Abstract
Two novel structural types of tyrosyl-DNA phosphodiesterase 1 (TDP1) inhibitors with hexahydroisobenzofuran 11 and 3-oxabicyclo [3.3.1]nonane 12 scaffolds were discovered. These monoterpene-derived compounds were synthesized through preliminary isomerization of (+)-3-carene to (+)-2-carene followed by reaction with heteroaromatic aldehydes. All the compounds inhibit the TDP1 enzyme at micro- and submicromolar levels, with the most potent compound having an IC50 value of 0.65 μM. TDP1 is an important DNA repair enzyme and a promising target for the development of new chemosensitizing agents. A panel of isogenic clones of the HEK293FT cell line knockout for the TDP1 gene was created using the CRISPR-Cas9 system. Cytotoxic effects of topotecan (Tpc) and non-cytotoxic compounds of the new structures were investigated separately and jointly in the TDP1 gene knockout cells. For two TDP1 inhibitors, 11h and 12k, a synergistic effect was observed with Tpc in the HEK293FT cells but was not found in TDP1 −/− cells. Thus, it is likely that the synergistic effect is caused by inhibition of TDP1. Synergy was also found for 11h in other cancer cell lines. Thus, sensitizing cancer cells using a non-cytotoxic drug can enhance the efficacy of currently used pharmaceuticals and, concomitantly, reduce toxic side effects.
Keywords: monoterpene, carene, topotecan, tyrosyl-DNA phosphodiesterase 1, synergy, TDP1 gene knockout cells, inhibitor
1. Introduction
There are a number of unresolved issues regarding chemotherapeutic treatment for oncological diseases, for example, low efficiency, resistance of malignant tumors, side effects, and high toxic load. The cytotoxic effect of chemotherapy is caused by DNA damage, and the ability of cancer cells to repair DNA lesions results in resistance. Therefore, the development of DNA repair enzyme inhibitors is a promising strategy to improve the efficacy of DNA damaging agents in clinical use.
Tyrosyl-DNA-phosphodiesterase 1 (TDP1) is a DNA repair enzyme and a promising therapeutic target to enhance established cancer treatment [1,2,3,4]. TDP1 plays a key role in the removal of DNA lesions, in particular those caused by DNA topoisomerase I (TOP1) inhibitors. TOP1 is an essential enzyme that regulates DNA topology by reducing DNA supercoiling. TOP1 introduces a transient single-strand break in DNA, enabling the broken strand to rotate around the TOP1-bound DNA during fundamental cellular events, such as replication, transcription, and repair [5]. TOP1 inhibitors, for example, topotecan (Tpc) and irinotecan, both important chemotherapeutic agents, stabilize the cleavage complex TOP1–DNA, leading to cell death [6]. However, TDP1 removes the stalled TOP1-DNA complex, reducing the efficacy of the drugs and leading to resistance [7,8,9]. As has been confirmed in gene knockout studies in mice and human cell lines, TDP1 contributes to the development of drug resistance in a number of cancers (see [3] and references therein). Indeed, human cell lines with mutant or missing TDP1 as well as TDP1-knockout mice are both hypersensitive to the well-established TOP1 inhibitor camptothecin and its derivatives. Conversely, at high TDP1 expression levels, camptothecin and etoposide cause limited DNA damage [3]. Thus, a promising approach to increase the selectivity and potency of TOP1 poisons to cancer cells is to combine them with TDP1 inhibitors, which can significantly increase the chemotherapeutic efficacy.
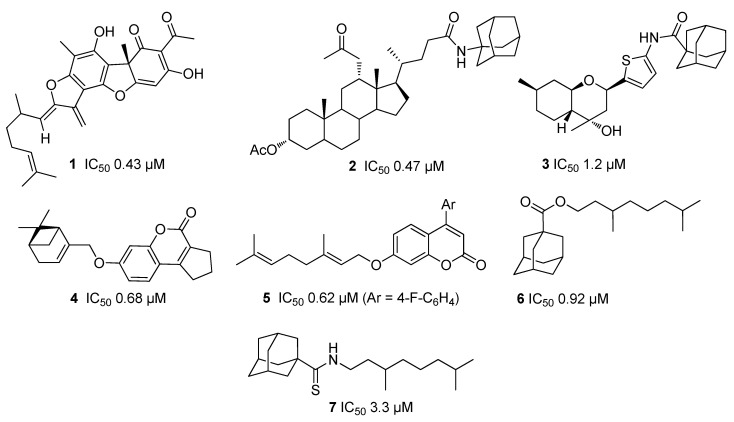
In recent years, TDP1 inhibitors of various chemical types have been discovered [1,2,3,4], including natural products, such as usnic [10,11,12] and bile acids [13] (for example, compounds 1 and 2, Figure 1).
Figure 1.
Structures of known TDP1 inhibitors and their half maximal inhibitory concentration (IC50) values.
Monoterpenes, which have a unique diverse structure and are inexpensive, available and often enantiomerically pure, is an attractive renewable material for the development of physiologically active agents [14]. One of the approaches to utilize various monoterpenes, such as pinenes and p-menthenes, is their capability to react with carbonyl compounds, mainly aldehydes, resulting in heterocyclic compounds of different structural types [14,15]. Some of these derivatives exhibit analgesic, antiviral, neuroprotective, and antitumor properties [14]. Moreover, new classes of TDP1 inhibitors with submicromolar half maximal inhibitory concentration (IC50) values have been developed that contain monoterpene moieties; for example, coumarin [16], 4-arylcoumarin [17], and adamantane [18,19,20,21] derivatives as well as octahydro-2H-chromenes 3 (see Figure 1) synthesized from p-menthane monoterpenoid (-)-isopulegol [22]. Their ability to enhance the Tpc cytotoxic potential was demonstrated for several of these derivatives. For example, 7-hydroxycoumarin derivative 4 (Figure 1) with a pinene-type fragment reduced the CC50 (median cytotoxic concentration causing 50% cell death) of camptothecin eight-fold for the human breast adenocarcinoma cell line MCF-7 [16] while the hybrid molecule 5, comprising of (4-fluorophenyl)-coumarin and geraniol moieties, showed a significant increase in the antitumor effect of Tpc on Krebs-2 ascites in an in vivo tumor model [17]. Among the adamantane derivatives, strong synergism was found for 6 and 7 (Figure 1), both containing the acyclic monoterpene substituents, when lung adenocarcinoma cells A549 were treated in combination with Tpc [18,20].
Monoterpene (+)-3-carene 8 (Scheme 1) is one of the main components of turpentine. Nevertheless, this compound is very rarely considered as a molecular platform for the development of new therapeutics and never considered for TDP1 inhibitors. In the current work, we studied the anti-TDP1 activity of (+)-3-carene derivatives for the first time. Two sets of (+)-3-carene derivatives with hexahydroisobenzofuran and 3-oxabicyclo [3.3.1]nonane skeletons were synthesized through preliminary isomerization of 3-carene to 2-carene followed by reaction with aldehydes. As a result, new structural types of potent TDP1 inhibitors were discovered. Furthermore, to elucidate the inhibitors’ mechanism of action on the cellular level, a panel of HEK293FT TDP1 knockout isogenic clones was created using the CRISPR-Cas9 approach and the cytotoxic potentials were measured.
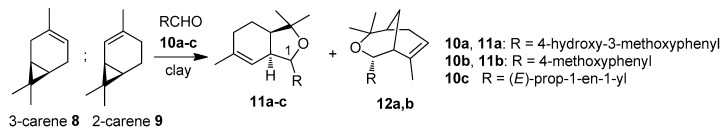
Scheme 1.
Reaction of carenes with aldehydes 10a–c [23].
2. Results and Discussion
2.1. Chemistry
It has been shown that carene can be successfully reacted with carbonyl compounds [23,24]. Previously, we showed that condensation of 2-carene 9 with aldehydes 10a–c in the presence of montmorillonite clays led to the formation of hexahydroisobenzofurans 11a–c as a mixture of two isomers at the C(1) position and compounds with 3-oxabicyclo[3.3.1]nonane structure 12 as minor products (Scheme 1). It was found that hexahydroisobenzofuran 11a obtained with condensation of 2-carene 9 with 4-hydroxy-3-methoxybenzaldehyde 10a demonstrated potent neuroprotective activity in an in vivo Parkinson model combined with low acute toxicity [24]. The reactions of 3-carene 8 with aldehydes gave the same products 11 and 12 as it did for 2-carene 9 but in much lower yields (only 3% for the reaction with aldehyde 10a), since 2-carene 9 is more reactive due to the conjugation of the double bond with the cyclopropane ring [23].
Interestingly, 2-carene 9 is widely found in nature albeit in small concentrations. The isolation of 9 from essential oils, or from reaction mixtures after 3-carene 8 isomerization, is difficult, making 9 expensive and therefore not attractive for use in organic synthesis [25,26,27,28,29]. Recently, we developed a method for the isomerization of readily available 3-carene 8 to 2-carene 9 using montmorillonite clays as a catalyst [30]. It was demonstrated that a mixture of carenes, limonene, and terpinenes containing 10% 2-carene can be effectively used as a starting material instead of pure 2-carene in a clay-catalyzed reaction with 4-hydroxy-3-methoxybenzaldehyde 10a, resulting in chiral isobenzofurans 11a. The largest yield of 11a was obtained with the 2-carene-containing mixture (60%) [31], which is much higher than using pure 2-carene 9 (33%). At the same time, condensation of carenes with aldehydes has not yet been systematically studied. Here, we synthesized heterocyclic compounds using 2-carene from a mixture containing carenes, limonene, and terpinenes with 2-carene comprising 10%, by condensation with heteroaromatic furan and thiophene aldehydes. The choice of this aldehyde was based on the presence of furan and thiophene moieties in many bioactive compounds, including TDP1 inhibitors [32,33,34,35], as well as preliminary molecular modelling that indicated favourable binding to TDP1.
For the synthesis of heterocyclic compounds, we used a mixture of terpenes obtained by the isomerization of 3-carene 8 in the presence of montmorillonite K10 [31], which consists of 10% 2-carene, 30% 3-carene, 20% m- and p-cymols, 12% terpinenes, 5% limonene, and a number of dimeric compounds (m/z = 272) (according to gas chromatography-mass spectrometry (GC-MS) data). The reaction pathways for 3-carene isomerization are shown in Scheme S1, Supplementary Material.
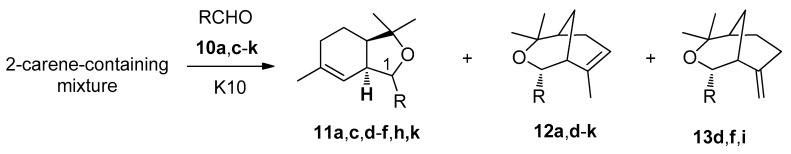
At first, we carried out the reactions with 4-hydroxy-3-methoxybenzaldehyde 10a and crotonaldehyde 10c previously studied using the 2-carene-containing mixture instead of neat 2-carene 9 [23]. In the present work, we used montmorillonite clay K10 as a catalyst, and the reaction was carried out without solvent at room temperature for 20 h. As a result of the reaction of 4-hydroxy-3-methoxybenzaldehyde 10a and the 2-carene-containing mixture, isobenzofurans 11a were obtained, which are diastereomers at position C(1) ((S)-/(R)- = 1:1, total yield 81%). In addition, compound 12a (11%) was isolated (Scheme 2, Table 1).
Scheme 2.
Reaction of 2-carene-containing mixture with aldehydes 10a, c-k.
Table 1.
Results of reaction of 2-carene-containing mixture with aldehides 10a, c–k.
| R | RCHO | Product 11 (Yield, (S)-/(R) Ratio) | Products 12 and 13 (Total Yield, %, 12/13) |
|---|---|---|---|
| 4-Hydroxy-3-methoxyphenyl | 10a | 11a (81%, (S)-/(R) = 1:1) | 12a (11%) |
| (E)-prop-1-en-1-yl | 10c | 11c (89%, (S)-/(R) = 1.3:1) | |
| Thiophen-2-yl | 10d | 11d (78%, (S)-isomer) | 12d, 13d (5%, 7:1) |
| 3-Methylthiophen-2-yl | 10e | 11e (86%, (S)-isomer) | 12e (2%) |
| 5-Methylthiophen-2-yl | 10f | 11f (85%, (S)-isomer) | 12f, 13f (2%, 3:1) |
| 4-Bromothiophen-2-yl | 10g | - | 12g (20%) |
| 5-Bromothiophen-2-yl | 10h | 11h (73%, (S)-isomer) | 12h (15%) |
| 5-Nitrothiophen-2-yl | 10i | - | 12i, 13i (18%, 5:1) |
| 5-Nitrofuran-2-yl | 10j | - | 12j (18%) |
| Thiophen-3-yl | 10k | 11k (91%, (S)-/(R) = 1.5:1) | 12k (21%) |
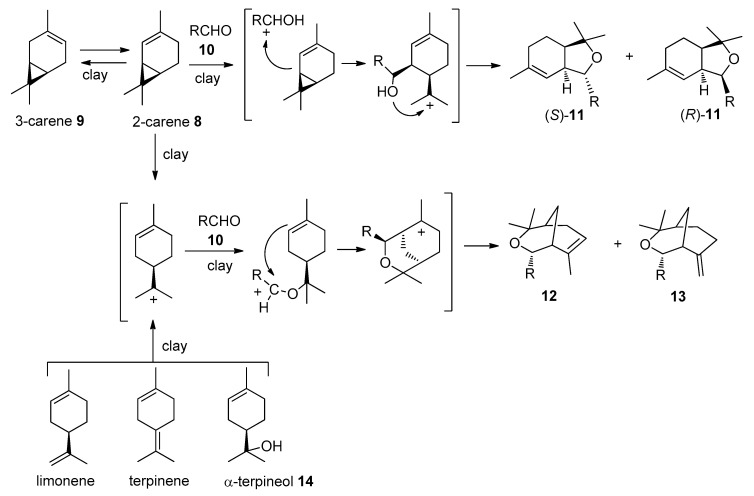
The reactants for the products with the 3-oxabicyclo[3.3.1]nonane framework 12a in reactions of the 2-carene-containing mixture with aldehydes can be both 2- and 3-carene as well as limonene and possibly terpinenes [15,23]; a proposed reaction mechanism for compounds 11, 12, and 13 is shown in Scheme 3. The yields for 11, 12, and 13 were calculated based on the 2-carene concentration in the mixture and on the total concentration of carenes, limonene, and terpinenes.
