Abstract
Cannabis is widely used as a therapeutic drug, especially by patients suffering from psychiatric and neurodegenerative diseases. However, the complex interplay between phytocannabinoids and their targets in the human receptome remains largely a mystery, and there have been few investigations into the relationship between the chemical composition of medical cannabis and the corresponding biological activity. In this study, we investigated 59 cannabis samples used by patients for medical reasons. The samples were subjected to extraction (microwave and supercritical carbon dioxide) and chemical analyses, and the resulting extracts were assayed in vitro using the CB1 and CB2 receptors. Using a partial least squares regression analysis, the chemical compositions of the extracts were then correlated to their corresponding cannabinoid receptor activities, thus generating predictive models that describe the receptor potency as a function of major phytocannabinoid content. Using the current dataset, meaningful models for CB1 and CB2 receptor agonism were obtained, and these reveal the insignificant relationships between the major phytocannabinoid content and receptor affinity for CB1 but good correlations between the two at CB2 receptors. These results also explain the anomalies between the receptor activities of pure phytocannabinoids and cannabis extracts. Furthermore, the models for CB1 and CB2 agonism in cannabis extracts predict the cannabinoid receptor activities of individual phytocannabinoids with reasonable accuracy. Here for the first time, we disclose a method to predict the relationship between the chemical composition, including phytocannabinoids, of cannabis extracts and cannabinoid receptor responses.
Keywords: medical cannabis, tetrahydrocannabinol, cannabidiol, cannabinoid receptors, chemoinformatics, partial least squares analysis, quantitative structure-activity relationship
1. Introduction
In recent years, cannabis has arguably become one of the most popular natural products across the globe. Cannabis sativa, a member of the herbaceous Cannabaceae family of plants, produces more than 568 unique compounds, of which more than 100 belong to the unique class of phytocannabinoids [1,2]. These are organic molecules with a polyphenolic structure. Phytocannabinoids such as Δ9-tetrahydrocannabinol (Δ9-THC), cannabidiol (CBD), Δ9-tetrahydrocannabinolic acid (Δ9-THCA), and cannabidiolic acid (CBDA) (Figure 1) exhibit a wide array of pharmacodynamic interactions in the human receptome [3]. Cannabinoids primarily interact with cannabinoid receptor types 1 and 2 (CB1 and CB2, respectively), but they can also interact with other receptors such as serotonin receptors and vanilloid-type transient receptor potential (TRPV) channels, which result in the physiological effects of cannabis, including psychoactivity or euphoria, motor impairment, analgesia, and immunomodulation [4,5,6].
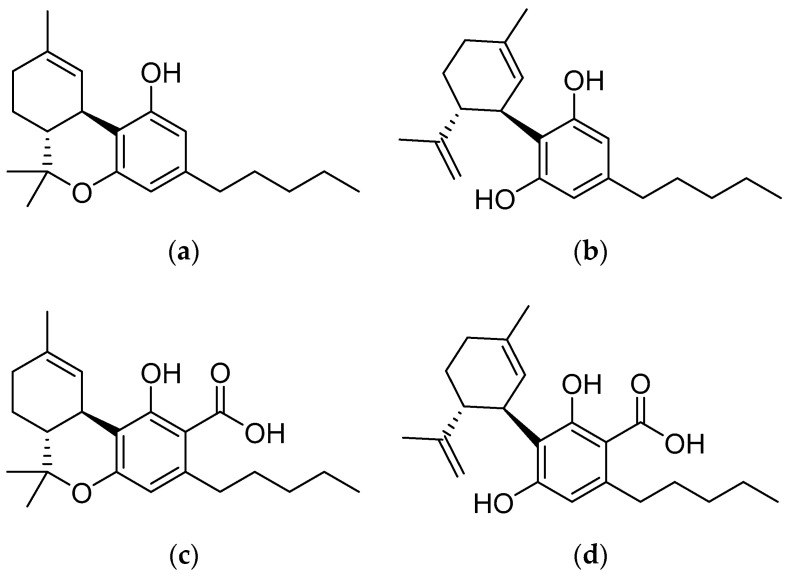
Figure 1.
Major phytocannabinoids present in C. sativa: (a) Δ9-tetrahydrocannabinol, (b) cannabidiol, (c) Δ9-tetrahydrocannabinolic acid, and (d) cannabidiolic acid.
The medicinal properties of cannabis and its derivative products make them potential treatments for managing the symptoms of several neurological and psychiatric disorders; they are also increasingly popular alternatives to opioids and are used by patients suffering from nausea, epilepsy, neuropathic pain, and post-traumatic stress disorder (PTSD) [7,8,9]. In most jurisdictions around the world, medical cannabis is accessible to patients, although it does not possess a drug identification number or a similar authorization by relevant drug control authorities [10,11]. There are hundreds of varieties of medical cannabis available in the market and these can be obtained in the forms of dried flowers, cannabis extracts, and infused oils [12,13]. The composition of phytocannabinoids in these cannabis varieties can vary significantly, leading to a range of biological and physiological responses; for example, the Δ9-THC content in cannabis samples can range from 0.14% to more than 25%, and, depending on the concentration of other phytocannabinoids, the resulting biological activities vary [12].
Interestingly, the variety (based on chemical composition), dose, and the route of administration of medical cannabis are largely determined by patients through self-titration with healthcare professionals playing an advisory role [7,11]. In fact, there have been no systematic studies investigating the relationship between the chemical constituents of different medical cannabis varieties and their corresponding combined therapeutic effects and receptor activities. Nevertheless, it is important to understand the differences between the natural product (which is a complex mixture of bioactive compounds) and individual bioactive compounds because the pharmacological responses produced by whole cannabis or cannabis extracts are different from those of single phytocannabinoids [13,14].
Here, through a series of experiments comprising chemical extractions, quantitative analyses, and CB1/CB2 receptor activity profiling, we have undertaken a comprehensive investigation of 59 cannabis samples, their chemical compositions, and receptor activities and identified if there are any relationships. In doing so, we hope to uncover any relationships between the chemical makeup and receptor activity of the medical cannabis samples. Such relationships will help explain differences in physiological effects caused by the different chemical compositions of cannabis samples. This may also lead to evidence of synergistic/antagonistic effects between the various phytocannabinoids present in medical cannabis and their relative concentrations.
2. Results
Fifty-nine samples of cannabis were collected from patients consuming cannabis for medical purposes. Among these samples, forty-five were dried cannabis samples, whereas the remaining fourteen samples were cannabis oils or resins. The concentrations of the four tested phytocannabinoids CBD, Δ9-THC, CBDA, and Δ9-THCA in all 59 cannabis samples varied significantly (Table 1 and Table 2). In our previous work, we showed that such variations can be a result of the different extraction methods employed, and, thus, extraction methods are important for quality control and quality assurance purposes [15]. Here, our focus is to investigate the effects of variations in phytocannabinoid concentrations on receptor responses and determine if there are quantitative correlations between the chemical composition and biological activities. In these cannabis extracts, the CBD, Δ9-THC, CBDA, and Δ9-THCA concentrations of a weight-by-weight (w/w) basis were 0–51.7%, 0–73.1%, 0–60.3%, and 0–76.1%, respectively (Table 1). Samples extracted using microwave assisted extraction (MAE) primarily contained decarboxylated phytocannabinoids, i.e., CBD and Δ9-THC, which was expected because of the high temperature produced using this extraction procedure, whereas samples obtained via supercritical fluid extraction (SFE) contained higher proportions of CBDA and Δ9-THCA. These differences contributed to a diverse sample set for the receptor activity evaluation. The cannabis oil samples, as received from various sources, are diluted solutions of phytocannabinoids; thus, the CBD, Δ9-THC, CBDA, and Δ9-THCA concentrations (w/v) varied from 0–8.9%, 0–4.6%, 0–31.7%, and 0–15.3%, respectively (Table 2). All but one of the cannabis oil samples contained only CBD and Δ9-THC.
Table 1.
