Abstract
In the present study, the structural characteristics that impart antibacterial activity to C16 alkynoic fatty acids (aFAs) was further investigated. The syntheses of hexadecynoic acids (HDA) containing triple bonds at C-6, C-8, C-9, C-10, and C-12 were carried out in four steps and with an overall yield of 34–78%. In addition, HDA analogs containing a sulfur atom at either C-4 or C-5 were also prepared in 69–77% overall yields, respectively. Results from this study revealed that the triple bond at C-2 is pivotal for the antibacterial activity displayed by 2-HDA, while the farther the position of the triple bond from the carbonyl group, the lower its bactericidal activity against Gram-positive bacteria, including clinical isolates of methicillin-resistant Staphylococcus aureus (CIMRSA) strains. The potential of 2-HDA as an antibacterial agent was also assessed in five CIMRSA strains that were resistant to Ciprofloxacin (Cipro) demonstrating that 2-HDA was the most effective treatment in inhibiting their growth when compared with either Cipro alone or equimolar combinations of Cipro and 2-HDA. Moreover, it was proved that the inhibition of S. aureus DNA gyrase can be linked to the antibacterial activity displayed by 2-HDA. Finally, it was determined that the ability of HDA analogs to form micelles can be linked to their decreased activity against Gram-positive bacteria, since critical micellar concentrations (CMCs) between 50 and 300 μg/mL were obtained.
Keywords: Alkynoic fatty acids, Ciprofloxacin-resistant S. aureus, Critical micelle concentration, DNA gyrase, MRSA, Susceptibility tests
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is considered by the Centers for Disease Control and Prevention (CDC) as a serious concern that requires prompt attention and sustained action (CDC, 2013). Some illnesses such as skin and wound infections, pneumonia, and bloodstream infections are attributed to MRSA (CDC, 2013). Serious attention is required since MRSA are gaining resistance to the current antibiotics through genetic mechanisms including mutation and acquisition of new DNA (McManus, 1997). This situation causes that bacteria resist antibiotics through the inactivation of the drugs with beta-lactamases, acetylases, adenylases, and phosphorylases (McManus, 1997). These enzymes reduce the drugs access to their active sites altering the target of these agents that results in the development of tolerance (McManus, 1997).
Ciprofloxacin (Cipro) is a fluoroquinolone that is commonly prescribed to treat MRSA infections (Piercy et al., 1989; Spetalnick et al., 1990). This broad-spectrum antibiotic exerts its bactericidal activity by inhibition of DNA gyrase (LeBel, 1988), a molecular target that is pivotal in the replication of DNA (LeBel, 1988). However, several MRSA strains have acquired resistance towards Cipro through mutations of gyrA/gyrB genes (Campion et al., 2004), which encode the subunits of DNA gyrase (Ito et al., 1994). For this reason, there is an urgent need to develop new antibiotics to replace those that are not effective.
Several studies have reported the potential of unsaturated fatty acids as antibacterial agents (Carballeira et al., 2002; Carballeira et al., 1998; Carballeira et al., 2017; Carballeira et al., 2006; Konthikamee et al., 1982; Morbidoni et al., 2006; Sanabria-Rios et al., 2014; Sanabria-Rios et al., 2015). For example, in a study performed by Carballeira et al., 1998, the (Z)-2-methoxy-5-hexadecenoic and (Z)-2-methoxy-6-hexadecenoic acids were biologically active against S. aureus and Streptococcus faecalis at minimal inhibitory concentrations (MICs) of 0.35 μM. In a more recent study, it was demonstrated that the (±)-2-methoxy-6-octadecynoic acid inhibited MRSA at MIC values ranging between 31.3 and 62.5 μg/mL (Carballeira et al., 2017).
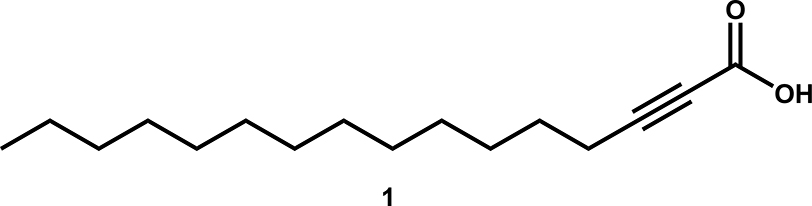
In 2014, Sanabria-Ríos and collaborators carried out the synthesis of a series of 2-alkynoic fatty acids (aFAs) and other synthetic analogs, such as 2-tetrahydropyranyl protected alkynols and 2-alkynols, to establish a structure activity relationship (SAR) with these compounds aimed at finding the most active compound against both Gram-positive and Gram-negative bacteria (Sanabria-Rios et al., 2014). There, it was demonstrated that the 2-hexadecynoic acid (1, Fig. 1) was the most active FA towards commercially available S. aureus and MRSA as well as towards clinical isolates of MRSA (CIMRSA) displaying MICs values that ranged from 3.9 to 15.6 μg/mL. In another study, Sanabria-Ríos and his research group chemically connected 1 to C5-Curcumin (C5-Curc) to enhance the antibacterial activity of C5-Curc (Sanabria-Rios et al., 2015). Specifically, they demonstrated that the presence of 1 in the C5-Curc-2-HDA conjugate increased 4–8-fold the antibacterial activity of C5-Curc against eight MRSA strains. Moreover, in the latter study it was suggested that the antibacterial activity of 1 can be due, in part, to the inhibition of the S. aureus DNA gyrase.
Fig. 1.
Chemical structure of 2-hexadecynoic acid (1).
In addition to the traditional antibacterial activity displayed by 1, the 2-HDA also displayed conjugation inhibitory (COIN) activity with Escherichia, Salmonella, Pseudomonas, and Acinetobacter spp (Getino et al., 2015). The use of 1 avoided the spreading of a depressed IncF plasmid into a recipient population, which demonstrates the feasibility of eliminating the dissemination of antimicrobial resistance by blocking bacterial conjugation (Getino et al., 2015). In a study performed by Ripoll-Rozada and collaborators, they identified that TrwD, a Type IV secretion traffic ATPase, is involved in the 1-mediated conjugation inhibition. Moreover, 1, which is also a potent COIN, displayed powerful inhibitory ATPase activity of TrwD as well, demonstrating that TrwD is the molecular target of 1 (Garcia-Cazorla et al., 2018; Ripoll-Rozada et al., 2016).
Despite the fact that several studies addressing the antibacterial activity of 2-aFAs exist (Carballeira et al., 2017; Carballeira et al., 2006; Morbidoni et al., 2006; Sanabria-Rios et al., 2014; Vilchèze et al., 2016), further studies are needed to better understand the chemical and structural characteristics required to improve the antibacterial activity of aFAs as well as to understand their mechanism in Gram-positive bacteria. In the present study, the synthesis of several analogs of 1 containing a triple bond at either C-3, C-6, C-8, C-9, C-10, and C-12 was undertaken to determine the effect of the triple bond position in the antibacterial activity towards clinical isolates of MRSA (CIMRSA). In addition, it was explored if the presence of a sulfur atom at C-4 and C-5 in the carbon chain of 2-HDA analogs influence their antibacterial activity. Since the antibacterial activity of 1 against CIMRSA can be related to its inhibitory activity towards S. aureus DNA gyrase (Sanabria-Rios et al., 2015), it was further investigated whether the analogs of 1 are able to inhibit the supercoiling activity of the above-mentioned enzyme. Moreover, a preliminary combinatorial study involving both 1 and Cipro was undertaken to determine if this combination enhances the antibacterial activity against CIMRSA as well as the inhibition of S. aureus DNA gyrase.
Materials and Methods
Instrumentation
Both 1H-NMR and 13C-NMR spectra were recorded on a Bruker Avance 500 spectrophotometer. 1H-NMR chemical shifts are reported relative to internal CDCl3 signal (7.26 ppm) and 13C-NMR chemical shifts are reported in parts per million (ppm) relative to CDCl3 signal (77.00 pm). FT-IR data were gathered on a Thermo Scientific Nicolet iS5 FT-IR spectrophotometer. Mass spectra data was acquired by using a chromatography-mass spectrometer (GC-MS, Agilent 5975C MS ChemStation; Agilent, Palo Alto, CA, USA) at 70 eV equipped with a 30 m x 0.25 mm special performance capillary column (HP-5MS) of polymethysiloxane cross-linked with 5% phenyl methylpolysiloxane. UV/Vis data were obtained on a Thermo Scientific Genesys 10S UV-Vis spectrophotometer (Cambridge, United Kingdom). MIC values were spectrophotometrically determined by using a Multiskan FC microplate reader (Fisher Scientific) at 620 nm.
Microorganisms
Staphylococcus aureus (ATCC 29213) and Escherichia coli (ATCC 25922) were purchased from the American Type Culture Collection (Manassas, VA, USA). Clinical isolates of MRSA (CIMRSA) were kindly donated by a community hospital in San Juan, Puerto Rico (USA). Stock cultures were kept on blood agar (TSA with 5% sheep blood, Remel and Oxoid Microbiology Products, Lenexa, KS). Inoculation of a single colony in 5 mL of Trypticase Soy Broth (TSB, BD Diagnostic Systems, Franklin Lakes, NJ) was performed to prepare suspension cultures that were incubated for 16–18 h at 37 °C. Prior to preparation of susceptibility assays, bacteria cells were re-suspended in TSB and visually standardized by using 0.5 McFarland standard solution providing an equivalent concentration of 1.0 × 108 colony-forming units (CFU)/mL.
