Abstract
Laryngeal cancer is a common head and neck cancer that effects the quality of life of those affected. Early diagnosis and treatment are vital to minimize the harmful effects of laryngeal cancer, which can improve the survival rate of patients following surgery and retain the voice function of the larynx. The purpose of the present study was to explore the molecular mechanism of the development of laryngeal cancer and to determine the biomarker for the diagnosis and treatment of laryngeal cancer. Reverse transcription-quantitative PCR (RT-qPCR) and The Cancer Genome Atlas database analysis were used to confirm high expression of TMEM158 in laryngeal cancer. The effects of TMEM158 and miR-548ac was investigated through in vitro and in vivo assays (MTT assay, colony-formation assay, flow cytometry assay, western blotting and tumor xenograft assay). Luciferase reporter assay, western blotting and RTq-PCR were used to confirm that miR-548 directly targeted the 3′-untranslated region of TMEM158 and inhibited TMEM158 expression. Taken together, the present results suggest that miR-548ac functions as a crucial cancer suppressor in laryngeal cancer, which induces apoptosis in laryngeal cancer cells by suppressing TMEM158. Thus, miR-548ac may be a potential target for the treatment of laryngeal cancer.
Keywords: transmembrane protein 158, microRNA-548ac, apoptosis, laryngeal cancer
Introduction
Laryngeal cancer is a common primary malignant tumor in the head and neck (1). The incidence of laryngeal cancer is 5.7–7.6% of total body tumors (2). It is estimated that there were 177,422 cases of laryngeal cancer and 94,711 mortalities worldwide in 2012 (3). Currently, there is no exact cause of laryngeal cancer, which may be caused by several factors, including smoking, alcohol consumption, air pollution and viral infection (4,5). Thus, the key to improving the prognosis of laryngeal cancer is to determine the biological characteristics of laryngeal cancer cells.
Transmembrane protein 158 (TMEM158), also known as RIS-1 protein, is recognized for its increased expression when Ras protein induces senescence of haploid fibroblasts infected with RAS V12 lentivirus (6). The expression and biological function of TMEM158 have been reported in different types of cancer (7–9); however, its role in laryngeal cancer remains unknown, thus, the present study aimed to investigate the regulatory mechanism and biological function of TMEM158 in laryngeal cancer.
MicroRNAs (miRNAs/miRs) are small non-coding RNA molecules that play key roles in the regulation of gene expression (10). Translation inhibition or silencing of target mRNAs is induced following complementary binding to the 3′-untranslated region (UTR) (11,12). miR-548 is a large and poorly conserved primate-specific miRNA gene family that consists of 68 members, and several members of this family have been reported to be associated with cancer (13). For example, miR-548b-3p functions as a tumor suppressor in lung cancer (14) and as an antioncogenic regulator in breast cancer (15).
The purpose of the present study is to explore the molecular mechanism of the development of laryngeal cancer and to identify the biomarker for diagnosis and treatment of laryngeal cancer. Reverse transcription-quantitative PCR (RT-qPCR) and the analysis of The Cancer Genome Atlas (TCGA) database were used to demonstrate overexpression of TMEM158 in laryngeal cancer. The carcinogenic effect of TMEM158 and the anticancer effect of miR-548ac was investigated through in vitro and in vivo assays (MTT assay, colony formation assay, flow cytometry assay, western blotting and tumor xenograft assay). Luciferase reporter assay, western blotting and RT-qPCR were used to demonstrate miR-548 directly targeted the 3′-untranslated region of TMEM158 and inhibited TMEM158 expression. These results suggest that miR-548ac functions as a crucial cancer suppressor in laryngeal cancer, which induces apoptosis in laryngeal cancer cells by suppressing TMEM158. The present findings enrich the pathogenesis of laryngeal cancer and provide a new target and new ideas for the treatment on laryngeal cancer.
Materials and methods
Cell lines and culture
Human LSC-1 cells and NP69 nasopharyngeal epithelial cells were purchased from the Institute of Oncology, Chinese Academy of Medical Sciences. Cells were maintained in DMEM medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (cat. no. 10100147; Thermo Fisher Scientific, Inc.), penicillin (100 IU/ml) and streptomycin (100 mg/ml) at 37°C in 5% CO2. LSC-1 cells were transfected with 10 µM miR-548ac agomir (LSC-1-miR-548ac) or 10 µM miR-548ac agomir negative control (NC; LSC-1-NC; Guangzhou RiboBio Co., Ltd.) using Lipofectamine® RNAiMAX reagent (cat. no. 13778030; Thermo Fisher Scientific, Inc.). The short hairpin (sh)RNA targeting positions 1520–1540 (5′-AAATGACCAAATCCTGTGTAT-3′; Sangon Biotech Co. Ltd.) of human TMEM158 mRNA was cloned into a lentiviral vector (PLKO.1). A non-specific scrambled shRNA sequence (5′-CCTAAGGTTAAGTCGCCCTCG-3′; Sangon Biotech Co. Ltd.) was used as the NC. 1 µg (50 pmol) shRNA was added into Opti-MEM (cat. no. 31985062; Thermo Fisher Scientific, Inc.) to prepare RNA diluent, and the final volume was 25 µl. LSC-1 cells were subsequently transfected with lentiviral packaging vectors (LSC-1-NC or LSC-1-shTMEM158) using Lipofectamine® 3000 reagent (cat. no. L3000001; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. Subsequent experiments began at 24 h after transfection of LSC-1 cells with shRNA. The LSC-1 cells stably transfected with NC, miR-548ac and shTMEM158 were constructed respectively.
Tumor xenograft model
A total of 30 4-week-old BALB/cA female nude mice (14–18 g) were purchased from the Beijing Vital River Laboratory Animal Technologies Co. Ltd. The present study was approved by the Ethics Committee of Tianjin Union Medical Center (Tianjin, China; approval no. 2018-269). All nude mice were kept at 28°C and SPF level with light/dark cycle. Mice received pathogen-free food and water. All food and water were sterilized and then sent into the animal isolator through a sterile entrance chamber to ensure that the nude mice were in a sterile living state. All animal experiments were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH Publications no. 8023, revised in 1978).
