Riniker S., Landrum G.A. Open-source platform to benchmark fingerprints for ligand-based virtual screening. J. Cheminform. 2013;5:26. doi: 10.1186/1758-2946-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Hagan S., Kell D.B. Consensus rank orderings of molecular fingerprints illustrate the ‘most genuine’ similarities between marketed drugs and small endogenous human metabolites, but highlight exogenous natural products as the most important ‘natural’ drug transporter substrates. ADMET & DMPK. 2017;5:85–125. [Google Scholar]
- 26.Dickens D., Rädisch S., Chiduza G.N., Giannoudis A., Cross M.J., Malik H., Schaeffeler E., Sison-Young R.L., Wilkinson E.L., Goldring C.E., et al. Cellular uptake of the atypical antipsychotic clozapine is a carrier-mediated process. Mol. Pharm. 2018;15:3557–3572. doi: 10.1021/acs.molpharmaceut.8b00547. [DOI] [PubMed] [Google Scholar]
- 27.Weininger D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1988;28:31–36. doi: 10.1021/ci00057a005. [DOI] [Google Scholar]
- 28.Rumelhart D.E., McClelland J.L. The PDP Research Group. Parallel Distributed Processing. Experiments in the Microstructure of Cognition. M.I.T. Press; Cambridge, MA, USA: 1986. [Google Scholar]
- 29.Goodacre R., Kell D.B., Bianchi G. Rapid assessment of the adulteration of virgin olive oils by other seed oils using pyrolysis mass spectrometry and artificial neural networks. J. Sci. Food Agric. 1993;63:297–307. doi: 10.1002/jsfa.2740630306. [DOI] [Google Scholar]
- 30.Goodacre R., Timmins É.M., Burton R., Kaderbhai N., Woodward A.M., Kell D.B., Rooney P.J. Rapid identification of urinary tract infection bacteria using hyperspectral whole-organism fingerprinting and artificial neural networks. Microbiology UK. 1998;144:1157–1170. doi: 10.1099/00221287-144-5-1157. [DOI] [PubMed] [Google Scholar]
- 31.Tetko I.V., Gasteiger J., Todeschini R., Mauri A., Livingstone D., Ertl P., Palyulin V., Radchenko E., Zefirov N.S., Makarenko A.S., et al. Virtual computational chemistry laboratory—Design and description. J. Comput. Aided Mol. Des. 2005;19:453–463. doi: 10.1007/s10822-005-8694-y. [DOI] [PubMed] [Google Scholar]
- 32.O’Boyle N., Dalke A. Deepsmiles: An Adaptation of Smiles for use in Machine-learning of Chemical Structures. [(accessed on 29 July 2020)];ChemRxiv. 2018 :7097960.v7097961. Available online: https://chemrxiv.org/articles/preprint/DeepSMILES_An_Adaptation_of_SMILES_for_Use_in_Machine-Learning_of_Chemical_Structures/7097960.
- 33.Segler M.H.S., Kogej T., Tyrchan C., Waller M.P. Generating focussed molecule libraries for drug discovery with recurrent neural networks. ACS Central Sci. 2017;4:120–131. doi: 10.1021/acscentsci.7b00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin W., Barzilay R., Jaakkola T. Junction Tree Variational Autoencoder for Molecular Graph Generation. arXiv. 20181802.04364v04362 [Google Scholar]
- 35.Kajino H. Molecular Hypergraph Grammar with its Application to Molecular Optimization. arXiv. 201802745v02741 [Google Scholar]
- 36.Panteleev J., Gao H., Jia L. Recent applications of machine learning in medicinal chemistry. Bioorg. Med. Chem. Lett. 2018;28:2807–2815. doi: 10.1016/j.bmcl.2018.06.046. [DOI] [PubMed] [Google Scholar]
- 37.Jaeger S., Fulle S., Turk S. Mol2vec: Unsupervised machine learning approach with chemical intuition. J. Chem. Inf. Model. 2018;58:27–35. doi: 10.1021/acs.jcim.7b00616. [DOI] [PubMed] [Google Scholar]
- 38.Shibayama S., Marcou G., Horvath D., Baskin I.I., Funatsu K., Varnek A. Application of the mol2vec technology to large-size data visualization and analysis. Mol. Inform. 2020;39:e1900170. doi: 10.1002/minf.201900170. [DOI] [PubMed] [Google Scholar]