Susceptibility Testing
Susceptibility tests were performed as previously described (Sanabria-Rios et al., 2014). Briefly, aFA’s stock solutions were prepared using 100 % DMSO and serially diluted with sterile TSB. A 100 μL of each dilution were transferred to flat-bottomed microplate wells previously inoculated with 10 μL of TSB solution containing 4–5 × 105 CFU. Each well was inspected spectrophotometrically at 620 nm using both a positive control well (containing the bacterial inoculated TSB but not the aFA treatment) and a negative control well (containing sterile TSB without aFA solution) for further comparisons. The minimum inhibitory concentration (MIC) was the concentration at which the test compound prevented turbidity in the well after incubation for 16–18 h at 37 °C.
Bacterial Growth Assays
CIMRSA XIII strain was grown to optical density of 1.0 (at 600 nm) and diluted 30,000-fold in TSB. A 10 μL aliquot of diluted bacteria was added to each well of a flat-bottom 96-well plate containing 100 μL of TSB with either Palmitic Acid, 1, or Cipro at an appropriated concentration of (4 × MIC). All FA dilutions contained 1% DMSO. TSB medium containing 1% DMSO was used as control with no treatment. The plate was incubated at 37 °C for 20 h in a Thermo Scientific Varioskan Lux plate reader and then read at 600 nm afterwards.
Effect of 1 on the S. aureus Fatty Acid Profile using Gas Chromatography-Mass Spectrometry (GC-MS)
CIMRSA XIII was grown in TSB medium in the presence or absence of 1 (0.16 μg/mL) at 37 °C for 18–20 h. Bacterial cells were centrifuged (Sorvall ST 16R, Thermo Scientific, Germany) at 5,000 rpm at 4 °C for 5 min and the resulting pellet was washed three times with 1 X PBS (HyClone, USA). Then, bacterial pellets were homogenized (PowerLyzer™, MO BIO Laboratories, Inc., USA) for 20 min at 2,500 rpm, sonicated in an Ultrasonic Cleaner (Fisher Brand FB11201, Germany) at 37 kHz (100 % power, 390 W) for 20 min at rt, and centrifugated (Sorvall Legend Micro 17, Thermo Scientific, Germany) at rt at 17,000 × g for 20 min. The resulting supernatant was chromatographed using silica gel column and diethyl ether as mobile phase. Subsequently, the organic extracts containing FAs were roto-evaporated (Büchi, Rotavapor R-114, Switzerland) and refluxed in 3 mL of methanol and 150 μl of 12 M HCl for 3 h. Once the transesterification reaction was completed, the methanolic extracts containing FAs were roto-evaporated and freeze dried at −80 °C (Millrock Technology, USA). The fatty acid methyl esters (FAME) were analyzed using a GC/MS-QP2010 (Shimadzu) equipped with a fused-silica FAMEWAX capillary column (30 m × 0.32 mm i.d. × 0.25μm film thickness) (Restek) and an auto-injector (Shimadzu, AOC-20i). The selected temperature method was: 130 °C initial temperature, 4° C/min temperature rate, and 250 °C final temperature using helium as the carrier gas. The split ratio was set at 15:1 for all analyses. Mass spectra data were obtained after electron impact ionization (EI, 70 eV, ion source temperature 200 °C) in full scan mode between 50 and 600 amu. The temperatures of the injection and the detector were set at 250 °C. All FAs were identified based on their molecular ion, base peak, fragmentation pattern, retention time, and equivalent chain length (ECL) values and then expressed as a percentage of total FAs. Experiments were performed in three biological replicates.
Determination of the Inhibitory Effect of aFAs on the Supercoiling Activity of S. aureus DNA Gyrase
The inhibitory effect of aFAs on the supercoiling activity of DNA gyrase was assessed using DNA gyrase from S. aureus (TopoGen, USA, 1 unit will supercoil 0.05 μg of DNA in 60 min at 37 °C) and 0.20 μg of relaxed pHOT1 plasmid DNA. AFas were dissolved in 100% DMSO and tested at different concentrations ranging from 1.9 to 1000 μg/mL. Reaction (final volume 20 μL) mixtures were incubated for 60 min at 37 °C, and subsequently treated with 2 μL of 10% sodium dodecyl sulfate (SDS). Incubation with proteinase K (0.05 mg/mL) for 30 min at 37 °C was performed to digest bounded proteins. Reactions were stopped by adding 4 μL of electrophoresis universal stop and loading buffer. Products were separated by electrophoresis in a 1% agarose gel (70 V/105 min). After electrophoresis, the gel was stained with ethidium bromide for 25 min and distained for 10–15 min in distilled water to visualize the reaction products. DNA bands were detected in a Min BIS bioimaging system (model 241016P1, Israel). MIC is defined at the lowest concentration in which supercoiled DNA band is practically not observable.
Determination of the Inhibitory Effect of 1 on the activity of Glyceraldehyde 3 Phosphate Dehydrogenase (G3PD)
Non-treated CIMRSA XIII, 1 -treated CIMRSA (0.16 μg/mL), and CIMRSA XIII containing 1% DMSO were grown in a shaker/incubator (Labnet International, Inc., USA) and centrifugated at 5,000 rpm at 4 °C for 18–20 h. TSB medium was removed, and the bacterial pellet was washed with 1 X PBS. Then, bacteria were resuspended in 1,000 μL of ice cold GAPDH Assay Buffer, homogenized (PowerLyzerTM, MO BIO Laboratories, Inc., USA) for 20 min at 2,500 rpm and sonicated in an Ultrasonic Cleaner (Fisher Brand FB11201, Germany) at 37 kHz (100 % power, 390 W) for 20 min in an ice bath. A final centrifugation at 10,000 × g for 20 min at 4 °C, allowed us to obtain bacterial lysates for the colorimetric assays involving G3PD. The effect of 1 on the activity of G3PD was determined as recommended by the manufacturer (Abcam, USA). After 30 min of incubation at 37 °C, the optical densities of orange-colored products were read at 450 nm. Experiments were performed in three biological replicates (N=3).
Synthesis of 3-, 6-, 8-, 9-, 10-, and 12-HDA
The syntheses of 3-, 6-, 8-, 9-, 10-, and 12-HDA were performed following the experimental procedures previously reported in the literature. The purity of the final products was confirmed by 1H-NMR and 13C-NMR spectroscopy as well as by the comparison of the MS spectral data previously reported (Arsequell et al., 1992; Camps et al., 1992; Carballeira et al., 2006; Fu et al., 2010; Ohashi et al., 2003; Tasdemir et al., 2010).
Synthesis of Alkythio-aFAs
Preparation of Alkyl (2-alkynyl) Sulfanes
A procedure described in the literature was modified to prepare alkyl (2-alkynyl) sulfanes (Vilchèze et al., 2016). The resulting crude was purified using vacuum distillation (120 °C, 10 min, 995 mBar).
2-Propynyl-1-(dodecyl)sulfane (15a)
Sulfane 15a (0.5 g, 2.1 mmol) was obtained as a yellowish oil in 90 % yield. IR (neat) vmax: 3312, 2921, 2852, 1463, 1231, 719, 630, 459 cm−1; 1H-NMR (300MHz, CDCl3) δ 3.24–3.23 (2H, d, J = 2.6 Hz), 2.70–2.65 (2H, t, J = 7.3 Hz), 2.22–2.21 (1H, t, J = 2.6 Hz), 1.66–1.57 (2H, m), 1.42–1.26 (18H, s), 0.90–0.86 (3H, t, J = 6.5 Hz); 13C NMR (75 MHz, CDCl3) δ 80.35 (s), 70.86 (s), 32.06 (t), 31.82 (t), 29.80 (t), 29.77 (t), 29.74 (t), 29.66 (t), 29.49 (t), 29.35 (t), 29.12 (t), 28.99 (t), 22.83 (t), 19.32 (t), 14.29 (q). GC/MS (70 eV) m/z (relative intensity): 240 (M+, 0.01), 201 (100), 171 (0.01), 159 (0.01), 145 (0.01), 127 (1), 111 (4), 97 (10), 83 (13), 69 (19), 55 (21).
3-Butynyl-1-(undecyl)sulfane (15b)
Compound 15b (0.6 g, 2.5 mmol) was obtained as a yellowish oil in a 91 % yield. IR (neat) vmax: 3312, 2921, 2852, 1463, 1377, 1223, 721, 630 cm−1; 1H-NMR (300MHz, CDCl3) δ 2.72–2.66 (2H, t, J = 7.7 Hz), 2.58–2.53 (2H, t, J = 7.3 Hz), 2.50–2.44 (3H, td, J = 7.4 Hz, J = 2.6 Hz), 2.03–2.01 (1H, t, J = 2.6 Hz), 1.61–1.26 (18H, m), 0.90–0.86 (3H, t, J = 6.4 Hz); 13C NMR (75 MHz, CDCl3) δ 82.5 (s), 69.41 (d), 32.37 (t), 30.04 (t), 29.78 (t), 29.73 (t), 29.71 (t), 29.65 (t), 29.46 (t), 29.37 (t), 29.02 (t), 22.82 (t), 20.08 (t), 14.23 (q). GC/MS (70 eV) m/z (relative intensity): 240 (M+, 2), 225 (3), 211 (6), 201 (25), 187 (5), 169 (3), 155 (5), 141 (4), 128 (2), 114 (7), 97 (21), 86 (100), 69 (32), 55 (42).
Synthesis of 4-(Dodecylthio)-2-butynoic acid (4S-2-HDA) and 5-(Undecylthio)-2-pentynoic acid (5S-2-HDA)
A procedure described in the literature was modified to prepare both 4S-2-HDA and 5S-2-HDA (Vilchèze et al., 2016). Once the desired products were obtained, the reaction crude was washed with a 1M NaOH (2 × 10 mL), extracted with DCM (2 × 15 mL), and the aqueous phase acidified using 1M HCl (10 mL). The acidified aqueous solution was extracted using diethyl ether (4 × 15 mL), and then the organic phase was dried over MgSO4 and filtered. The organic phase was removed in vacuo and the obtained brownish tar was purified using silica gel column chromatography eluting either 4S-2-HDA or 5S-2-HDA with hexane/ether (6:4).