Scheme 3.
The proposed reaction mechanism for compounds 11, 12, and 13.
The reaction of crotonaldehyde 10c with the 2-carene-containing mixture on K10 resulted in isobenzofurans 11c ((S)/(R) = 1.3:1, total yield of 89%). The reaction yielded more 11c than the 2-carene’s concentration in the mixture, with a higher product yield than for the reaction only using 2-carene 9. It is therefore clear that the mixture can be successfully used without isolating the 2-carene 9. We also studied the reactions of the 2-carene-containing mixture with the heteroaromatic aldehydes 10d-k (thiophenecarboxaldehyde, furaldehyde, and their derivatives). Compound (S)-11d with a hexahydroisobenzofuran moiety was synthesized in a 78% yield, reacting the 2-carene-containing mixture with 2-thiophenecarboxaldehyde 10d catalyzed by montmorillonite K10. In addition, a mixture of two compounds 12d and 13d with a 3-oxabicyclo[3.3.1]nonane skeletone was formed, which are isomers depending on the double bond position (12d/13d = 7:1, total yield 5%) (Scheme 2, Table 1). It should be noted that compound 11d, in contrast to the previously studied reactions with aromatic aldehydes, the crotonaldehyde, formed exclusively as an (S)-isomer [23]. Moreover, although Prins cyclization in reactions of terpenoids with aldehydes, leading to the formation of heterocycles, with the 3-oxabicyclo[3.3.1]nonane framework is well studied [15,23], the only example of isolating a compound with an exocyclic double bond is the reaction of α-terpineol 14 with benzaldehyde in the presence of BF3∗Et2O, leading to 13d [36].
The reaction of the 2-carene-containing mixture with methyl-substituted thiophene-2-carboxaldehydes 10e and 10f is similar to the previously mentioned reactions. Thus, starting from 3-methylthiophene-2-carboxaldehyde 10e, compounds 11e and 12e were synthesized with yields of 86% and 2%, respectively; compound 13e was formed only in trace amounts. Compound 11f (85%) and a mixture of 12f and 13f (12f/13f = 3:1, 2%) were obtained in the reaction with 5-methylthiophene-2-carboxaldehyde 10f. Compounds 11h and 12h were synthesized with yields of 73% and 35%, respectively, using 5-bromothiophen-2-carboxaldehyde 10h. The reaction of the 2-carene-containing mixture with 4-bromothiophen-2-carboxaldehyde 10g proceeded quite differently, as the only product of this reaction was compound 12g with the 3-oxabicyclo[3.3.1]nonane framework with a 20% yield.
When 5-nitro-substituted heteroaromatic aldehydes were reacted with the 2-carene-containing mixture, compounds with the 3-oxabicyclo[3.3.1]nonane framework were only obtained. This can be explained by the low ability for protonation by these aldehydes, leading to high-energy barriers for the first stage of the isobenzofuran formation path (Scheme 3). Thus, by the reaction with 5-nitrothiophene-2-carboxaldehyde 13i and 5-nitro-2-furaldehyde 13j, compounds 12i, 13i (18%, 12i/13i = 5:1), and 12j (18%) were synthesized.
Interestingly, reaction of the 2-caren-containing mixture with 3-thiophenecarboxaldehyde 11k, the isomer of 2-thiophenecarboxaldehyde 10d, proceeded in a non-stereoseletive way with the formation of 11k as a pair of diastereomers at the substituent position at C(1) in the ratio (S)-11k/(R)-11k = 1.5:1, with a total yield of 91%. Additionally, compound 12k with 3-oxabicyclo[3.3.1]nonane skeletone (21%) was isolated from the reaction mixture.
It should be noted that in the case of the 2-carene-containing mixture reactions with aldehydes 10, the main or even the only way of isobenzofuran synthesis was the formation of (S)-isomers (excluding the interaction with 4-hydroxy-3-methoxybenzaldehyde 10a). Predominant formation of the (S)-isomer of isobenzofuran was clearly observed for the condensation of 2-carene and anisaldehyde [37]. According to denstity functional theory (DFT) calculations, the formation of (S)-isomer is energetically more favorable (by 6.8 kJ/mol) compared to formation of the (R)-isomer [37]. Moreover, the lower stability of the (R)-isomer in comparison with the (S)-isomer due to secondary transformations, leading to resinofication under the reaction conditions compared with (S)-isomer, was also shown [37].
Reactions of 2- and 3-furaldehyde as well as pyrrole-2-carboxaldehyde with the 2-carene-containing mixture did not lead to the formation of the target heterocyclic compounds. According to GC-MS data, after the reactions, 2-carene remained unconverted, but limonene was completely consumed and traces of compounds with m/z values corresponding to the intermolecular products, most likely with the 3-oxabicyclo[3.3.1]nonane skeleton, were observed. Apparently, the intermolecular products formed during the reaction are unstable and undergo degradation in the presence of an acid catalyst.
It should be noted that unreacted monoterpenes and other substances from the 2-carene-containing mixture are easily separated from the more polar condensation products by column chromatography using hexane as an elute. Moreover, compounds with the 3-oxabicyclo[3.3.1]nonane sceletone 12 can also be synthesized based on more available limonene [15]. For example, condensation of limonene with 2-thiophenecarboxaldehyde 10d catalyzed by montmorillonite K10 without solvent at room temperature after 2 h led to the formation of compounds 12d and 13d (total yield 64%, 12d/13d = 14:1).
Thus, we obtained a wide range of hydrogenated isobenzofuran and 3-oxabicyclo[3.3.1]nonane heterocycles, including compounds with an exocyclic double bond.
2.2. Biology
2.2.1. Structure–Activity Relationship Analysis
The compounds synthesized starting from (+)-3-carene 8 were tested for their TDP1 inhibitory properties by measuring their IC50 values using a real-time fluorescent oligonucleotide biosensor [38]. This biosensor demonstrated high sensitivity and specificity for the detection of TDP1 activity. The commercially available TDP1 inhibitor furamidine, a bisbenzamidine derivative belonging to the diamidines family [34], was used as a positive control (IC50 1.2 ± 0.3 μM). The results for all the derivatives are summarized in Table 2.
Table 2.
TDP1 inhibition potency of the 3-carene derivatives.
| R | Compound 11 | IC501, μM | Compound 12 | IC501, μM |
|---|---|---|---|---|
| 4-Hydroxy-3-methoxyphenyl | 11a | >20 | 12a | >20 |
| (E)-prop-1-en-1-yl | 11c | >20 | - | - |
| Thiophen-2-yl | 11d | 4.85 ± 1.06 | 12d | 3.35 ± 1.06 |
| 3-Methylthiophen-2-yl | 11e | 3.6 ± 1.7 | 12e | 2.25 ± 0.63 |
| 5-Methylthiophen-2-yl | 11f | 4.7 ± 2.0 | - | - |
| 4-Bromothiophen-2-yl | - | - | 12g | 0.65 ± 0.22 |
| 5-Bromothiophen-2-yl | 11h | 0.75 ± 0.07 | 12h | 1.75 ± 0.78 |
| 5-Nitrothiophen-2-yl | - | - | 12i | 14 ± 1 |
| 5-Nitrofuran-2-yl | - | - | 12j | 28 ± 10 |
| Thiophen-3-yl | 11k | 1.60 ± 0.56 | 12k | 1.20 ± 0.14 |
| Furamidine | 1.2 ± 0.3 |
1 IC50—half maximal inhibitory concentration.
Compounds 11a, 12a, and 11c with aromatic and alkyl substituents at position C(1) do not exhibit activity against TDP1, while the IC50 values for substances with heterocyclic substituents varied from 0.65 (12g) to 28 μM (12j) (Table 2).
Both 11 and 12, with the thiophen-2-yl moiety, give good inhibition (IC50 values 4.85 μM (11d), 3.35 μM (12d)). The addition of a methyl group at the 3-position (compounds 11e, 12e) as well as at the 5-position (compound 11f) on the thiophene ring had no effect.
The addition of bromine onto the heterocycle significantly increased the inhibition potency of the ligands. Thus, compound 11h with a bromine atom at the 5-position of the thiophene ring had the best activity in the submicromolar range of the hexahydroisobenzofurans (IC50 0.75 μM). From the compounds with the 3-oxabicyclo[3.3.1]nonane skeleton, the most active was 4-bromine-substituted compound 12g (IC50 0.65 μM). A bromine shift from position 4 to 5 from 12g to 12h decreased the inhibition potency (IC50 1.75 μM); nevertheless, 12h is approximately two-fold more active than 12d, which does not have substituents on the thiophene ring.
Higher activity of the 3-substituted thiophenes 11k and 12k in comparison with 2-substituted thiophenes 11d and 12d was also observed. Interestingly, the 5-nitro-substituted thiophen- (12i) and furan (12j) with a 3-oxabicyclo[3.3.1]nonane skeleton did not exhibit high activity against TDP1.
2.2.2. Cell Growth and Viability
Topotecan Cytotoxicity on HEK293FT Wild Type and TDP1−/− Cells
TDP1 plays an essential role in the resistance of cancer cells to antitumour TOP1-inhibiting drugs. It was shown previously for HEK293 [39] and other cell lines that cells lacking TDP1 expression exhibit hypersensitivity to camptothecin and its derivatives, such as Tpc [40]. To examine the potential impact of TDP1 knockout on cell survival and dosing with the synthesized compounds, we generated HEK293FT TDP1-deficient (TDP1−/−) cell line. Using a paired gRNA CRISPR-Cas9 strategy and polymerase chain reaction (PCR) screening of cell clones, we obtained three clones containing deletions in the first protein coding the exon of the TDP1 gene. PCR products obtained from cell clones containing deletions were further cloned in a T-vector and 10 independent plasmid clones were sequenced by the Sanger method. Alignments of these deletions with the wild-type TDP1 gene sequence showed that modifications in all three clones lead to a shift in the reading frame of the gene and should lead to impaired synthesis of TDP1 (Figure S1). Clone B2 was found to have a 197-bp deletion in one of the TDP1 alleles, as well as a 16-bp deletion and T nucleotide insertion in the other allele; clone B5 carried 198-bp and 196-bp deletions; and the E3 clone had deletions of 197bp and 200bp. We treated TDP1-proficient (wild-type, WT) and TDP1-deficient (TDP1−/−) cells with increasing concentrations of Tpc. The results for all three clones are presented in Figure S2A. Clone E3 was selected for further work as it has a homozigous deletion causing an open reading frame shift in both alleles of the TDP1 gene and demonstrated sensitivity to Tpc. Clone E3 TDP1 mRNA was confirmed to have a 197/200-bp deletion causing the open reading frame shift (Figures S1, S2B). There is a deletion at the level of genomic DNA (Figure S2B, lane 4) and only the mutant form was transcribed (Figure S2B, lane 7). The clone E3 was subsequently screened by the biochemical assay for 3′-phosphotyrosyl cleavage activity to identify detectable TDP1 activity in the cell extract (Figure S2C). None was found in contrast to the control WT cell extract and purified TDP1.