Phytocannabinoid concentrations and corresponding receptor potencies of cannabis extracts, as well as pure phytocannabinoids and reference compounds.
| Sample | Phytocannabinoid Concentration (% w/w) | Receptor Potency (μM) | Normalized Receptor Potency (nM) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a EC50 (Agonist) | b IC50 (Antagonist) | Log(EC50) | Log(IC50) | |||||||||
| CBD | Δ9-THC | CBDA | Δ9-THCA | CB1 | CB2 | CB1 | CB2 | CB1 | CB2 | CB1 | CB2 | |
| CP-55940 3 | - | - | - | - | 0.0025 ± 0.0009 | 0.0026 ± 0.0004 | - | - | 0.398 | 0.415 | - | - |
| SR-141716 3 | - | - | - | - | - | - | 0.024 ± 0.003 | - | - | - | 1.380 | - |
| AM-630 3 | - | - | - | - | - | - | - | 0.146 ± 0.084 | - | - | - | 2.164 |
| CBD 3 | 100 | - | - | - | 10 ± 2 | >10 | >10 | 7 ± 2 | 4.000 | - | - | 3.845 |
| Δ9-THC 3 | - | 100 | - | - | 0.015 ± 0.001 | >10 | >10 | 1.4 ± 0.3 | 1.176 | - | - | 3.146 |
| CBDA | - | - | 100 | - | 17 ± 3 | 17 ± 4 | >10 | >10 | 4.230 | 4.230 | - | - |
| Δ9-THCA 3 | - | - | - | 100 | 1.8 ± 0.7 | 30 ± 15 | >10 | >10 | 3.255 | 4.477 | - | - |
| 1 | 0.4 ± 0.04 | 49 ± 0.9 | 0 | 0 | 0.05 ± 0.02 | >100 | >1 | >10 | 1.675 | - | - | - |
| 2 | 1.05 ± 0.02 | 29 ± 0.2 | 0 | 0 | 0.013 ± 0.008 | >7 | >10 | >1 | 1.114 | - | - | - |
| 3 | 0.169 ± 0.007 | 39.2 ± 0.8 | 0 | 0.036 ± 0.005 | 1.3 ± 0.5 | >100 | >1 | >100 | 3.117 | - | - | - |
| 4 | 27.9 ± 0.3 | 11.5 ± 0.2 | 0 | 0 | 30 ± 10 | >100 | 0.13 ± 0.06 | >10 | 4.479 | - | 2.117 | - |
| 5 | 33.0 ± 0.8 | 0 | 0 | 0 | 0.2 ± 0.1 | >10 | >1 | >1 | 2.332 | - | - | - |
| 6 | 0.163 ± 0.006 | 39 ± 1 | 0 | 0 | 0.1 ± 0.1 | >33 | >1 | >10 | 2.152 | - | - | - |
| 7 | 0.135 ± 0.003 | 35.9 ± 0.3 | 0 | 0 | >10 | >10 | >1 | >10 | - | - | - | - |
| 8 | 10.5 ± 0.3 | 23.4 ± 0.9 | 0 | 0 | >10 | >10 | >1 | 2.5 ± 0.8 | - | - | - | 3.401 |
| 9 | 0.163 ± 0.003 | 70.0 ± 0.9 | 0 | 0 | 0.2 ± 0.1 | >2 | >2 | >19 | 2.332 | - | - | - |
| 10 | 0.153 ± 0.002 | 73.1 ± 0.9 | 0 | 0 | 0.4 ± 0.3 | >13 | 6 ± 2 | >19 | 2.651 | - | 3.779 | - |
| 11 | 0.159 ± 0.010 | 72.5 ± 0.3 | 0 | 0 | 0.7 ± 0.3 | >13 | >2 | >0.2 | 2.846 | - | - | - |
| 12 | 0 | 41.8 ± 0.3 | 0 | 0 | 6 ± 2 | >10 | >1 | >10 | 3.803 | - | - | - |
| 13 | 21.9 ± 0.7 | 36.2 ± 0.5 | 0 | 0 | 9 ± 6 | >48 | >1 | >10 | 3.978 | - | - | - |
| 14 | 0 | 42.2 ± 0.6 | 0 | 0 | 0.5 ± 0.4 | >100 | >1 | >10 | 2.696 | - | - | - |
| 15 | 51.7 ± 0.4 | 0 | 0 | 0 | 0.8 ± 0.4 | >100 | >10 | >1 | 2.926 | - | - | - |
| 16 | 17.6 ± 0.5 | 41.1 ± 0.7 | 0 | 0 | 0.2 ± 0.1 | >99 | >1 | >10 | 2.318 | - | - | - |
| 17 | 0 | 28.2 ± 0.4 | 0 | 0 | 0.2 ± 0.1 | >100 | >1 | 1.3 ± 0.8 | 2.365 | - | - | 3.102 |
| 18 | 0 | 17.4 ± 0.9 | 0 | 0 | 0.2 ± 0.1 | >100 | >1 | >10 | 2.362 | - | - | - |
| 19 | 23.9 ± 0.3 | 11.57 ± 0.10 | 0 | 0 | 0.11 ± 0.06 | >10 | >1 | >1 | 2.060 | - | - | - |
| 20 | 18.1 ± 0.4 | 10.7 ± 0.6 | 0 | 0 | >0.1 | >10 | >1 | >1 | - | - | - | - |
| 21 | 0 | 33 ± 1 | 0 | 0 | >0.1 | >10 | >1 | >10 | - | - | - | - |
| 22 | 0 | 65.6 ± 0.5 | 0 | 0 | 0.19 ± 0.04 | >100 | >1 | >10 | 2.279 | - | - | - |
| 23 | 0 | 39.4 ± 0.1 | 0 | 0 | 0.04 ± 0.04 | >100 | >1 | 1.6 ± 0.9 | 1.646 | - | - | 3.202 |
| 24 | 0 | 47.4 ± 0.5 | 0 | 0 | 0.15 ± 0.05 | >100 | >10 | >10 | 2.187 | - | - | - |
| 25 | 0 | 54.9 ± 0.3 | 0 | 0 | 0.20 ± 0.09 | >100 | >10 | >10 | 2.292 | - | - | - |
| 26 | 48.9 ± 0.6 | 0 | 0 | 0 | 0.5 ± 0.3 | >100 | >10 | >1 | 2.676 | - | - | - |
| 27 | 0 | 48.6 ± 0.7 | 9.7 ± 0.3 | 7.6 ± 0.4 | 0.15 ± 0.08 | 3 ± 2 | >1 | >10 | 2.108 | 3.477 | - | - |
| 28 | 0.30 ± 0.01 | 55 ± 2 | 0 | 0 | 0.20 ± 0.08 | 0.2 ± 0.1 | >10 | >10 | 2.307 | 2.304 | - | - |
| 29 | 0.37 ± 0.02 | 44 ± 2 | 0 | 0 | 0.20 ± 0.09 | >114 | >11 | >11 | 2.291 | - | - | - |
| 30 | 0 | 43.2 ± 0.1 | 0 | 0 | 0.19 ± 0.08 | 0.16 ± 0.06 | >11 | >10 | 2.290 | 2.215 | - | - |
| 31 | 0.37 ± 0.01 | 56 ± 1 | 0 | 0 | 0.3 ± 0.3 | >100 | >10 | >1 | 2.442 | - | - | - |
| 32 | 0.378 ± 0.008 | 46 ± 1 | 0 | 0 | >0.012 | >12 | >12 | >12 | - | - | - | - |
| 33 | 0.282 ± 0.009 | 46 ± 1 | 0 | 0 | 0.12 ± 0.05 | 0.10 ± 0.07 | >10 | >10 | 2.079 | 2.015 | - | - |
| 34 | 0.44 ± 0.02 | 38.7 ± 0.8 | 0 | 0 | 0.3 ± 0.2 | >100 | >100 | >100 | 2.402 | - | - | - |
| 35 | 0.77 ± 0.03 | 43 ± 1 | 0 | 0 | 0.15 ± 0.08 | >10 | >10 | >0.1 | 2.166 | - | - | - |
| 36 | 0 | 0 | 49 ± 1 | 32.0 ± 0.3 | 3 ± 1 | 18 ± 7 | >10 | >10 | 3.544 | 4.262 | - | - |
| 37 | 0 | 0 | 0.092 ± 0.007 | 51.2 ± 0.6 | 3 ± 1 | 21 ± 11 | >93 | >0.09 | 3.490 | 4.323 | - | - |
| 38 | 0 | 0 | 28.3 ± 0.4 | 27.6 ± 0.2 | 0.08 ± 0.03 | 13 ± 5 | >1 | >11 | 1.878 | 4.098 | - | - |
| 39 | 0 | 0 | 0.050 ± 0.003 | 47.8 ± 0.8 | 1.3 ± 0.2 | 10 ± 2 | >88 | >7 | 3.109 | 3.999 | - | - |
| 40 | 0 | 0 | 60 ± 1 | 0 | 0.8 ± 0.3 | 15 ± 6 | >9 | >89 | 2.920 | 4.183 | - | - |
| 41 | 0 | 0 | 0.232 ± 0.005 | 61.5 ± 0.4 | 5 ± 3 | 9 ± 2 | >103 | >10 | 3.708 | 3.935 | - | - |
| 42 | 0 | 0 | 0.094 ± 0.010 | 62.2 ± 0.5 | 0.6 ± 0.2 | 6 ± 2 | >94 | >9 | 2.767 | 3.796 | - | - |
| 43 | 0 | 0 | 0.076 ± 0.002 | 64 ± 1 | 0.5 ± 0.2 | 46 ± 19 | >10 | >10 | 2.724 | 4.663 | - | - |
| 44 | 0 | 0 | 0 | 76.1 ± 0.3 | 0.5 ± 0.1 | 73 ± 59 | >10 | >10 | 2.738 | 4.860 | - | - |
| 45 | 0 | 0 | 52 ± 1 | 15.7 ± 0.02 | 2 ± 1 | 13 ± 5 | >10 | >10 | 3.358 | 4.100 | - | - |
| 46 | 0 | 0 | 0 | 54 ± 1 | 0.7 ± 0.4 | 12 ± 4 | >10 | >10 | 2.825 | 4.096 | - | - |
| 47 | 0 | 0 | 0 | 73 ± 3 | 4 ± 1 | >10 | >10 | >10 | 3.564 | - | - | - |
a Half-maximal effective concentrations. b Half-maximal inhibitory concentrations. 3 [15].