4S-2-HDA (16a)
Compound 16a (0.5 g, 1.8 mmol) was obtained as a yellow oil in an 86% yield (Vilchèze et al., 2016). IR (neat) vmax: 2921, 2852, 2237, 1704, 1463, 1226, 907, 733 cm−1; 1H-NMR (300MHz, CDCl3) δ 3.35 (2H, s), 2.70–2.67 (2H, t, J = 7.3 Hz), 1.64–1.26 (20H, m), 0.90 (3H, t, J = 6.8 Hz); 13C NMR (75 MHz, CDCl3) δ 157.19 (s), 86.60 (s), 74.31 (s), 32.18 (t), 32.06 (t), 29.79 (t), 29.77 (t), 29.73 (t), 29.64 (t), 29.49 (t), 29.33 (t), 29.01 (t), 28.88 (t), 22.83 (t), 19.23 (t), 14.26 (s).
5S-2-HDA (16b)
Compound 16b (0.5 g, 1.8 mmol) was obtained as a white solid in a 76% yield. M. p.: 62–64 ˚C; IR (neat) vmax: 3310, 2955–2857, 2236, 1686, 1461, 1379, 1226, 908, 730, 633 cm−1; 1H-NMR (300MHz, CDCl3) δ 2.78–2.61 (4H, m), 2.58–2.53 (2H, t, J = 7.3 Hz), 1.61–1.26 (18H, m), 0.90–0.86 (3H, t = 6.4 Hz); 13C NMR (75 MHz, CDCl3) δ 158.87 (s), 90.00 (s), 73.42 (s), 32.48 (t), 32.05 (t), 29.88 (t), 29.74 (t), 29.73 (t), 29.71 (t), 29.66 (t), 29.47 (t), 29.36 (t), 28.99 (t), 22.83 (t), 20.52 (t), 14.25 (q).
Determination of the Critical Micelle Concentration (CMC)
The CMC of all the aFAs tested in this study were determined as described before (Sanabria-Rios et al., 2014).
Statistical analysis
The GI50 values were determined by plotting dose-response curves. All dose-response curves were carried out by using GraphPad Prism® (v 6.01) biostatistics software (San Diego, CA). The GI50 was defined as the concentration in which an aFA inhibits 50% of the bacterial growth.
Results
Synthesis of aFAs
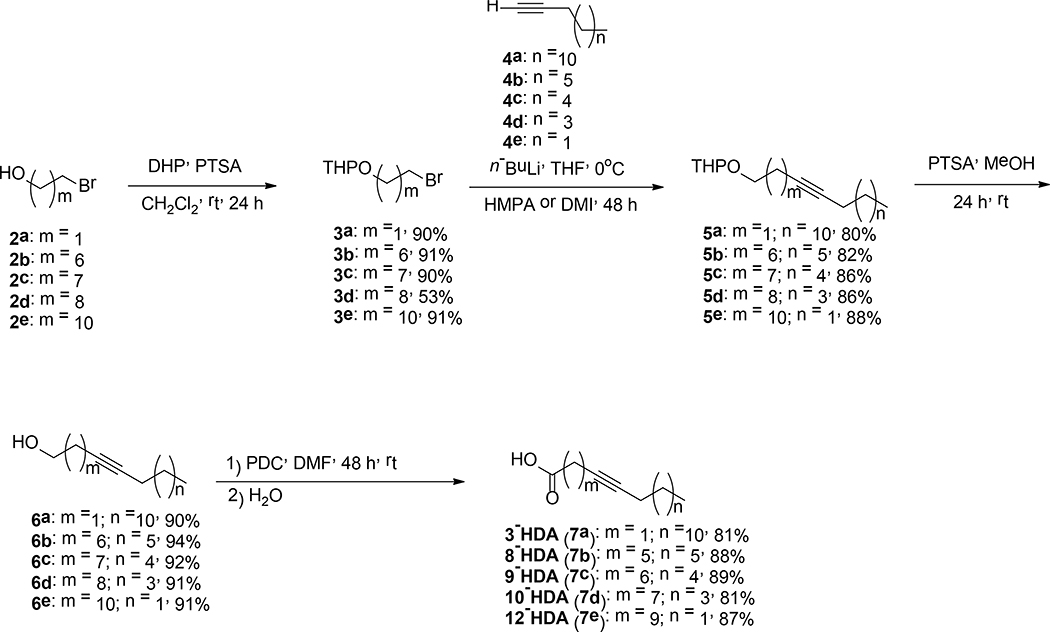
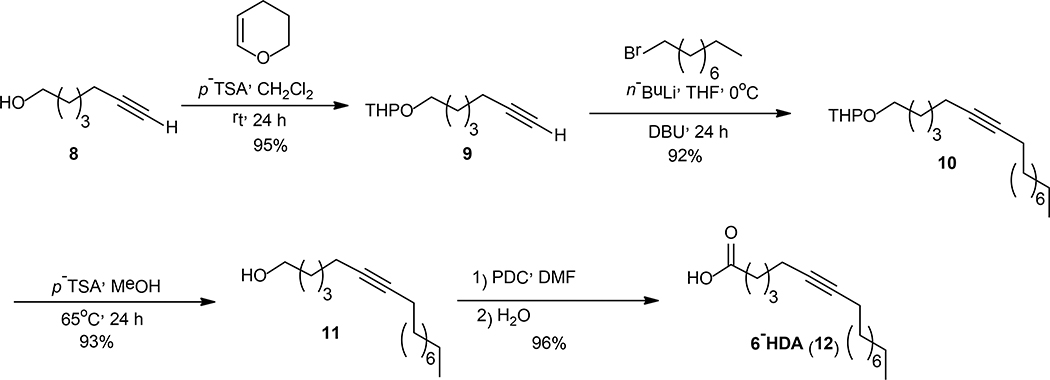
In this study a series of aFAs were synthesized as shown in Schemes 1, 2, and 3. In Scheme 1, the syntheses of 3-, 8-, 9-, 10-, and 12-HDA are displayed. These four-step syntheses started with the protection of 2 with 3,4-dihydro-2H-pyran (DHP) in dichloromethane (DCM) followed by an acetylenic coupling reaction using n-BuLi in dry tetrahydrofuran (THF) combined with either hexamethylphosphoramide (HMPA) or 1,3-dimethyl-2-imidazolidinone (DMI) to yield compounds 5a-e in 80–88% yields. The resulting tetrahydropyranyl protected alkynols 5a-e, were treated with catalytic amounts of p-toluenesulfonic acid (PTSA) in DCM at rt to afford 6a-e in 90–94% yields. Alkynols 6a-e were oxidized using PDC in DMF to produce the aFAs 7a-e in 81–89% yields. Compounds 7a-d were obtained in 34–63% overall yields. The synthesis displayed in Scheme 2, is different when compared to those displayed in Scheme 1. The difference consists in the protection of alkynol 8 and the alkylation of 9 to afford 10 in 93% yield. For the four synthetic steps, acid 12 was obtained in 78% overall yield.
Scheme 1.
Synthesis of 3– 8–, 9–, 10–, and 12-HDA.
Scheme 2.
Synthesis of 6-HDA.
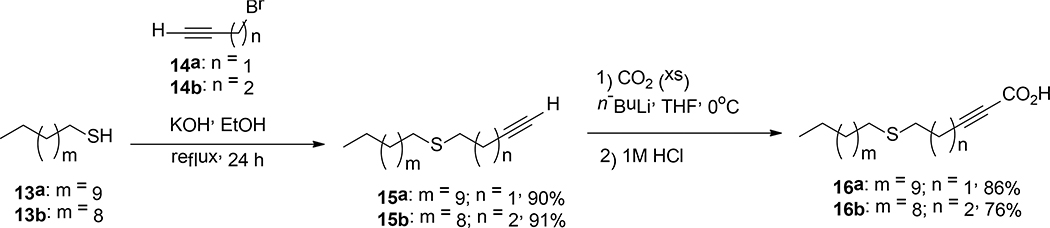
Scheme 3.
Synthesis of 4S-2-HDA (16a) and 5S-2-HDA (16b).
In a study performed by Vilchèze et al., it was reported for the first time, the synthesis of 16a, an aFA that contains a sulfur atom at C-4 in its carbon chain (Vilchèze et al., 2016). They demonstrated that 16a had better bactericidal activity than 1 against Mycobacterium bovis BCG (Vilchèze et al., 2016), which inspired us to synthesize compound 16a and its isomer 16b to determine whether these compounds displayed enhanced inhibitory activity against S. aureus. The synthetic strategy towards compounds 16a and 16b is displayed in Scheme 3. This two-step synthesis started with an acid-base reaction between thiols 13a-b and KOH to form sulfur nucleophiles, which were reacted with the bromoalkynes 14a-b via a SN2-like reaction to afford compounds 15a-b in 90–91% yields. Finally, aFAs 16a-b were obtained in 76–86% yield through the reaction between 15a-b and n-BuLi in THF and subsequent quenching of the lithium acetylide with carbon dioxide followed by an acidic workup with 1M HCl.
Antibacterial Activity of aFAs against CIMRSA Strains
Before starting biological studies involving CIMRSA, these bacteria were characterized using three microbiological techniques; the mannitol, coagulase, and non-staining KOH tests (Sanabria-Ríos et al., 2014, 2015). In both mannitol and coagulase tests, all CIMRSA strains used in this study were positive to the above-mentioned tests. In the case of the non-staining KOH test, all CIMRSA strains were not able to form a gel-like suspension, which agrees with the coagulase and mannitol tests.