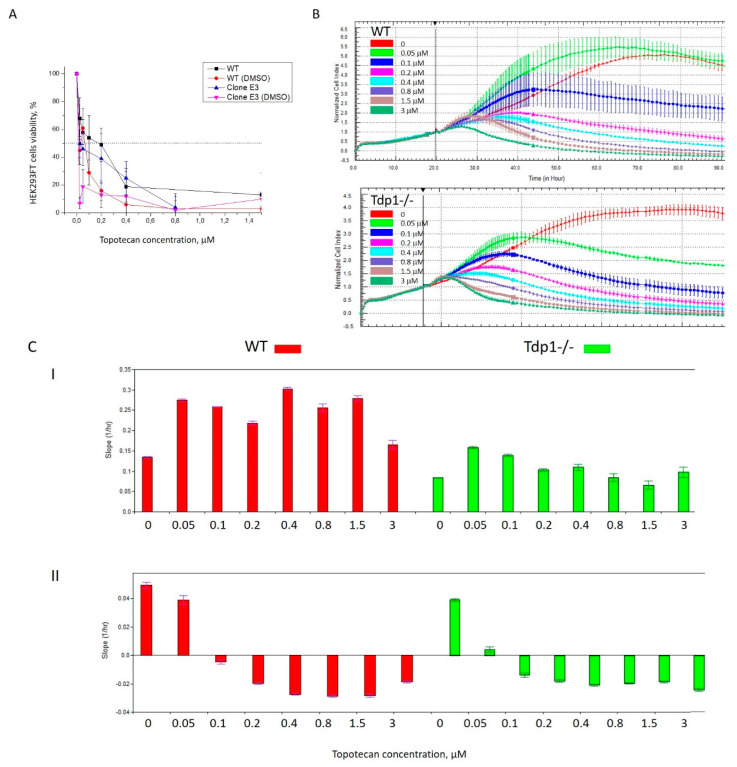
We analyzed, using a colorimetric test, the relative amount of viable cells after treating HEK293FT WT and TDP1-deficient (TDP1−/−) cells with increasing concentrations of Tpc for 72 h. TDP1 knockout reduced cell viability, where the IC50 values in H2O were 150 nM for TDP1−/− and 250 nM for WT cells (Figure 2A); and in DMSO, they were 30 nM for TDP1−/− and 200 nM for WT cells. Next, we studied WT and mutant cells’ live activity after Tpc treatment on the xCELLigence System (ACEA Biosciences, USA) by the impedance-based assay. HEK293FT TDP1−/− cells were more sensitive to Tpc (Figure 2B). After Tpc administration to the cells, an increase of the slope was seen, characterizing the velocity of the cell growth with Tpc as compared to the control both for WT (red columns) and mutant cells (green columns) (Figure S1, Supplementary Materials). The analysis of the averaged slope for the time period of ~70 h after Tpc treatment showed that the slope decreased depending on the Tpc concentration (Figure 2CII). The sharper decrease of the slope for the mutant cells than for the WT (compare the histogram for the 0 and 0.05 μM Tpc concentration, ~1.5 times for WT cells (red) and ~6 times for TDP1−/−cells (green)) also characterizes the higher sensitivity of the mutant to the TOP1 inhibitor. The Tpc IC50 values determined by the impedance-based assay were 60 nM for WT and 20 nM for TDP1−/− cells in H2O. There was a three-fold difference for these IC50 values, which is nearly the same as for the colorimetric assay. The absolute values slightly differed for these methods and solvents, but still, HEK293FT TDP1−/− cells showed a higher sensitivity to the TOP1 inhibitor Tpc, thus verifying the contribution of TDP1 on cell survival.
Figure 2.
Topotecan (Tpc) cytotoxicity on HEK293FT WT and TDP1−/− knockout cells (A). Dose-dependent action of Tpc by the colorimetric test (B). Dose-dependent action of Tpc on cell viability by the impedance-based real-time assay on the xCELLigence System (C). The slope characterizing the velocity of cell growth with Tpc compared to the control for WT (red columns) and mutant cells (green columns): I) After the Tpc addition, II) 70 h after Tpc treatment. Real-time cell analysis demonstrates sensitivty to Tpc. Cell proliferation and viability are affected by different Tpc concentrations in the growth medium.
TDP1 Inhibitors’ Cytotoxicity on HEK293FT TDP1−/− Cells
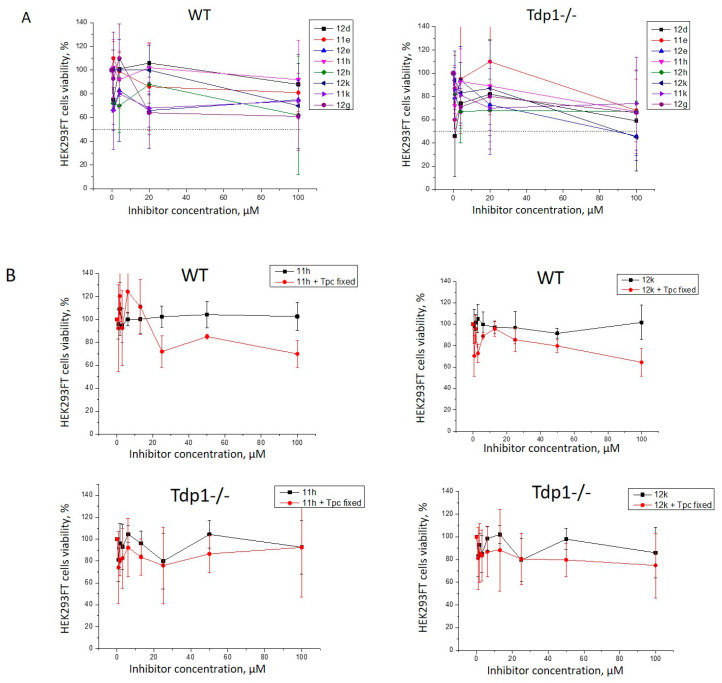
An analysis of the intrinsic cytotoxicity of the synthesized compounds was performed on HEK293FT WT and TDP1-deficient (TDP1−/−) cell lines by a colorimetric test. We tested eight compounds with the highest TDP1 inhibitory activity: 11e (3.6 μM), 11h (0.75 μM), 11k (1.6 μM), 12d (IC50 3.35 μM), 12e (2.25 μM), 12g (0.65 μM), 12h (1.75 μM), and 12k (1.2 μM). Interestingly, cytotoxicity was absent, or insignificant, in the range of concentrations (0.08–100 µM) for all the compounds in both cell lines (Figure 3A). The absence of toxicity is very important since it will not lead to undesirable side effects.
Figure 3.
The TDP1 inhibitors’ intrinsic cytotoxicity on HEK293FT WT and TDP1−/− cells (A). Dose-dependent action of the derivatives (B). Activity with topotecan (Tpc) against HEK293FT cells in combination with 11h and 12k. The Tpc concentration was 30 nM for TDP1−/− and 200 nM for the WT cells.
Next, we checked the influence of the eight most potent TDP1 inhibitors on the cytotoxic effect of Tpc on the WT and mutant cell lines. We chose 30 and 700 nM Tpc concentrations for the TDP1−/− and WT cells, respectively, which are close to the IC50 values for Tpc in DMSO in the colorimetric assay (Figure 2A). Then, we selected two candidates (11h and 12k) for subsequent studies based on their ability to inhibit TDP1 activity and low cytotoxicity. We tested the HEK293FT cells’ viability after treatment with 11h and 12k and in conjunction with Tpc in a concentration close to its IC50 value (30 nM for TDP1−/− and 200 nM for WT cells). Suppressed cell growth was displayed with the TDP1 inhibitors and Tpc on the WT cells; however, no effect was seen for Tpc in TDP1-deficient cells (Figure 3B). Thus, the synergistic action of Tpc in conjunction with 11h and 12k on HEK293FT WT cells is most likely due to the TDP1 inhibition making it the main target of action. The initial increase in cell proliferation on the graphs up to the 20 µM concentration can be observed because we diluted the inhibitor with the medium before adding it to the well plate. Thus, a stimulation effect at low inhibitor concentrations could be observed due to the addition of fresh medium.
Activity of 11h and 12k with Topotecan against Tumor Cells
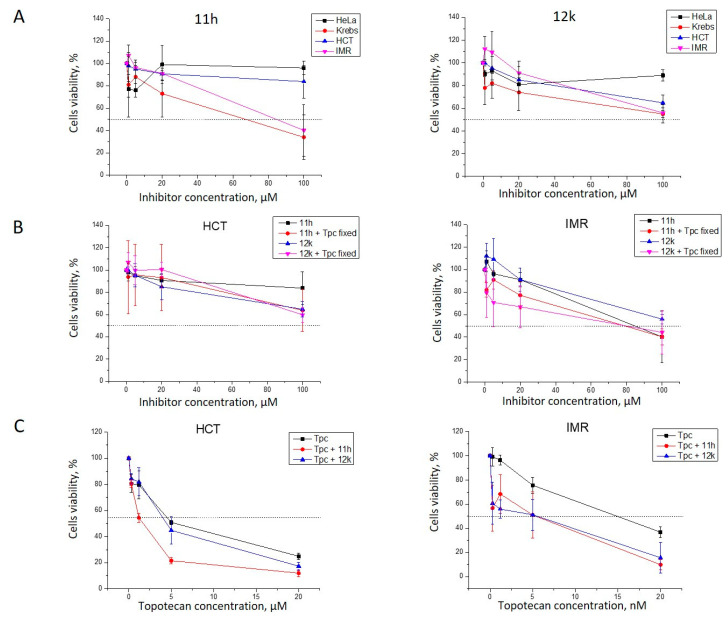
Next, we checked the cytotoxicity of the combination of Tpc with 11h and 12k against several cancer cell lines and the results are shown in Figure 4.
Figure 4.
Activity of 11h and 12k with topotecan (Tpc) against tumor cells (A). The intrinsic cytotoxicity of 11h and 12k derivatives on the cancer cell lines (B). Dose-dependent action of 11h and 12k in combination with Tpc (C). Dose-dependent action of Tpc in conjunction with 11h and 12k.
The intrinsic cytotoxicity of both 11h and 12k on HeLa (cervical carcinoma), HCT116 (human colon cancer), IMR-32 (human neuroblastoma cell line), and Krebs-2 carcinoma cells was low (Figure 4A). Due to their low cytotoxicity, these derivatives were accepted as being suitable for further study on their cytotoxic effect with Tpc (Figure 4B,C). Both compounds showed no enhancement of the Tpc cytotoxicity for HeLa cells (data not shown). We compared HCT and IMR-32 cells’ viability after treatment with 11h and 12k alone and in the presence of Tpc in a concentration close to their IC50 values (900 nM, HCT; and 8 nM, IMR-32 cells). We observed suppressed cell growth in the joint presence of Tpc with 11h or 12k on the IMR-32 cells and no effect of Tpc on the HCT cell line (Figure 4B). We tested HCT and IMR-32 cells’ viability after Tpc treatment in the absence and presence of 11h/12k (Figure 4C). Both 11h and 12k demonstrated synergistic action with Tpc on IMR-32 cell growth. 11h also enhanced Tpc’s potency on HCT116 cells. These compounds show promise as candidates for anticancer therapy in conjunction with Tpc.
2.2.3. Chemical Space
The calculated molecular descriptors MW (molecular weight), log P (water-octanol partition coefficient), HD (hydrogen bond donors), HA (hydrogen bond acceptors), PSA (polar surface area), and RB (rotatable bonds)) are given in Table S2. The molecular weight of the ligands lies between 206.3 and 327.3 g mol−1, falling into lead- and drug-like chemical spaces. The log P values range from 3.2 and 5.1, lying mostly in the drug-like but reaching into known drug space whilst the HD, HA, RB, and PSA values are within the lead-like space (for the definition of lead-like, drug-like, and known drug space (KDS) regions (see [41], Table S3).
The known drug index (KDI) of each ligand was calculated to gauge the balance of the molecular descriptors of the ligands. This method is based on the statistical analysis of drugs in clinical use (KDS) and a weighted index for each molecular descriptor. Both the summation (KDI2a) and multiplication (KDI2b) methods were used [42]; as shown for KDI2a in Equation (1) and for KDI2b in Equation (2). Finally, the numerical results are given in Table S4 in the Supplementary Material:
| KDI2a = IMW + Ilog P + IHD+ IHA + IRB + IPSA, | (1) |
| KDI2b = IMW × Ilog P × IHD× IHA × IRB × IPSA. | (2) |
The KDI2a values range from 4.15 to 5.15, with a theoretical maximum of 6 and an average of 4.08 for known drugs. KDI2b ranges from 0.10 to 0.39, with a theoretical maximum of 1 and with a KDS average of 0.18. This indicates that the majority of the ligands are well balanced. 11h has KDI2a of 4.25 and KDI2b of 0.11 and 12g has similar values, with KDI2a of 4.16 and KDI2b of 0.10. The KDI2a of both compounds is above the average index value for known drugs; however, the KDI2b is below the average mainly due to the low PSA values. Nevertheless, these ligands can be considered reasonably well balanced in terms of their molecular descriptors.