Table 2.
Phytocannabinoid concentrations and corresponding receptor potencies of cannabis oil samples.
| Sample | Phytocannabinoid Concentration (% w/v) | Receptor Potency (μM) | Normalized Receptor Potency (nM) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a EC50 (Agonist) | b IC50 (Antagonist) | Log(EC50) | Log(IC50) | |||||||||
| CBD | Δ9-THC | CBDA | Δ9-THCA | CB1 | CB2 | CB1 | CB2 | CB1 | CB2 | CB1 | CB2 | |
| 1 | 2.05 ± 0.03 | 0.158 ± 0.007 | 0 | 0 | >2 | >2 | >2 | >0.02 | - | - | - | - |
| 2 | 0.0072 ± 0.0002 | 1.815 ± 0.010 | 0 | 0 | 0.03 ± 0.03 | >2 | >0.2 | >0.2 | 1.513 | - | - | - |
| 3 | 1.168 ± 0.009 | 0.290 ± 0.007 | 0 | 0 | 0.04 ± 0.02 | >1 | >37 | 0.024 ± 0.008 | 1.642 | - | - | 1.385 |
| 4 | 0.427 ± 0.004 | 0.512 ± 0.006 | 0 | 0 | 0.03 ± 0.02 | >0.8 | >0.8 | >0.08 | 1.516 | - | - | - |
| 5 | 1.71 ± 0.02 | 0.112 ± 0.001 | 0 | 0 | 0.2 ± 0.2 | >2 | >2 | >2 | 2.287 | - | - | - |
| 6 | 3.79 ± 0.09 | 0.199 ± 0.006 | 0 | 0 | 0.20 ± 0.08 | >13 | >13 | >13 | 2.296 | - | - | - |
| 7 | 8.9 ± 0.2 | 0 | 0 | 0 | 0.5 ± 0.3 | >10 | >10 | >10 | 2.711 | - | - | - |
| 8 | 7.5 ± 0.1 | 4.6 ± 0.1 | 31.7 ± 0.6 | 15.3 ± 0.3 | 0.3 ± 0.2 | >0.001 | >10 | >10 | 2.304 | - | - | - |
| 9 | 0.0133 ± 0.0003 | 4.1 ± 0.2 | 0 | 0 | 0.3 ± 0.2 | >0.009 | >0.9 | >9 | 2.401 | - | - | - |
| 10 | 5.4 ± 0.1 | 0 | 0 | 0 | >0.1 | >0.001 | >1 | >1 | - | - | - | - |
| 11 | 2.73 ± 0.02 | 0.173 ± 0.003 | 1.34 ± 0.03 | 0.017 ± 0.003 | 0.04 ± 0.02 | >3 | >30 | >3 | 1.391 | - | - | - |
| 12 | 1.258 ± 0.008 | 0.909 ± 0.006 | 0.035 ± 0.001 | 0.0017 ± 0.0001 | >0.001 | 0.0015 ± 0.0007 | >4 | >4 | - | 0.162 | - | - |
a Half-maximal effective concentrations. b Half-maximal inhibitory concentrations.
The agonist and antagonist activities for all 59 cannabis samples, the pure cannabinoids (Δ9-THC, CBD, Δ9-THCA, and CBDA), and the reference compounds (CP55940, SR-141716, and AM-630) at CB1 and CB2 were evaluated, and the results are presented in Table 1 and Table 2. The receptor activities (half-maximal effective concentrations (EC50) or half-maximal inhibitory concentrations (IC50), for the characterization of agonist and antagonist effects, respectively) for each cannabis sample were calculated using the concentration of the phytocannabinoid present in the highest proportion (i.e., the major phytocannabinoid) in the corresponding cannabis sample. Notably the receptor activities of several extracts were poor or required concentrations of the major phytocannabinoids in excess of 10 µM, resulting in solubility issues and being physiologically impractical. These cannabis extracts were, thus, considered inactive.
Subsequently, we performed chemoinformatic analyses using partial least squares (PLS) methods, which are widely used in quantitative structure-activity relationship (QSAR) analyses to relate the phytocannabinoid concentrations and receptor responses. In a traditional QSAR, a dependent variable, such as receptor response, is correlated with physicochemical descriptors, such as octanol/water partition coefficients or Hammett constants derived for a set of molecules, to generate mathematical models. These models can explain the relevance of various molecular features to the observed biological activities and may enable the prediction of the physicochemical properties of untested compounds [16]. Using a similar strategy, we correlated the CB1 and CB2 receptor potencies of the medical cannabis samples to the four cannabinoid concentrations using PLS regression analysis to obtain QSAR-like prediction models (Table S2).
The concentrations of the four phytocannabinoids quantified in the 59 cannabis samples were fit against the corresponding agonist and antagonist activities of each cannabis sample at CB1 and CB2. Three subsets emerged with significant statistical power, containing sufficient data points for the generation of the corresponding mathematical models (Equations (1)–(3)).
The first relationship, modeled by Equation (1), is between the linear terms of the concentrations of CBD, Δ9-THC, and CBDA, and the quadratic term of the Δ9-THCA concentration, describing the corresponding agonist activities at the CB2 receptor (EC50):
| Log(EC50) = −2.967 CBD − 1.649 × 10−2Δ9-THC + 5.143 × 10−3 CBDA + 1.363 × 10−4Δ9-THCA2 + 3.765 | (1) |
where n = 15, r2 = 0.842, Q2 = 0.784, RMSE = 0.338, Components = 1. The second relationship between the phytocannabinoid composition of the cannabis oil samples and their CB1 receptor agonism is described by Equation (2):
| Log(EC50) = 0.142 CBD + 5.089 × 10−2Δ9-THC2 − 2.356 × 10−2 CBDA − 3.833 × 10−2Δ9-THCA + 1.510 | (2) |
where n = 9, r2 = 0.679, Q2 = 0.304, RMSE = 0.260, Components = 3. The third meaningful relationship between the phytocannabinoid composition of the cannabis extracts and their CB1 receptor potencies as agonists is described by Equation (3):
| Log(EC50) = 5.479 × 10−3 CBD − 6.151 × 10−3Δ9-THC + 1.085 × 10−4 CBDA2 + 5.882 × 10−3Δ9-THCA + 2.681 | (3) |
where n = 42, r2 = 0.204, Q2 = 0.140, RMSE = 0.598, Components = 1.