Once the bacterial characterization tests were performed, microdilution susceptibility tests were carried out as described before (Carballeira et al., 2017; Sanabria-Rios et al., 2014; Sanabria-Rios et al. 2015). Table 1 shows the antibacterial activity of aFAs against MRSA ATCC 43300 and three CIMRSA strains. Results reveal that the presence of a triple bond is key for the antibacterial activity of these FA, since palmitic acid did not display antibacterial activity towards neither Gram-positive nor Gram-negative bacteria. However, it can be appreciated that the position of the triple bond also played an important role in the antibacterial activity of these acids. For example, it can be noted that the triple bond at C-9 in 9-HDA decreases 8- to 16-fold the antibacterial activity as compared to where the triple bond is at C-2 in acids such as 2-HDA. A similar trend was observed when 8-HDA was compared to 2-HDA. Also, it can be noted that the replacement of a carbon atom by a sulfur atom at either C-4 or C-5 in a 16-carbon length chain significantly decreased the antibacterial activity of 4S-2-HDA and 5S-2-HDA. Therefore, the following valuable information can be extracted from Table 1: i) presence of a triple bond in a C16-FA increases the antibacterial activity against MRSA, ii) a triple bond at either C-6, C-8, C-9, C-10, or C-12 did not significantly increase the antibacterial activity of the aFAs, and iii) the position of the triple bond at C-2 in aFA is pivotal for its antibacterial activity.
Table 1.
Antibacterial activity of HDA analogs against multidrug-resistant bacteria.a
| Compounds | MIC/GI50 ± SEM, μg/mL | CMC, μg/mL | |||||
|---|---|---|---|---|---|---|---|
| Bacterial Strains | |||||||
| S. aureus ATCC 29213 | MRSA ATCC 43300 | CIMRSA I | CIMRSA II | CIMRSA IX | E. coli ATCC 25922 | ||
| 6-HAD | 125/75.7 ± 0.3 | 250/168.7 ± 1.1 | 125/68.1 ± 1.4 | 62.5/32.9 ± 0.5 | 250/162.9 ± 6.5 | >1,000 | 100–300 |
| 8-HAD | 125/76.5 ± 0.7 | 125/73.2 ± 0.2 | 125/91.0 ± 2.7 | 62.5/43.3 ± 2.3 | 62.5/47.6 ± 1.1 | >1,000 | 100–300 |
| 9-HAD | 62.5/39.7 ± 1.8 | 62.5/31.7 ± 4.9 | 125/69.3 ± 2.3 | 62.5/29.6 ± 0.7 | 62.5/30.2 ± 1.4 | >1,000 | 100–300 |
| 10-HAD | 125/76.6 ± 1.5 | 125/70.2 ± 3.5 | 250/130.8 ± 1.1 | 250/104.6 ± 4.1 | 250/ 127.6 ± 2.4 | >1,000 | 100–300 |
| 12-HAD | 62.5/39.0 ± 0.7 | 250/178.6 ± 0.7 | 250/130.9 ± 4.7 | 1,000/522.8 ± 5.8 | 500/264.5 ± 2.0 | >1,000 | 100–300 |
| 4S-2-HAD | 1,000/514.4 ± 8.9 | 250/118.9 ± 9.6 | 1,000/509.2 ± 4.8 | 250/119.25 ± 0.5 | 250/ 121.6 ± 3.0 | >1,000 | 50–70 |
| 5S-2-HAD | >1,000 | 250/139.3 ± 5.6 | >1,000 | >1,000 | >1,000 | >1,000 | 50–70 |
| 2-HAD | 7.8/ 3.3 ± 0.3 | 7.8/5.1 ± 0.1 | 7.8/4.2 ± 0.1 | 7.8/5.0 ± 0.5 | 7.8/5.1 ± 0.2 | >1,000 | 90–100 |
| Palmitic Acid | >1,000 | >1,000 | >1,000 | >1,000 | >1,000 | >1,000 | >1,0000 |
| Ciprob | 0.14 ± 0.01 | 0.20 ± 0.02 | 15.6/9.1 ± 0.2 | 3.9/2.22 ± 0.02 | 7.8/5.3 ± 0.5 | <0.03 | N.D.c |
Experiments were performed in triplicate (N =3). GI50 values were obtained from dose-response curves.
Ciprofloxacin (Cipro) was used as a positive control and palmitic acid was used as negative control.
N.D. = Not determined.
Antibacterial Effect of 3-HDA in CIMRSA Strains
In a mechanistic study performed by Morbidoni et al., it was demonstrated that toxic concentrations of 1 resulted in the accumulation of 3-HDA, which inhibits the FA degradation in M. smegmatis (Morbidoni et al., 2006). Although the metabolism of FAs in M. smegmatis does not necessarily follow the same pathway as in S. aureus, the similarity of enzymes participating in their core fatty acid biosynthesis suggests that a similar metabolism of FA could be taking place in S. aureus. Therefore, the synthesis of 3-HDA was carried out to determine if the antibacterial activity of the above-mentioned FA is like 1 in S. aureus. It was hypothesized that exogenous 3-HDA will exhibit a similar inhibitory activity as 1 in the case that 1 isomerizes into 3-HDA in the S. aureus FA metabolic pathway. Results from this study are shown in Table 2. It can be noted that 3-HDA displayed decreased cytotoxicity towards S. aureus as compared to 1. It was also observed that 3-HDA is 32- to 255-fold less active than 1 in all S. aureus strains displayed in Table 2.
Table 2.
Comparison of the antibacterial activities of both 3-HDA and 1 in six CIMRSA strainsa
| Compounds | MIC/GI50 ± SEM, μg/mL | |||||||
|---|---|---|---|---|---|---|---|---|
| Bacterial Strains | ||||||||
| S. aureus ATCC 29213b | CIMRSA XI | CIMRSA XII | CIMRSA XIII | CIMRSA XIV | CIMRSA XV | CIMRSA XVI | E. coli ATCC 25922 b | |
| 1 | 7.8/ 3.3 ± 0.3 | 0.49/0.30 ± 0.03 | 0.49/0.25 ± 0.03 | 0.24/0.16 ± 0.02 | 0.49/0.34 ± 0.03 | 0.49/0.27 ± 0.01 | 0.49/0.30 ± 0.03 | >1,00 |
| 3-HDA (7a) | 250/107.8± 5.4 | 62.5/43.4 ± 1.1 | 125/89.2 ± 0.9 | 500/200.6 ± 1.1 | 62.5/35.1 ± 3.5 | 15.6/8.7 ± 0.9 | 62.5/37.9 ± 3.8 | >1,000 |
Experiments were performed in triplicate (N =3). GI50 values were obtained from dose-response curves.
Both S. aureus ATCC 29213 and E. coli ATCC 25922 were included in the experimental design for comparative purposes.
Antibacterial Effect of the Combination of 2-HDA and Cipro on the Growth of CIMRSA Strains
Since 1 displayed the best antibacterial activity among the HDA, this acid was combined with Cipro and tested against six CIMRSA strains. Results in Table 3 demonstrate that 1 enhanced the antibacterial activity of Cipro, but Cipro decreases the bioactivity of 1 against these CIMRSA strains. For example, when 1 was combined with Cipro and tested against CIMRSA XI, a 2-fold increase was observed in MIC values when compared with Cipro alone, while a 16-fold change was observed in MIC values when CIMRSA XI was treated with 1. In the case of CIMRSA XII, CIMRSA XIII, and CIMRSA XVI strains treated with the combination of 1 and Cipro, an 8- to 16-fold change was observed when compared with Cipro alone. However, when the MIC values of 1 in CIMRSA XII, CIMRSA XIII, and CIMRSA XVI strains were compared with those of Cipro, a 64-, 260-, and 16-fold change was observed, respectively. From all CIMRSA strains tested, CIMRSA XIV did not show any significant change in MIC values when this bacterial strain was treated with either Cipro or 1. Only a 2-fold change was observed when this strain was treated with the combination of Cipro and 1 when compared with Cipro alone. On the other hand, CIMRSA XV was susceptible to both 1 and the combination of Cipro and 1 since their MICs values were the same. For this MRSA strain, only a 16-fold change was observed when both 1 and a combination of 1 and Cipro were compared with Cipro alone. It can be appreciated as well that CIMRSA XII and CIMRSA XIII are the most resistant MRSA strains to Cipro, and 1 was the most effective treatment in inhibiting their growth.
Table 3.
Antibacterial activity of 1 and Cipro against Ciprofloxacin-resistant S. aureus strains.a
| Compounds | MIC/GI50 ± SEM, μg/mL | |||||
|---|---|---|---|---|---|---|
| Bacterial Strains | ||||||
| CIMRSA XI | CIMRSA XII | CIMRSA XIII | CIMRSA XIV | CIMRSA XV | CIMRSA XVI | |
| 2-HDA (1) | 0.49/0.30 ± 0.03 | 0.49/0.25 ± 0.03 | 0.24/0.16 ± 0.02 | 0.49/0.34 ± 0.03 | 0.49/0.27 ± 0.01 | 0.49/0.30 ± 0.03 |
| CIPROb | 7.8/5.7 ± 1.1 | 31.3/17.2 ± 1.0 | 62.5/30.4 ± 1.4 | 0.49/0.35 ± 0.04 | 7.8/4.7± 1.1 | 7.8/2.3 ± 1.1 |
| CIPRO/2-HDA (1) | 3.9/1.8 ± 0.2 | 3.9/2.1 ± 0.2 | 3.9/1.7 ± 0.2 | 0.98/0.64 ± 0.06 | 0.49/0.28 ± 0.03 | 0.98/0.60 ± 0.06 |
Experiments were performed in triplicate (N =3). GI50 values were obtained from dose-response curves.
According to Raviglione et al. Ciprofloxacin-resistant S. aureus strains show MIC values higher than 2 μg/mL (Raviglione et al., 1990).