3. Materials and Methods
3.1. Chemistry
All commercially available compounds and solvents were reagent grade and used without further treatment unless otherwise noted. We used montmorillonite K10 clay (Sigma-Aldrich, St. Louis, MO, USA) as the catalyst. The clay was calcinated at 105 °C for 3 h immediately before use. CH2Cl2 was passed through calcined Al2O3. The 2-carene-containing mixture was synthesized according to Sidorenko et al. [31] by isomerization of 3-carene (Acros Organics, 98%). Column chromatography (CC): silica gel (SiO2; 60–200 μ; Macherey-Nagel, Dueren, Germany); hexane/Et2O 100:0 → 90:10, analysis of fractions composition after column chromatography: GC on an Agilent 7820A, HP-5 quartz column, 30,000 × 0.25 mm, flame-ionization detector, He (1 atm) as the carrier gas. GC/MS (purity control and products analysis): Agilent 7890A with a quadrupole mass spectrometer Agilent 5975C as a detector, HP-5MS quartz column, 30,000 × 0.25 mm, He (1 atm) as the carrier gas. Optical rotation: polAAr 3005 spectrometer, CHCl3 soln. HR-MS: DFS-Thermo-Scientific spectrometer in the full scan mode (15–500 m/z, 70 eV electron-impact ionization, direct sample introduction). 1H and 13C NMR: Bruker DRX-500 apparatus at 500.13 MHz (1H) and 125.76 MHz (13C) and Bruker Avance-III 600 apparatus at 600.30 MHz (1H) and 150.95 MHz (13C) in CDCl3; chemical shifts δ in ppm rel. to residual CHCl3 (δ (H) 7.24, δ (C) 76.90 ppm), J in Hz; structure determinations by analyzing the 1H NMR spectra, including 1H – 1H double resonance spectra and 1H – 1H 2D homonuclear correlation (COSY, NOESY); J-modulated 13C NMR spectra (JMOD), and 13C – 1H 2D heteronuclear correlation with one-bond and long-range spin-spin coupling constants (C – H COSY, 1J(C,H) = 135 Hz; HSQC, 1J(C,H) = 145 Hz; COLOC, 2,3J(C,H) = 10 Hz; HMBC, 2,3J(C,H) = 7 Hz). All the target compounds reported have a purity of ≥95%. Numeration of atoms in the compounds (see Supplementary Material) is given for assigning the signals in the NMR spectra and does not coincide with that for the names according to the nomenclature of compounds. Spectral and analytical studies were carried out at the Collective Chemical Service Center of the Siberian Branch of the Russian Academy of Sciences.
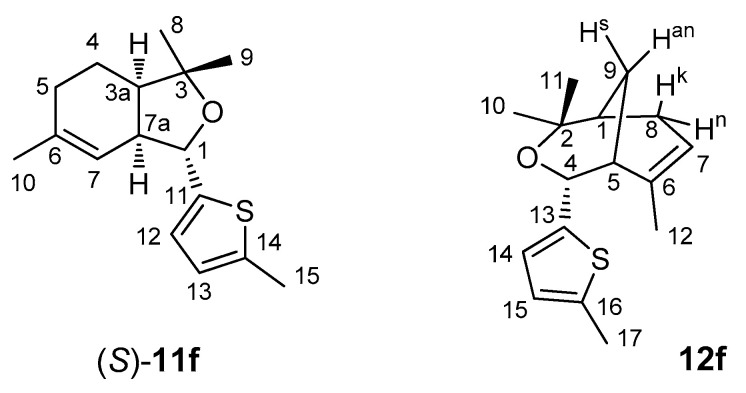
Numeration for the carbon atoms used for assignment in NMR spectra is presented in Figure 5 with compound (S)-11f and 12f as an example.
Figure 5.
Numeration of carbon atoms for NMR spectra.
3.1.1. General Procedure (GP)
To the suspension of the K10 clay (3.0 g) in methylene chloride (15 mL) solution of 0.40 g aldehyde in methylene chloride (10 mL) and 1.0 g of a 2-carene containing mixture (10 wt.% of 2-carene., 57 wt.% of the sum of 2- and 3-carenes, limonene, and menthadienes., obtained by isomerization of 3-carene according to [30,31], were added. The solvent was distilled off, and the reaction mixture was kept at rt for 20 h. Thereafter, ethyl acetate was added, the catalyst was filtered off, and the filtrate was evaporated. Isolation of the reaction products was carried out by column chromatography (silica gel (17 g), hexane/Et2O 100:0 → 90:10). Compounds 11 were eluted with a 4–6% solution of Et2O in hexane, and compounds 12 and 13 with a 0.5–2% solution of Et2O in hexane. The yields of compounds 11, 12, and 13 were calculated based on the 2-carene content and on the sum of 2- and 3-carenes, limonene, and menthadienes, respectively.
3.1.2. Reaction of 2-Carene-Containing Mixture and 4-hydroxy-3-methoxybenzaldehyde 10a
According to the GP, the reaction of the 2-carene containing mixture and 4-hydroxy-3-methoxybenzaldehyde 10a gave isobenzofurans 11a (0.172 g, total yield 81%, (S)-/(R)- = 1:1), and compound 12a (0.135 g, 11%) was isolated. The spectrum of the substance (S)-11a, (R)-11a, and 12a corresponds to the literature [23].
3.1.3. Reaction of 2-Carene-Containing Mixture and Crotonaldehyde 10c
According to the GP, the reaction of the 2-carene containing mixture and crotonaldehyde 10c gave isobenzofurans 11c (0.135 g, total yield of 89%, (S)/(R) = 1.3: 1). The spectrum of the substance (S)-11c and (R)-10c corresponds to the literature [23].
3.1.4. Reaction of 2-Carene-Containing Mixture and 2-Thiophenecarbaldehyde 10d
According to the GP, the reaction of the 2-carene containing mixture and 2-thiophenecarbaldehyde 10d gave (1S,3aR,7aS)-3,3,6-trimethyl-1-(thiophen-2-yl)-1,3,3a,4,5,7a-hexahydroisobenzofuran 11d (0.143 g, 78%) and the mixture of (1R,4R,5R)-2,2,6-trimethyl-4-(thiophen-2-yl)-3-oxabicyclo[3.3.1]non-6-ene 12d and (1R,4R,5R)-2,2-dimethyl-6-methylene-4-(thiophen-2-yl)-3-oxabicyclo[3.3.1]nonane 13d (0.050 g, 5%, 7:1).
11d: NMR 1H (600 MHz, CDCl3, δ, ppm, J/Hz): 1.28 (s, 3H, Me-9), 1.39 (s, 3H, Me-8), 1.51–1.60 (m, 1H, Ha-4), 1.68 (br.s, 3H, Me-10), 1.74 (dm, 1H, 2J = 13.0, He-4), 1.95–2.01 (m, 3H, 2H-5, H-3a), 2.83–2.88 (m, 1H, H-7a), 4.85 (d, 1H, J1,7a = 10.0, H-1), 5.27–5.29 (m, 1H, H-7), 6.93 (dd, 1H, J13,14 = 5.0, J13,12 = 3.5, H-13), 6.96 (dm, 1H, J12,13 = 3.5, H-12), 7.22 (dd, 1H, J14,13 = 5.0, J14,12 = 1.2, H-14). NMR 13C (150 MHz, CDCl3, δ, ppm): 81.33 (d, C-1), 83.05 (c, C-3), 46.38 (d, C-3a), 21.83 (t, C-4), 29.98 (t, C-5), 136.55 (c, C-6), 118.41 (d, C-7), 48.68 (d, C-7a), 30.92 (q, C-8), 24.00 (q, C-9), 23.49 (q, C-10), 146.76 (s, C-11), 123.89 (d, C-12), 126.42 (d, C-13), 124.26 (d, C-14). HR-MS: m/z calcd. for C15H20OS: 248.1229. Found: 248.1227. = 120 (c = 0.3, CHCl3).
The NMR spectra of 12d and 13d were recorded for their mixture (≈ 7:1).
12d: NMR 1H (600 MHz, CDCl3, δ, ppm, J/Hz): 1.00–1.03 (m, 3H, Me-12), 1.30 (s, 3H, Me-10), 1.37 (s, 3H, Me-11), 1.51–1.56 (m, 1H, H-1), 1.71 (ddd, 1H, 2J = 12.5, J9an,1 = J9an,5 = 3.2, H-9an), 2.08 (dm, 1H, J8k,8n = 18.8, H-8k), 2.21 (ddd, 1H, J5,9s = J5,9an = 3.2, J5,4 = 2.3, H-5), 2.30 (dddd, 1H, 2J = 12.5, J9s,1 = J9s,5 = 3.2, J9s,8n = 1.2, H-9s), 2.38 (br.d, 1H, 2J = 18.8, H-8n), 5.14 (d, 1H, J4,5 = 2.3, H-4), 5.46–5.49 (m, 1H, H-7), 6.89 (dm, 1H, J14,15 = 3.5, H-14), 6.91 (dd, 1H, J15,16 = 5.0, J15,14 = 3.5, H-15), 7.12 (dd, 1H, J16,15 = 5.0, J16,14 = 1.3, H-16). NMR 13C (150 MHz, CDCl3, δ, ppm): 33.86 (d, C-1), 75.89 (c, C-2), 71.94 (d, C-4), 42.31 (d, C-5), 133.04 (c, C-6), 123.46 (d, C-7), 27.55 (t, C-8), 27.74 (t, C-9), 28.44 (q, C-10), 23.87 (q, C-11), 23.97 (q, C-12), 146.97 (c, C-13), 121.54 (d, C-14), 126.04 (d, C-15), 123.05 (d, C-16). HR-MS: m/z calcd. for C15H20OS: 248.1229. Found: 248.1230.
13d: NMR 1H (600 MHz, CDCl3, δ, ppm, J/Hz): 1.36 (s, 3H, Me-10), 1.40 (s, 3H, Me-11), 1.50–1.53 (m, 1H, H-1), 1.72–1.77 (m, 1H, H-9s), 2.86–2.93 (m, 1H, Ha-7), 4.31 (dd, 1H, 2J = 2.3, J12,5 = 2.3, H-12), 4.56 (dd, 1H, 2J = 2.3, J12′,5 = 2.3, H-12′), 5.22 (br.d, 1H, J4,5 = 1.9, H-4), 6.87 (dm, 1H, J14,15 = 3.5, H-14), 7.07–7.10 (m, 1H, H-15), 7.11–7.13 (m, 1H, H-16). The remaining proton signals are overlapped by the signals of the main isomer 12a. NMR 13C (150 MHz, CDCl3, δ, ppm): 35.10 (d, C-1), 75.99 (c, C-2), 73.46 (d, C-4), 46.00 (d, C-5), 148.29 (c, C-6), 31.08 (t, C-7), 28.92 (t, C-8), 32.05 (t, C-9), 27.85 (q, C-10), 24.29 (q, C-11), 109.69 (t, C-12), 148.29 (c, C-13), 122.40 (d, C-14), 125.66 (d, C-15), 129.45 (d, C-16). HR-MS: m/z calcd. for C15H20OS: 248.1229. Found: 248.1230.
3.1.5. Reaction of 2-Carene-Containing Mixture and 3-methylthiophene-2-carbaldehyde 10e
According to the GP, the reaction of the 2-carene-containing mixture and 3-methylthiophene-2-carbaldehyde 10e gave (1S,3aR,7aS)-3,3,6-trimethyl-1-(3-methylthiophen-2-yl)-1,3,3a,4,5,7a-hexahydroisobenzofuran 11e (0.167 g, 86%) and (1R,4R,5R)-2,2,6-trimethyl-4-(3-methylthiophen-2-yl)-3-oxabicyclo[3.3.1]non-6-ene 12e (0.025 g, 2%).
11e: NMR 1H (600 MHz, CDCl3, δ, ppm, J/Hz): 1.28 (s, 3H, Me-9), 1.41 (s, 3H, Me-8), 1.57–1.65 (m, 1H, Ha-4), 1.67 (br.s, 3H, Me-10), 1.74 (dm, 1H, 2J = 12.9, He-4), 1.96–2.02 (m, 3H, 2H-5, H-3a), 2.17 (s, 3H, Me-15), 2.84–2.90 (m, 1H, H-7a), 4.92 (d, 1H, J1,7a = 10.1, H-1), 5.21–5.24 (m, 1H, H-7), 6.74 (d, 1H, J13,14 = 5.0, H-13), 7.13 (d, 1H, J14,13 = 5.0, H-14).
NMR 13C (150 MHz, CDCl3, δ, ppm): 79.14 (d, C-1), 82.64 (c, C-3), 46.40 (d, C-3a), 21.92 (t, C-4), 30.02 (t, C-5), 136.38 (c, C-6), 118.47 (d, C-7), 49.18 (d, C-7a), 31.14 (q, C-8), 24.03 (q, C-9), 23.45 (q, C-10), 139.46 (s, C-11), 134.13 (s, C-12), 129.79 (d, C-13), 123.04 (d, C-14), 13.88 (q, C-15). HR-MS: m/z calcd. for C16H22OS: 262.1386. Found: 262.1388. = 100 (c = 0.6, CHCl3).
12e: NMR 1H (600 MHz, CDCl3, δ, ppm, J/Hz): 0.97–1.00 (m, 3H, Me-12), 1.29 (s, 3H, Me-10), 1.37 (s, 3H, Me-11), 1.51–1.55 (m, 1H, H-1), 1.70 (ddd, 1H, 2J = 12.5, J9an,1 = J9an,5 = 3.1, H-9an), 2.04–2.09 (m, 1H, H-8k), 2.20 (s, 3H, Me-17), 2.20–2.22 (m, 1H, H-5), 2.28 (dddd, 1H, 2J = 12.5, J9s,1 = J9s,5 = 3.1, J9s,8n = 1.2, H-9s), 2.39 (br.d, 1H, 2J = 18.4, H-8n), 5.10 (d, 1H, J4,5 = 2.5, H-4), 5.46–5.49 (m, 1H, H-7), 6.71 (d, 1H, J15,16 = 5.0, H-15), 7.04 (d, 1H, J16,15 = 5.0, H-16).
