Abstract
The pandemic coronavirus disease 2019 (COVID‐19), caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS‐CoV‐2), has affected millions of people worldwide. To date, there are no proven effective therapies for this virus. Efforts made to develop antiviral strategies for the treatment of COVID‐19 are underway. Respiratory viral infections, such as influenza, predispose patients to co‐infections and these lead to increased disease severity and mortality. Numerous types of antibiotics such as azithromycin have been employed for the prevention and treatment of bacterial co‐infection and secondary bacterial infections in patients with a viral respiratory infection (e.g., SARS‐CoV‐2). Although antibiotics do not directly affect SARS‐CoV‐2, viral respiratory infections often result in bacterial pneumonia. It is possible that some patients die from bacterial co‐infection rather than virus itself. To date, a considerable number of bacterial strains have been resistant to various antibiotics such as azithromycin, and the overuse could render those or other antibiotics even less effective. Therefore, bacterial co‐infection and secondary bacterial infection are considered critical risk factors for the severity and mortality rates of COVID‐19. Also, the antibiotic‐resistant as a result of overusing must be considered. In this review, we will summarize the bacterial co‐infection and secondary bacterial infection in some featured respiratory viral infections, especially COVID‐19.
Keywords: antibiotic, bacterial co‐infection, COVID‐19, SARS‐CoV‐2, viral infection
Abbreviations
- AMPs
antimicrobial peptides
- CCL2
C‐C Motif Chemokine Ligand 2
- COVID‐19
coronavirus disease 2019
- HCoV
human coronavirus
- HCoV‐NL63
Human coronavirus NL63
- ICAM‐1
intercellular adhesion molecule 1
- IFNs
interferons
- IL
interleukin
- IP‐10
interferon‐γ‐inducible protein‐10
- IRF‐3
interferon regulatory factor 3
- MAVS
mitochondrial antiviral‐signaling
- MCP‐1
monocyte chemoattractant protein‐1
- MDA5
RIG‐1/melanoma differentiation‐associated gene 5
- MERS
Middle East Respiratory Syndrome‐related coronavirus
- NADPH
nicotinamide adenine dinucleotide phosphate
- NTHI
non‐typeable H. influenzae
- PERK
PKR‐like endoplasmic reticulum kinase
- PKR
protein kinase R
- REG3B
Regenerating Islet Derived Protein 3 Beta
- RIG‐I
retinoic acid‐inducible gene 1
- SARS
severe acute respiratory syndrome
- SARS‐CoV‐2
Severe Acute Respiratory Syndrome Coronavirus 2
- STAT1
signal transducer and activator of transcription 1
- TANK
TRAF Family Member Associated NFKB Activator
- TB
tuberculosis
- TGF‐β
transforming growth factor‐beta
- TLR‐3
Toll‐like receptor 3
- TRAF3
TNF Receptor Associated Factor 3
- T4P
Type 4 pilus
- WGMs
whole‐genome metagenomics
1. INTRODUCTION
The coronavirus disease 2019 (COVID‐19) is an infectious disease caused by the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) that was first identified in December 2019 in Wuhan, China, and is currently circulating throughout the world. 1 By July 5, 2020, more than 11,125,245 million cases have been diagnosed in 216 countries, and more than 528,204 deaths have been reported. 2 The ongoing COVID‐19 pandemic highlights the critical need for rapid development of vaccines and antiviral treatments to reduce the number of hospitalizations and deaths caused by this new dangerous coronavirus. 3 Co‐infections and superinfections are common in respiratory viral infections. 4 , 5 According to the laboratory, clinical, and epidemiological studies, secondary or bacterial co‐infections with other viruses can significantly increase the mortality rate in patients infected with viral infections. 6 , 7 It has previously documented that the mortality rate of viral infections can be influenced by different factors, such as bacterial co‐infection. 8 , 9 , 10 For instance, influenza‐related bacterial infections contribute to severe illness and mortality during the epidemic and seasonal influenza outbreaks. 11 Some influenza‐related bacterial species include Streptococcus pyogenes, Neisseria meningitidis, Moraxella catarrhalis, Streptococcus pneumoniae, Haemophilus influenzae, and Staphylococcus aureus. 11 , 12 , 13 , 14 , 15 , 16 , 17 , 18 The mechanisms of severe complications caused by influenza‐bacterial co‐infections mainly include a lack of effective immune response as well as pathogenic synergy. 5 Although multiple microbial agents can cause acute lower respiratory tract infections, in most cases, the disease is caused by viruses and bacteria at the same time. 19 Secondary and bacterial co‐infections with pandemics and viral epidemics have irreversible consequences, especially in high‐risk groups, including those with immunodeficiency or immunosuppression. 20