Effect of Exogenous 1 on the Growth of CIMRSA XIII
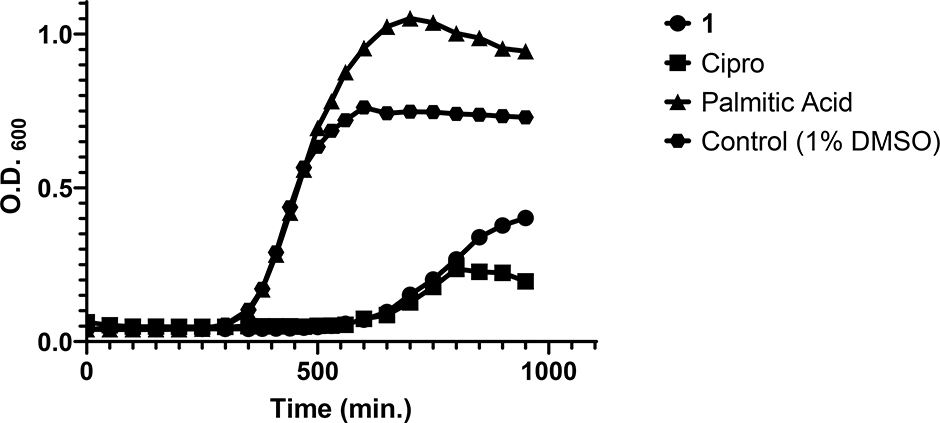
Because 1 displayed better cytotoxicity than Cipro in different S. aureus strains, kinetic growth assays were performed to determine whether CIMRSA XIII is adapted to metabolize 1 (Fig. 2). CIMRSA XIII was selected in this study, because this MRSA demonstrated to be the most resistant strain to Cipro (see Table 3). Results in Fig. 2 reveal that palmitic acid is not cytotoxic against CIMRSA XIII, since its growth curve is like the control that only contains 1% DMSO. The only difference between the palmitic acid curve and the control curve is that the former has higher O.D.600 values, due to the slight turbidity that was generated when palmitic acid was added to the TSB. In the case of 1 and Cipro, these compounds arrested the growth of CIMRSA XIII within 600 minutes of its addition. It can be appreciated from the 1 curve that 960 ng/mL of 1 in CIMRSA XIII was toxic when compared to the control, while Cipro displayed similar cytotoxicity to 1 but at 250 μg/mL. As observed in palmitic acid, 1 has a higher O.D.600 than Cipro due to the slight turbidity of 1 in TSB. Altogether, these results, recapitulated those findings obtained in Tables 1 and 3 and demonstrate once again the potency of 1 as an antibacterial agent in ciprofloxacin-resistant S. aureus.
Fig. 2.
CIMRSA XIII growth curve showing its log-phase in TSB when treated with 1 (960 ng/mL), Cipro (250 μg/mL), and palmitic acid (4 mg/mL). CIMRSA XIII treated with 1% DMSO was used as control. Experiments were performed in three biological replicates (N =3).
The Effect of 1 on the FA Profile of S. aureus
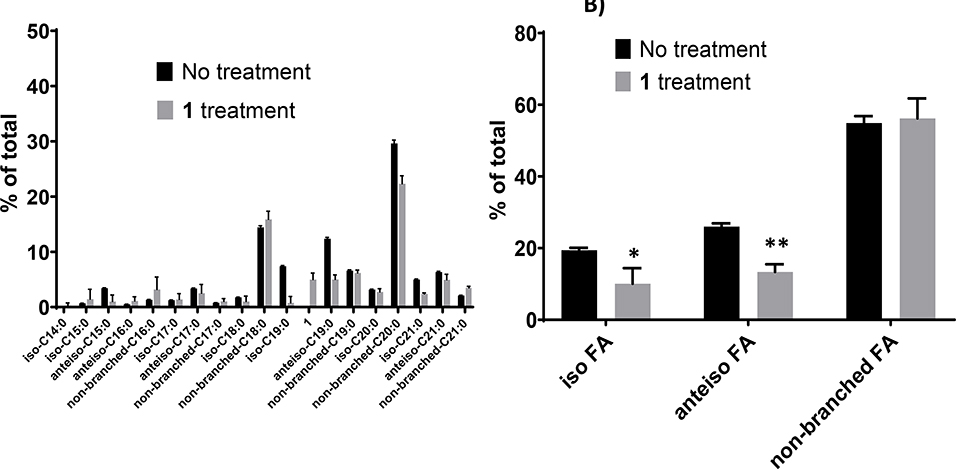
To further understand the mechanism of action of 1 in multidrug-resistant S. aureus, GC-MS experiments were performed aimed at identifying metabolites that might be generated from exogenous 1. Results from these experiments are displayed in Fig. 3. Results in Fig. 3A clearly show the presence of saturated branched FA consisting of both iso- and anteiso-branched FA. These findings are consistent with previous studies that have reported that the presence of iso- and anteiso-branched FA are common in Gram-positive bacteria, including S. aureus (Bisignano et al., 2019). One interesting finding that was obtained from our study is the high abundance of straight-chain FA such as n-18:0 and n-20:0. Literature explains that S. aureus growth in TSB led to reduction of branched FA and increase of straight-chain FA (Sen et al., 2016). The above-mentioned phenomenon could also be occurring in our study. In Fig. 3B, it can be observed that 1 decreased 2-fold the composition of both iso- and anteiso-branched FA in CIMRSA XIII, while straight chain FA in CIMRSA XIII did not change their composition after exposure to 1. Another aspect that needs to be highlighted is that S. aureus did not produce 3-HDA as a possible metabolite derived from the isomerization of 1, since GC-MS did not detect the presence of this product.
Fig. 3.
A) FA profiles of both non-treated and 1-treated CIMRSA XIII. CIMRSA XIII cells were grown at 37 °C for 18–20 h in TSB containing 0.16 μg/mL 1 (treated), or in its absence (untreated). FA were eluted, converted to methyl-esters and analyzed by GC-MS, as described in materials and methods. Bars represent the percentage of different FAs obtained from three separated experiments (mean ± SD). FA nomenclature indicates the type of branched FA, the number of carbons followed by the number of double bonds. Acid 1 refers to 2-hexadecynoic acid. B) Comparison of iso-, anteiso-, and straight-FA acids that are present in 1-treated- and non-treated-CIMRSA XIII. Histograms were constructed by classifying all branched FA and non-branched FA from three independent experiments (mean ± SD) that were previously identified based on their molecular ion, base peak, fragmentation pattern, retention time, and ECL values. Multiple t test analyses were performed using GraphPad Prism® v.6.01 and revealed statistical difference between iso-FA and anteiso-FA, since p = 0.0422 (*) and p = 0.0031 (**) were obtained.
Inhibitory Effect of aFAs on the Supercoiling Activity of S. aureus DNA Gyrase
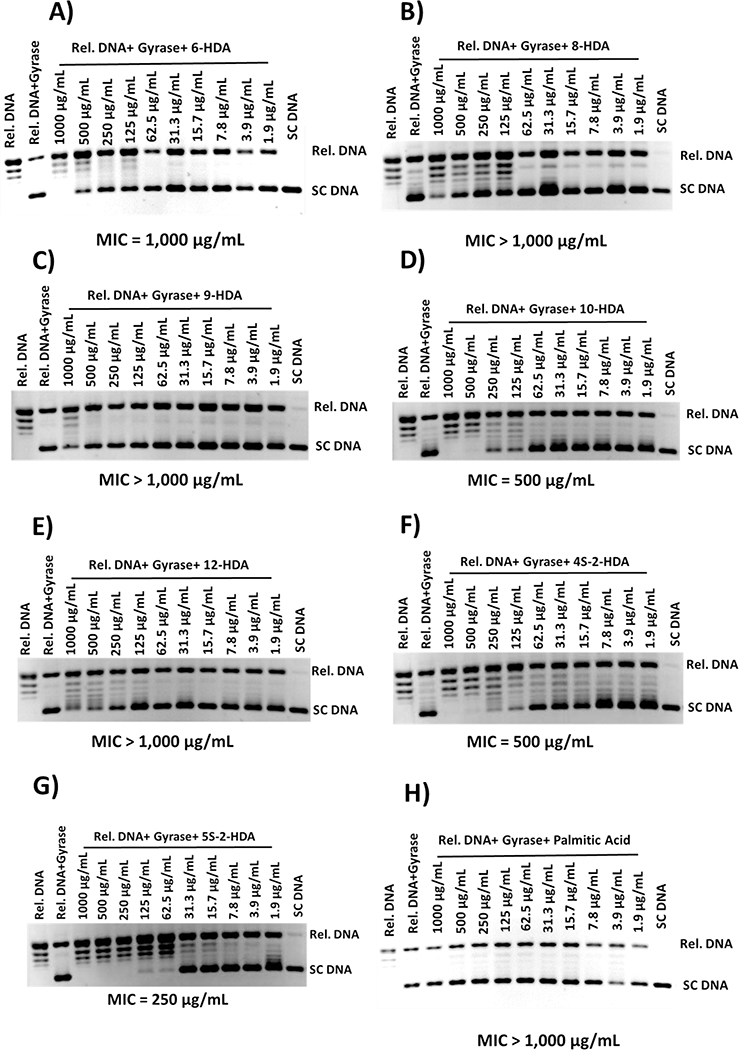
In a previous study, Sanabria-Ríos et al. demonstrated that 1 inhibited the supercoiling activity of S. aureus DNA gyrase suggesting that the inhibition of this enzyme can be implied in the mechanism of action of this aFA (Sanabria-Rios et al., 2015). When all aFAs in Table 1 were compared to 1, it was observed that the presence of a triple bond at C-6, C-8, C-9, C-10, or C-12 decreases the antibacterial activity as well as the presence of sulfur atoms at either C-4 or C-5 in the carbon chain. Results from S. aureus gyrase inhibitory tests displayed in Fig. 4 revealed that a triple bond at C-6, C-8, C-9, C-10, or C-12 or the presence of a sulfur atom at either C-4 or C-5 did not improve their inhibitory activity towards the above-mentioned enzyme, which supports the findings observed in Table 1. Only 4S-2-HDA and 5S-2-HDA displayed some inhibitory activity at MIC values of 500 μg/mL and 250 μg/mL, respectively, but these values are not better than 1 (MIC = 31 μg/mL) (Sanabria-Rios et al., 2015).