LSC-1 cells (2×106 cells/mouse) were inoculated into the back of the nude mice following light anesthesia with 350 mg/kg chloral hydrate. The mice were randomly divided into four groups (6 mice/group), as follows: i) LSC-1 cells with a blank vector; ii) LSC-1 cells with shTMEM158; ii) LSC-1 cells with miR-548ac NC and iv) LSC-1 cells with miR-548ac agomir. The experimental procedures lasted for 3 weeks. The health and behavior of all animals were monitored every day and the individuals with poor health were terminated in time. The nude mice were free from stress, discomfort, pain and injury. All mice were sacrificed via intraperitoneal injection with 200 mg/kg pentobarbital sodium. Mortality was confirmed by checking for a heartbeat and examining the pupil dilation of the nude mice. The maximum diameter of the tumor was 0.87 cm. The tumors were subsequently removed using resection, photographed using a camera (model, D3600; Nikon Corporation) and weighed. Tumor volume was calculated using the following formula: V=(length × width2)/2.
MTT assay
The cell suspensions (LSC-1-NC, LSC-1- miR-548ac; LSC-1-NC or LSC-1-shTMEM158) were added into 96-well plates at a density of 6,000 cells/well. After 24 h of cell attachment, the culture medium was discarded. Cells were subsequently incubated with serum-free DMEM (100 µl) and 10 µl MTT (5 mg/ml; cat. no. ST316; Beyotime Institute of Biotechnology, Inc.) for 4 h at 37°C. Following the MTT incubation, the purple formazan crystals were dissolved using 100 µl dimethyl sulfoxide and shaking vigorously for 10 min using a vortex oscillator (model, MixMax; AIBENSEN, Co. Ltd.). Cell viability was subsequently analyzed at a wavelength of 570 nm. All experiments were performed in triplicate and three biological repeats were conducted.
Colony formation assay
The cell suspensions (LSC-1-NC, LSC-1-miR-548ac; LSC-1-NC or LSC-1-shTMEM158) were added into 6-well plates at a density of 300 cells/well. Cells were maintained in DMEM medium supplemented with 10% fetal bovine serum (cat. no. 10100147; Thermo Fisher Scientific, Inc.) for 7–10 days at 37°C. After ~70 cells appeared in each colony, each pore was fixed with 1 ml methanol for 15 min at room temperature. Cell colonies were subsequently stained with 0.5% crystal violet for 10 min at room temperature, and excess stain was discarded using ultrapure water. Cell colonies were observed and recorded using a camera (model, D3600; Nikon Corporation).
Annexin V-propidium iodide (PI) assay
Cells (LSC-1-NC, LSC-1-miR-548ac; LSC-1-NC or LSC-1-shTMEM158) were digested with trypsin-EDTA solution (0.05% trypsin and 0.02% EDTA; cat. no. C0202; Beyotime Institute of Biotechnology, Inc.) and collected by centrifugation at 161 × g and room temperature. Cells were washed three times with sterile PBS and subsequently resuspended in 500 µl Annexin V-PI binding solution. PI (10 µl) and annexin V-fluorescein isothiocyanate (5 µl) (cat. no. C1062S; Beyotime Institute of Biotechnology, Inc.) were added to the cell suspensions of each group and manually gently mixed. The whole reaction was 515 µl at room temperature and dark for 30 min. Apoptotic cells were subsequently detected using a flow cytometer.
Flow cytometric analysis of cell cycle distribution
Cells (LSC-1-NC, LSC-1-miR-548ac; LSC-1-NC or LSC-1-shTMEM158) at 1×106 cells/ml were collected following digestion with trypsin-EDTA solution (cat. no. C0202; Beyotime, Inc.), washed twice with PBS, and resuspended and fixed with 70% ethanol overnight at −20°C. The cell suspension was re-washed twice with PBS and stained with 400 µl PI at 4°C for 30 min in the dark. Cell cycle distribution was subsequently detected via flow cytometric analysis.
Western blotting
Total protein from cells (LSC-1-NC, LSC-1-miR-548ac; LSC-1-NC or LSC-1-shTMEM158) was extracted using RIPA lysis buffer (cat. no. R0010; Beijing Solarbio Science & Technology Co., Ltd.). Detection of protein concentration was used BCA assay. Equal amounts (15 µg) of protein were separated via SDS-PAGE (12% separating gel and 5% concentrating gel) and subsequently transferred onto polyvinylidene difluoride (PVDF) membranes. The PVDF membrane was blocked in 5% skimmed milk at room temperature for 2 h. The membranes were incubated with primary antibodies against: Bax (1:1,000; cat. no. ab32503), B-cell lymphoma-2 (Bcl-2; 1:1,000; cat. no. ab8227), TMEM158 (1:1,000; cat. no. ab98335) and β-actin (1:2,000; cat. no. ab98335) (all purchased from Abcam) at room temperature for 2 h. Following the primary incubation, membranes were incubated with secondary antibodies (HRP goat anti-Rabbit; 1:5,000; cat. no. A0208; Beyotime Institute of Biotechnology, Inc.) at room temperature for 1 h. Protein bands were visualized using enhanced chemiluminescence buffer and detected using the multifunctional gel imaging system (model, Gel Doc XR+; Bio-Rad Laboratories, Inc.). Protein expression was calculated via the gray value.