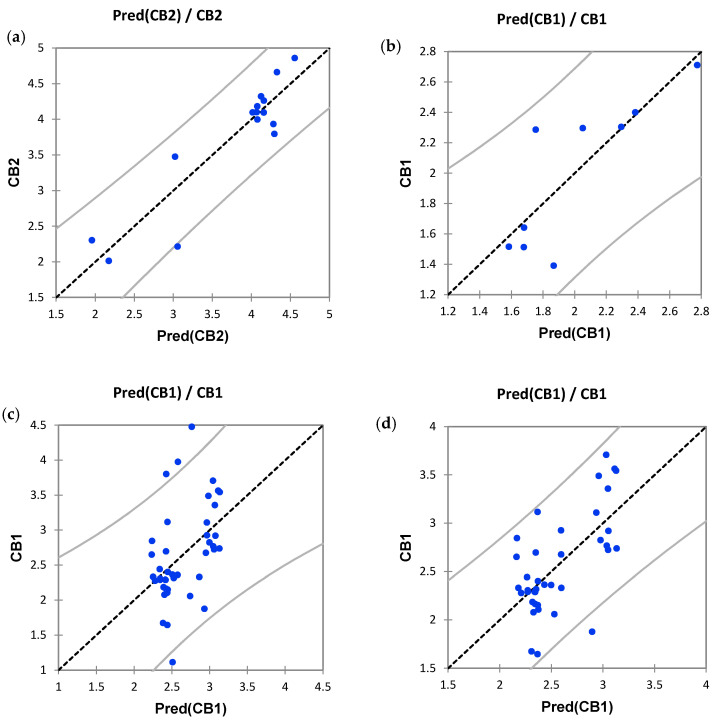
Figure 2 illustrates the distributions of the experimental and predicted receptor potencies for the three models (Equations (1)–(3) and Figure 2a–c, respectively).
Figure 2.
Distribution of calculated versus experimental receptor potencies for the models described by Equations (1)–(3) (above) (a–c, respectively) and Equation (4) (vide infra, d).
3. Discussion
Individual phytocannabinoids exert different physiological effects on the human endocannabinoid system and other receptors, albeit with different potencies at each receptor. For example, Δ9-THC acts as a potent partial agonist whereas CBD is known as a moderately potent antagonist at both the CB1 and CB2 receptors [15,17,18]. Similarly, Δ9-THCA exhibits agonist activity at CB1 and CB2, although its affinities for these receptors are several orders of magnitude lower than those of Δ9-THC and CBD. In contrast, CBDA shows little to no binding at either cannabinoid receptor [15,19]. One of the interesting findings in research into cannabis and cannabis-derived extracts is that the combined biological and physiological actions of these phytocannabinoids when consumed as extracts are quite different to those of the pure compounds, even when considering cannabis extracts enriched in a particular phytocannabinoid. Here, using 59 cannabis samples consumed by patients, we were interested in understanding the collective response of the phytocannabinoids (and potentially other bioactive compounds) in these cannabis extracts. We initially limited our efforts to the four most abundant and studied phytocannabinoids, Δ9-THC, Δ9-THCA, CBD, and CBDA, to gain an understanding of potential relationships with routinely measurable receptor activities using PLS methods.
One of the main advantages of PLS regression is its ability to account for multi-collinearity among the independent variables, i.e., the concentrations of the phytocannabinoids in the samples. This is especially pertinent with regards to medical cannabis, for which the phytocannabinoids are produced through a multi-branched biosynthetic pathway [2]. The four phytocannabinoids in the cannabis samples considered in this study (Δ9-THCA, CBDA, Δ9-THC, and CBD) are synthesized through a combination of enzyme catalysis and exposure to environmental heat, and undergo various changes during processing and storage until they are consumed [20,21,22,23,24]. CBDA and Δ9-THCA, and by extension CBD and Δ9-THC, lie on separate branches of the phytosynthetic pathway originating from a common precursor, cannabigerolic acid (CBGA). A negative correlation, thus, can be observed between the total Δ9-THC content and total CBD content in the cannabis samples. Indeed, regression of our quantitative data revealed strong negative correlations between total Δ9-THC and total CBD contents in both dried cannabis and cannabis oil sample groups (Table 3A,B, respectively). Further breakdown of the cannabis samples reveals that a stronger correlation exists between the CBD and Δ9-THC concentrations when extracted with MAE, as well as a stronger correlation between CBDA and Δ9-THCA concentrations of SFE samples (Table 3C,D, respectively).
Table 3.
Intra-variable correlation (Pearson’s correlation coefficients) between major cannabinoid concentrations in the cannabis extracts from dried cannabis (A), cannabis oils (B), cannabis extracts using MAE (C) and SFE (D). Note: Total Δ9-THC = Δ9-THC + Δ9-THCA Content; Total CBD = CBD + CBDA Content.
| CBD | Δ9-THC | CBDA | Δ9-THCA | Total Δ9-THC | Total CBD | |
|---|---|---|---|---|---|---|
| A | ||||||
| CBD | 1 | |||||
| Δ9 -THC | −0.330 | 1 | ||||
| CBDA | −0.137 | −0.344 | 1 | |||
| Δ9 -THCA | −0.230 | −0.613 | 0.056 | 1 | ||
| Total Δ9-THC | 0.615 | −0.513 | 0.697 | −0.122 | 1 | |
| Total CBD | −0.637 | 0.443 | −0.328 | 0.437 | −0.722 | 1 |
| B | ||||||
| CBD | 1 | |||||
| Δ9 -THC | 0.002 | 1 | ||||
| CBDA | 0.489 | 0.675 | 1 | |||
| Δ9 -THCA | 0.489 | 0.680 | 0.999 | 1 | ||
| Total Δ9-THC | 0.681 | 0.568 | 0.972 | 0.971 | 1 | |
| Total CBD | 0.383 | 0.819 | 0.976 | 0.978 | 0.923 | 1 |
| C | ||||||
| CBD | 1 | |||||
| Δ9 -THC | −0.750 | 1 | ||||
| CBDA | −0.091 | 0.094 | 1 | |||
| Δ9 -THCA | −0.091 | 0.094 | 1.000 | 1 | ||
| Total Δ9-THC | 0.993 | −0.742 | 0.025 | 0.024 | 1 | |
| Total CBD | −0.750 | 0.998 | 0.159 | 0.159 | −0.734 | 1 |
| D | ||||||
| CBD | 1 | |||||
| Δ9 -THC | N/A | 1 | ||||
| CBDA | N/A | N/A | 1 | |||
| Δ9 -THCA | N/A | N/A | −0.910 | 1 | ||
| Total Δ9-THC | N/A | N/A | 1 | −0.910 | 1 | |
| Total CBD | N/A | N/A | −0.910 | 1 | −0.910 | 1 |
The first model (Equation (1)) suggests that increasing the concentrations of CBD and Δ9-THC increases the potency of the cannabis extracts as CB2 receptor agonists, whereas increasing the CBDA and Δ9-THCA concentrations decreases the corresponding potency. The coefficient of determination (r2) and Q2 statistics, 0.842 and 0.784, respectively, for this model indicate a strong correlation between the dependent and independent variables, as well as good predictive power for the equation. Interestingly, CBD, a known antagonist at CB2 receptor, exhibits CB2 agonism when administered as part of a cannabis extract where the other three phytocannabinoids are in smaller proportions. Furthermore, the relative values of the variable coefficients in Equation (1) show that CBD has a higher CB2 receptor potency among the four phytocannabinoids—two orders of magnitude greater than that of Δ9-THC, which exhibits the highest individual CB2 receptor activity. Such pronounced pharmacodynamic differences may be due to synergistic and possibly antagonistic interplay between the ligands at the cannabinoid receptors, where simultaneous occupation of the orthosteric and allosteric sites by multiple ligands in the mixture of compounds leads to unique pharmacological responses [15]. This phenomenon of inter-modulation between phytocannabinoids observed in medical cannabis and cannabis formulations has been collectively termed the entourage effect [13]. Here, in the context of CB2 receptor agonism, we stress that the combined effects of complex natural products, such as cannabis extracts, can be described by mathematical models with high correlation using the concentrations of individual quantifiable compounds, as shown in this study.
The model described by Equation (1) also lends weight to the hypothesis that medical cannabis and cannabinoid-based products show efficacy for neuropathic pain management [25,26]. In contrast to the CB1 receptor, which is mainly found in the central nervous system (CNS), the CB2 receptor is primarily expressed in peripheral tissues, especially those belonging to the immune system [27]. Activation of the CB2 receptor pathway has been shown to modulate nociception in animal models [28,29,30]. Although none of the four phytocannabinoids quantified in the current study are known to behave as CB2 agonists individually, phytocannabinoid synergy at the CB2 receptor may result in CB2 agonism and, in turn, the subsequent anti-nociceptive effects observed clinically. This may explain the anti-nociceptive effects observed in cannabis plant extracts [31,32,33], as well as CBD-containing cannabinoid mixtures, such as nabiximols, unlike CBD alone [34].