Emerging evidence suggests that the number of patients with COVID‐19 diagnosed with bacterial co‐infections during hospitalization periods is increasingly raised. 21 , 22 , 23 The source and specific nature of these infections are yet to be fully explored, but there is some evidence suggesting that multidrug‐resistant bacteria are among the pathogens that are thought to be responsible for the development of these infections. 21 , 22 , 23 Patients vulnerable to viral lung infections, such as influenza, severe acute respiratory syndrome (SARS), and COVID‐19 are the greatest risk to be co‐infected with superbugs. 21 , 22 , 23 , 24 , 25 For example, the 2009 H1N1 influenza pandemic caused approximately 300,000 deaths around the world in which 30–55% of cases die of bacterial pneumonia. 26 , 27 It is now known that viral infections can weaken the host immunity, paving the way for the development of viral‐bacterial co‐infection. 28 , 29 The new coronavirus, COVID‐19, is another example of this fact as most of the hospitalized patients with COVID‐19 acquired a secondary bacterial infection. 30 , 31 , 32 In some severe form of SARS‐CoV‐2, patients exhibited increased levels of infection‐related biomarkers and inflammatory cytokines, suggesting potential bacterial co‐infection as a result of the dysregulated immune system. 33 Besides, the emergence of antibiotic resistance could be an additional burden for the health care system as co‐infection with coronavirus and pneumonia stretches health care units beyond their capabilities and resources. Understanding the mechanism underlying the synergy between Covid‐19 and bacteria paves the way for the discovery of novel therapeutic agents to prevent the mortality rate in patients co‐infected with COVID‐19 and bacteria. In the current situation, appropriate and systematic analysis of COVID‐19 patients diagnosed with bacterial co‐infection should be implemented to choose proper antibiotics to increase the survival of patients and limit the spread of drug‐resistant bacteria. The use of rapid diagnostic tools and methods promoting the prescription of effective narrow‐spectrum antibiotics should be taken into account. In this review, we will summarize the current data available for bacterial infections in patients with COVID‐19.
1.1. Bacterial co‐infection with viral respiratory infections
Viral pneumonia and lower respiratory tract infections are well characterized in adult patients, including those diagnosed with severe forms of viral infection. 34 Most viral lower respiratory tract infections seem to be acquired in the community and considered a leading cause of infection in patients who undergo mechanical ventilation. 34 The most common cases diagnosed with bacterial co‐infection with viral infections are seen in those infected with influenza virus. 27 The oldest report of bacterial infections that occurred simultaneously or shortly after influenza is related to the 1918 Influenza pandemic, in which most deaths occurred as a result of co‐infection with infectious bacteria. 35 Also, the H1N1 Influenza pandemic in 2009 was complicated by bacterial pneumonia in 4–33% of hospitalized patients. 36 , 37 , 38 Bacterial‐viral co‐infection is not restricted to influenza and also caused by other respiratory viruses, such as parainfluenza virus, respiratory syncytial virus, adenovirus, rhinovirus, and human metapneumovirus. 34 , 39 , 40 Despite the discovery of antibiotics and viral vaccines in 1918–1957, the mortality rate, resulting from secondary bacterial pneumonia remained a major problem. The mortality rate seems to be still growing mostly because of the rapid rate of aging in the human population. 41 , 42
Although viruses are commonly responsible for the development of acute upper and lower respiratory infections, in most cases patients may be infected by both bacterial and viral pathogens; however, the clinical manifestations at the early stages of the disease would not be nosologically distinguishable for physicians to differentially diagnose viral from a bacterial infection. 43 Recently, a group of respiratory emerging viruses has been identified, such as human coronavirus (HCoV), NL63, human bocavirus, influenza viruses' type H1N1 and H5N1, SARS, Middle East Respiratory Syndrome‐related coronavirus (MERS), and Covid‐19. 44 , 45 , 46 , 47 , 48 In children, atypical bacterial pathogens, such as Legionella pneumophila, Mycoplasma pneumoniae, and Chlamydophila pneumoniae include the majority of infectious agents that cause mild, moderate, or even severe forms of acute respiratory infections. 49 , 50 Bacterial co‐infections with respiratory viral pathogens are very common, often through synergistic interaction among viruses such as influenza virus, and bacterial pathogens and the host immune system of the human being; nevertheless, the interaction between viruses and unusual bacteria is not yet fully understood. 50 , 51 These secondary infections predominantly involve a specific group of bacterial pathogens, such as S. aureus, S. pneumoniae, S. pyogenes, and H. influenzae. 11 , 12 , 13 , 14 , 15 , 16 , 17 , 18 A complete list of bacterial co‐infections with viral pathogens is depicted in Table 1.