NMR 13C (150 MHz, CDCl3, δ, ppm): 33.79 (d, C-1), 75.79 (c, C-2), 71.15 (d, C-4), 39.12 (d, C-5), 133.11 (c, C-6), 123.51 (d, C-7), 27.44 and 27.49 (2t, C-8, C-9), 28.47 (q, C-10), 23.89 (q, C-11), 23.61 (q, C-12), 140.20 (c, C-13), 129.93 (s, C-14), 129.03 (d, C-15), 122.72 (d, C-16), 14.09 (q, C-17). HR-MS: m/z calcd. for C16H22OS: 262.1386. Found: 262.1384. = 0 (c = 0.3, CHCl3).
3.1.6. Reaction of 2-Carene-Containing Mixture and 5-methylthiophene-2-carbaldehyde 10f
According to the GP, the reaction of the 2-carene-containing mixture and 5-methylthiophene-2-carbaldehyde 10f gave (1S,3aR,7aS)-3,3,6-trimethyl-1-(5-methylthiophen-2-yl)-1,3,3a,4,5,7a-hexahydroisobenzofuran 11f (0.163 g, 85%) and mixture of (1R,4R,5R)-2,2,6-trimethyl-4-(5-methylthiophen-2-yl)-3-oxabicyclo[3.3.1]non-6-ene 12f and (1R,4R,5R)-2,2-dimethyl-6-methylene-4-(5-methylthiophen-2-yl)-3-oxabicyclo[3.3.1]nonane 13f (0.025 g, 2%, 3:1).
11f: NMR 1H (600 MHz, CDCl3, δ, ppm, J/Hz): 1.27 (s, 3H, Me-9), 1.38 (s, 3H, Me-8), 1.51–1.59 (m, 1H, Ha-4), 1.68 (br.s, 3H, Me-10), 1.73 (dm, 1H, 2J = 12.9, He-4), 1.94–2.01 (m, 3H, 2H-5, H-3a), 2.44 (d, 3H, J15,13 = 0.8, Me-15), 2.82–2.87 (m, 1H, H-7a), 4.75 (d, 1H, J1,7a = 10.0, H-1), 5.27–5.30 (m, 1H, H-7), 6.56 (dq, 1H, J13,12 = 3.5, J13,15 = 0.8, H-13), 6.74 (d, 1H, J12,13 = 3.5, H-12). NMR 13C (150 MHz, CDCl3, δ, ppm): 81.45 (d, C-1), 82.78 (c, C-3), 46.31 (d, C-3a), 21.81 (t, C-4), 29.97 (t, C-5), 136.34 (c, C-6), 118.54 (d, C-7), 48.41 (d, C-7a), 30.92 (q, C-8), 23.98 (q, C-9), 23.45 (q, C-10), 144.02 (s, C-11), 124.15 (d, C-12), 124.38 (d, C-13), 138.92 (s, C-14), 15.29 (q, C-15). HR-MS: m/z calcd. for C16H22OS: 262.1386. Found: 262.1382. = 119 (c = 0.4, CHCl3).
The NMR spectra of 12f and 13f were recorded for their mixture (≈ 3:1).
12f: NMR 1H (600 MHz, CDCl3, δ, ppm, J/Hz): 1.10–1.12 (m, 3H, Me-12), 1.28 (s, 3H, Me-10), 1.35 (s, 3H, Me-11), 1.50–1.53 (m, 1H, H-1), 1.69 (ddd, 1H, 2J = 12.5, J9an,1 = J9an,5 = 3.2, H-9an), 2.06 (dm, 1H, J8k,8n = 18.8, H-8k), 2.17 (ddd, 1H, J5,9s = J5,9an = 3.2, J5,4 = 2.0, H-5), 2.27 (dddd, 1H, 2J = 12.5, J9s,1 = J9s,5 = 3.2, J9s,8n = 1.2, H-9s), 2.36 (br.d, 1H, 2J = 18.8, H-8n), 2.39 (br.s, 3H, Me-17), 5.04 (br.d, 1H, J4,5 = 2.0, H-4), 5.45–5.48 (m, 1H, H-7), 6.53–6.55 (m, 1H, H-15), 6.65 (dd, 1H, J14,15 = 3.3, J14,4 = 1.0, H-14). NMR 13C (150 MHz, CDCl3, δ, ppm): 33.87 (d, C-1), 75.78 (c, C-2), 71.93 (d, C-4), 42.22 (d, C-5), 133.29 (c, C-6), 123.28 (d, C-7), 27.56 (t, C-8), 27.74 (t, C-9), 28.46 (q, C-10), 23.87 (q, C-11), 24.20 (q, C-12), 144.32 (c, C-13), 121.31 (d, C-14), 124.03 (d, C-15), 137.35 (s, C-16), 15.06 (q, C-17). HR-MS: m/z calcd. for C16H22OS: 262.1386. Found: 262.1387. = 0 (c = 0.2, CHCl3).
13f: NMR 1H (600 MHz, CDCl3, δ, ppm, J/Hz): 1.33 (s, 3H, Me-10), 1.38 (s, 3H, Me-11), 1.47–1.50 (m, 1H, H-1), 2.09–2.16 (m, 2H, H-7e, H-8e), 2.36–2.46 (m, 2H, H-9an, H-5), 2.39 (br.s, 3H, Me-17), 2.86–2.93 (m, 1H, Ha-7), 4.37 (dd, 1H, 2J = 2.4, J12,5 = 2.4, H-12), 4.61 (dd, 1H, 2J = 2.4, J12′,5 = 2.4, H-12′), 5.11 (br.d, 1H, J4,5 = 1.9, H-4), 6.53–6.55 (m, 1H, H-15), 6.65 (dd, 1H, J14,15 = 3.3, J14,4 = 1.0, H-14). The remaining proton signals are overlapped by the signals of the main isomer 12f. NMR 13C (150 MHz, CDCl3, δ, ppm): 35.12 (d, C-1), 75.88 (c, C-2), 73.52 (d, C-4), 45.79 (d, C-5), 148.56 (c, C-6), 31.17 (t, C-7), 29.02 (t, C-8), 32.21 (t, C-9), 27.88 (q, C-10), 24.28 (q, C-11), 109.72 (t, C-12), 143.30 (c, C-13), 122.50 (d, C-14), 123.98 (d, C-15), 137.68 (s, C-16), 15.12 (q, C-17). HR-MS: m/z calcd. for C16H22OS: 262.1386. Found: 262.1387.
3.1.7. Reaction of 2-Carene-Containing Mixture and 4-bromothiophene-2-carbaldehyde 10g
According to the GP, the reaction of the 2-carene-containing mixture and 4-bromothiophene-2-carbaldehyde 10g gave (1R,4R,5R)-2,2,6-trimethyl-4-(4-bromothiophen-2-yl)-3-oxabicyclo[3.3.1]non-6-ene 12g (0.273 g, 20%).
12g: NMR 1H (600 MHz, CDCl3, δ, ppm, J/Hz): 1.06–1.08 (m, 3H, Me-12), 1.28 (s, 3H, Me-10), 1.34 (s, 3H, Me-11), 1.50–1.54 (m, 1H, H-1), 1.70 (ddd, 1H, 2J = 12.5, J9an,1 = J9an,5 = 3.2, H-9an), 2.06 (dm, 1H, 2J = 18.8, H-8k), 2.16–2.19 (m, 1H, H-5), 2.27 (dm, 1H, 2J = 12.5, H-9s), 2.35 (br.d, 1H, 2J = 18.8, H-8n), 5.05 (dd, 1H, J4,5 = 2.5, J4,14 = 0.7, H-4), 5.46–5.49 (m, 1H, H-7), 6.81 (dd, 1H, J14,16 = 1.5, J14,4 = 0.7, H-14), 7.03 (d, 1H, J16,14 = 1.5, H-16). NMR 13C (150 MHz, CDCl3, δ, ppm): 33.73 (d, C-1), 76.18 (c, C-2), 71.49 (d, C-4), 42.10 (d, C-5), 132.46 (c, C-6), 123.93 (d, C-7), 27.49 and 27.62 (2t, C-8, C-9), 28.33 (q, C-10), 23.85 (q, C-11), 24.12 (q, C-12), 148.37 (c, C-13), 124.37 (d, C-14), 108.51 (s, C-15), 120.68 (d, C-16). HR-MS: m/z calcd. for C15H19OBrS: 326.0335. Found: 326.0334. = 0 (c = 1.3, CHCl3).
3.1.8. Reaction of 2-Carene-Containing Mixture and 5-bromothiophene-2-carbaldehyde 10h
According to the GP, the reaction of the 2-carene-containing mixture and 5-bromothiophene-2-carbaldehyde 10h gave (1S,3aR,7aS)-3,3,6-trimethyl-1-(5-bromothiophen-2-yl)-1,3,3a,4,5,7a-hexahydroisobenzofuran 11h (0.176 g, 73%) and (1R,4R,5R)-2,2,6-trimethyl-4-(5-bromothiophen-2-yl)-3-oxabicyclo[3.3.1]non-6-ene 12h (0.199 g, 15%).
11h: NMR 1H (600 MHz, CDCl3, δ, ppm, J/Hz): 1.26 (s, 3H, Me-9), 1.36 (s, 3H, Me-8), 1.47–1.56 (m, 1H, Ha-4), 1.68 (br.s, 3H, Me-10), 1.72 (dm, 1H, 2J = 12.9, He-4), 1.93–2.00 (m, 3H, 2H-5, H-3a), 2.75–2.81 (m, 1H, H-7a), 4.74 (d, 1H, J1,7a = 9.9, H-1), 5.24–5.27 (m, 1H, H-7), 6.69 (dd, 1H, J12,13 = 3.7, J12,3a = 0.8, H-12), 6.86 (d, 1H, J13,12 = 3.7, H-13). NMR 13C (150 MHz, CDCl3, δ, ppm): 81.56 (d, C-1), 83.42 (c, C-3), 46.27 (d, C-3a), 21.73 (t, C-4), 29.92 (t, C-5), 136.95 (c, C-6), 117.95 (d, C-7), 48.56 (d, C-7a), 30.83 (q, C-8), 23.92 (q, C-9), 23.49 (q, C-10), 148.65 (s, C-11), 124.00 (d, C-12), 129.22 (d, C-13), 110.99 (s, C-14). HR-MS: m/z calcd. for C15H19OBrS: 326.0335. Found: 326.0330. = 34 (c = 1.2, CHCl3).
12h: NMR 1H (600 MHz, CDCl3, δ, ppm, J/Hz): 1.10–1.12 (m, 3H, Me-12), 1.27 (s, 3H, Me-10), 1.34 (s, 3H, Me-11), 1.50–1.54 (m, 1H, H-1), 1.69 (ddd, 1H, 2J = 12.5, J9an,1 = J9an,5 = 3.2, H-9an), 2.06 (dm, 1H, J8k,8n = 18.8, H-8k), 2.15–2.18 (m, 1H, H-5), 2.26 (dddd, 1H, 2J = 12.5, J9s,1 = J9s,5 = 3.2, J9s,8n = 1.1, H-9s), 2.35 (br.d, 1H, 2J = 18.8, H-8n), 5.03 (br.d, 1H, J4,5 = 2.1, H-4), 5.47–5.50 (m, 1H, H-7), 6.63 (dd, 1H, J14,15 = 3.7, J14,4 = 0.8, H-14), 6.85 (d, 1H, J15,14 = 3.7, H-15). NMR 13C (150 MHz, CDCl3, δ, ppm): 33.74 (d, C-1), 76.09 (c, C-2), 72.07 (d, C-4), 42.00 (d, C-5), 132.61 (c, C-6), 123.87 (d, C-7), 27.49 (t, C-8), 27.55 (t, C-9), 28.35 (q, C-10), 23.84 (q, C-11), 24.24 (q, C-12), 148.84 (c, C-13), 121.61 (d, C-14), 128.76 (d, C-15), 109.96 (s, C-16). HR-MS: m/z calcd. for C15H19OBrS: 326.0335. Found: 326.0331. = 0 (c = 0.9, CHCl3).
3.1.9. Reaction of 2-Carene-Containing Mixture and 5-nitrothiophene-2-carbaldehyde 10i
According to the GP, the reaction of the 2-carene-containing mixture and 5-nitrothiophene-2-carbaldehyde 10i gave a mixture of (1R,4R,5R)-2,2,6-trimethyl-4-(5-methylthiophen-2-yl)-3-oxabicyclo[3.3.1]non-6-ene 12i and (1R,4R,5R)-2,2-dimethyl-6-methylene-4-(5-methylthiophen-2-yl)-3-oxabicyclo[3.3.1]nonane 13i (0.221 g, 18%, 5.4:1).
The NMR spectra of 12i and 13i were recorded for their mixture (≈ 5.4:1).