The second model, represented by Equation (2), shows that increasing CBD and Δ9-THC concentrations decreases their potency as CB1 receptor agonists, whereas increasing CBDA and Δ9-THCA concentrations increases the potency of cannabis oils. Similar to the trend observed in model 1, CBD concentration exerts the largest effect among the four variables on receptor affinity; additionally, among the three models the third is the only model that attributes receptor activity solely to the cannabinoid acids (CBDA and Δ9-THCA), which show weaker or no CB1 affinity. It is noteworthy that Equation (3) describes a similar relationship with cannabis extracts, rather than cannabis oils, and projects a different result whereby CB1 receptor activity is entirely attributed to the concentration of Δ9-THC. Although arguments can be made for the entourage effect, it is unlikely that a change in the form of dosing (from extracts to oils) alone could lead to such drastic differences in cannabinoid pharmacodynamics at the CB1 receptor [35,36]. Instead, this is likely the result of model inadequacy, brought about by the small sample size (9) that was used to construct Equation (2). Indeed, although the coefficient of determination (r2 = 0.679) indicates moderate correlation, the Q2 parameter are much smaller (Q2 = 0.304), and this difference is indicative of the poor predictive power of the model. As such, the incorporation of additional data from the remaining patients may serve to reveal the true nature of this relationship.
As mentioned above, the third model represented by Equation (3) shows that increasing Δ9-THC concentration increases potency, whereas increasing concentrations of CBD, CBDA, and Δ9-THCA lower the combined potency of cannabis extracts as agonists at the CB1 receptor. This relationship is consistent with the individual pharmacological behavior of the four phytocannabinoids at the CB1 receptors. This also corroborates observations that CBD modulates the acute psychoactive properties of Δ9-THC in vivo, as well as having potential for psychosis, after cannabis consumption [27,37,38]. However, the coefficient of determination (r2 = 0.204) suggests a poor correlation at best between these two variables. Furthermore, the leave-one-out (LOO) cross-validated r2 value (Q2), was also relatively low (Q2 = 0.140). Although the sample size is large (n = 42), this model yielded the poorest correlation between cannabinoid receptor agonist activity and the concentrations of the four phytocannabinoids in the corresponding cannabis samples. However, the closeness of the r2 and Q2 parameters suggests that the model may have high predictive quality.
Comparison between predicted and experimental receptor potencies for Equation (3) (Figure 2c) revealed four outliers corresponding to samples #2, #4, #12, and #13 (Table 1). Exclusion of these outliers generated the following model (Equation (4)) for the CB1 agonism of cannabis extracts:
| Log(EC50) = −1.684 × 10−4 CBD − 6.018 × 10−3Δ9-THC + 1.240 × 10−4 CBDA2 + 6.966 × 10−3Δ9-THCA + 2.602, | (4) |
where n = 38, r2 = 0.422, Q2 = 0.367, RMSE = 0.387, Components = 1.
In addition to an improvement in correlation strength, the pharmacodynamics of CBD are reversed in the new model, where increasing CBD concentrations is shown to improve CB1 agonist potency, albeit to a lesser extent than Δ9-THC. Similar to the results revealed by Equation (1), the pharmacology of CBD when administered as part of a cannabis extract is once again pronouncedly different from that of CBD alone. As shown by its Q2 value, this model maintains good predictive quality. Among the outliers, samples #2 and #12 were from high-THC cannabis samples, whereas samples #4 and #13 contained significant amounts of CBD and Δ9-THC. The chemical profiles of the four quantified phytocannabinoids are not anomalous in these four outliers. However, it is possible that potentially high concentrations of yet-to-be-quantified phytochemicals may have influenced the affinities of these four outliers for the CB1 receptors.
To evaluate the utility of the above models, we simulated the EC50 values of the four quantified phytocannabinoids at the CB1 receptor (Equations (2)–(4)) and CB2 receptor (Equation (1)) as agonists. Thus, the 100% concentration of each test phytocannabinoid along with null concentrations of the remaining three phytocannabinoids yielded the predicted receptor potencies (Table 4). These predictions were then compared against experimental EC50 values determined in the same assays [13]. The predicted EC50 values from Equation (1) for Δ9-THCA and CBDA, Equation (3) for Δ9-THC, CBD, Δ9-THCA, and CBDA and Equation (4) for CBD, Δ9-THCA, and CBDA are within an order of magnitude from their experimental counterparts. In particular, the predicted EC50 values for Δ9-THCA at CB1 (Equations (3) and (4)) and for CBDA at CB2 (Equation (1)) differ by <10% from their experimental values. Thus, with the incorporation of additional training data, these models can be further optimized to estimate the in vitro activity of any cannabis extract with accuracy.
Table 4.
Predicted and experimental EC50 values for four major phytocannabinoids at CB1 and CB2 receptors. Generally, a difference of less than 0.2 between r2 and Q2 is indicative of good predictive power for models. Note: Pred—Predicted, Exp—Experimental.
| Cannabinoid | EC50, CB2 Agonism (μM) | EC50, CB1 Agonism (μM) | ||||||
|---|---|---|---|---|---|---|---|---|
| Equation (1) (r2 = 0.842, Q2 = 0.784) |
Equation (2) (r2 = 0.679, Q2 = 0.304) |
Equation (3) (r2 = 0.204, Q2 = 0.140) |
Equation (4) (r2 = 0.422, Q2 = 0.367) |
|||||
| Pred | Exp | Pred | Exp | Pred | Exp | Pred | Exp | |
| CBD | <10−290 | >10 | >1012 | 10 ± 2 | 1.7 | 10 ± 2 | 0.4 | 10 ± 2 |
| Δ9-THC | 0.13 | >10 | >10500 | 0.015 ± 0.001 | 0.12 | 0.015 ± 0.001 | 0.100 | 0.015 ± 0.001 |
| CBDA | 18.79 | 17 ± 4 | 1.4 × 10−4 | 17 ± 3 | 5.9 | 17 ± 3 | 7 | 17 ± 3 |
| Δ9-THCA | 131.83 | 30 ± 15 | 4.8 × 10−6 | 1.8 ± 0.7 | 1.9 | 1.8 ± 0.7 | 2 | 1.8 ± 0.7 |
However, predictions from these models may be limited by the trends and the size of the training datasets. For example, Equations (1), (3) and (4) are applicable to extracts obtained from cannabis flowers, in which the concentration of the most abundant phytocannabinoid consistently exceeds 25% (w/w). On the other hand, Equation (2) may be used for cannabis derivative oil products, in which the phytocannabinoid content generally does not exceed 10% (w/v) for any quantified phytocannabinoid. Additionally, Schüürmann et al. noted that the derivation for Q2 in the LOO methodology (i.e., where Q2 is calculated based on the training set rather than the test set) tends to overestimate Q2 when observations lie mostly at the two extremes of the dependent variable range, a situation that may apply to Equation (1) [39,40].
Another caveat when interpreting the models lies in their constant terms, which are up to four magnitudes of order larger than the coefficients of the independent variables. This captures the effects of unquantified factors on receptor potency, including minor cannabinoids and non-cannabinoid phytochemicals in this case. The rationale for selecting the CBD and Δ9-THC compound families as independent variables lies in their relative abundance because total CBD and Δ9-THC concentrations in the cannabis plant are generally many times higher than those of other phytocannabinoids [41] and are, thus, thought to be the primary determinants of cannabinoid receptors activities. However, complex natural products such as cannabis can contain up to hundreds of unique compounds, and it remains unknown how less abundant phytocannabinoids, such as cannabichromene (CBC) and cannabigerol (CBG), affect the activity of cannabis extracts at cannabinoid receptors [42,43].
4. Materials and Methods
4.1. General Information
Milli-Q purified water and high-performance liquid chromatography (HPLC) grade methanol were used for chromatographic analyses. All other commercial solvents and reagents were used without further processing. Liquid CO2 (SFE grade) for supercritical fluid extraction was purchased from Praxair® (Scarborough, ON, Canada). Analytical standards for Δ9-THCA, CBDA, Δ9-THC, CBD, and Δ9-THC-d3 were purchased from Sigma-Aldrich® Canada (Oakville, ON, Canada) as Certified Reference Standards. Tissue culture media, fetal bovine serum, and trypsin were obtained from Gibco™ (ThermoFischer Scientific, Ottawa, ON, Canada). HitHunter cAMP assay kit was obtained from DiscoverX Corporation (Fremont, CA, USA). All other chemicals were obtained from Sigma-Aldrich and were of analytical grade and used as such.