TABLE 1.
Common respiratory viral‐bacterial coinfections and their associated clinical infections in human
| Viral infection | Bacterial coinfection | Clinical infection | References |
|---|---|---|---|
| Influenza | Staphylococcus aureus, MRSA | Community‐acquired pneumonia, | 18, 52, 53, 54, 55, 56, 57 |
| Streptococcus pneumoniae | Pneumococcal pneumonia, sepsis, meningitis, otitis media | 58 | |
| Streptococcus pyogenes (group A streptococci) | Sepsis, pleural empyema | 15 | |
| Haemophilus influenzae | Pneumonia | 59 | |
| Moraxella catarrhalis | Pneumonia and bacteremia | 60, 61 | |
| Neisseria meningitidis | Meningococcemia | 62, 63 | |
| Chlamydophila pneumoniae | Pneumonia | 64 | |
| Mycoplasma pneumoniae | Pneumonia | 64 | |
| Legionella pneumophila, Klebsiella pneumoniae, Pseudomonas aeruginosa, Acinetobacter baumannii, Burkholderia cepacia, Enterobacter aerogenes | Pneumonia | 57 | |
| Metapneumovirus | Haemophilus influenzae, enterococcus spp, N. meningitidis group B, Brucella spp, Streptococcus pyogenes, Streptococcus pneumoniae | Acute otitis media, pneumonia | 65 |
| Respiratory syncytial virus | Pseudomonas aeruginosa | Respiratory infections in cystic fibrosis patients | 66, 67 |
| Adenovirus | Non‐typeable Haemophilus influenzae, Chlamydia trachomatis | Pneumonia or acute otitis media | 68 |
| Parainfluenza |
Streptococcus pneumonia, Streptococcus agalactiae, Haemophilus influenza |
Acute otitis media, pneumonia | 69, 70 |
| Rhinovirus |
Streptococcus pneumonia, Mycoplasma |
Pneumonia | 50 |
| Staphylococcus aureus | Respiratory complications | 71 | |
| SARS |
Chlamydophila pneumonia Mycoplasma pneumonia |
Pneumonia | 72 |
| MRSA | Pneumonia | 73, 74 | |
| MERS | Mycobacterium tuberculosis | Immune suppression and augment the infection of each other | 75 |
|
Mycoplasma spp. Legionella Chlamydia spp. |
Not reported | 76 |
Abbreviations: MERS, Middle East respiratory syndrome; MRSA, methicillin‐resistant Staphylococcus aureus; SARS, severe acute respiratory syndrome.
1.2. Viral predisposition to bacterial co‐infection in the respiratory tract
Commonly, viral infection can destroy histologically and functionally the respiratory tract of individuals upon viral spread. 42 Depending on the type of the virus, the histopathological outcomes could be relatively different from mild types to severe ones. These detrimental changes include altered mucus secretion, cell death, hyperplasia, decreased mucosal clearance, reduced oxygen exchange, and impaired surfactant secretion. 42 , 77 Each of these effects is caused by various molecular mechanisms, depending on the virus, bacterial species, as well as the degree of the host immune reaction to either a bacterium or virus. 42 It has been noted that viral infections promote bacterial colonization of the airway through a variety of mechanisms. 42 Peltola and colleagues found that influenza viruses can enhance the colonization of the nasopharynx by S. pneumoniae bacterium, however, only particular subtypes were found to mediate the development of bacterial otitis media and sinusitis. 78 These data explain why the rate of bacterial infection is high in influenza seasons. 42 The neuraminidase enzyme of the influenza virus has been found to be presented on the host cell receptors, and they are employed for the adherence of bacteria due to its sialidase ability that changes the carbohydrate moieties on the host epithelial cells. 77 , 79 This enzyme is also capable of increasing the possibility of bacterial adherence to the host cells through the stimulation of transforming growth factor‐beta (TGF‐β) which triggers the up‐regulation of integrins and fibronectin. Integrins and fibronectin have been shown to act as receptors for bacteria. 80 Besides, interferons (IFNs) induction of by influenza virus can cause reduced C‐C Motif Chemokine Ligand 2 (CCL2) expression resulting in failed macrophages recruitment that necessary for clearance of pneumococcal cells thereby enhance the colonization of S. pneumoniae in vivo. 81 Also, it has been found that the influenza virus predisposes the host to develop pneumonia caused by S. aureus where viral and bacterial loads are increased during co‐infection. 52 It has been hypothesized that viral load is increased following bacterial co‐infection because of increased shedding rate of the virus from infected host cells; however, bacterial loads would be elevated as a result of impaired function of alveolar macrophages. 82