12i: NMR 1H (600 MHz, CDCl3, δ, ppm, J/Hz): 1.03–1.05 (m, 3H, Me-12), 1.27 (s, 3H, Me-10), 1.338 (s, 3H, Me-11), 1.53–1.56 (m, 1H, H-1), 1.73 (ddd, 1H, 2J = 12.5, J9an,1 = J9an,5 = 3.2, H-9an), 2.07 (dm, 1H, 2J = 19.0, H-8k), 2.23–2.26 (m, 1H, H-5), 2.28 (dddd, 1H, 2J = 12.5, J9s,1 = J9s,5 = 3.2, J9s,8n = 1.2, H-9s), 2.35 (br.d, 1H, 2J = 19.0, H-8n), 5.06 (br.d, 1H, J4,5 = 2.4, H-4), 5.48–5.51 (m, 1H, H-7), 6.84 (dd, 1H, J14,15 = 4.2, J14,4 = 1.0, H-14), 7.75 (d, 1H, J15,14 = 4.2, H-15). NMR 13C (150 MHz, CDCl3, δ, ppm): 33.49 (d, C-1), 76.60 (c, C-2), 71.74 (d, C-4), 41.75 (d, C-5), 131.38 (c, C-6), 124.82 (d, C-7), 27.35 (t, C-8), 27.45 (t, C-9), 28.14 (q, C-10), 23.77 (q, C-11), 24.12 (q, C-12), 157.31 (c, C-13), 120.69 (d, C-14), 128.02 (d, C-15), 150.16 (br.s, C-16). HR-MS: m/z calcd. for C15H19O3NS: 293.1080. Found: 293.1076.
13i: NMR 1H (600 MHz, CDCl3, δ, ppm, J/Hz): 1.342 (s, 3H, Me-10), 1.37 (s, 3H, Me-11), 1.47–1.53 (m, 1H, H-8a), 1.76 (dm, 1H, 2J = 13.0, H-9s), 2.07–2.12 (m, 2H, H-7e, H-8e), 2.36–2.41 (m, 1H, H-9a), 2.43–2.46 (m, 1H, H-5), 2.67–2.75 (m, 1H, H-7a), 4.30 (dd, 1H, 2J = 2.4, J12,5 = 2.4, H-12), 4.54 (dd, 1H, 2J = 2.4, J12′,5 = 2.4, H-12′), 5.16 (br.d, 1H, J4,5 = 2.5, H-4), 6.73 (dd, 1H, J14,15 = 4.2, J14,4 = 1.0, H-14), 7.75 (d, 1H, J15,14 = 4.2, H-15). The H-1 signal is overlapped by the H-1 signal of the main isomer 12i. NMR 13C (150 MHz, CDCl3, δ, ppm): 34.76 (d, C-1), 76.71 (c, C-2), 73.30 (d, C-4), 45.30 (d, C-5), 146.62 (c, C-6), 30.73 (t, C-7), 28.46 (t, C-8), 31.39 (t, C-9), 27.53 (q, C-10), 24.23 (q, C-11), 110.58 (q, C-12), 156.23 (c, C-13), 121.10 (d, C-14), 128.22 (d, C-15), ≈150 (br.s, C-16). Broad singlet signal of C-16 could not be determined accurately due to low concentrations of 13i. HR-MS: m/z calcd. for C15H19O3NS: 293.1080. Found: 293.1076.
3.1.10. Reaction of 2-Carene-Containing Mixture and 5-nitrofuran-2-carbaldehyde 10j
According to the GP, the reaction of the 2-carene-containing mixture and 5-nitrofuran-2-carbaldehyde 10j gave (1R,4R,5R)-2,2,6-trimethyl-4-(5-nitrofuran-2-yl)-3-oxabicyclo[3.3.1]non-6-ene 12j (0.215 g, 18%).
12j: NMR 1H (600 MHz, CDCl3, δ, ppm, J/Hz): 1.08–1.09 (m, 3H, all J < 2.5 Hz, Me-12), 1.28 (s, 3H, Me-10), 1.35 (s, 3H, Me-11), 1.54–1.58 (m, 1H, H-1), 1.75 (ddd, 1H, 2J = 12.6, J9an,1 = J9an,5 = 3.2, H-9an), 2.09 (dm, 1H, J8k,8n = 18.9, H-8k), 2.25 (dm, 1H, 2J = 12.6, H-9s), 2.34 (dm, 1H, 2J = 18.9, H-8n), 2.46–2.49 (m, 1H, H-5), 4.90 (br.d, 1H, J4,5 = 2.2, H-4), 5.48–5.51 (m, 1H, H-7), 6.39 (dd, 1H, J14,15 = 3.7, J14,4 = 1.2, H-14), 7.24 (d, 1H, J15,14 = 3.7, H-15). NMR 13C (150 MHz, CDCl3, δ, ppm): 33.75 (d, C-1), 76.38 (c, C-2), 69.63 (d, C-4), 38.69 (d, C-5), 131.49 (c, C-6), 124.56 (d, C-7), 27.47 (t, C-8), 27.00 (t, C-9), 28.23 (q, C-10), 23.65 (q, C-11), 23.07 (q, C-12), 160.14 (c, C-13), 108.81 (d, C-14), 112.97 (d, C-15), 150.96 (s, C-16). HR-MS: m/z calcd. for C15H19O4N: 277.1309. Found: 277.1306.
3.1.11. Reaction of 2-Carene-Containing Mixture and thiophene-3-carbaldehyde 10k
According to the GP, the reaction of the 2-carene-containing mixture and thiophene-3-carbaldehyde 10k gave (1S(R),3aR,7aS)-3,3,6-trimethyl-1-(thiophen-3-yl)-1,3,3a,4,5,7a-hexahydroisobenzofuran 11k (0.165 g, 91%, S/R = 1.5:1) and (1R,4R,5R)-2,2,6-trimethyl-4-(thiophen-3-yl)-3-oxabicyclo[3.3.1]non-6-ene 12k (0.213 g, 21%).
The NMR spectra of compound 11k were recorded for the mixture (S)-11k and (R)-11k isomers (1.3:1).
(S)-11k: NMR 1H (500 MHz, CDCl3, δ, ppm, J/Hz): 1.28 (s, 3H, Me-9), 1.37 (s, 3H, Me-8), 1.53–1.61 (m, 1H, Ha-4), 1.68 (br.s, 3H, Me-10), 1.69–1.76 (m, 1H, He-4), 1.88–2.02 (m, 3H, 2H-5, H-3a), 2.73–2.80 (m, 1H, H-7a), 4.68 (d, 1H, J1,7a = 10.1, H-1), 5.22–5.25 (m, 1H, H-7), 7.09 (dd, 1H, J12,13 = 5.0, J12,14 = 1.3, H-12), 7.18 (ddd, 1H, J14,13 = 3.0, J14,13 = 1.3, J14,1 = 0.7, H-14), 7.22 (dd, 1H, J13,12 = 5.0, J13,14 = 3.0, H-13). NMR 13C (125 MHz, CDCl3, δ, ppm): 81.53 (d, C-1), 82.61 (c, C-3), 46.20 (d, C-3a), 21.82 (t, C-4), 29.99 (t, C-5), 136.40 (c, C-6), 118.65 (d, C-7), 47.37 (d, C-7a), 30.95 (q, C-8), 24.02 (q, C-9), 23.53 (q, C-10), 143.91 (s, C-11), 125.64 (d, C-12), 125.68 (d, C-13), 120.75 (d, C-14). HR-MS: m/z calcd. for C15H20OS: 248.1229. Found: 248.1230.
(R)-11k: NMR 1H (500 MHz, CDCl3, δ, ppm, J/Hz): 1.30 (s, 3H, Me-8), 1.33 (s, 3H, Me-9), 1.50 (br.s, 3H, Me-10), 1.69–1.76 (m, 1H, He-4), 1.81–1.85 (m, 2H, 2H-5), 1.89–1.97 (m, 1H, H-3a), 3.17–3.24 (m, 1H, H-7a), 4.89–4.93 (m, 1H, H-7), 5.22 (d, 1H, J1,7a = 9.5, H-1), 6.89 (dd, 1H, J12,13 = 5.0, J12,14 = 1.2, H-12), 7.08 (dm, 1H, J14,13 = 3.0, H-14), 7.17 (dd, 1H, J13,12 = 5.0, J13,14 = 3.0, H-13). NMR 13C (125 MHz, CDCl3, δ, ppm): 77.91 (d, C-1), 81.72 (c, C-3), 44.94 (d, C-3a), 22.45 (t, C-4), 28.70 (t, C-5), 134.35 (c, C-6), 119.72 (d, C-7), 42.83 (d, C-7a), 26.98 (q, C-8), 23.76 (q, C-9), 23.49 (q, C-10), 142.92 (s, C-11), 126.92 (d, C-12), 124.49 (d, C-13), 121.07 (d, C-14). HR-MS: m/z calcd. for C15H20OS: 248.1229. Found: 248.1230.
12k: NMR 1H (500 MHz, CDCl3, δ, ppm, J/Hz): 0.95–0.98 (ddd, 3H, J12,7 = 2.4, J12,8k = 2.1, J12,8n = 1.8, Me-12), 1.29 (s, 3H, Me-10), 1.36 (s, 3H, Me-11), 1.52–1.55 (m, 1H, H-1), 1.71 (ddd, 1H, 2J = 12.5, J9an,1 = J9an,5 = 3.2, H-9an), 2.07 (dm, 1H, J8k,8n = 18.8, H-8k), 2.17–2.19 (m, 1H, H-5), 2.30 (dddd, 1H, 2J = 12.5, J9s,1 = J9s,5 = 3.2, J9s,8n = 1.2, H-9s), 2.37 (br.d, 1H, 2J = 18.8, H-8n), 4.95 (d, 1H, J4,5 = 2.1, H-4), 5.42–5.45 (m, 1H, H-7), 6.98 (dd, 1H, J14,15 = 5.0, J14,16 = 1.2, H-14), 7.09 (ddd, 1H, J16,15 = 3.0, J16,14 = 1.2, J16,4 = 0.8, H-16), 7.19 (dd, 1H, J15,14 = 5.0, J15,16 = 3.0, H-15). NMR 13C (125 MHz, CDCl3, δ, ppm): 33.97 (d, C-1), 75.31 (c, C-2), 72.06 (d, C-4), 40.97 (d, C-5), 133.23 (c, C-6), 123.14 (d, C-7), 27.61 (t, C-8), 27.92 (t, C-9), 28.58 (q, C-10), 23.87 (q, C-11), 23.68 (q, C-12), 144.27 (c, C-13), 125.62 (d, C-14), 119.47 (d, C-15), 124.69 (d, C-16). HR-MS: m/z calcd. for C15H20OS: 248.1229. Found: 248.1230. = 0 (c = 1.8, CHCl3).
3.1.12. Reaction of Limonene and 2-thiophenecarbaldehyde 10d
To the suspension of the K10 (1.5 g) in methylene chloride (15 mL), a solution of 0.40 g aldehyde in methylene chloride (5 mL) and 0.50 g of limonene was added. The solvent was distilled off, and the reaction mixture was kept at room temperature for 2 h. Thereafter, ethyl acetate was added, the catalyst was filtered off, and the filtrate was evaporated. Isolation of the reaction products was carried out by column chromatography to give 11d and 12d (0.582 g, 64%, 14:1).
3.2. Real-Time Detection of TDP1 Activity
A fluorophore quencher-coupled oligonucleotide biosensor was used for TDP1 enzyme activity real-time fluorescence detection [38]. The biosensor 5′-FAM-AAC GTC AGG GTC TTC C- BHQ1-3′ is a 16-mer single-stranded oligonucleotide with a 5′- fluorophore (FAM), and a 3′-quencher (BHQ1). Recombinant protein TDP1 was expressed in Escherichia coli (pET 16B plasmid containing TDP1 cDNA was provided by Dr. K.W. Caldecott, University of Sussex, United Kingdom) and isolated as described by Lebedeva et al. [43]. The reaction mixture contained TDP1 reaction buffer (50 mM Tris-HCl, 50 mM NaCl, 7 mM β-mercaptoethanol), 50 nM oligonucleotide substrate, varied concentrations of the tested compounds, and 1.5 nM purified TDP1 in a final volume of 200 μL.
The TDP1 reaction mixture was incubated at rt in a POLARstar OPTIMA fluorimeter (BMG LABTECH, GmbH). Fluorescence intensity was measured every 1 min (Ex485/Em520 nm). The efficiency of TDP1 inhibition was evaluated by comparing the rate of increase in the fluorescence of the biosensor in the presence of the compound to that of DMSO (1.5%) control wells. IC50 values were determined using an 11-point concentration–response curve. The data were imported into MARS Data Analysis 2.0 program (BMG LABTECH) and the slope during the linear phase was calculated. The IC50 measurements were carried out in at least three independent experiments.
3.3. TDP1 Activity by Gel-Based Assay
A fluorophore-labeled oligonucleotide 5′- FAM-AAC GTC AGG GTC TTC C-tyrosine-3′ was used as a standard gel-based method for the indication of TDP1 enzyme activity. The oligonucleotide containing a natural phosphotyrosine adduct at the 3′-end is a substrate for TDP1 [33]. The reaction mixture contained reaction buffer (50 mM Tris-HCl, 50 mM NaCl, 7 mM β-mercaptoethanol), 50 nM oligonucleotide substrate, 5 nM purified TDP1, or 1 μg of cell extract in a final volume of 20 μL. The reaction was conducted at 37 °C for 20 min. The reaction products were separated by electrophoresis in a 20% denaturing polyacrylamide gel with 7 M urea. A Typhoon FLA 9500 phosphorimager (GE Healthcare) was used for gel scanning and imaging, and the data were analyzed with QuantityOne 4.6.7 software.