4.2. Cannabis Samples
Samples were collected from patients consuming cannabis for medical reasons. These patients were recruited as a part of two clinical studies investigating the use of medical cannabis in chronic pain (University Health Network REB#: 16-6375) and PTSD (University Health Network REB#: 17-5180.0; Center for Addiction and Mental Health REB#: 036/2017). A study visit was arranged for eligible participants who had passed clinical study screening. During the visit, a sample of medical cannabis products consumed by each participant (200 mg of dried medical cannabis or 0.2 mL of cannabis oil) was collected for analysis. The cannabis samples were transported to a designated secure area by qualified personnel for further processing and analysis.
4.3. Extraction of Cannabis for Cannabinoids
Cannabis samples in their oil form were analyzed for their chemical content without further processing. Cannabis samples collected as dried plant material were extracted using either supercritical fluid extraction (SFE) or microwave assisted extraction (MAE) following previously described procedures [2,15,44] and the extracts were subjected to chemical analyses. A total of 59 cannabis samples are included in this investigation: 45 dried flower samples, two resins, and 12 oil samples. Samples 1 and 2 were obtained as resins and analyzed as received, samples 3–35 were processed using MAE, and samples 36–47 were processed using SFE (Table 1).
4.4. Chemical Analysis
The cannabis extracts were analyzed by liquid chromatography-mass spectrometry (LC-MS) on an ACQUITY UPLC H-Class system (Waters® Limited, Mississauga, ON, Canada) equipped with Quaternary Solvent Manager and Sample Manager FTN. A Waters® MS 3100 mass spectrometer was used to monitor the samples. Chromatographic separation was achieved on an Acquity UPLC® BEH column (2.1 × 50 mm, C18, 1.7 µm). The sample injection plate and the column were maintained at 15 and 40 °C, respectively. The injection volume was 10 µL. Δ9-THC-d3 was added to samples as an internal standard to monitor the sensitivity of the UPLC-MS system. Working stock solutions of the extracts and cannabis oil samples were prepared in ethanol and methanol, respectively.
Analytical samples were prepared by diluting defined amounts of stock solutions in H2O/MeOH (3:7) with 0.1% formic acid (mobile phase I), filtering (Millex-GV® Syringe Filters, 0.22 µm; EMD Millipore, Oakville, ON, Canada), and further diluting with mobile phase I as needed. The internal standard was then added before analytical samples were injected into the UPLC system. The mobile phase for LC-MS consisted of H2O (A) and MeOH (B), with 0.1% formic acid. The gradient was programmed as follows: 0–4.5 min (30% A/70% B to 0% A/100% B, linear gradient), 4.5–5.0 min (0% A/100% B, isocratic), 5.0–5.2 min (0% A/100% B to 30% A/70% B, linear gradient), and 5.2–6 min (30% A/70% B, isocratic). The flow rate was 0.6 mL/min, and all samples were analyzed in triplicate. The mass scan in the range of 150–500 m/z, and single ion recordings (SIRs) in positive and negative modes (+ve = 287.20, 311.20, 315.23, 317.25, 318.24, 331.47, 345.45; −ve = 329.46, 343.45, 357.21, 359.22 m/z) were monitored.
When quantifying the phytocannabinoids present at relatively low abundances (i.e., <5% w/w and 1% w/v in extracts and oils, respectively), analytical samples were prepared by diluting defined amounts of stock solutions in H2O/MeOH (4:6) with 0.1% formic acid (mobile phase II), filtering (Millex-GV® Syringe Filters, 0.22 µm; EMD Millipore), and further diluting with mobile phase II as needed. The internal standard was then added before analytical samples were injected into the UPLC system. The mobile phase for LC-MS consisted of H2O (A) and MeOH (B), with 0.1% formic acid. The gradient was programmed as follows: 0–13.5 min (40% A/60% B to 0% A/100% B, linear gradient), 13.5–14.0 min (0% A/100% B, isocratic), 14.0–14.2 min (0% A/100% B to 40% A/60% B, linear gradient), and 14.2–15 min (30% A/70% B, isocratic). The flow rate was 0.5 mL/min, and all samples were analyzed in triplicate. Single ion recordings (SIRs) in positive and negative modes (+ve = 287.20, 311.20, 315.23, 317.25, 318.24; −ve = 329.46, 357.21, 359.22 m/z) were monitored.
All SIR chromatograms were analyzed using Waters® Empower3®, and the concentration of each phytocannabinoid was determined using its corresponding standard curve plotted in Grafit® 5.0.10 (Erithacus Software Limited, East Grinstead, West Sussex, United Kingdom) using the following expression:
| % (w/w) or % (w/v) Phytocannabinoid = [(Concentration) × (Dilution Factor) × 100]/(Sample Stock Solution Concentration). |
All injections were performed in triplicate, and all reported values are averaged values. The lower limits of detection (LLD) and lower limits of quantitation (LLQ) for the quantified phytocannabinoids are reported in Table S1.
4.5. CB1 and CB2 Receptor Assays
The cannabis samples extracts and cannabis oils were assayed against CB1 and CB2 receptors on a cAMP reporter platform using CHO-K1-CB1 and CHO-K1-CB2 cell lines, respectively (DiscoverX Corporation), according to previously described methods [15]. The efficacies of the cannabis extracts were calculated as the percentage inhibition of forskolin-stimulated cAMP production. CP-55940 was used as a reference agonist at CB1 and CB2 receptors, whereas SR-141716 and AM630 were used as reference antagonists at CB1 and CB2 receptors, respectively. The data were analyzed according to previously described methods [15]. Graphpad® Prism 7 (GraphPad Software, San Diego, CA, USA) was used to derive dose-response curves for each sample. Concentration of the major phytocannabinoid in each cannabis extract (as shown in Table 1 and Table 2) was used to fit and derive EC50 and IC50 for each cannabis sample. For example, for cannabis sample 1, Δ9-THC concentration was used to derive the dose-response profile and the corresponding EC50 or IC50 value.
4.6. Statistical Analysis
A multivariate PLS analysis was employed to evaluate potential relationships between receptor responses (agonism and antagonism at the CB1 and CB2 receptors) and chemical composition (Δ9-THCA, CBDA, Δ9-THC, and CBD). Normalized potencies of the extracts at each receptor (log(EC50) or log(IC50), nM) were correlated against the concentrations of four major phytocannabinoids (% w/w or % w/v) on the XLSTAT® 2019 platform (Addinsoft, Paris, France) for Microsoft Excel® (Microsoft Corporation, Redmond, WA, USA) Because of differences in measurement units, the dried plant samples (concentrations in % w/w) and oil samples (concentrations in % w/v) were correlated separately. Before analysis, the data were categorized into eight subsets based on sample type (dried plant/cannabis oil), receptor subtype (CB1/CB2) and assay mode (agonism/antagonism) (Table S2). Extracts that were inactive in the receptor assays or whose EC50/IC50 values could not be precisely derived were excluded from the analysis. Groups with n < 5 samples were deemed to not be statistically significant and excluded from the analysis. For each remaining group, prediction models were generated using a combination of the linear explanatory variables Δ9-THCA, CBDA, Δ9-THC, and CBD, as well as the quadratic variables Δ9-THCA2, CBDA2, Δ9-THC2, and CBD2, where models were optimized to maximize r2 while minimizing root mean square error (RMSE) and the value of the constant term (Tables S3–S5). All models were also cross-validated using the LOO technique, allowing for the evaluation of predictive quality through the Q2 statistic, which is the LOO-cross-validated r2 value [40]. In general, a difference of less than 0.2 between the r2 and Q2 parameters is indicative of good predictive power for a mathematical model [45]. PLS regression captures factors that describe most of the relationship between the dependent and independent variables while preventing the incorporation of superfluous factors that lower the predictive power of models [46,47].