Reverse transcription-quantitative (RT-q)PCR
Total RNA was extracted from cells (LSC-1-NC, LSC-1-miR-548ac; LSC-1-NC or LSC-1-shTMEM158) using TRIzol® reagent (cat. no. 5301100; SIMGEN) according to the manufacturer's instructions, and quantified using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Inc.). Total RNA was reverse transcribed into cDNA using the TransScript RT Reagent kit (cat. no. AH411-02; TransGen Biotech Co., Ltd.). The reverse transcription reaction conditions were 25°C, 10 min (miRNA skip); 42°C, 30 min; 85°C, 5 min. qPCR was subsequently performed using the 2×SYBR Green qPCR mix (cat. no. 153301; Beijing BLKW Biotechnology Co. Ltd.) and a ABI 7500 system (Thermo Fisher Scientific, Inc.). The qPCR reaction conditions were 1 cycle at 94°C for 2 min; 40 cycles (94°C for 15 sec; 55°C for 15 sec; and 72°C for 20 sec). The following primer sequences were used for qPCR: TMEM158 forward, 5′-TGCCCAACGGCATGGAACA-3′ and reverse, 5′-AGGAGCGGAGCGGGTCACTT-3′; GAPDH forward, 5′-ATGACATCAAGAAGGTGGTGAAGCAGG-3′ and reverse, GCGTCAAAGGTGGAGGAGTGGGT; miR-548ac forward, 5′-CAAAAACCGGCAATTACTTTTG-3′ and reverse, 5′-CTCAACTGGTGTCGTGGA-3′; and U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ andreverse, 5′-CTCAACTGGTGTCGTGGA-3′. Relative expression levels were calculated using the 2−ΔΔCq method (16) and normalized to the internal reference genes GAPDH and U6, respectively.
Dual-luciferase reporter assay
The wild (WT)/mutant (MUT) 3′-UTR fragments of TMEME158 were amplified via PCR using 2X PCR Master mix (cat. no. D7228; Beyotime Institute of Biotechnology, Inc.). The PCR reaction conditions were: 1 cycle at 94°C for 5 min; 35 cycles (94°C for 30 sec; 60°C for 1 min; 72°C at 1 min); and 1 cycle (72°Cfor 10 min). Th fragments were cloned into the Dual-Luciferase miRNA targeted expression vector (Promega Corporation) to synthesize WT/MUT-TMEM158-3′-UTR. LSC-1 cells with 70% confluence were transfected with WT-TMEM158-3′-UTR or MUT-TMEM158-3′-UTR and miR-548ac agomirs or miR-548ac agomirs NC/miR-548ac antagonists or miR-548ac antagonists NC(Guangzhou RiboBio Co., Ltd.) usingLipofectamine®3000 reagent (cat. no. L3000008; Thermo Fisher Scientific, Inc.). Following incubation at 37°Cfor 48 h, Luciferase activity was measured using a multimode microplate reader (BioTek Instruments, Inc.) and normalized with Renilla luciferase activity.
Bioinformatics analysis
The mRNA and miRNA sequencing data and the corresponding clinical information of the patients with laryngeal cancer were downloaded from the TCGA data portal (http://cancergenome.nih.gov). The data were screened and laryngeal cancer samples and non-tumor samples, which had detailed clinical information (laryngeal cancer, 116 cases; normal, 11 cases), were selected. The clinical features of the patients are shown in Table I. Patients with head and neck cancer were divided into four stages, according to tumor-node-metastasis (TNM) (17) and differences in TMEM158 expression at different Tumor-Node-Metastasis stages were analyzed. RNAInter software (http://www.rna-society.org/raid/; Department of Bioinformatics, Southern Medical University) was used to predict the target association between miR-548ac and TMEM158. Univariate Cox proportional hazards regression analysis was performed to determine the association between TMEM158 expression and overall survival rate. Kaplan-Meier survival analysis and log-rank test were used to assess the association between TMEM158 expression and overall survival time. A total of 38 cases of TMEM158 with high or low expression were selected.
Statistical analysis
Statistical analyses were performed using SPSS 18.0 software (SPSS, Inc.). All experiments were performed in triplicate and data are presented as the mean ± standard deviation. Unpaired Student's two-tailed t-test was used to compare differences between two independent samples, whilst one-way analysis of variance and Bonferroni's correction were used to compare differences between multiple groups (Figs. 1B, 2F, 3G, 4D and 5B and C). P<0.05 was considered to indicate a statistically significant difference.
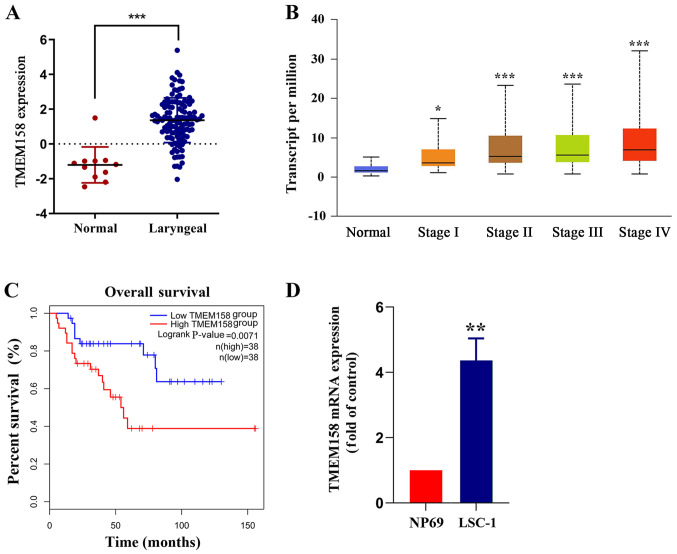
Figure 1.
TMEM158 expression using data downloaded from TCGA database. (A) TMEM158 expression was assessed in laryngeal carcinoma tissues and adjacent normal tissues using mRNA sequencing data from the TCGA database. Red represents cancer tissues and blue represents adjacent normal tissues. ***P<0.001, normal tissue vs. laryngeal cancer. (B) TMEM158 expression at different tumor-node-metastasis stages. *P<0.05 and ***P<0.001, stage I/II/III/IV vs. normal. (C) Kaplan-Meier survival analysis of patients with laryngeal cancer based on TMEM158 expression. ***P<0.001, lowTMEM158 group vs. highTMEM158 group. (D) Reverse transcription-quantitative PCR was performed to assess TMEM158 mRNA expression in human laryngeal squamous cell carcinoma cells and NP69 nasopharyngeal epithelial cells. **P<0.01, LSC-1 cells vs. NP69 cells. TMEM158, transmembrane protein 158; TCGA, The Cancer Genome Atlas.