- 39.Duvenaud D., Maclaurin D., Aguilera-Iparraguirre J., Gómez-Bombarelli R., Hirzel T., Aspuru-Guzik A., Adams R.P. Convolutional networks on graphs for learning molecular fingerprints. Adv. NIPS. 2015;2:2224–2232. [Google Scholar]
- 40.Kearnes S., McCloskey K., Berndl M., Pande V., Riley P. Molecular graph convolutions: Moving beyond fingerprints. J. Comput. Aided Mol. Des. 2016;30:595–608. doi: 10.1007/s10822-016-9938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gupta A., Müller A.T., Huisman B.J.H., Fuchs J.A., Schneider P., Schneider G. Generative recurrent networks for de novo drug design. Mol. Inform. 2018;37:1700111. doi: 10.1002/minf.201700111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schneider G. Generative models for artificially-intelligent molecular design. Mol. Inf. 2018;37:188031. doi: 10.1002/minf.201880131. [DOI] [PubMed] [Google Scholar]
- 43.Grisoni F., Schneider G. De novo molecular design with generative long short-term memory. Chimia. 2019;73:1006–1011. doi: 10.2533/chimia.2019.1006. [DOI] [PubMed] [Google Scholar]
- 44.Arús-Pous J., Blaschke T., Ulander S., Reymond J.L., Chen H., Engkvist O. Exploring the gdb-13 chemical space using deep generative models. J. Cheminform. 2019;11:20. doi: 10.1186/s13321-019-0341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jørgensen P.B., Schmidt M.N., Winther O. Deep generative models for molecular science. Mol. Inf. 2018;37:1700133. doi: 10.1002/minf.201700133. [DOI] [PubMed] [Google Scholar]
- 46.Li Y., Hu J., Wang Y., Zhou J., Zhang L., Liu Z. Deepscaffold: A comprehensive tool for scaffold-based de novo drug discovery using deep learning. J. Chem. Inf. Model. 2020;60:77–91. doi: 10.1021/acs.jcim.9b00727. [DOI] [PubMed] [Google Scholar]
- 47.Lim J., Hwang S.Y., Moon S., Kim S., Kim W.Y. Scaffold-based molecular design with a graph generative model. Chem. Sci. 2020;11:1153–1164. doi: 10.1039/C9SC04503A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moret M., Friedrich L., Grisoni F., Merk D., Schneider G. Generative molecular design in low data regimes. Nat. Mach. Intell. 2020;2:171–180. doi: 10.1038/s42256-020-0160-y. [DOI] [Google Scholar]
- 49.van Deursen R., Ertl P., Tetko I.V., Godin G. Gen: Highly efficient smiles explorer using autodidactic generative examination networks. J. Cheminform. 2020;12:22. doi: 10.1186/s13321-020-00425-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walters W.P., Murcko M. Assessing the impact of generative ai on medicinal chemistry. Nat Biotechnol. 2020;38:143–145. doi: 10.1038/s41587-020-0418-2. [DOI] [PubMed] [Google Scholar]
- 51.Yan C., Wang S., Yang J., Xu T., Huang J. Re-balancing Variational Autoencoder Loss for Molecule Sequence Generation. arXiv. 20191910.00698v00691 [Google Scholar]
- 52.Winter R., Montanari F., Noé F., Clevert D.A. Learning continuous and data-driven molecular descriptors by translating equivalent chemical representations. Chem. Sci. 2019;10:1692–1701. doi: 10.1039/C8SC04175J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Samanta B., De A., Ganguly N., Gomez-Rodriguez M. Designing Random Graph Models using Variational Autoencoders with Applications to Chemical Design. arXiv. 20181802.05283 [Google Scholar]
- 54.Krenn M., Häse F., Nigam A., Friederich P., Aspuru-Guzik A. Self-Referencing Embedded Strings (selfies): A 100% Robust Molecular String Representation. arXiv. 20191905.13741 [Google Scholar]
- 55.Sattarov B., Baskin I.I., Horvath D., Marcou G., Bjerrum E.J., Varnek A. De novo molecular design by combining deep autoencoder recurrent neural networks with generative topographic mapping. J. Chem. Inf. Model. 2019;59:1182–1196. doi: 10.1021/acs.jcim.8b00751. [DOI] [PubMed] [Google Scholar]
- 56.Bengio Y., Courville A., Vincent P. Representation learning: A review and new perspectives. IEEE Trans. Patt. Anal. Mach. Intell. 2013;35:1798–1828. doi: 10.1109/TPAMI.2013.50. [DOI] [PubMed] [Google Scholar]