5. Conclusions
A first-of-its-kind QSAR-like quantitative analysis was performed on 59 samples of cannabis extracts and cannabis oils to elucidate the relationships between the concentrations of phytocannabinoids in the cannabis extracts and the corresponding biological activities at the CB1 and CB2 receptors. To achieve this, we employed PLS regression analyses. These models can be useful for predicting the potency of (1) CB1 and (2) CB2 agonism in cannabis extracts; however, (3) CB1 agonism in cannabis oils will likely require a larger training dataset to generate accurate predictions. Our analysis of the statistical parameters suggest insignificant relationships between the major phytocannabinoid content and receptor affinity for CB1 but good correlative models at CB2 receptors with the current dataset. The models disclosed here for the CB1 and CB2 agonism of cannabis extracts reliably predicted the potencies of single phytocannabinoids, which were comparable to the experimentally derived potencies in turn validating the models. It must also be noted that some phytocannabinoids such as CBD are considered “safe” by many people although any chemical compound including CBD [48] can exhibit adverse effects leading to toxicities; data and results presented in this study do not consider any such effects. Note that one can include other compounds present at physiologically significant concentrations from the cannabis extracts in the description of the chemical composition in each equation, and future work will include such expansion. This is the first study disclosing a relationship between the major phytocannabinoids concentrations in cannabis extracts and the corresponding cannabinoid receptors activities. The strategies described in this work and further refinement utilizing larger sample sets and additional variables may further improve the accuracy to understand the effects of cannabis extracts on cannabinoid receptors.
Acknowledgments
We thank Mahmood Tazari for useful discussions on PLS and statistical models. We would like to thank Editage (www.editage.com) for language editing.
Supplementary Materials
The following are available online, Table S1: LLD and LLQ values for the phytocannabinoids quantified in this study; Table S2: Categorization of data prior to regression analysis; Table S3: Preliminary prediction models generated using all combinations of linear and quadratic explanatory variables, describing the relationship between chemical composition and potency at CB2 as agonists for extracts obtained from dried plant samples; Table S4: Preliminary prediction models generated using all combinations of linear and quadratic explanatory variables, describing the relationship between chemical composition and potency at CB1 as agonists for cannabis-derivative samples; Table S5: Preliminary prediction models generated using all combinations of linear and quadratic explanatory variables, describing the relationship between chemical composition and potency at CB1 as agonists for extracts obtained from dried plant samples; Figure S1: Dose-response curves for CBD, CBDA, Δ9-THC and Δ9-THCA as agonists and antagonists at CB1 receptor; Figure S2: Dose-response curves for CBD, CBDA, Δ9-THC and Δ9-THCA as agonists and antagonists at CB2 receptor.
Author Contributions
Conceptualization, L.P.K.; methodology, Y.Y., M.L.-B., R.V.; validation, Y.Y., R.V., M.L.-B.; resources, L.P.K., H.A.C., A.H.C.W.; data curation, Y.Y.; writing—original draft preparation, Y.Y., R.V., L.P.K.; writing—review and editing, Y.Y., L.P.K.; supervision, L.P.K.; project administration, L.P.K.; funding acquisition, L.P.K., H.A.C., A.H.C.W. All authors have read and agreed to the published version of the manuscript.
Funding
Authors gratefully acknowledge the financial support from Canada Foundation for Innovation (Grant No. CFI 32350 to LPK), and University Health Network.
Conflicts of Interest
There are no conflicts.
Footnotes
Sample Availability: Samples of the cannabis extracts cannot be made available from the authors due to regulatory restrictions.
References
- 1.Owens B. Drug development: The treasure chest. Nature. 2015;525:S6–S8. doi: 10.1038/525S6a. [DOI] [PubMed] [Google Scholar]
- 2.Lewis M., Yang Y., Wasilewski E., Clarke H.A., Kotra L.P. Chemical profiling of medical cannabis extracts. ACS Omega. 2017;2:6091–6103. doi: 10.1021/acsomega.7b00996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pertwee R.G., editor. Handbook of Cannabis. Oxford University Press; Oxford, UK: 2014. [Google Scholar]
- 4.Newmeyer M.N., Swortwood M.J., Abulseoud O.A., Huestis M.A. Subjective and physiological effects, and expired carbon monoxide concentrations in frequent and occasional cannabis smokers following smoked, vaporized, and oral cannabis administration. Drug Alcohol Depend. 2017;175:67–76. doi: 10.1016/j.drugalcdep.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 5.Herreros I., Miquel L., Blithikioti C., Nuno L., Ballester B.R., Grechuta K., Gual A., Balcells-Olivero M., Verschure P. Motor Adaptation Impairment in Chronic Cannabis Users Assessed by a Visuomotor Rotation Task. J. Clin. Med. 2019;8:1049. doi: 10.3390/jcm8071049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Howlett A.C., Abood M.E. CB1 and CB2 Receptor Pharmacology. Adv. Pharmacol. 2017;80:169. doi: 10.1016/bs.apha.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pettinato M. Medicinal cannabis: A primer for nurses. Nursing. 2017;47:40–46. doi: 10.1097/01.NURSE.0000521022.07638.35. [DOI] [PubMed] [Google Scholar]
- 8.Haroutounian S., Ratz Y., Ginosar Y., Furmanov K., Saifi F., Meidan R., Davidson E. The Effect of Medicinal Cannabis on Pain and Quality-of-Life Outcomes in Chronic Pain: A Prospective Open-label Study. Clin. J. Pain. 2016;32:1036–1043. doi: 10.1097/AJP.0000000000000364. [DOI] [PubMed] [Google Scholar]
- 9.Suraev A.S., Todd L., Bowen M.T., Allsop D.J., McGregor I.S., Ireland C., Lintzeris N. An Australian nationwide survey on medicinal cannabis use for epilepsy: History of antiepileptic drug treatment predicts medicinal cannabis use. Epilepsy Behav. 2017;70:334–340. doi: 10.1016/j.yebeh.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Lintzeris N., Driels J., Elias N., Arnold J.C., McGregor I.S., Allsop D.J. Medicinal cannabis in Australia, 2016: The Cannabis as Medicine Survey (CAMS-16) Med. J. Aust. 2018;209:211–216. doi: 10.5694/mja17.01247. [DOI] [PubMed] [Google Scholar]
- 11.Information for Health Care Professionals: Cannabis (Marihuana, Marijuana) and the Cannabinoids. October 2018, Health Canada. [(accessed on 20 March 2020)]; Available online: https://www.canada.ca/en/health-canada/services/drugs-medication/cannabis/information-medical-practitioners/information-health-care-professionals-cannabis-cannabinoids.html.