3.4. Obtainment of TDP1 Knockout HEK293FT Clones
3.4.1. Plasmid Construction for Human TDP1 Gene Knockout
The sgRNAs design was performed using the Benchling CRISPR tool (https://www.benchling.com/). Two protospacers (PAM sequences in brackets) were selected for the DNA sequence deletion in the first protein-coding exon of the human TDP1 gene exon 3 (third exon in mRNA, NM_001008744.2): TDP1-gRNA1 AGACGAGTATGAGACATCAG(GGG) and TDP1-gRNA2 GCAGAAAAGCGGTTCCCAGG(AGG). Corresponding oligonucleotides were cloned in plasmid pSpCas9(BB)-2A-GFP (PX458) (the plasmid was a gift from Feng Zhang (Addgene plasmid #48138; http://n2t.net/addgene:48138; RRID:Addgene_48138)) as previously described [44]. Transfection-grade plasmid DNA was isolated using the PureLink HiPure Plasmid Miniprep Kit (ThermoFisher Scientific, Waltham, MA, USA).
3.4.2. Knockout HEK293FT Clone Generation
First, 5 × 105 HEK293FT cells were plated into a well of a 12-well plate and cotransfected with the constructed plasmids TDP1-gRNA1 and TDP1-gRNA2 (0.25 µg of each) using Lipofectamine 3000 Reagent (Thermofisher Scientific). Growth medium contained DMEM/F12 (Gibco, Co Dublin, Ireland) 1:1, 10% fetal bovine serum (FBS) (Gibco), 100 U/mL penicillin-streptomycin (Gibco), and 1x GlutaMAX (Gibco). Then, 48 h after transfection, cells were detached using TrypLE Express (TrypLE, Gibco), and the GFP-positive cell population was enriched by cell sorting using a BD FACSAria III Cell Sorter (BD Biosciences, East Rutherford, NJ, USA). Transfected cells were plated onto a 96-well plate, one cell per well. Single-cell clones grew for two weeks before they were replicated to another 96-well plate, so we obtained two equal 96-well plates with cell clones: One plate was used for PCR analysis of the deletion in the TDP1 gene while the other plate was used for the mutant cell clone multiplicatioin.
3.4.3. Analysis of CRISPR/Cas9-Mediated Deletions in the TDP1 Gene
Genome DNA was extracted from cells on one of two 96-well plates using 50 μL of QuickExtract™ DNA Extraction Solution (Epicentre) per well. The DNA extracts were diluted with 200 μL of mQ water. Two microliters of diluted DNA extract were used for PCR amplification of the target region with the primers: TDP1-scF 5′-TCAGGAAGGCGATTATGGGAG-3′ and TDP1-scR 5′-TTGATGTGGAGGGCTCCAG-3′. Reactions were run on a S1000 Thermal Cycler (Bio-Rad, Hercules, CA ,USA) using BioMaster HS-Taq PCR-Color (2×) (Biolabmix, Novosibirsk Oblast, Russia) with the following program: 95 °C 3 min; 35 cycles: 95 °C 30 s, 68 °C 30 s, 72 °C 10 s; 72 °C 3 min. Products of the reactions were resolved in 1.5% agarose gel stained with ethidium bromide. PCR products with a lower molecular weight than the wild type were selected for subsequent analysis. PCR products were gel purified and cloned in the pGEM-T Easy vector (Promega, Madison, WI, USA). Ten independent plasmid clones were sequenced using M13 universal primers. Products of Sanger sequencing reactions were analyzed using the ABI 3130xl Genetic Analyzer (SB RAS Genomics Core Facility, http://www.niboch.nsc.ru/doku.php/corefacility). Three clones containing deletions in the TDP1 gene were found: Clone B2 Δ197bp/Δ16bp+InT; clone B5 Δ198bp/Δ196bp; and clone E3 Δ197bp/Δ200bp. To study the expression of the mutant form of TDP1 in clone E3, total RNA was isolated from cells using TRIZOL reagent as per the manufacturer’s protocols. The synthesis of the first cDNA strand was performed using SuperScript ™ III Reverse Transcriptase (ThermoFisher Scientific) according to the manufacturer’s recommendations. To control DNA contamination of the RNA sample, a reaction was performed without the addition of reverse transcriptase (RT-).
3.5. Cell Culture Assay
The cytotoxicity of the compounds was examined against human cell lines HEK293FT (human embryonic kidney), WT and TDP1-deficient (TDP1-/-), HCT116 (human colon cancer), IMR-32 (human neuroblastoma cell line), HeLa (cervical cancer), and Krebs-2 carcinoma ascites using MTT and EZ4U colorimetric tests or an xCELLigence DP Real-Time Cell Analyzer (ACEA Biosciences, San Diego, CA, USA) for HEK293FT in cells exposed to the compounds. The cells were grown in DMEM/F12 or IMDM (for IMR-32 and Krebs-2) medium (Gibco), with 1x GlutaMAX (Gibco), 50 IU/mL penicillin, 50 μg/mL streptomycin (MP Biomedicals, Irvine, CA, USA) and in the presence of 10% fetal bovine serum (Biolot). The cells (~20,000 cells per well) were incubated for 24 h at 37 °C in 5% CO2, and then treated with the tested compounds (the volume of the added reagents was 1/100 of the total volume of the culture medium, the amount of DMSO was 1% of the final volume as indicated). Control cells were grown in the presence of 1% DMSO where indicated. After 72 h of cell incubation, the relative amount of living cells was determined using a colorimetric test of the amount of formazan converted from 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT test) or EZ4U Cell Proliferation and Cytotoxicity Assay (Biomedica, Austria), as per the manufacturer’s protocols. The real-time detection of the living cells was done by an xCELLigence DP Real-Time Cell Analyzer (ACEA Biosciences). The measurements were carried out in three parallel experiments.
3.6. Calculation of Molecular Descriptors
The QikProp 3.2 [45] software was used to calculate the molecular descriptors of the ligands. The reliability of QikProp is established for the calculated descriptors [46]. The known drug indexes (KDI) were calculated from the molecular descriptors as described by Eurtivong and Reynisson [42]. For application in Excel, columns for each property were created and the following equations used do derive the KDI numbers for each descriptor: KDI MW: =EXP(-((MW-371.76)2)/((2 × 112.76)2)), KDI Log P: =EXP(-((LogP-2.82)2)/((2 × 2.21)2)), KDI HD: =EXP(-((HD-1.88)2)/((2 × 1.7)2)), KDI HA: =EXP(-((HA-5.72)2)/((2 × 2.86)2)), KDI RB- =EXP(-((RB-4.44)2)/((2 × 3.55)2)), and KDI PSA: =EXP(-((PSA-79.4)2)/((2 × 54.16)2)). These equations could simply be copied into Excel and the descriptor name (e.g., MW) substituted with the value in the relevant column. In order to derive KDI2A, this equation was used: = (KDI MW + KDI LogP + KDI HD + KDI HA + KDI RB + KDI PSA) and for KDI2B: = (KDI MW × KDI LogP × KDI HD × KDI HA × KDI RB × KDI PSA).
4. Conclusions
A new class of effective TDP1 inhibitors was discovered with activity in the low micromolar to nanomolar concentrations. All the tested derivatives exhibited low intrinsic cytotoxicity when tested in a panel of cancer cell lines. The most promising compounds with the best combination of TDP1 inhibition, low toxicity, and enhancement of Tpc efficacy were compounds 11h and 12k, with IC50 values of 0.75 and 1.20 μM, respectively. Thus, these new inhibitors are promising candidates for adjuvant therapy, mainly due to the absence of an additional toxic load.
Furthermore, a panel of isogenic clones of the HEK293FT cell line knockout for TDP1 was created using the CRISPR-Cas9 system. Clone E3 with the Δ197bp/Δ200bp deletion in the TDP1 gene was selected for the experiments. We investigated the cytotoxic effect of the Tpc and the TDP1 inhibitors 11h and 12k separately and jointly. HEK293FT TDP1-/- cells were more sensitive to Tpc compared to WT cells. For both 11h and 12k, we observed cell growth suppression in the presence of Tpc only for WT cells but not for the TDP1 knockout cells. It can therefore be stated that the synergistic effect with Tpc on HEK293FT cell growth is only caused by TDP1 activity inhibition by 11h and 12k, with minimal off-target effects. These results make a significant contribution to our understanding of the mechanism of action for the non-cytotoxic 3-carene-derived TDP1 inhibitors. In conclusion, we demonstrated the existence of TDP1 inhibitors, which do not modulate other biomolecular targets, which in turn makes the prospect of developing viable drug candidates more realistic since off-target toxicity often leads to undesirable side effects [47].
Acknowledgments
The authors would like to acknowledge the Multi-Access Chemical Research Center SB RAS for their assistance with the spectral and analytical measurements. Cell lines were obtained from the Russian Cell Culture Collection (RCCC) Institute of Cytology RAS, St. Petersburg, Russia.
Supplementary Materials
Supplementary Materials can be found online.
Author Contributions
These authors contributed equally: I.V.I., N.S.D. Chemistry investigation, I.V.I., N.S.L.-Z., A.Y.S., D.V.K. and K.P.V.; In vitro investigation, N.S.D., A.L.Z.; A.A.C., O.D.Z., E.S.I.; TDP1 knockout clones generation, A.A.M., S.P.M., S.M.Z.; Modeling, J.R., R.C., D.M.A.-T.; Methodology, N.F.S. and O.I.L.; Project administration, K.P.V.; Supervision, K.P.V.; Writing—original draft, N.S.D., I.V.I., A.L.Z. and S.P.M.; Writing—review and editing, K.P.V., J.R., S.M.Z., N.F.S., O.I.L. All authors have read and agreed to the published version of the manuscript.
Funding
Study of TDP1 inhibition was funded by the Russian Science Foundation (grant no. 19-13-00040); generation of cell line knockout for TDP1 was funded by the Russian Foundation for Basic Research (grant no. 19-415-540002).
Conflicts of Interest
The authors report no conflicts of interest.
Footnotes
Sample Availability: Samples of the compounds of structural types 11 and 12 are available from the authors.