Fig. 4.
Ethidium bromide-stained agarose electrophoresis gels showing the inhibitory effect of HDA analogs on the supercoiling activity of S. aureus DNA gyrase. The gels shown above are representative gels from experiments that were carried out in triplicate (N=3). Reaction products were separated by electrophoresis in 1% agarose gels.
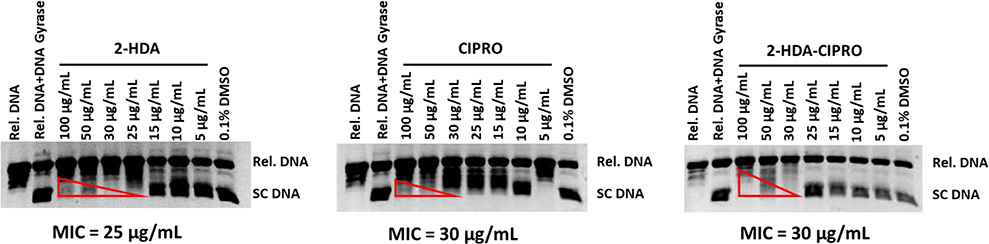
Given that 1 showed a better inhibitory activity against S. aureus DNA gyrase than the other aFAs, several S. aureus DNA gyrase inhibitory assays were performed aimed to corroborate the findings displayed in Table 3. In these assays, an equimolar stock solution containing both 1 and Cipro was prepared. Different reactions containing S. aureus DNA gyrase, relaxed pHOT1 plasmid DNA, various concentrations of either 1 in combination with Cipro, 1 alone, or Cipro alone were prepared to determine the effect of these treatments on the supercoiling activity of S. aureus DNA gyrase. Results in Fig. 5 show that 1 was able to inhibit the supercoiling activity of S. aureus DNA gyrase, since this compound displayed similar MIC values than Cipro, a well-known DNA gyrase inhibitor (LeBel, 1988). Also, it was observed that the equimolar combination of 1 and Cipro did not increase the DNA gyrase inhibitory activity of neither 1 nor Cipro, which is consistent with findings reported in Table 3.
Fig. 5.
Ethidium bromide-stained gel showing the effect of the equimolar combination of 1 and Cipro on the supercoiling activity of S. aureus DNA gyrase. The gels shown above are representative gels from experiments that were carried out in triplicate (N=3). Reaction products were separated by electrophoresis in 1% agarose gels.
Inhibitory Effect of 1 on the Activity of G3PD
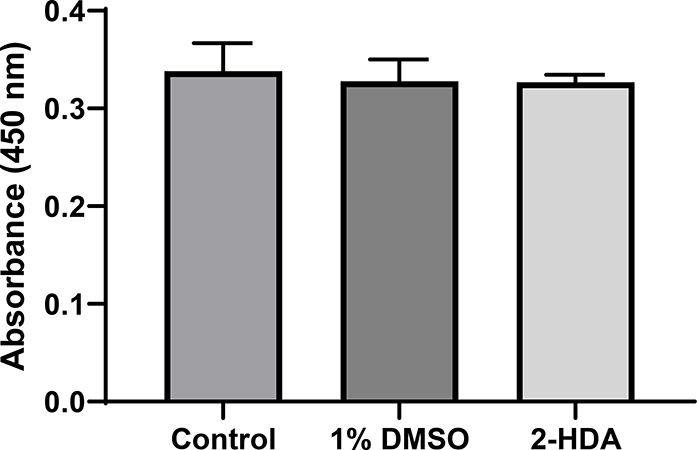
The participation of G3PD in the glycolytic pathway of S. aureus, is needed for the pathogen’s subsistence. G3PD is responsible for the conversion of glyceraldehyde-3-phosphate (G3P) into 1,3-bisphosphoglycerate (1,3-BPG) and it consumes inorganic phosphate to produce NADH (Mukherjee et al., 2010). Therefore, the study of the inhibitory effect of 1 on the activity of G3PD was performed to discard any physical effect of this FA in S. aureus. To do this, we used the Abcam G3PD Assay Kit™ to monitor the transformation of G3P into 1,3-BPG and intermediate, which reacts with a developer to form an orange-colored product that absorbs maximally at 450 nm. Results from this biochemical assay are shown in Fig. 6. It can be appreciated that CIMRSA XIII treated with 1% DMSO did not affect the activity of G3PD when compared to the control (CIMRSA XIII with no treatment). The same behavior is observed when 1 is compared to the control. In fact, one-way ANOVA did not detect any significant difference among means (i.e. control, 1% DMSO, and 1), which implies that 1 does not affect the glycolytic pathway of CIMRSA XIII.
Fig. 6.
GAPDH specific activity from lysates of CIMRSA XIII without treatment (control), CIMRSA XIII treated with 1% DMSO, and CIMRSA treated with 0.16 μg/mL of 1. Experiments were performed in three biological replicates (N =3). One-way ANOVA followed by Dunnett’s test were performed using GraphPad Prism V. 6.01. Non-statistical differences between mean values were obtained since p values higher than 0.05 were obtained. 2-HDA = 1.
Discussion
In a previous study, it was determined that 1 was effective in inhibiting the growth of Gram-positive bacteria including MRSA and CIMRSA strains when compared to other aFAs containing different carbon chain lengths as well as different functional groups (Sanabria-Rios et al. 2014). This study claims that the triple bond position at C-2, as well as the presence of a carboxylic acid moiety, are critical for the antibacterial activity of 1. Although the findings above are important for understanding the antibacterial properties of aFAs, further studies were needed to determine those characteristics that are fundamental in aFAs to display antibacterial activity. One of these characteristics that needed exploration was the position of a triple bond in a C16 aFA. In the present study, C16 aFAs containing triple bonds at either C-3, C-8, C-9, C-10, or C-12 were synthesized. These unsaturation positions were selected because several reports have claimed that they have an important role in the biological activity of aFAs against cancer cells (Ohashi et al., 2003), mycobacteria (Morbidoni et al.), fungi (Carballeira et al., 2006), parasites (Tasdemir et al., 2010), and enzymes derived from plants (Arsequell et al., 1992). Results displayed in Table 1 once again confirm that a triple bond at C-2 in a C16 aFA is fundamental for the antibacterial activity of HDAs towards Gram-positive bacteria, including MRSA and CIMRSA strains. These findings are in agreement with other studies that have shown that 1 was the most effective aFA in inhibiting the biological activity of Plasmodial FAS-II enzymes (Tasdemir et al., 2010) or inhibiting the plasmid-mediated horizontal gene transfer in E. coli (Getino et al., 2015).
Results in Table 1 also demonstrate that the replacement of a carbon atom by a sulfur atom at either C-4 or C-5 in a C16 aFA decreased the antibacterial activity of these aFAs when compared to 1. These results are quite different to those findings reported by Vilchèze et al., where they showed that 16a has a better anti-mycobacterial activity towards M. bovis BCG (Vilchèze et al., 2016). According to Morbidoni and collaborators, the antimycobacterial activity displayed by 1 towards M. smegmatis is because 1 acts as a prodrug that is metabolized into 3-HDA and 3-ketohexadecanoic acid, which inhibits fatty acid degradation and fatty acid biosynthesis, respectively (Morbidoni et al., 2006). With respect to S. aureus, some unsaturated fatty acids (e.g. linoleic acid) display antibacterial activity against the above mentioned bacteria through the inhibition of the bacterial enoyl-acyl carrier protein reductase (FabI), an essential component of bacterial fatty acid synthesis (Parsons et al., 2011; Zheng et al., 2005). However, the results in Table 1 support the hypothesis that other resistance-related mechanisms such as multidrug pump efflux (Jang, 2016) and detoxification pathways (Newton et al., 2012) may be involved in the lack of activity of the HDA analogs, including 16a and 16b. Results also show that 9-HDA displayed moderate inhibitory activity against MRSA (ATCC 43300) and CIMRSA strains. This is not surprising since 9-HDA was also antiparasitic against Plasmodium falciparum blood stage (BS) and P. yoelii liver stage (LS) parasites (Tasdemir et al., 2010) as well as against Cryptococcus neoformans (Carballeira et al., 2006), implying that the triple bond position at C-9 has an important role in its antimicrobial properties. Moreover, 9-HDA is inhibitory towards both PfFabI and PfFabZ enzymes, which have an important function in the Plasmodium FAS II system (Carballeira, 2008; Tasdemir et al., 2010).
Another aspect that was addressed in this study is the ability of aFAs to form micelles. This is important because a possible relationship between the CMC and the biological activities of FA has been reported in the past (Courtney et al., 1986). Table 1 shows that the decreased antibacterial activity of HDA analogs depend on their ability to form micelles. Only one exception was observed, where the antibacterial activity of 1 depends on its free monomeric form in aqueous media, which is in agreement with previous studies (Sanabria-Rios et al., 2014).
The cytotoxic effect of synthetic 3-HDA on the growth of CIMRSA was also assessed using broth dilution susceptibility tests to gain more knowledge regarding the mode of action of 1. It was evidenced that 1 is metabolized into toxic 3-HDA to inhibit different lipid synthesis pathways in M. smegmatis (Morbidoni et al., 2006). However, results from susceptibility tests suggest that isomerization of 1 into 3-HDA is not taking place here since treatment of seven S. aureus strains with 3-HDA resulted to be 32- to 255-fold less active than 1 (see Table 2).