Additionally, other upper respiratory tract viruses increase the adherence ability of bacterial pathogens to primary and immortalized epithelial cells with particular differences. Such differences are determined by the types of epithelial cells and their response to parainfluenza virus‐3, respiratory syncytial virus, and/or influenza virus. 83 Novotny et al 84 showed that adenovirus and respiratory syncytial virus stimulated the expression of intercellular adhesion molecule 1 (ICAM‐1) by primary respiratory tract epithelial cells. ICAM1 acts as a receptor for Type 4 pilus (T4P) of non‐typeable H. influenzae (NTHI), thus promoting the binding of this pathogen to cells expressing this molecule. Also, respiratory syncytial virus infection increases the binding ability of P. aeruginosa to normal epithelial cells, as well as cells affected by cystic fibrosis. Such phenomena have been frequently employed by other bacteria to increase their virulence to infect the cells. 66 Studies have indicated that, following the infection with the respiratory syncytial virus, the rate of the binding of S. pneumoniae serotypes epithelial cells is increased by 2–10 folds. 85 Similarly, the antibody titers against S. pneumoniae are elevated in the nasopharynx when the cells co‐infected with the respiratory syncytial virus, rhinovirus, and community‐acquired pneumonia. 86 The higher degree of colonization of nasopharynx with S. pneumoniae is also found when the individuals are co‐infected with viral upper respiratory tract infection or human immunodeficiency virus infection. 87
Bacterial and viral co‐infection can alter some properties of the host mucosal immunity, leading to the failure in controlling the replication of bacteria in this site. 88 , 89 Some key findings are discussed here, including the influence of viral infection on phagocytic activity. The reduction of alveolar macrophages by the influenza virus facilitates bacterial co‐infection. 88 , 90 Several lines of evidence demonstrated that 90% of resident alveolar macrophages were lost in the early weeks after influenza infection through tracking dye‐labeled alveolar macrophages, 42 whereas 95% of the initial bacterial inoculum was eliminated during 3 hr by alveolar macrophages in non‐influenza inoculated hosts. Notably, in those cells co‐infected with influenza virus about 50% of the bacterial inoculum remained recoverable. 42 Also, it has been suggested that phagocyte activity, along with the cell proliferation could be influenced by viral infection. The infection of alveolar macrophages with influenza causes a marked decrease in the level of cytokines and chemokines, leading to decreased rates of recruitment and stimulation of neutrophils. 88 It may also suppress the phagocytic bacterial clearance mediated to nicotinamide adenine dinucleotide phosphate (NADPH), thereby increasing the susceptibility to secondary bacterial infection. 91 As mentioned earlier, the dysregulation of pro‐inflammatory cytokine‐induced by viral infection has been shown to play an essential role in the susceptibility of the cells to secondary bacterial infection. It is now known that type 1 IFNs have antiviral and immune‐stimulatory properties and could have detrimental effects on human cells when their expression is inappropriate and excessive. It has been reported that IFNs play a fundamental role in the production of anti‐inflammatory cytokines, such as interleukin (IL)‐10 and IL‐6, as well as the inhibition of proinflammatory cytokines, linking the innate immunity to adaptive immune responses, such as IL‐17 and IL‐23. They also decrease the activity of macrophages, dendritic cells, natural killer cells, along with the number of CD4‐ and CD8‐positive T cells, leading to the impaired eradication of bacterial co‐infection. 79 , 92 , 93 , 94
Also, there are a number of mechanisms that are independent of the phagocytosis process by which viral host infection can predispose the human body to secondary bacterial co‐infection. The production of antimicrobial peptides (AMPs), such as lipocalin‐2, cathelicidin, Regenerating Islet Derived Protein 3 Beta (REG3B), calprotectin, may be dysregulated by upper respiratory tract viruses. 94 The respiratory syncytial virus infection is able to diminish the expression of human β‐defensin‐3 orthologue named chinchilla beta‐defensin 1 when used in vivo. Of note, con‐infection with the respiratory syncytial virus is capable of stimulating the viral load of Nontypeable Haemophilus influenzae (NTHi) in the nasopharynx by 10–100 folds. 95 The intranasal delivery of anti‐ chinchilla beta‐defensin 1, human β‐defensin 3, or the recombinant form of chinchilla beta‐defensin 1 showed that the disruption of the availability of even a single innate immune effector could have a great impact on bacterial load since the viral infection has a critical in the loading of H. influenzae within the host airway. 95 Some mechanisms by which viral respiratory infections may predispose patients to bacterial infections to include failure immune response, viral‐induced changes in epithelial cells, and the increased bacterial colonization 34 and summarize of the potential mechanisms responsible for bacterial coinfection with viral respiratory infections is depicted in Table 2.