- 57.Bousquet O., Gelly S., Tolstikhin I., Simon-Gabriel C.-J., Schoelkopf B. From Optimal Transport to Generative Modeling: The Vegan Cookbook. arXiv. 20171705.07642 [Google Scholar]
- 58.Husain H., Nock R., Williamson R.C. Adversarial Networks and Autoencoders: The Primal-dual Relationship and Generalization Bounds. arXiv. 20191902.00985 [Google Scholar]
- 59.Goodfellow I.J., Pouget-Abadie J., Mirza M., Xu B., Warde-Farley D., Ozairy S., Courville A., Bengio Y. Generative adversarial nets. arXiv. 20141406.2661v1401 [Google Scholar]
- 60.Polykovskiy D., Zhebrak A., Vetrov D., Ivanenkov Y., Aladinskiy V., Mamoshina P., Bozdaganyan M., Aliper A., Zhavoronkov A., Kadurin A. Entangled conditional adversarial autoencoder for de novo drug discovery. Mol. Pharm. 2018;15:4398–4405. doi: 10.1021/acs.molpharmaceut.8b00839. [DOI] [PubMed] [Google Scholar]
- 61.Arjovsky M., Chintala S., Bottou L. Wasserstein gan. arXiv. 20171701.07875v07873 [Google Scholar]
- 62.Goodfellow I. Generative adversarial networks. arXiv. 20171701.00160v00161 [Google Scholar]
- 63.Foster D. Generative Deep Learning. O’Reilly; Sebastopol, CA, USA: 2019. [Google Scholar]
- 64.Langr J., Bok V. Gans in Action. Manning; Shelter Island, NY, USA: 2019. [Google Scholar]
- 65.Prykhodko O., Johansson S.V., Kotsias P.C., Arús-Pous J., Bjerrum E.J., Engkvist O., Chen H.M. A de novo molecular generation method using latent vector based generative adversarial network. J. Cheminform. 2019;11:74. doi: 10.1186/s13321-019-0397-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao J.J., Kim Y., Zhang K., Rush A.M., LeCun Y. Adversarially Regularized Autoencoders for Generating Discrete Structures. arXiv. 20171706.04223v04221 [Google Scholar]
- 67.Kingma D., Welling M. Auto-encoding variational bayes. arXiv. 20141312.6114v1310 [Google Scholar]
- 68.Rezende D.J., Mohamed S., Wierstra D. Stochastic Backpropagation and Approximate Inference in Deep Generative Models. arXiv. 20141401.4082v1403 [Google Scholar]
- 69.Doersch C. Tutorial on Variational Autoencoders. arXiv. 20161606.05908v05902 [Google Scholar]
- 70.Benhenda M. Chemgan Challenge for Drug Discovery: Can ai Reproduce Natural Chemical Diversity? arXiv. 20171708.08227v08223 [Google Scholar]
- 71.Griffiths R.-R., Hernández-Lobato J.M. Constrained Bayesian Optimization for Automatic Chemical Design. arXiv. 2017 doi: 10.1039/c9sc04026a.1709.05501v05505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Aumentado-Armstrong T. Latent Molecular Optimization for Targeted Therapeutic Design. arXiv. 20181809.02032 [Google Scholar]
- 73.Blaschke T., Olivecrona M., Engkvist O., Bajorath J., Chen H.M. Application of generative autoencoder in de novo molecular design. Mol. Inform. 2018;37:1700123. doi: 10.1002/minf.201700123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gómez-Bombarelli R., Wei J.N., Duvenaud D., Hernández-Lobato J.M., Sánchez-Lengeling B., Sheberla D., Aguilera-Iparraguirre J., Hirzel T.D., Adams R.P., Aspuru-Guzik A. Automatic chemical design using a data-driven continuous representation of molecules. ACS Cent. Sci. 2018;4:268–276. doi: 10.1021/acscentsci.7b00572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tschannen M., Bachem O., Lucic M. Recent Advances in Autoencoder-based Representation Learning. arXiv. 20181812.05069v05061 [Google Scholar]
- 76.Kingma D.P., Welling M. An Introduction to Variational Autoencoders. arXiv. 20191906.02691v02691 [Google Scholar]
- 77.Rezende D.J., Viola F. Taming vaes. arXiv. 20181810.00597v00591 [Google Scholar]
- 78.Hutson M. Core progress in ai has stalled in some fields. Science. 2020;368:927. doi: 10.1126/science.368.6494.927. [DOI] [PubMed] [Google Scholar]