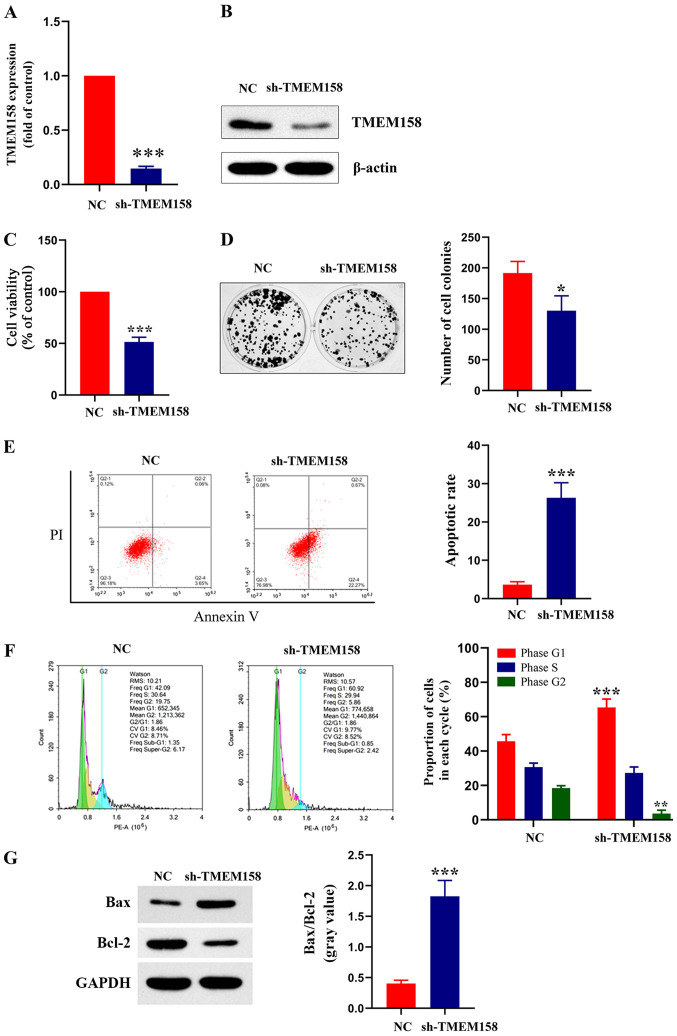
Figure 2.
TMEM158 affects viability, formation of colonies, cell cycle distribution and apoptosis of laryngeal cancer cells. (A) Reverse transcription-quantitative PCR and (B) Western blot analyses were performed to assess mRNA and protein expression of TMEM158 in LSC-1 cells transfected with sh-TMEM158 or NC, respectively. ***P<0.001, shTMEM158 vs. NC. (C) The MTT assay was performed to assess cell viability in LSC-1 cells transfected with sh-TMEM158 or NC. ***P<0.001, shTMEM158 vs. NC. (D) Effect of TMEM158 on colony formation of laryngeal cancer cells. Representative photos (left) and relative quantification analysis of colonies (right). *P<0.05, shTMEM158 vs. NC. (E) Flow cytometric analysis of apoptosis. ***P<0.001, shTMEM158 vs. NC. (F) Flow cytometric analysis of cell cycle distribution. Histograms depict the proportion of LSC-1 cells in the G1, S and G2 phases. ***P<0.001, shTMEM158 (G1) vs. NC (G1); **P<0.01, shTMEM158 (G2) vs. NC (G2). (G) Western blot analysis was performed to assess protein expression levels of Bax and Bcl-2. Histograms depict the gray value of Bax/Bcl-2. ***P<0.001, shTMEM158 vs. NC. TMEM158, transmembrane protein 158; LSC-1, laryngeal squamous cell carcinoma; sh, short hairpin; NC, negative control; Bcl-2, B-cell lymphoma 2; PI, propidium iodide.
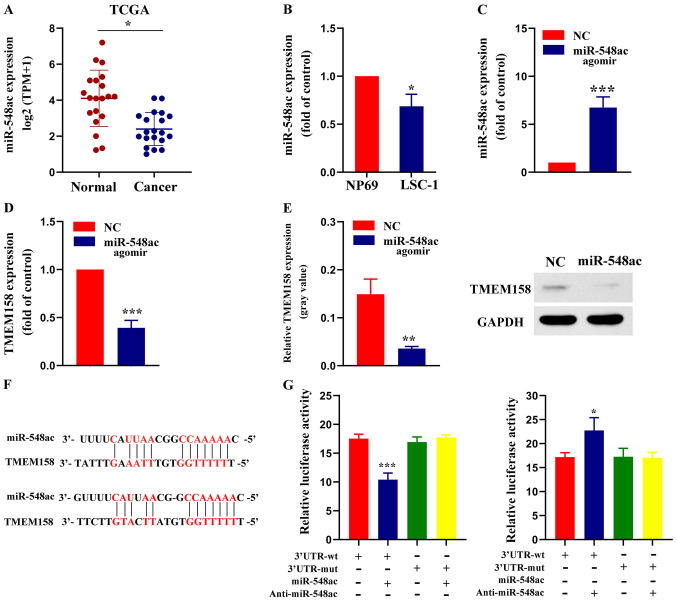
Figure 3.
miR-548ac suppresses TMEM158 expression by directly binding to its 3′-UTR. (A) miR-548ac expression was assessed in laryngeal carcinoma tissues and adjacent normal tissues using miRNA sequencing data from TCGA database. *P<0.05 normal group vs. tumor group. (B) RT-qPCR analysis was performed to assess miR-548ac expression in NP69 and LSC-1 cells. *P<0.05, LSC-1 cells vs. NP69 cells. (C) RT-qPCR assay was performed to assessmiR-548ac expression in LSC-1 cells transfected with miR-548ac agomir or NC. ***P<0.001, miR-548ac agomirs vs. NC. (D) RT-qPCR analysis was performed to assess TMEM158 expression in LSC-1 cells transfected with miR-548ac agomir or NC. ***P<0.001, miR-548ac agomirs vs. NC. (E) Western blot analysis was performed to assess TMEM158 protein expression in LSC-1 cells transfected with miR-548ac agomir or NC. Histograms depict the gray value of TMEM158/GAPDH. **P<0.01, miR-548ac agomirs vs. NC. (F) Sequence alignment of miR-548ac and its predicted binding sites (red) in TMEM158 3′-UTR. (G) Luciferase reporter assay. ***P<0.001, 3′UTR-WT (+miR-548ac) vs. 3′UTR-WT; *P<0.05, 3′UTR-WT (+Anti-miR-548ac) vs. 3′UTR-WT. miR, microRNA; TMEM158, transmembrane protein 158; UTR, untranslated region; TCGA, The Cancer Genome Atlas; RT-qPCR, reverse transcription-quantitative PCR; LSC-1, laryngeal squamous cell carcinoma; NC, negative control.