Kinetic growth assays were conducted to determine whether CIMRSA XIII are adapted to metabolize 1 (Fig. 2). Results from this study suggests that 1 is a potent growth inhibitor displaying a similar growth pattern as palmitoleate (16:1 Δ9), a FA that is cytotoxic against S. aureus by rapid membrane depolarization, disruption of all major branches of macromolecular synthesis, and the release of key proteins in the medium (Parsons et al., 2012). The findings displayed in Fig. 2 support the idea that 1 can be disrupting and releasing important proteins that are necessary for the bacterial subsistence.
The GC-MS experiments performed were aimed to determine the effect of 1 on the FA profile of S. aureus (Fig. 3). Results from Fig. 3A were interesting since GC-MS did not detect the presence of 3-HDA after exposure of CIMRSA XIII to 1 validating those bioactivity results displayed in Table 2. The fact that 3-HDA is not present in 1-treated CIMRSA XIII strain is indicative that isomerization of 1 into 3-HDA is not occurring in S. aureus. Another relevant information that can be interpreted is that the percentage of total branched FAs 2-fold decreased after exposure of S. aureus to 1 (Fig. 3B). These results may imply that 1 is affecting the synthetic pathways of both iso- and anteiso-branched FAs, by inhibiting the activity of key enzymes such as branched-chain aminotransferases (BATs), branched-chain α-ketoacid dehydrogenases (BKDs), NAD(P)H-dependent trans-2-enoyl-ACP reductases FabI and FabG, FabF condensation enzymes, and dehydrases FabZ (Heath et al., 2004; Madsen et al., 2002; Schiebel et al., 2012).
Since the presence of a triple bond at C-3, C-6, C-8, C-9, C-10, or C-12 did not increase the antibacterial activity of aFAs in S. aureus when compared to 1 (Table 1) coupled to the fact that previous studies suggest that 1 is able to inhibit the supercoiling activity of S. aureus DNA gyrase (Sanabria-Rios et al., 2015), biochemical assays aimed at determining if the aFAs displayed in Table 1 are able to inhibit the activity of S. aureus DNA gyrase was carried out. Results in Fig. 4 demonstrate that none of the aFAs tested showed a better anti-topoisomerase activity than 1. These results confirm the findings displayed in Table 1 and they strongly support the hypothesis that DNA gyrase can be involved in the mechanism of the antibacterial activity of 1 towards Gram-positive bacteria. Moreover, the results displayed in Fig. 4 demonstrate that the triple bond position at C-2 in a C16-aFA is key for the inhibitory activity against DNA gyrase.
The potential of 1 as an antibacterial agent was also assessed in six CIMRSA strains, where five of these strains were resistant to Cipro (Table 3), since MIC values higher than 2 μg/mL were determined (Raviglione et al., 1990). Results in Table 3 are revealing because: 1) 2-HDA displayed better antibacterial activity than Cipro and 2) the antibacterial activity of the combination of 1 and Cipro is not better than 1 alone towards these CIMRSA strains. In terms of cytotoxicity, it was previously reported that 1 displayed an IC50 of 57.2 μg/mL in peripheral blood mononuclear cells (PBMC) (Sanabria-Ríos et al., 2014), which allowed to determine that 1 is 168- to 356-fold more selective towards the CIMRSA strains displayed in Table 3 than PBMC. Because Cipro is a well-known inhibitor of S. aureus DNA gyrase (LeBel, 1988) and five of the six CIMRSA strains that were tested in this study resulted to be resistant to Cipro and susceptible to 1, it is suggested that the inhibition of the supercoiling activity of S. aureus DNA gyrase is another viable mechanism that can be involved in the antibacterial activity of 1. In fact, results in Fig. 5 clearly demonstrate that 1 has similar inhibitory effect on the supercoiling activity of S. aureus DNA gyrase than Cipro alone. However, when both compounds (1 and Cipro) were combined in equimolar concentrations, no improvement in their inhibitory activity against the above-mentioned enzyme was observed. The results in Fig. 5 also show that both 1 and Cipro inhibit the activity of S. aureus DNA gyrase, but the way in which these compounds inhibit the enzyme are not necessarily the same. In the case of Cipro, it is known that this compound inhibits the supercoiling activity of DNA gyrase by the formation of a fluoroquinolone-enzyme-DNA complex in which the DNA is broken (Fàbrega et al., 2009; Mustaev et al., 2014). This “cleaved complex” is characterized by the presence of DNA breaks, which have been crystallized and found to have the fluoroquinolone C-7 ring system facing the GyrB subunit (Mustaev et al., 2014). In the case of 1, it was demonstrated that this compound displayed COIN activity in E. coli by the inhibition of TrwD ATPase (Garcia-Cazorla et al., 2018; Getino et al., 2015; Ripoll-Rozada et al., 2016). It was shown that 1 binds to the N-terminal domain (NTD) and the linker region of TrwD ATPase resulting in the restriction of the movement of the NTD over the catalytic domain (CTD), which would result in a reduction of the TrwD ATPase activity (Ripoll-Rozada et al., 2016). These findings are revealing because S. aureus DNA gyrase contains a GyrB subunit, an enzymatic region that has ATPase activity (Eakin et al., 2012; Fàbrega et al., 2009), which could explain the observed inhibitory activity of 1 towards the S. aureus DNA gyrase. In addition, it was determined that 1 is not inhibitory against G3PD implying that 1 does not affect the glycolytic pathway of S. aureus (Fig. 6). The fact that 1 is not active in G3PD, discard physical effects of the above-mentioned FA and provides an appropriated control system to this study.
In conclusion, we further investigated how the triple bond position and the presence of a sulfur atom in a C16 aFA influence the antibacterial activity of aFAs. Results from this study confirm that the presence of a triple bond at C-2 is pivotal for the observed antibacterial activity against Gram-positive bacteria, including clinical isolates of methicillin-resistant S. aureus that are also resistant to Cipro. Moreover, it was outlined, for the first time, that the inhibition of S. aureus DNA gyrase may be implicated in the antibacterial activity of 1. Results from this study also demonstrated that the decreased antibacterial activity of HDAs depend on their ability to form micelles in aqueous media, while the antibacterial activity of 1 is not mediated by micellar aggregates formation. This study will surely impact the understanding regarding the antibacterial properties of aFAs and highlight 1 as a novel agent against nosocomial bacterial infections.
Acknowledgements
This project was supported by the National Center for Research Resources and the National Institute of General Medical Sciences of the National Institutes of Health through Grant Number P20GM103475. Dr. D.J. Sanabria-Ríos would like to thank the IAUPR-MC Seed Money for sponsoring this project and the Metabolomics Research Core funded through the PR-INBRE program (NIGMS P20 GM103475) for the use of the GC-MS in the lipidomic component of this project. Special thanks to the Materials Characterization Center (MCC) for the use of NMR and Ms. Denisse Alequín-Torres, Ms. Aleni De Jesús, and Mr. José R. León for assisting in the biological, synthetic, and chemical characterization part of this study. Also, Dr. Sanabria-Ríos thanks Dr. Charles O. Rock (St. Jude Children’s Research Hospital) for his advice in the design of kinetic growth experiments that were carried out in this study. Dr. Christian Morales-Guzmán, Mr. Joseph Mooney, and Mr. Tomás Pereles-De-León acknowledge the support of the PR-INBRE program for the graduate and undergraduate research fellowships provided. Dr. Christian Morales-Guzmán also thanks to the RISE Program (Grant No. 5R25GM061151–17) for the graduate fellowship provided.