- 79.Burgess C.P., Higgins I., Pal A., Matthey L., Watters N., Desjardins G., Lerchner A. Understanding disentangling in β-vae. arXiv. 20181804.03599 [Google Scholar]
- 80.Taghanaki S.A., Havaei M., Lamb A., Sanghi A., Danielyan A., Custis T. Jigsaw-vae: Towards Balancing Features in Variational Autoencoders. arXiv. 20202005.05496 [Google Scholar]
- 81.Caterini A., Cornish R., Sejdinovic D., Doucet A. Variational Inference with Continuously-Indexed Normalizing Flows. arXiv. 20202007.05426 [Google Scholar]
- 82.Nielsen D., Jaini P., Hoogeboom E., Winther O., Welling M. Survae flows: Surjections to bridge the Gap between Vaes and Flows. arXiv. 20202007.02731 [Google Scholar]
- 83.Li Y., Yu S., Principe J.C., Li X., Wu D. Pri-vae: Principle-of-relevant-information Variational Autoencoders. arXiv. 20202007.06503 [Google Scholar]
- 84.Wolpert D.H., Macready W.G. No free lunch theorems for optimization. IEEE Trans. Evol. Comput. 1997;1:67–82. doi: 10.1109/4235.585893. [DOI] [Google Scholar]
- 85.Vaswani A., Shazeer N., Parmar N., Uszkoreit J., Jones L., Gomez A.N., Kaiser L., Polosukhin I. Attention is All You Need. arXiv. 20171706.03762 [Google Scholar]
- 86.Devlin J., Chang M.-W., Lee K., Toutanova K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv. 20181810.04805 [Google Scholar]
- 87.Dai B., Wipf D. Diagnosing and Enhancing vae Models. arXiv. 20191903.05789v05782 [Google Scholar]
- 88.Asperti A., Trentin M. Balancing Reconstruction Error and Kullback-leibler Divergence in Variational Autoencoders. arXiv. 20202002.07514v07511 [Google Scholar]
- 89.Goodacre R., Pygall J., Kell D.B. Plant seed classification using pyrolysis mass spectrometry with unsupervised learning: The application of auto-associative and kohonen artificial neural networks. Chemometr. Intell. Lab. Syst. 1996;34:69–83. doi: 10.1016/0169-7439(96)00021-4. [DOI] [Google Scholar]
- 90.Yao X. Evolving artificial neural networks. Proc. IEEE. 1999;87:1423–1447. [Google Scholar]
- 91.Floreano D., Dürr P., Mattiussi C. Neuroevolution: From architectures to learning. Evol. Intell. 2008;1:47–62. doi: 10.1007/s12065-007-0002-4. [DOI] [Google Scholar]
- 92.Vassiliades V., Christodoulou C. Toward nonlinear local reinforcement learning rules through neuroevolution. Neural Comput. 2013;25:3020–3043. doi: 10.1162/NECO_a_00514. [DOI] [PubMed] [Google Scholar]
- 93.Stanley K.O., Clune J., Lehman J., Miikkulainen R. Designing neural networks through neuroevolution. Nat. Mach. Intell. 2019;1:24–35. doi: 10.1038/s42256-018-0006-z. [DOI] [Google Scholar]
- 94.Iba H., Noman N. Deep Neural Evolution: Deep Learning with Evolutionary Computation. Springer; Berlin, Germany: 2020. [Google Scholar]
- 95.Le Cun Y., Denker J.S., Solla S.A. Optimal brain damage. Adv. Neural Inf. Proc. Syst. 1990;2:598–605. [Google Scholar]
- 96.Dietterich T.G. Ensemble methods in machine learning. LNCS. 2000;1857:1–15. [Google Scholar]
- 97.Hinton G.E., Srivastava N., Krizhevsky A., Sutskever I., Salakhutdinov R.R. Improving neural networks by preventing co-adaptation of feature detectors. arXiv. 20121207.0580 [Google Scholar]
- 98.Keskar N.S., Mudigere D., Nocedal J., Smelyanskiy M., Tang P.T.P. On large-batch training for deep learning: Generalization gap and sharp minima. arXiv. 20171609.04836v04832 [Google Scholar]