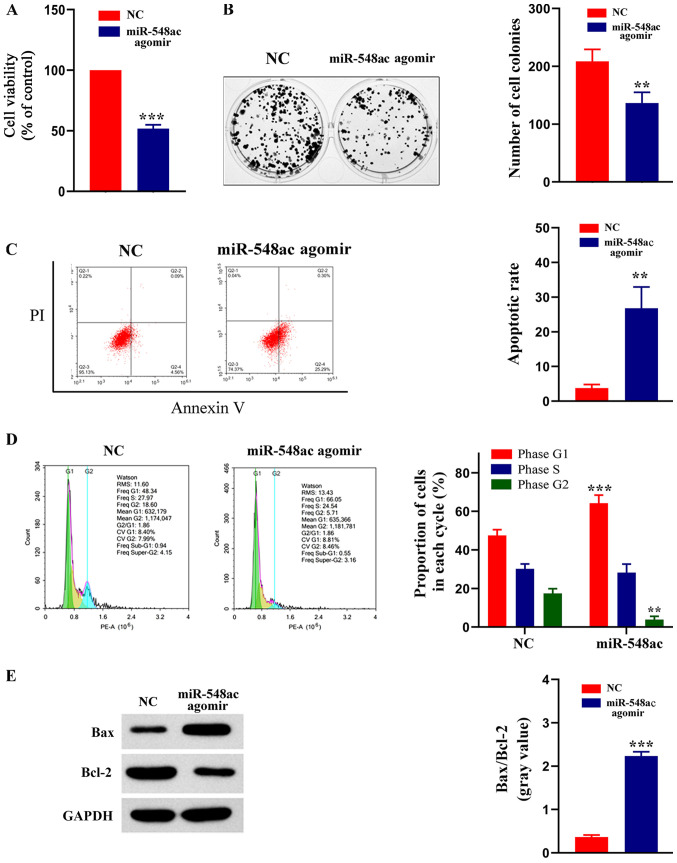
Figure 4.
miR-548ac affects viability, formation of colonies, cell cycle distribution and apoptosis of laryngeal cancer cells. (A) MTT assay was performed to assess cell viability in LSC-1 cells transfected with miR-548ac antagomir or NC. ***P<0.001, miR-548ac agomir vs. NC. (B) Effect of miR-548ac on colony formation of laryngeal cancer cells. Representative micrographs (left) and relative quantification analysis of colonies (right). **P<0.01, miR-548ac agomir vs. NC. (C) Flow cytometric analysis of apoptosis. **P<0.01, miR-548ac agomir vs. NC. (D) Flow cytometric analysis of cell cycle distribution. Histograms depict the proportion of LSC-1 cells in the G1, S and G2 phases. ***P<0.001, miR-548ac agomir (G1) vs. NC (G1); **P<0.01, miR-548ac agomir (G2) vs. NC (G2). (E) Western blot analysis was performed to assess protein expression levels of Bax and Bcl-2. Histograms depict the gray value of Bax/Bcl-2. ***P<0.001, miR-548ac agomir vs. NC. miR, microRNA; LSC-1, laryngeal squamous cell carcinoma; NC, negative control; Bcl-2, B-cell lymphoma 2; PI, propidium iodide.
Figure 5.
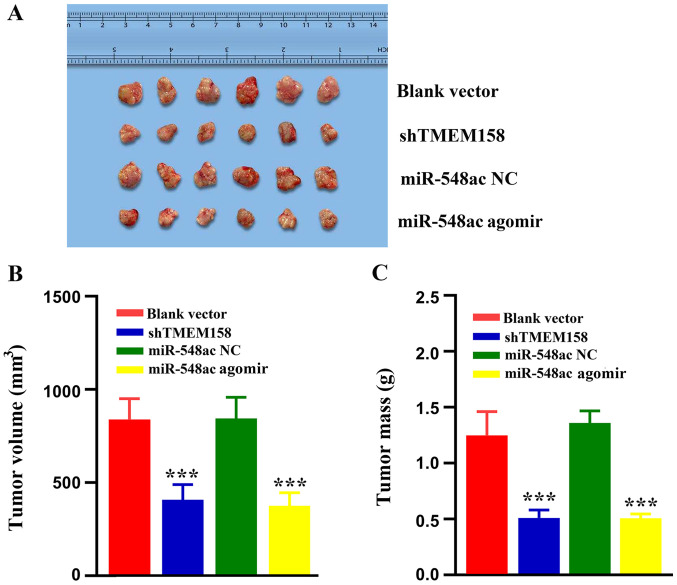
Effects of TMEM158 and miR-548ac on tumor growth in vivo. (A) Representative images of tumors derived from nude mice in the blank vector, shTMEM158, miR-548ac NC and miR-548ac agomir groups. (B) Tumor volumes in the different groups. (C) Tumor masses in the different groups. ***P<0.001 vs. blank vector group and miR-548ac NC group. TMEM158, transmembrane protein 158; miR, microRNA; sh, short hairpin; NC, negative control.