References
- 1.Huang S.-Y.N., Pommier Y., Marchand C. Tyrosyl-DNA Phosphodiesterase 1 (Tdp1) inhibitors. Expert Opin. Ther. Patents. 2011;21:1285–1292. doi: 10.1517/13543776.2011.604314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laev S.S., Salakhutdinov N.F., Lavrik O.I. Tyrosyl-DNA phosphodiesterase inhibitors: Progress and potential. Bioorganic Med. Chem. 2016;24:5017–5027. doi: 10.1016/j.bmc.2016.09.045. [DOI] [PubMed] [Google Scholar]
- 3.Zakharenko A., Dyrkheeva N., Lavrik O.I. Dual DNA topoisomerase 1 and tyrosyl-DNA phosphodiesterase 1 inhibition for improved anticancer activity. Med. Res. Rev. 2019;39:1427–1441. doi: 10.1002/med.21587. [DOI] [PubMed] [Google Scholar]
- 4.Kawale A.S., Povirk L.F. Tyrosyl-DNA phosphodiesterases: Rescuing the genome from the risks of relaxation. Nucleic Acids Res. 2018;46:520–537. doi: 10.1093/nar/gkx1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Interthal H., Pouliot J.J., Champoux J.J. The tyrosyl-DNA phosphodiesterase Tdp1 is a member of the phospholipase D superfamily. Proc. Natl. Acad. Sci. USA. 2001;98:12009–12014. doi: 10.1073/pnas.211429198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pommier Y. Topoisomerase I inhibitors: Camptothecins and beyond. Nat. Rev. Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 7.Dexheimer T.S., Antony S., Marchand C., Pommier Y. Tyrosyl-DNA phosphodiesterase as a target for anticancer therapy. Anti-Cancer Agents Med. Chem. 2008;8:381–389. doi: 10.2174/187152008784220357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beretta G.L., Cossa G., Gatti L., Zunino F., Perego P. Tyrosyl-DNA Phosphodiesterase 1 Targeting for Modulation of Camptothecin-Based Treatment. Curr. Med. Chem. 2010;17:1500–1508. doi: 10.2174/092986710790979971. [DOI] [PubMed] [Google Scholar]
- 9.Ledesma F.C., El Khamisy S.F., Zuma M.C., Osborn K., Caldecott K.W. A human 5′-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature. 2009;461:674–678. doi: 10.1038/nature08444. [DOI] [PubMed] [Google Scholar]
- 10.Filimonov A.S., Chepanova A.A., Luzina O.A., Zakharenko A.L., Zakharova O.D., Ilina E.S., Dyrkheeva N.S., Kupryushkin M.S., Kolotaev A.V., Khachatryan D.S., et al. New Hydrazinothiazole Derivatives of Usnic Acid as Potent Tdp1 Inhibitors. Molecules. 2019;24:3711. doi: 10.3390/molecules24203711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zakharenko A., Luzina O., Sokolov D.N., Kaledin V.I., Nikolin V., Popova N., Patel J., Zakharova O., Chepanova A., Zafar A., et al. Novel tyrosyl-DNA phosphodiesterase 1 inhibitors enhance the therapeutic impact of topotecan on in vivo tumor models. Eur. J. Med. Chem. 2019;161:581–593. doi: 10.1016/j.ejmech.2018.10.055. [DOI] [PubMed] [Google Scholar]
- 12.Dyrkheeva N., Luzina O., Filimonov A., Zakharova O., Ilina E., Zakharenko A., Kuprushkin M., Nilov D., Gushchina I., Švedas V., et al. Inhibitory Effect of New Semisynthetic Usnic Acid Derivatives on Human Tyrosyl-DNA Phosphodiesterase 1. Planta Medica. 2018;85:103–111. doi: 10.1055/a-0681-7069. [DOI] [PubMed] [Google Scholar]
- 13.Salomatina O., Popadyuk I.I., Zakharenko A.L., Zakharova O.D., Fadeev D.S., Komarova N.I., Reynisson J., Arabshahi H.J., Chand R., Volcho K.P., et al. Novel Semisynthetic Derivatives of Bile Acids as Effective Tyrosyl-DNA Phosphodiesterase 1 Inhibitors. Molecules. 2018;23:679. doi: 10.3390/molecules23030679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salakhutdinov N.F., Volcho K.P., Yarovaya O.I. Monoterpenes as a renewable source of biologically active compounds. Pure Appl. Chem. 2017;89:1105–1117. doi: 10.1515/pac-2017-0109. [DOI] [Google Scholar]
- 15.Patrusheva O., Volcho K., Salakhutdinov N. Approaches to the synthesis of oxygen-containing heterocyclic compounds based on monoterpenoids. Russ. Chem. Rev. 2018;87:771–796. doi: 10.1070/RCR4810. [DOI] [Google Scholar]
- 16.Khomenko T., Zakharenko A., Odarchenko T., Arabshahi H.J., Sannikova V., Zakharova O., Korchagina D., Reynisson J., Volcho K., Salakhutdinov N. New inhibitors of tyrosyl-DNA phosphodiesterase I (Tdp 1) Bioorg. Med. Chem. 2016;24:5573–5581. doi: 10.1016/j.bmc.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 17.Khomenko T.M., Zakharenko A.L., Chepanova A.A., Ilina E.S., Zakharova O.D., I. Kaledin V., Nikolin V.P., A. Popova N., Korchagina D.V., Reynisson J., et al. Promising New Inhibitors of Tyrosyl-DNA Phosphodiesterase I (Tdp 1) Combining 4-Arylcoumarin and Monoterpenoid Moieties as Components of Complex Antitumor Therapy. Int. J. Mol. Sci. 2019;21:126. doi: 10.3390/ijms21010126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mozhaitsev E.S., Zakharenko A.L., Suslov E.V., Korchagina D.V., Zakharova O.D., Vasil’eva I.A., Chepanova A.A., Black E., Patel J., Chand R., et al. Novel Inhibitors of DNA Repair Enzyme TDP1 Combining Monoterpenoid and Adamantane Fragments. Anti-Cancer Agents Med. Chem. 2019;19:463–472. doi: 10.2174/1871520619666181207094243. [DOI] [PubMed] [Google Scholar]
- 19.Mozhaitsev E., Suslov E.V., Demidova Y., Korchagina D., Volcho K.P., Zakharenko A., Vasil’eva I., Kupryushkin M., Chepanova A., Ayine-Tora D.M., et al. The Development of Tyrosyl-DNA Phosphodyesterase 1 (TDP1) Inhibitors Based on the Amines Combining Aromatic/Heteroaromatic and Monoterpenoid Moieties. Lett. Drug Des. Discov. 2019;16:597–605. doi: 10.2174/1570180816666181220121042. [DOI] [Google Scholar]
- 20.Chepanova A.A., Mozhaitsev E.S., Munkuev A.A., Suslov E.V., Korchagina D.V., Zakharova O.D., Zakharenko A.L., Patel J., Ayine-Tora D.M., Reynisson J., et al. The Development of Tyrosyl-DNA Phosphodiesterase 1 Inhibitors. Combination of Monoterpene and Adamantine Moieties via Amide or Thioamide Bridges. Appl. Sci. 2019;9:2767. doi: 10.3390/app9132767. [DOI] [Google Scholar]
- 21.Ponomarev K.Y., Suslov E.V., Zakharenko A.L., Zakharova O.D., Rogachev A.D., Korchagina D.V., Zafar A., Reynisson J., Nefedov A.A., Volcho K.P., et al. Aminoadamantanes containing monoterpene-derived fragments as potent tyrosyl-DNA phosphodiesterase 1 inhibitors. Bioorganic Chem. 2018;76:392–399. doi: 10.1016/j.bioorg.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 22.Li-Zhulanov N.S., Zakharenko A.L., Chepanova A.A., Patel J., Zafar A., Volcho K.P., Salakhutdinov N.F., Reynisson J., Leung I.K.H., Lavrik O.I. A Novel Class of Tyrosyl-DNA Phosphodiesterase 1 Inhibitors That Contains the Octahydro-2H-chromen-4-ol Scaffold. Molecules. 2018;23:2468. doi: 10.3390/molecules23102468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Il’ina I.V., Volcho K.P., Korchagina D.V., Salnikov G.E., Genaev A.M., Karpova E.V., Salakhutdinov N.F. Unusual reactions of (+)-2- and (+)-3-carene with aldehydes on K10 clay. Helv. Chim. Acta. 2010;93:2135–2150. doi: 10.1002/hlca.201000145. [DOI] [Google Scholar]
- 24.Pavlova A., Il’Ina I., Morozova E., Korchagina D., Kurbakova S., Sorokina I., Tolstikova T., Volcho K.P., Salakhutdinov N. Potent Neuroprotective Activity of Monoterpene Derived 4-[(3aR,7aS)- 1,3,3a,4,5,7a-Hexahydro-3,3,6-trimethylisobenzofuran-1-yl]-2-methoxyphenol in MPTP Mice Model. Lett. Drug Des. Discov. 2013;11:611–617. doi: 10.2174/1570180811666131210000316. [DOI] [Google Scholar]
- 25.Acharya S.P., Brown H.C. Hydroboration of Terpenes. III. Isomerization of (+)-3-Carene to (+)-2-Carene. Hydroboration of (+)-2-Carene ([UNK]4-Carene). Nuclear Magnetic Resonance Spectra with Absolute Configurational and Conformational Assignments for the 2-Caranols and 2-Caranones. J. Am. Chem. Soc. 1967;89:1925–1932. doi: 10.1021/ja00984a030. [DOI] [Google Scholar]
- 26.Meyer U., Hoelderich W. Application of basic zeolites in the decomposition reaction of 2-methyl-3-butyn-2-ol and the isomerization of 3-carene. J. Mol. Catal. A: Chem. 1999;142:213–222. doi: 10.1016/S1381-1169(98)00289-1. [DOI] [Google Scholar]
- 27.Julianto T.S., Jumina, Sastrohamidjojo H., Mustofa Solvent-free isomerization of 3-carene to 2-carene using Na/o-chlorotoluene catalyst in trans-isolimonene production. Orient. J. Chem. 2017;33:3107–3111. doi: 10.13005/ojc/330652. [DOI] [Google Scholar]
- 28.Eswaramoorthy M., Krishnasamy V. Influence of coke on the aromatization of 3-carene in the vapour phase over zeolites. Indian J. Chem. 2001;40:264–269. [Google Scholar]
- 29.Krishnasamy V., Yeddanapalli L.M. Vapour phase catalytic transformations of terpene hydrocarbons in the C10H16 series. III. Dehydrogenation of Δ3-carene over modified chromia and chromia–alumina catalysts. Can. J. Chem. 1976;54:3458–3463. doi: 10.1139/v76-496. [DOI] [Google Scholar]
- 30.Sidorenko A., Aho A., Ganbaatar J., Batsuren D., Utenkova D., Sen’Kov G., Wärnå J., Murzin D., Agabekov V. Catalytic isomerization of α-pinene and 3-carene in the presence of modified layered aluminosilicates. Mol. Catal. 2017;443:193–202. doi: 10.1016/j.mcat.2017.10.014. [DOI] [Google Scholar]
- 31.Sidorenko A., Il’Ina I., Kravtsova A., Aho A., Ardashov O.V., Li-Zhulanov N., Volcho K.P., Salakhutdinov N., Murzin D.Y., Agabekov V. Preparation of chiral isobenzofurans from 3-carene in the presence of modified clays. Mol. Catal. 2018;459:38–45. doi: 10.1016/j.mcat.2018.07.025. [DOI] [Google Scholar]
- 32.Pommier Y., Marchand C., Thibaut L. Diamidine derivatives as inhibitors of human tyrosyl-DNA-phosphodiesterase (Tdp1) 60/786,604. USA Patent. 2006 Mar 27;
- 33.Zakharova O., Luzina O., Zakharenko A., Sokolov D., Filimonov A., Dyrkheeva N., Chepanova A., Ilina E., Ilyina A., Klabenkova K., et al. Synthesis and evaluation of aryliden- and hetarylidenfuranone derivatives of usnic acid as highly potent Tdp1 inhibitors. Bioorganic Med. Chem. 2018;26:4470–4480. doi: 10.1016/j.bmc.2018.07.039. [DOI] [PubMed] [Google Scholar]
- 34.Antony S., Marchand C., Stephen A.G., Thibaut L., Agama K.K., Fisher R.J., Pommier Y. Novel high-throughput electrochemiluminescent assay for identification of human tyrosyl-DNA phosphodiesterase (Tdp1) inhibitors and characterization of furamidine (NSC 305831) as an inhibitor of Tdp1. Nucleic Acids Res. 2007;35:4474–4484. doi: 10.1093/nar/gkm463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Majumdar P., Bathula C., Basu S.M., Das S.K., Agarwal R., Hati S., Singh A., Sen S., Das B.B. Design, synthesis and evaluation of thiohydantoin derivatives as potent topoisomerase I (Top1) inhibitors with anticancer activity. Eur. J. Med. Chem. 2015;102:540–551. doi: 10.1016/j.ejmech.2015.08.032. [DOI] [PubMed] [Google Scholar]
- 36.Volcho K.P., Tatarova L.E., Korchagina D.V., Salakhutdinov N.F., Aul’chenko I.S., Ione K.G., Barkhash V.A. Cycloaddition of carbonyl compounds to olefins on aluminosilicate catalysts. Russ. J. Org. Chem. 1994;30:641–653. [Google Scholar]
- 37.Sidorenko A., Kravtsova A., Mäki-Arvela P., Aho A., Sandberg T., Il’Ina I., Li-Zhulanov N., Korchagina D., Volcho K., Salakhutdinov N., et al. Synthesis of isobenzofuran derivatives from renewable 2-carene over halloysite nanotubes. Mol. Catal. 2020;490:110974. doi: 10.1016/j.mcat.2020.110974. [DOI] [Google Scholar]
- 38.Zakharenko A., Khomenko T., Zhukova S., Koval O.A., Zakharova O., Anarbaev R., Lebedeva N., Korchagina D., Komarova N., Vasiliev V., et al. Synthesis and biological evaluation of novel tyrosyl-DNA phosphodiesterase 1 inhibitors with a benzopentathiepine moiety. Bioorganic Med. Chem. 2015;23:2044–2052. doi: 10.1016/j.bmc.2015.03.020. [DOI] [PubMed] [Google Scholar]
- 39.Li J., Summerlin M., Nitiss K.C., Nitiss J.L., Hanakahi L. TDP1 is required for efficient non-homologous end joining in human cells. DNA Repair. 2017;60:40–49. doi: 10.1016/j.dnarep.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 40.Brettrager E.J., Segura I.A., Van Waardenburg R.C., Waardenburg V. Tyrosyl-DNA Phosphodiesterase I N-Terminal Domain Modifications and Interactions Regulate Cellular Function. Genes. 2019;10:897. doi: 10.3390/genes10110897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu F., Logan G., Reynisson J. Wine Compounds as a Source for HTS Screening Collections. A Feasibility Study. Mol. Informatics. 2012;31:847–855. doi: 10.1002/minf.201200103. [DOI] [PubMed] [Google Scholar]
- 42.Eurtivong C., Reynisson J. The Development of a Weighted Index to Optimise Compound Libraries for High Throughput Screening. Mol. Informatics. 2018;38:1800068. doi: 10.1002/minf.201800068. [DOI] [PubMed] [Google Scholar]
- 43.Lebedeva N.A., Rechkunova N.I., Lavrik O.I. AP-site cleavage activity of tyrosyl-DNA phosphodiesterase 1. FEBS Lett. 2011;585:683–686. doi: 10.1016/j.febslet.2011.01.032. [DOI] [PubMed] [Google Scholar]
- 44.Ran F.A., Hsu P.D., Wright J., Agarwala V., A. Scott D., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.QikProp: Schrödinger, LLC, New York NY. [(accessed on 11 May 2020)];2020 Available online: https://www.schrodinger.com/citations.
- 46.Ioakimidis L., Thoukydidis L., Mirza A., Naeem S., Reynisson J. Benchmarking the Reliability of QikProp. Correlation between Experimental and Predicted Values. QSAR Comb. Sci. 2008;27:445–456. doi: 10.1002/qsar.200730051. [DOI] [Google Scholar]
- 47.Liu S., Kurzrock R. Toxicity of targeted therapy: Implications for response and impact of genetic polymorphisms. Cancer Treat. Rev. 2014;40:883–891. doi: 10.1016/j.ctrv.2014.05.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.