- 99.O’Hagan S., Swainston N., Handl J., Kell D.B. A ‘rule of 0.5′ for the metabolite-likeness of approved pharmaceutical drugs. Metabolomics. 2015;11:323–339. doi: 10.1007/s11306-014-0733-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.O’Hagan S., Kell D.B. Understanding the foundations of the structural similarities between marketed drugs and endogenous human metabolites. Front. Pharmacol. 2015;6:105. doi: 10.3389/fphar.2015.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.O’Hagan S., Kell D.B. Metmaxstruct: A tversky-similarity-based strategy for analysing the (sub)structural similarities of drugs and endogenous metabolites. Front. Pharmacol. 2016;7:266. doi: 10.3389/fphar.2016.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.O’Hagan S., Kell D.B. Analysis of drug-endogenous human metabolite similarities in terms of their maximum common substructures. J. Cheminform. 2017;9:18. doi: 10.1186/s13321-017-0198-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.O’Hagan S., Kell D.B. Analysing and navigating natural products space for generating small, diverse, but representative chemical libraries. Biotechnol. J. 2018;13:1700503. doi: 10.1002/biot.201700503. [DOI] [PubMed] [Google Scholar]
- 104.O’Hagan S., Kell D.B. Structural Similarities between Some Common Fluorophores used in Biology and Marketed drugs, Endogenous Metabolites, and Natural Products. [(accessed on 29 July 2020)];bioRxiv. 2019 :834325. doi: 10.3390/md18110582. Available online: https://www.biorxiv.org/content/10.1101/834325v1.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Samanta S., O’Hagan S., Swainston N., Roberts T.J., Kell D.B. Vae-sim: A novel Molecular Similarity Measure Based on a Variational Autoencoder. [(accessed on 29 July 2020)];bioRxiv. 2020 :172908. doi: 10.3390/molecules25153446. Available online: https://www.biorxiv.org/content/10.1101/2020.06.26.172908v1.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dai H., Tian Y., Dai B., Skiena S., Song L. Syntax-Directed Variational Autoencoder for Structured data. arXiv. 20181802.08786v08721 [Google Scholar]
- 107.Kusner M.J., Paige B., Hernández-Lobato J.M. Grammar Variational Autoencoder. arXiv. 20171703.01925v01921 [Google Scholar]
- 108.Kingma D.P., Ba J.L. Adam: A Method for Stochastic Optimization. arXiv. 20151412.6980v1418 [Google Scholar]
- 109.Glorot X., Bengio Y. Understanding the difficulty of training deep feedforward neural networks. Proc. AISTATs. 2010;9:249–256. [Google Scholar]
- 110.O’Hagan S., Kell D.B. The knime workflow environment and its applications in genetic programming and machine learning. Genetic Progr. Evol. Mach. 2015;16:387–391. doi: 10.1007/s10710-015-9247-3. [DOI] [Google Scholar]
- 111.McInnes L., Healy J., Melville J. Umap: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv. 20181802.03426v03422 [Google Scholar]
- 112.McInnes L., Healy J., Saul N., Großberger L. Umap: Uniform manifold approximation and projection. J. Open Source Software. 2018 doi: 10.21105/joss.00861. [DOI] [Google Scholar]
- 113.Citraro R., Leo A., Aiello R., Pugliese M., Russo E., De Sarro G. Comparative analysis of the treatment of chronic antipsychotic drugs on epileptic susceptibility in genetically epilepsy-prone rats. Neurotherapeutics. 2015;12:250–262. doi: 10.1007/s13311-014-0318-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Thorn C.F., Muller D.J., Altman R.B., Klein T.E. Pharmgkb summary: Clozapine pathway, pharmacokinetics. Pharmacogenet. Genomics. 2018;28:214–222. doi: 10.1097/FPC.0000000000000347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hopkins A.L., Mason J.S., Overington J.P. Can we rationally design promiscuous drugs? Curr. Opin. Struct. Biol. 2006;16:127–136. doi: 10.1016/j.sbi.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 116.Mestres J., Gregori-Puigjané E., Valverde S., Solé R.V. The topology of drug-target interaction networks: Implicit dependence on drug properties and target families. Mol. Biosyst. 2009;5:1051–1057. doi: 10.1039/b905821b. [DOI] [PubMed] [Google Scholar]