Results
TMEM158 is upregulated in laryngeal cancer tissues and cell lines
The Cancer Genome Atlas (TCGA) database was used to compare mRNA sequencing data between laryngeal carcinoma tissues and adjacent normal tissues. The results demonstrated that TMEM158 expression was significantly higher in laryngeal carcinoma tissues compared with adjacent normal tissues (Fig. 1A). TMEM158 expression according to different tumor-node-metastasis (TNM) stages was also assessed. The results demonstrated that TMEM158 expression increased in accordance with the increase of the TNM stage (Fig. 1B). The prognostic value of TMEM158 in laryngeal carcinoma was determined via overall survival analysis. The results demonstrated that patients with low TMEM158 expression had a significantly longer survival time than that of patients with high TMEM158 expression (Fig. 1C). TMEM158 mRNA expression was further assessed in NP69 and LSC-1 cells. The results demonstrated that TMEM158mRNA expression was 4.35±0.56 times greater in LSC-1 cells compared with NP69 cells (P=0.0052; Fig. 1D). Taken together, these results suggest that upregulated TMEM158 expression may play a critical role in the development and progression of laryngeal cancer.
Effect of TMEM158 on the viability of laryngeal cancer cells
Normal LSC-1 cells were used as the control group, while cells transfected with sh-TMEM158 were used as the experimental group. RT-qPCR analysis demonstrated that TMEM158 mRNA expression was significantly lower in LSC-1 cells transfected with sh-TMEM158 compared with LSC-1 cells transfected with NC (14.67±2.07% of the control; P<0.001; Fig. 2A). The results of western blot analysis were consistent with these findings (Fig. 2B). Similarly, the results of the MTT assay demonstrated that cell viability was significantly lower in LSC-1 cells transfected with sh-TMEM158 compared with the control group (51.49±4.51% of the control; P<0.0001; Fig. 2C). The results of the colony formation assay demonstrated that the number of cell colonies was significantly lower in the experimental group compared with the control group (67.62±4.72% of the control; P=0.025; Fig. 2D). The results of the Annexin V-PI assay indicated that the apoptotic rate was significantly higher in the experimental group compared with the control group (7.3±1.33 of the control; P=0.00062; Fig. 2E). The cell cycle distribution assay demonstrated a higher proportion of cells transfected with sh-TMEM158 in the G1 phase compared with the control group (Fig. 2F). The protein expression levels of Bax and Bcl-2 were determined via western blot analysis, and the gray value was used for relative quantification. The results demonstrated that Bax and Bcl-2 expression levels were significantly higher in the experimental group compared with the control group (4.53±0.03 of the control; P=0.00072; Fig. 2G). Collectively, these results indicate that silencing TMEM158 significantly inhibits the viability of laryngeal cancer cells.
miR-548ac is downregulated in laryngeal cancer tissues and cell lines
RNAInter software analysis demonstrated that miR-548ac has a high probability of binding to TMEM158. TCGA database was used to compare miRNA sequencing data between laryngeal carcinoma tissues and adjacent normal tissues. The results demonstrated that miR-548ac expression was significantly lower in laryngeal carcinoma tissues compared with adjacent normal tissues (Fig. 3A). miR-548ac expression was further assessed inNP69 and LSC-1 cells. The results demonstrated that miR-548ac expression was significantly lower in LSC-1 cells compared with NP69 cells (68.67±10.21% of NP69 cells; P=0.027; Fig. 3B). Taken together, these results suggest that downregulation of miR-548ac may play a critical role in the progression of laryngeal cancer.
Association between miR-548ac and TMEM158
miR-548ac agomir (experimental group) or miR-548ac agomir NC (control group) were transfected into LSC-1 cells, respectively. RT-qPCR analysis demonstrated that miR-548ac expression was significantly higher in the experimental group compared with the control group (7.76±1.1-fold of the control; P<0.001; Fig. 3C). Furthermore, TMEM158 mRNA expression was significantly lower in the experimental group compared with the control group (39.33±6.34% of the control; P=0.00071; Fig. 3D). The results of western blot analysis were consistent with these findings (Fig. 3E). The results of the dual-luciferase reporter assay demonstrated that luciferase activity decreased in LSC-1 cells transfected with WT-TMEM158-3′-UTR and miR-548ac agomir compared with WT-TMEM158-3′-UTR alone (59.58±6.78% of WT-TMEM158-3′-UTR alone; P<0.001). However, the luciferase activity remained unchanged whenLSC-1 cells were transfected with MUT-TMEM158-3′-UTR and miR-548ac agomir. Luciferase activity increased whenLSC-1 cells were transfected withWT-TMEM158-3′-UTR and anti-miR-548ac agomir compared with WT-TMEM158-3′-UTR alone (1.32±0.07 of WT-TMEM158-3′-UTR alone; P=0.03). However, the luciferase activity remained unchanged whenLSC-1 cells were transfected with MUT-TMEM158-3′-UTR and anti-miR-548ac agomir compared with MUT-TMEM158-3′-UTR alone (Fig. 3G). Collectively, these results indicate that TMEM158 is a direct target gene of miR-548ac.
Effect of miR-548ac on the viability of laryngeal cancer cells
Normal LSC-1 cells were used as the control group, while cells transfected with miR-548ac agomir were used as the experimental group. The results of the MTT assay demonstrated that cell viability significantly decreased in the experimental group compared with the control group (51.86±3.11% of the control; P<0.0001; Fig. 4A). The results of the colony formation assay demonstrated that the number of cell colonies was significantly lower in the experimental group compared with the control group (65.44±4.34% of the control; P=0.0094; Fig. 4B). The results of the Annexin V-PI assay demonstrated that the apoptotic rate was significantly higher in the experimental compared with the control group (7.27±1.22-fold of the control; P=0.003; Fig. 4C). The cell cycle distribution assay demonstrated a higher proportion of cells transfected with miR-548ac agomir in the G1 phase compared with the control group (Fig. 4D). The protein expression levels of Bax and Bcl-2 were detected via western blot analysis, and the gray value was used for relative quantification. The western blots showed that Bcl-2 protein expression was higher and Bax protein expression was lower in NC group compared with the experimental group. Gray value ratio of Bax and Bcl-2 was significantly higher in the experimental group compared with the control group (6.19±0.46 fold of the control; P=0.00045; Fig. 4E). Collectively, these results indicate that overexpressing miR-548ac significantly inhibits the viability of laryngeal cancer cells. Thus, it is speculated that miR-548ac plays a cancer suppressive role in laryngeal cancer.