TABLE 2.
Summary of the potential mechanisms responsible for the bacterial coinfection with viral respiratory infections
| Mechanism | Description | References |
|---|---|---|
| Elevation in bacterial adherence due to viral infection | Virus can modulate surface membrane receptors, thereby enhancing bacterial adhesion | 16, 96, 97, 98 |
| Cell destruction by viral enzymes | Viral enzymes destroy mucosal glycoproteins, mainly those inhibiting bacterial attachment | 98, 99 |
| Reduction of mucociliary clearance | Virus can reduce mucociliary clearance leading to the decreased production of bactericidal materials | 100 |
| Reduction in chemotaxis | Virus can decrease the chemotactic factors, leading to the reduced cell response to attacking organisms | 101 |
| Direct effect on phagocytic and induction of post phagocytic alveolar macrophage functions | Virus hinders or modifies a number of immune functions, such as phagosome‐lysosome fusion and intracellular killing | 102, 103 |
| Induction of immature phagocytes | Virus can disrupt macrophages and probably replace them with immature phagocytes | 98, 104, 105 |
| Reduction of surfactant levels | Virus impairs the function of alveolar type‐2 pneumocyte | 98, 106, 107 |
| Induction of dysbiosis in lower respiratory tract microbiome | Microbiome dysbiosis can affect the immune response against respiratory viral infection | 108 |
| Dysregulation of the innate and adaptive immune responses | Virus decreases the number of alveolar macrophages through the development of apoptosis | 42, 88, 90, 91 |
| Modulation of apoptosis and inflammation | Autophagy and apoptosis facilitates secondary bacterial pneumonia after viral infection | 109 |
| Reduction of antibacterial immune function at the respiratory epithelium | Respiratory viral infection leads to the predisposition to secondary bacterial infection via the deviation of the respiratory tract immune status | 110, 111, 112, 113, 114, 115 |
| Dysregulation of nutritional immunity | Some viruses can subvert nutritional protection to promote bacterial infection | 116, 117, 118 |
| Immunosuppression | Immunosuppression is induced by several viruses such as HIV | 119, 120, 121 |
| Synergism during viral/bacterial co‐infections | Both viruses and bacteria play a role in the immunopathogenicity of co‐infection | 8, 122, 123 |
| Release of planktonic bacteria from biofilms | Viruses can manipulate many factors such as chemokines and hydrogen peroxide, thereby leading to the disruption of biofilm structure | 42, 124, 125, 126 |
Abbreviation: HIV, human immunodeficiency viruses.
1.3. Coronaviruses influence on host immune system
The human immune reaction to SARS‐CoV‐2 infection is a two‐step reaction that during the non‐severe infection phase, a particular adaptive immune reaction is necessary to viral eradicate and prevent the disease from progressing to a severe phase. 127 , 128 Hence, approaches to increase immune reactions such as using anti‐serum and Pegylated IFNα are crucial at this phase. 127 , 128 To provide a protective immune reaction in the early phase, the host must be in a good general health condition and have a genetic background to be able to exhibit an acceptable antiviral reaction. Genetic diversity is well known to involve in particular changes in the host immune response to various microbial pathogens.