- 117.Mestres J., Gregori-Puigjané E. Conciliating binding efficiency and polypharmacology. Trends Pharmacol. Sci. 2009;30:470–474. doi: 10.1016/j.tips.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 118.Oprea T.I., Bauman J.E., Bologa C.G., Buranda T., Chigaev A., Edwards B.S., Jarvik J.W., Gresham H.D., Haynes M.K., Hjelle B., et al. Drug repurposing from an academic perspective. Drug Discov. Today Ther. Strateg. 2011;8:61–69. doi: 10.1016/j.ddstr.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dimova D., Hu Y., Bajorath J. Matched molecular pair analysis of small molecule microarray data identifies promiscuity cliffs and reveals molecular origins of extreme compound promiscuity. J. Med. Chem. 2012;55:10220–10228. doi: 10.1021/jm301292a. [DOI] [PubMed] [Google Scholar]
- 120.Peters J.U., Hert J., Bissantz C., Hillebrecht A., Gerebtzoff G., Bendels S., Tillier F., Migeon J., Fischer H., Guba W., et al. Can we discover pharmacological promiscuity early in the drug discovery process? Drug Discov. Today. 2012;17:325–335. doi: 10.1016/j.drudis.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 121.Hu Y., Gupta-Ostermann D., Bajorath J. Exploring compound promiscuity patterns and multi-target activity spaces. Comput. Struct. Biotechnol. J. 2014;9:e201401003. doi: 10.5936/csbj.201401003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bajorath J. Molecular similarity concepts for informatics applications. Methods Mol. Biol. 2017;1526:231–245. doi: 10.1007/978-1-4939-6613-4_13. [DOI] [PubMed] [Google Scholar]
- 123.Eckert H., Bajorath J. Molecular similarity analysis in virtual screening: Foundations, limitations and novel approaches. Drug Discov. Today. 2007;12:225–233. doi: 10.1016/j.drudis.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 124.Medina-Franco J.L., Maggiora G.M. Molecular similarity analysis. In: Bajorath J., editor. Chemoinformatics for Drug Discovery. Wiley; Hoboken, NJ, USA: 2014. pp. 343–399. [Google Scholar]
- 125.Zhang B., Vogt M., Maggiora G.M., Bajorath J. Comparison of bioactive chemical space networks generated using substructure- and fingerprint-based measures of molecular similarity. J. Comput. Aided Mol. Des. 2015;29:595–608. doi: 10.1007/s10822-015-9852-5. [DOI] [PubMed] [Google Scholar]
- 126.Hornik K., Stinchcombe M., White H. Multilayer feedforward networks are universal approximators. Neural Netw. 1989;2:359–366. doi: 10.1016/0893-6080(89)90020-8. [DOI] [Google Scholar]
- 127.Hornik K. Approximation capabilities of multilayer feedforward networks. Neural Netw. 1991;4:251–257. doi: 10.1016/0893-6080(91)90009-T. [DOI] [Google Scholar]
- 128.Everitt B.S. Cluster Analysis. Edward Arnold; London, UK: 1993. [Google Scholar]
- 129.Jain A.K., Dubes R.C. Algorithms for Clustering Data. Prentice Hall; Englewood Cliffs, NJ, USA: 1988. [Google Scholar]
- 130.Kaufman L., Rousseeuw P.J. Finding Groups in Data. An Introduction to Cluster Analysis. Wiley; New York, NY, USA: 1990. [Google Scholar]
- 131.Handl J., Knowles J., Kell D.B. Computational cluster validation in post-genomic data analysis. Bioinformatics. 2005;21:3201–3212. doi: 10.1093/bioinformatics/bti517. [DOI] [PubMed] [Google Scholar]
- 132.MacCuish J.D., MacCuish N.E. Clustering in Bioinformatics And Drug Discovery. CRC Press; Boca Raton, FL, USA: 2011. [Google Scholar]
- 133.Hong S.H., Ryu S., Lim J., Kim W.Y. Molecular generative model based on an adversarially regularized autoencoder. J. Chem. Inf. Model. 2020;60:29–36. doi: 10.1021/acs.jcim.9b00694. [DOI] [PubMed] [Google Scholar]
- 134.Bozkurt A., Esmaeili B., Brooks D.H., Dy J.G., van de Meent J.-W. Evaluating Combinatorial Generalization in Variational Autoencoders. arXiv. 20191911.04594v04591 [Google Scholar]
- 135.Bozkurt A., Esmaeili B., Brooks D.H., Dy J.G., van de Meent J.-W. Can Vaes Generate novel Examples? arXiv. 20181812.09624v09621 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.