Effect of TMEM158 and miR-548ac on tumor growth in vivo
A xenograft model was used to determine the effects of TMEM158 and miR-548ac on tumor growth in vivo. As presented in Fig. 5A, the tumors were smaller in the shTMEM158 and miR-548ac agomir groups compared with the corresponding control groups. The tumor volume in the shTMEM158 group was 49.05±9.78% that of the blank vector group (P<0.001), while the tumor volume in the miR-548ac group was 45.36±10.54% that of the miR-548ac NC group (P<0.001) (Fig. 5B). The tumor mass in the shTMEM158 group was 42.21±11.47% that of the blank vector group (P<0.0001), while the tumor mass in the miR-548ac group was 37.49±4.43% that of the miR-548ac NC group (P<0.001) (Fig. 5C).
Discussion
miRNAs function in repressing translation of target mRNAs or degrading it, thus negatively regulating the expression of target genes following transcription (18). The regulatory mechanism of miRNAs to their target genes has been extensively studied and demonstrated to be involved in biological events, such as cell proliferation, apoptosis, differentiation, exercise and angiogenesis (19,20). It has been reported that the miR-548 family can inhibit the growth of several tumors. For example, Shi et al (15) reported that mir-548-3p inhibits the proliferation of breast cancer cells by regulating ECHS1 expression, suggesting its potential as a therapeutic target for breast cancer. Furthermore, Wang et al (14) demonstrated that mir-548-3p can effectively inhibit the proliferation of lung cancer cells by inhibiting the PI3K/Akt signaling pathway, indicating that miR-548b-3p has the potential to be developed as an antitumor target of lung cancer. The results of the present study demonstrated that LSC-1 cell viability significantly decreased following overexpression of miR-548ac, whereby some cells exhibited early apoptosis accompanied by activation of the apoptosis factor, Bax, and further cell cycle arrest at the G1 phase. Thus, it was speculated that overexpression of miR-548ac in laryngeal cancer cells significantly inhibits cell proliferation and induces apoptosis. Collectively, the results of the present study suggest that miR-548ac may function as a tumor suppressor in laryngeal cancer.
The expression and molecular mechanism of TMEM158 have been reported in different types of tumor. For example, Iglesias et al (21) suggested that TMEM158 acts as an oncogene in the development of colon cancer. In addition, Mohamed et al (8) demonstrated that TMEM158 interference significantly decreases the toxic effect exerted by cisplatin on NSCLC cells, suggesting that TMEM158 is associated with the sensitivity of chemotherapy drugs. Zirn et al (22) reported that TMEM158 is highly expressed in CTNNB1 mutant nephroblastoma, which may be associated with the Ras and Wnt signaling pathways. Furthermore, Liu et al (23) demonstrated that TMEM158 is a key regulatory factor of colorectal carcinogenesis and drug resistance, and thus may be a promising target for colorectal cancer treatment. The results of the present study demonstrated that TMEM158 was highly expressed in laryngeal carcinoma tissues from the TCGA database, which was verified via cell line experiments. The results demonstrated that LSC-1 cell viability significantly decreased following TMEM158 silencing, which induced early apoptosis, accompanied by activation of the apoptotic factors (Bax). Simultaneously, TMEM158 silencing resulted in cell arrest in the G1 phase. Collectively, these results suggest that TMEM158 may act as an oncogene, affecting the viability of laryngeal cancer cells.
miRNAs regulate the expression of ~30% of human protein-coding genes. Each miRNA can regulate hundreds of mRNA genes, and the same target gene can be regulated by multiple miRNAs, to regulate the expression of thousands of proteins directly or indirectly (24). miRNAs regulate gene expression by specifically pairing with the 3′-UTR region of their target gene, resulting in the degradation of the target mRNA or post transcriptional silencing, thus inhibiting protein synthesis (25,26). The results of the present study demonstrated that TMEM158 mRNA and protein expression levels significantly decreased following overexpression of miR-548ac in LSC-1 cells. The results of the dual-luciferase reporter assay confirmed that miR-548ac directly binds to the 3′-UTR of TMEM158, thus inhibiting TMEM158 expression. Consistent with previous findings, the results of the present study demonstrated that miR-548ac induces apoptosis in LSC-1 cells by targeting TMEM158.
The present results suggest that miR-548ac functions as a crucial cancer suppressor in laryngeal cancer, which induces apoptosis in laryngeal cancer cells by suppressing TMEM158. The present findings enrich the pathogenesis of laryngeal cancer and provide a new target and new insights for the treatment of laryngeal cancer. The limitation of this study is that there is no biological detection of TMEM158 and miR-548ac in clinical samples. At the same time, there is no detection on the influence of the follow-up signal pathway. These limitations will be addressed in future studies.
Acknowledgements
The authors would like to thank The Cancer Genome Atlas (TCGA) project for providing the data.
Funding
Support for the study was provided by the Tianjin Natural Science Foundation (No 20170842).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
FS designed the experiment and wrote the manuscript; YY contributed to animal and cell experiments; JL performed the statistical and bioinformatics analysis. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Tianjin Union Medical Center (Tianjin, China; approval no. 2018-269,). All animal experiments were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH Publications no. 8023, revised in 1978).