Abbreviations
- aFA
Alkynoic Fatty Acid
- ATCC
American Type Culture Collection
- CIMRSA
Clinical Isolates of Methicillin-Resistant Staphylococcus Aureus
- COIN
Conjugation Inhibition
- DMSO
Dimethyl Sulfoxide
- ECL
Equivalent Chain Length
- FAME
Fatty Acid Methyl Ester
- HAD
Hexadecynoic Acid
- MIC
Minimal Inhibitory Concentration
- MRSA
Methicillin-Resistant Staphylococcus Aureus
- NMR
Nuclear Magnetic Resonance
- PBMC
Peripheral Blood Mononuclear Cells
- SEM
Standard Error of the Mean
References
- Arsequell G, Fabriás G, Gosalbo L, & Camps F (1992). Synthesis of inhibitors of a delta-11 desaturase in the moth Spodoptera littoralis. Chem. Phys. Lipids, 63(1), 149–158. [Google Scholar]
- Bisignano Carlo, Ginestra Giovanna, Smeriglio Antonella, Camera La, Erminia Crisafi, Giuseppe Franchina, Tranchida Flavio A., Peter Q, Alibrandi Angela, Trombetta Domenico, Mondello Luigi, &Mandalari Giuseppina. (2019). Study of the Lipid Profile of ATCC and Clinical Strains of Staphylococcus aureus in Relation to Their Antibiotic Resistance. Molecules, 24(7), 1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campion JJ, McNamara PJ, & Evans ME (2004). Evolution of ciprofloxacin-resistant Staphylococcus aureus in in vitro pharmacokinetic environments. Antimicrob. Agents Chemother, 48(12), 4733–4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps F, Hospital S, Rosell G, Delgado A, & Guerrero A (1992). Synthesis of biosynthetic inhibitors of the sex pheromone of Spodoptera littoralis. Part II: acetylenic and cyclopropane fatty acids. Chem. Phys. Lipids, 61(2), 157–167. [DOI] [PubMed] [Google Scholar]
- Carballeira NM (2008). New advances in fatty acids as antimalarial, antimycobacterial and antifungal agents. Prog. Lipid Res, 47, 50–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballeira NM, Betancourt JE, Orellano EA, & González FA (2002). Total synthesis and biological evaluation of (5Z,9Z)-5,9-hexadecadienoic acid, an inhibitor of human topoisomerase I. J. Nat. Prod, 65(11), 1715–1718. [DOI] [PubMed] [Google Scholar]
- Carballeira NM, Emiliano A, Hernández-Alonso N, & González FA (1998). Facile total synthesis and antimicrobial activity of the marine fatty acids (Z)-2-methoxy-5-hexadecenoic acid and (Z)-2-methoxy-6-hexadecenoic acid. J. Nat. Prod, 61(12), 1543–1546. [DOI] [PubMed] [Google Scholar]
- Carballeira NM, Montano N, Morales C, Mooney J, Torres X, Diaz D, & Sanabria-Rios DJ (2017). 2-Methoxylated FA display unusual antibacterial activity towards clinical isolates of methicillin-resistant Staphylococcus aureus (CIMRSA) and Escherichia coli. Lipids, 52(6), 535–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballeira NM, Sanabria D, Cruz C, Parang K, Wan B, & Franzblau S (2006). 2,6-Hexadecadiynoic acid and 2,6-nonadecadiynoic acid: Novel synthesized acetylenic fatty acids as potent antifungal agents. Lipids, 41(5), 507–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC (2013). Antibiotic resistance threats in the United States, 2013. Atlanta, GA: Centers for Disease Control and Prevention [Google Scholar]
- Courtney HS, Simpson WA, & Beachey EH (1986). Relationship of critical micelle concentrations of bacterial lipoteichoic acids to biological activities. Infection and Immunity, 51(2), 414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eakin AE, Green O, Hales N, Walkup GK, Bist S, Singh A, Mullen G, Bryant J, Embrey K, Gao N, Breeze A, Timms D, Andrews B, Uria-Nickelsen M, Demeritt J, Loch JT 3rd, Hull K, Blodgett A, Illingworth RN, Prince B, Boriack-Sjodin PA, Hauck S, MacPherson LJ, Ni H, & Sherer B (2012). Pyrrolamide DNA gyrase inhibitors: fragment-based nuclear magnetic resonance screening to identify antibacterial agents. Antimicrob. Agents Chemother, 56(3), 1240–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fàbrega A, Madurga S, Giralt E, & Vila J (2009). Mechanism of action of and resistance to quinolones. Microb. Biotechnol, 2(1), 40–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Weng Y, Hong W, & Zhang Q (2010). Efficient Synthesis of Unsaturated 1-Monoacyl Glycerols for in meso Crystallization of Membrane Proteins. Synlett : accounts and rapid communications in synthetic organic chemistry, 2011(6), 809–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Cazorla Y, Getino M, Sanabria-Rios DJ, Carballeira NM, de la Cruz F, Arechaga I, & Cabezon E (2018). Conjugation inhibitors compete with palmitic acid for binding to the conjugative traffic ATPase TrwD, providing a mechanism to inhibit bacterial conjugation. J. Biol. Chem, 293(43), 16923–16930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getino M, Sanabria-Rios DJ, Fernandez-Lopez R, Campos-Gomez J, Sanchez-Lopez JM, Fernandez A, Carballeira NM, & de la Cruz F (2015). Synthetic fatty acids prevent pasmid-mediated horizontal gene transfer. mBio, 6(5), e01032–01015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath RJ, & Rock CO (2004). Fatty acid biosynthesis as a target for novel antibacterials. Curr. Opin. Investig. Drugs, 5(2), 146–153. [PMC free article] [PubMed] [Google Scholar]
- Ito H, Yoshida H, Bogaki-Shonai M, Niga T, Hattori H, & Nakamura S (1994). Quinolone resistance mutations in the DNA gyrase gyrA and gyrB genes of Staphylococcus aureus. Antimicrob. Agents Chemother, 38(9), 2014–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S (2016). Multidrug efflux pumps in Staphylococcus aureus and their clinical implications. Journal of Microbiology, 54(1), 1–8. [DOI] [PubMed] [Google Scholar]
- Konthikamee W, Gilbertson JR, Langkamp H, & Gershon H (1982). Effect of 2-alkynoic acids on in vitro growth of bacterial and mammalian cells. Antimicrob. Agents Chemother, 22(5), 805–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBel M (1988). Ciprofloxacin: Chemistry, mechanism of action, resistance, antimicrobial spectrum, pharmacokinetics, clinical trials, and adverse reactions. Pharmacotherapy, 8(1), 3–30. [DOI] [PubMed] [Google Scholar]
- Madsen S, Beck H, Ravn P, Vrang A, Hansen A, & Israelsen H (2002). Cloning and Inactivation of a branched-chain-amino-acid aminotransferase gene from Staphylococcus carnosus and characterization of the enzyme. Appl. Environ. Microb, 68, 4007–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus MC (1997). Mechanisms of bacterial resistance to antimicrobial agents. Am. J. Health Syst. Pharm, 54(12), 1420–1433. [DOI] [PubMed] [Google Scholar]
- Morbidoni HR, Vilchèze C, Kremer L, Bittman R, Sacchettini JC, & Jacobs WR (2006). Dual Inhibition of mycobacterial fatty acid biosynthesis and degradation by 2-alkynoic acids. Chem. Biol, 13(3), 297–307. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Dutta D, Saha B, & Das AK (2010). Crystal Structure of Glyceraldehyde-3-Phosphate Dehydrogenase 1 from Methicillin-Resistant Staphylococcus aureus MRSA252 Provides Novel Insights into Substrate Binding and Catalytic Mechanism. Journal of Molecular Biology, 401(5), 949–968. [DOI] [PubMed] [Google Scholar]
- Mustaev A, Malik M, Zhao X, Kurepina N, Luan G, Oppegard LM, Hiasa H, Marks KR, Kerns RJ, Berger JM, & Drlica K (2014). Fluoroquinolone-gyrase-DNA complexes: Two modes of drug binding. J. Biol. Chem, 289(18), 12300–12312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton GL, Fahey RC, & Rawat M (2012). Detoxification of toxins by bacillithiol in Staphylococcus aureus. Microbiology 158(Pt 4), 1117–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi K, Winarno H, Mukai M, & Shibuya H (2003). Preparation and cancer cell invasion inhibitory effects of C16-alkynoic fatty acids. Chem. Pharm. Bull, 51(4), 463–466. [DOI] [PubMed] [Google Scholar]
- Parsons JB, Frank MW, Subramanian C, Saenkham P, & Rock CO (2011). Metabolic basis for the differential susceptibility of Gram-positive pathogens to fatty acid synthesis inhibitors. Proc. Natl. Acad. Sci, 108(37), 15378–15383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons JB, Yao J, Frank MW, Jackson P, & Rock CO (2012). Membrane disruption by antimicrobial fatty acids releases low-molecular-weight proteins from Staphylococcus aureus. Journal of Bacteriology, 194(19), 5294–5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piercy EA, Barbaro D, Luby JP, & Mackowiak PA (1989). Ciprofloxacin for methicillin-resistant Staphylococcus aureus infections. Antimicrob. Agents Chemother, 33(1), 128–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raviglione MC, Boyle JF, Mariuz P, Pablos-Mendez A, Cortes H, & Merlo A (1990). Ciprofloxacin-resistant methicillin-resistant Staphylococcus aureus in an acute-care hospital. Antimicrob. Agents Chemother, 34(11), 2050–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripoll-Rozada J, Garcia-Cazorla Y, Getino M, Machon C, Sanabria-Rios D, de la Cruz F, Cabezon E, & Arechaga I (2016). Type IV traffic ATPase TrwD as molecular target to inhibit bacterial conjugation. Mol. Microbiol, 100(5), 912–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanabria-Rios DJ, Rivera-Torres Y, Maldonado-Dominguez G, Dominguez I, Rios C, Diaz D, Rodriguez JW, Altieri-Rivera JS, Rios-Olivares E, Cintron G, Montano N, & Carballeira NM (2014). Antibacterial activity of 2-alkynoic fatty acids against multidrug-resistant bacteria. Chem. Phys. Lipids, 178, 84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanabria-Rios DJ, Rivera-Torres Y, Rosario J, Gutierrez R, Torres-Garcia Y, Montano N, Ortiz-Soto G, Rios-Olivares E, Rodriguez JW, & Carballeira NM (2015). Chemical conjugation of 2-hexadecynoic acid to C5-curcumin enhances its antibacterial activity against multi-drug resistant bacteria. Bioorg Med Chem Lett, 25(22), 5067–5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiebel J, Chang A, Lu H, Baxter M V., Tonge, P. J., & Kisker, C. (2012). Staphylococcus aureus FabI: Inhibition, substrate recognition, and potential implications for in vivo essentiality. Structure, 20(5), 802–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen S, Sirobhushanam S, Johnson SR, Song Y, Tefft R, Gatto C, & Wilkinson BJ (2016). Growth-environment dependent modulation of Staphylococcus aureus branched-chain to straight-chain fatty acid ratio and incorporation of unsaturated fatty acids. PloS One, 11a(10), e0165300- e0165300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spetalnick BM, & Powers JS (1990). Eradication of Methicillin-Resistant Staphylococcus aureus Colonization with Ciprofloxacin. J. Am. Geriatr. Soc, 38(3), 389–390. [DOI] [PubMed] [Google Scholar]
- Tasdemir D, Sanabria D, Lauinger IL, Tarun A, Herman R, Perozzo R, Zloh M, Kappe SH, Brun R, & Carballeira NM (2010). 2-Hexadecynoic acid inhibits plasmodial FAS-II enzymes and arrests erythrocytic and liver stage Plasmodium infections. Bioorg. Med. Chem, 18(21), 7475–7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilchèze C, Leung LW, Bittman R, & Jacobs WR Jr (2016). Synthesis and biological activity of alkynoic acids derivatives against mycobacteria. Chem. Phys. Lipids, 194, 125–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng CJ, Yoo JS, Lee TG, Cho HY, Kim YH, & Kim WG (2005). Fatty acid synthesis is a target for antibacterial activity of unsaturated fatty acids. FEBS Lett, 579(23), 5157–5162. [DOI] [PubMed] [Google Scholar]