- 12.Mammen G., Freitas L., Rehm J., Rueda S. Cannabinoid concentrations in Canada’s regulated medical cannabis industry. Addiction. 2017;112:730–732. doi: 10.1111/add.13738. [DOI] [PubMed] [Google Scholar]
- 13.Russo E.B. The Case for the Entourage Effect and Conventional Breeding of Clinical Cannabis: No “Strain”, No Gain. Front. Plant Sci. 2019;9:1969. doi: 10.3389/fpls.2018.01969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Russo E.B. Taming THC: Potential cannabis synergy and phytocannabinoid-terpenoid entourage effects. Br. J. Pharmacol. 2011;163:1344–1364. doi: 10.1111/j.1476-5381.2011.01238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis-Bakker M.M., Yang Y., Vyawahare R., Kotra L.P. Extractions of Medical Cannabis Cultivars and the Role of Decarboxylation for Optimal Receptor Responses. Cannabis Cannabinoid Res. 2019;4:183–194. doi: 10.1089/can.2018.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gini G. QSAR Methods. In Silico Methods for Predicting Drug Toxicity. In: Benfenati E., editor. Methods in Molecular Biology. Vol. 1425 Humana Press; New York, NY, USA: 2016. [Google Scholar]
- 17.Hua T., Vemuri K., Pu M., Qu L., Han G.W., Wu Y., Zhao S., Shui W., Li S., Korde A., et al. Crystal Structure of the Human Cannabinoid Receptor CB1. Cell. 2016;167:750–762. doi: 10.1016/j.cell.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pertwee R.G. The diverse CB1 and CB2 receptor pharmacology of three plant cannabinoids: Δ9 tetrahydrocannabinol, cannabidiol and Δ9-tetrahydrocannabivarin. Br. J. Pharmacol. 2008;153:199–215. doi: 10.1038/sj.bjp.0707442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McPartland J.M., MacDonald C., Young M., Grant P.S., Furkert D.P., Glass M. Affinity and efficacy studies of tetrahydrocannabinolic acid A at cannabinoid receptor types one and two. Cannabis Cannabinoid Res. 2017;2:87–95. doi: 10.1089/can.2016.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Backer B., Maebe K., Verstraete A.G., Charlier C. Evolution of the content of THC and other major cannabinoids in drug-type cannabis cuttings and seedlings during growth of plants. J. Forensic Sci. 2012;57:918–922. doi: 10.1111/j.1556-4029.2012.02068.x. [DOI] [PubMed] [Google Scholar]
- 21.Morimoto S., Komatsu K., Taura F., Shoyama Y. Purification and characterization of cannabichromenic acid synthase from Cannabis sativa. Phytochemistry. 1998;49:1525–1529. doi: 10.1016/S0031-9422(98)00278-7. [DOI] [PubMed] [Google Scholar]
- 22.Shoyama Y., Fujita T., Yamauchi T., Nishioka I. Cannabichromenic acid, a genuine substance of cannabichromene. Chem. Pharm. Bull. 1968;16:1157–1158. doi: 10.1248/cpb.16.1157. [DOI] [PubMed] [Google Scholar]
- 23.Shoyama Y., Oku R., Yamauchi T., Nishioka I. Cannabis. VI. Cannabicyclolic Acid. Chem. Pharm. Bull. 1972;20:1927–1930. doi: 10.1248/cpb.20.1927. [DOI] [Google Scholar]
- 24.ElSohly M.A., Slade D. Chemical constituents of marijuana: The complex mixture of natural cannabinoids. Life Sci. 2005;78:539–548. doi: 10.1016/j.lfs.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 25.Notcutt W., Price M., Miller R., Newport S., Phillips C., Simmons S., Sansom C. Initial experiences with medicinal extracts of cannabis for chronic pain: Results from 34 ‘N of 1’ studies. Anesthesia. 2004;59:440–452. doi: 10.1111/j.1365-2044.2004.03674.x. [DOI] [PubMed] [Google Scholar]
- 26.Nugent S.M., Morasco B.J., O’Neil M.E., Freeman M., Low A., Kondo K., Elven C., Zakher B., Motu’apuaka M., Paynter R., et al. The effects of cannabis among adults with chronic pain and an overview of general harms: A systematic review. Ann. Intern. Med. 2017;167:319–331. doi: 10.7326/M17-0155. [DOI] [PubMed] [Google Scholar]
- 27.Svizenska I., Dubovy P., Sulcova A. Cannabinoid receptors 1 and 2 (CB1 and CB2), their distribution, ligands and functional involvement in nervous system structures—A short review. Pharmacol. Biochem. Behav. 2008;90:501–511. doi: 10.1016/j.pbb.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 28.Whiteside G.T., Lee G.P., Valenzano K.J. The role of the cannabinoid CB2 receptor in pain transmission and therapeutic potential of small molecule CB2 receptor agonists. Curr. Med. Chem. 2007;14:917–936. doi: 10.2174/092986707780363023. [DOI] [PubMed] [Google Scholar]
- 29.Malan T.P., Ibrahim M.M., Lai J., Vanderah T.W., Makriyannis A., Porreca F. CB2 cannabinoid receptor agonists: Pain relief without psychoactive effects? Curr. Opin. Pharmacol. 2003;3:62–67. doi: 10.1016/S1471-4892(02)00004-8. [DOI] [PubMed] [Google Scholar]
- 30.Luongo L., Palazzo E., Tambaro S., Giordano C., Gatta L., Scafuro M.A., Rossi F.s., Lazzari P., Pani L., de Novellis V., et al. 1-(2′,4′-dichlorophenyl)-6-methyl-N-cyclohexylamine-1,4-dihydroindeno[1,2-c]pyrazole-3-carboxamide, a novel CB2 agonist, alleviates neuropathic pain through functional microglial changes in mice. Neurobiol. Dis. 2009;37:177–185. doi: 10.1016/j.nbd.2009.09.021. [DOI] [PubMed] [Google Scholar]
- 31.Hegazy O., Platnick H. Cannabidiol (CBD) for Treatment of Neurofibromatosis-related Pain and Concomitant Mood Disorder: A Case Report. Curceus. 2019;11:e6312. doi: 10.7759/cureus.6312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Capano A., Weaver R., Burkman E. Evaluation of the effects of CBD hemp extract on opioid use and quality of life indicators in chronic pain patients: A prospective cohort study. Postgrad. Med. 2020;132:56–61. doi: 10.1080/00325481.2019.1685298. [DOI] [PubMed] [Google Scholar]
- 33.Cuñetti L., Manzo L., Peyraube R., Arnaiz J., Curi L., Orihuela S. Chronic Pain Treatment With Cannabidiol in Kidney Transplant Patients in Uruguay. Transpl. Proc. 2018;50:461–464. doi: 10.1016/j.transproceed.2017.12.042. [DOI] [PubMed] [Google Scholar]
- 34.Boyaji S., Merkow J., Elman R.N.M., Kaye A.D., Yong R.J., Urman R.D. The Role of Cannabidiol (CBD) in Chronic Pain Management: An Assessment of Current Evidence. Curr. Pain Headache Rep. 2020;24:4. doi: 10.1007/s11916-020-0835-4. [DOI] [PubMed] [Google Scholar]
- 35.Canada’s Medical Cannabis-Oil Extraction Process. [(accessed on 21 March 2020)]; Available online: https://www.cannimed.ca/pages/oil-extraction-process.
- 36.Cannabis Oil Canada. [(accessed on 21 March 2020)]; Available online: https://aphria.ca/cannabis-oil/
- 37.Mackie K. Mechanisms of CB1 receptor signaling: Endocannabinoid modulation of synaptic strength. Int. J. Obes. (Lond) 2006;30:S19–S23. doi: 10.1038/sj.ijo.0803273. [DOI] [PubMed] [Google Scholar]
- 38.Iseger T.A., Bossong M.G. A systematic review of the antipsychotic properties of cannabidiol in humans. Schizophr. Res. 2015;162:153–161. doi: 10.1016/j.schres.2015.01.033. [DOI] [PubMed] [Google Scholar]
- 39.Schüürmann G., Ebert R., Chen J., Wang B., Kühne R. External Validation and Prediction Employing the Predictive Squared Correlation Coefficient—Test Set Activity Mean vs Training Set Activity Mean. J. Chem. Inf. Model. 2008;48:2140–2145. doi: 10.1021/ci800253u. [DOI] [PubMed] [Google Scholar]
- 40.Consonni V., Ballabio D., Todeschini R. Comments on the Definition of the Q2 Parameter for QSAR Validation. J. Chem. Inf. Model. 2009;47:1669–1678. doi: 10.1021/ci900115y. [DOI] [PubMed] [Google Scholar]
- 41.Mudge E.M., Murch S.J., Brown P.N. Chemometric Analysis of Cannabinoids: Chemotaxonomy and Domestication Syndrome. Sci. Rep. 2018;8:13090–13099. doi: 10.1038/s41598-018-31120-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Udoh M., Santiago M., Devenish S., McGregor I.S., Connor M. Cannabichromene is a cannabinoid CB2 receptor agonist. Br. J. Pharmacol. 2019;176:4537–4547. doi: 10.1111/bph.14815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Navarro G., Varani K., Reyes-Resina I., de Sánchez Medina V., Rivas-Santisteban R., Sánchez-Carnerero Callado C., Vincenzi F., Casano S., Ferreiro-Vera C., Canela E.I., et al. Cannabigerol Action at Cannabinoid CB1 and CB2 Receptors and at CB1-CB2 Heteroreceptor Complexes. Front. Pharmacol. 2018;9:632. doi: 10.3389/fphar.2018.00632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang Y., Lewis M.M., Bello A.M., Wasilewski E., Clarke H.A., Kotra L.P. Cannabis sativa (Hemp) Seeds, Δ9-Tetrahydrocannabinol, and Potential Overdose. Cannabis Cannabinoid Res. 2017;2:274–281. doi: 10.1089/can.2017.0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eriksson L., Byrne T., Johansson E., Trygg J., Vikström C. Multi- and Megavariate Data Analysis Basic Principles and Applications. Vol. 1 Umetrics Academy; Umeå, Sweden: 2013. PLS. [Google Scholar]
- 46.Tobias R.D. Proceedings of the Twentieth Annual SAS Users Group International Conference. SAS Institute Inc.; Cary, NC, USA: 1995. An introduction to partial least squares regression; pp. 1250–1257. [Google Scholar]
- 47.Haenlein M., Kaplan A.M. A Beginner’s Guide to Partial Least Squares Analysis. Underst. Stat. 2004;3:283–297. doi: 10.1207/s15328031us0304_4. [DOI] [Google Scholar]
- 48.Huestis M.A., Solimini R., Pichini S., Pacifici R., Carlier J., Busardò F.P. Cannabidiol adverse effects and toxicity. Curr. Neuropharmacol. 2019;17:974–989. doi: 10.2174/1570159X17666190603171901. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.