Patients consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Beibei Y, Rong Y, Yunfei Y, Wenchao Z. Research progress regarding surgical margins, molecular margins, and prognosis of laryngeal carcinoma. Ear Nose Throat J 145561320903146. 2020 doi: 10.1177/0145561320903146. (Epubahead of print) [DOI] [PubMed] [Google Scholar]
- 2.Olaleye O, Siddiq S, Ekrikpo U, Kazi R. Regional differences in incidence and mortality trends in cancers of the larynx, thyroid, oral cavity and pharynx in England and Scotland: 1975–2002. Open Epidemiol J. 2013;3:70–78. doi: 10.4236/ojepi.2013.32011. [DOI] [Google Scholar]
- 3.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 4.Steuer CE, El-Deiry M, Parks JR, Higgins KA, Saba NF. An update on larynx cancer. CA Cancer J Clin. 2017;67:31–50. doi: 10.3322/caac.21386. [DOI] [PubMed] [Google Scholar]
- 5.Menicagli R, Bolla G, Menicagli L, Esseridou A. The possible role of diabetes in the etiology of laryngeal cancer. Gulf J Oncolog. 2017;1:44–51. [PubMed] [Google Scholar]
- 6.Barradas M, Gonos ES, Zebedee Z, Kolettas E, Petropoulou C, Delgado MD, Leon J, Hara E, Serrano M. Identification of a candidate tumor-suppressor gene specifically activated during Ras-induced senescence. Exp Cell Res. 2002;273:127–137. doi: 10.1006/excr.2001.5434. [DOI] [PubMed] [Google Scholar]
- 7.Cheng Z, Guo J, Chen L, Luo N, Yang W, Qu X. Overexpression of TMEM158 contributes to ovarian carcinogenesis. J Exp Clin Cancer Res. 2015;34:75. doi: 10.1186/s13046-015-0193-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mohammed Ael S, Eguchi H, Wada S, Koyama N, Shimizu M, Otani K, Ohtaki M, Tanimoto K, Hiyama K, Gaber MS, Nishiyama M. TMEM158 and FBLP1 as novel marker genes of cisplatin sensitivity in non-small cell lung cancer cells. Exp Lung Res. 2012;38:463–474. doi: 10.3109/01902148.2012.731625. [DOI] [PubMed] [Google Scholar]
- 9.Fu Y, Yao N, Ding D, Zhang X, Liu H, Ma L, Shi W, Zhu C, Tang L. TMEM158 promotes pancreatic cancer aggressiveness by activation of TGFβ1 and PI3K/AKT signaling pathway. J Cell Physiol. 2020;235:2761–2775. doi: 10.1002/jcp.29181. [DOI] [PubMed] [Google Scholar]
- 10.Hummel R, Maurer J, Haier J. MicroRNAs in brain tumors: A new diagnostic and therapeutic perspective? Mol Neurobiol. 2011;44:223–234. doi: 10.1007/s12035-011-8197-x. [DOI] [PubMed] [Google Scholar]
- 11.Rouleau S, Glouzon JS, Brumwell A, Bisaillon M, Perreault JP. 3′ UTR G-quadruplexes regulate miRNA binding. RNA. 2017;23:1172–1179. doi: 10.1261/rna.060962.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao X, Cao Y, Chen H. Profiling and characterization of microRNAs responding to sodium butyrate treatment in A549 cells. J Cell Biochem. 2018;119:3563–3573. doi: 10.1002/jcb.26547. [DOI] [PubMed] [Google Scholar]
- 13.Liang T, Guo L, Liu C. Genome-wide analysis of mir-548 gene family reveals evolutionary and functional implications. J Biomed Biotechnol. 2012;2012:679563. doi: 10.1155/2012/679563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z, Wu X, Hou X, Zhao W, Yang C, Wan W, Chen L. MiR-548b-3p functions as a tumor suppressor in lung cancer. Lasers Med Sci. 2020;35:833–839. doi: 10.1007/s10103-019-02865-7. [DOI] [PubMed] [Google Scholar]
- 15.Shi Y, Qiu M, Wu Y, Hai L. MiR-548-3p functions as an anti-oncogenic regulator in breast cancer. Biomed Pharmacother. 2015;75:111–116. doi: 10.1016/j.biopha.2015.07.027. [DOI] [PubMed] [Google Scholar]
- 16.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 17.Tashiro K, Kuroki N, Einama T, Iwasaki T, Miyata Y, Aosasa S, Inoue Y, Takahashi Y, Ogata S, Ueno H, et al. Prognostic significance of regional lymph node metastasis according to station in ampullary carcinoma. J Hepatobiliary Pancreat Sci. 2020 doi: 10.1002/jhbp.791. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 18.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mellis D, Caporali A. MicroRNA-based therapeutics in cardiovascular disease: Screening and delivery to the target. Biochem Soc Trans. 2018;46:11–21. doi: 10.1042/BST20170037. [DOI] [PubMed] [Google Scholar]
- 20.Qadir MI, Faheem A. MiRNA: A diagnostic and therapeutic tool for pancreatic cancer. Crit Rev Eukaryot Gene Expr. 2017;27:197–204. doi: 10.1615/CritRevEukaryotGeneExpr.2017019494. [DOI] [PubMed] [Google Scholar]
- 21.Iglesias D, Fernandez-Peralta AM, Nejda N, Daimiel L, Azcoita MM, Oliart S, Gonzalez-Aguilera JJ. RIS1, a gene with trinucleotide repeats, is a target in the mutator pathway of colorectal carcinogenesis. Cancer Genet Cytogenet. 2006;167:138–144. doi: 10.1016/j.cancergencyto.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 22.Zirn B, Samans B, Wittmann S, Pietsch T, Leuschner I, Graf N, Gessler M. Target genes of the WNT/beta-catenin pathway in Wilms tumors. Genes Chromosomes Cancer. 2006;45:565–574. doi: 10.1002/gcc.20319. [DOI] [PubMed] [Google Scholar]
- 23.Liu L, Zhang J, Li S, Yin L, Tai J. Silencing of TMEM158 inhibits tumorigenesis and multidrug resistance in colorectal cancer. Nutr Cancer. 2020;72:662–671. doi: 10.1080/01635581.2019.1650192. [DOI] [PubMed] [Google Scholar]
- 24.Kura B, Kalocayova B, LeBaron TW, Frimmel K, Buday J, Surovy J, Slezak J. Regulation of microRNAs by molecular hydrogen contributes to the prevention of radiation-induced damage in the rat myocardium. Mol Cell Biochem. 2019;457:61–72. doi: 10.1007/s11010-019-03512-z. [DOI] [PubMed] [Google Scholar]
- 25.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 26.Correia de Sousa M, Gjorgjieva M, Dolicka D, Sobolewski C, Foti M. Deciphering miRNAs' Action through miRNA Editing. Int J Mol Sci. 2019;20:6249. doi: 10.3390/ijms20246249. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.