There is few information about the initial events in the process of virus elimination and inflammatory reactions during SARS‐CoV infection. However, the innate immune reactions is mediated to the adaptive immunity development and disease severity in SARS‐CoV. 129 Of note, SARS‐CoV evolved escape mechanisms to avoid IFN responses in infected host cells. Besides, inflammatory reactions are regulated by inflammatory cytokines and chemokines such as IL‐6, interferon‐γ‐inducible protein‐10 (IP‐10), monocyte chemoattractant protein‐1(MCP‐1) as well as the penetration of inflammatory cells such as macrophages into infected host tissues. 129 The suppression of antiviral type I interferons is one of the marked characteristics of SARS‐CoV infection as well as other group II coronaviruses including mouse hepatitis virus. 130 It has been shown that the formation of type I interferons is impaired in host cells infected with SARS‐CoV, but pretreatment of cells with IFN suppresses the growth of SARS‐CoV. 131 , 132 This shows that these viruses have developed mechanisms to overcome IFN responses in infected cells. Type I interferons are rarely found in acute SARS patients, and SARS‐CoV is sensitive to Pegylated IFN‐α, as shown in in vivo murine models. 133 Studies indicated that type I interferon suppression in SARS‐CoV‐infected hosts is mediated by the inactivation of the IRF‐3 (interferon regulatory factor 3) protein, a transcriptional factor that controls the transcription of interferons. 132 Also, other SARS‐CoV accessory proteins act as powerful interferon antagonists through various strategies. 129 For instance, N proteins inhibit the expression of interferons, whereas open‐reading frame 3b and 6 proteins oppress the signaling pathway and expression of interferons. 129 Also, open‐reading frame 6 proteins can halt the translocation of signal transducer and activator of transcription 1 (STAT1). The open‐reading frame 3b protein is a shuttling protein, impeding the stimulation of type I interferon, which is triggered by retinoic acid‐inducible gene 1 (RIG‐I) and mitochondrial antiviral‐signaling (MAVS) protein. 134 Additionally, M proteins suppress the production of type I interferon by inhibiting the generation of the TNF Receptor Associated Factor 3 (TRAF3)–TRAF Family Member Associated NFKB Activator (TANK) – TANK binding kinase 1 (TBK1)/IκB kinase ε (IKKε) complex. 134 Besides, SARS‐CoV NSP1 proteins are able to halt the expression of IFN‐β in host cells by promoting the degradation of the host mRNA and the suppression of the translation process. 135 A polyprotein complex of SARS‐CoV named papain‐like protease is able to inhibit the phosphorylation and nuclear translocation of IRF‐3, resulting in disrupting the activation of type I IFN response via either Toll‐like receptor 3 (TLR‐3) or RIG‐1/melanoma differentiation‐associated gene 5 (MDA5) complex. 136 Also, infected cells with SARS‐CoV can stimulate the expression of protein kinase R (PKR) and PKR‐like endoplasmic reticulum kinase (PERK). 137
1.4. Bacterial co‐infection with coronaviruses and COVID‐19
Although numerous studies performed on viral and bacterial co‐infections, little information exists about human coronaviruses. In addition to seasonal influenza, it has been reported corona pathogens of pneumonia include coronavirus 229E, NL63, OC43, SARS, MERS, and SARS‐CoV‐2. These viruses can cause co‐infection in the setting of community‐acquired bacterial pneumonia. 138 , 139 , 140 , 141 Human coronavirus NL63 (HCoV‐NL63) has been recently discovered as a human respiratory pathogen with a high worldwide prevalence. 142 , 143 Arguably, HCoV‐NL63 is among the most clinically significant human coronaviruses and associated with upper and lower respiratory tract infections, frequently occurring in the winter and presenting more severe symptoms in children, the elderly, and immunocompromised patients. 142 , 143 , 144 In a study conducted by Golda et al., 143 they evaluated the impact of HCoV‐NL63 on bacterial adherence causing respiratory tract diseases. HCoV‐NL63 infection has been shown to enhance the adherence of S. pneumoniae to cells infected with the virus. 143 In one study, Zahariadis et al. 72 showed the coinfection of SARS patients with other pulmonary pathogens. They found that 30 and 9% of cases with SARS were co‐infected with C. pneumoniae or M. pneumonia, respectively. Additionally, Alfaraj et al. 75 reported the coinfection of MERS‐CoV with tuberculosis (TB) in two cases. In a study carried out by Wang et al., 145 they reported seven cases of SARS‐related deaths who developed a secondary bacterial infection.
The COVID‐19 pandemic caused a large number of immunocompromised individuals to be hospitalized and some reports indicated that some COVID‐19 patients were diagnosed with secondary infections. 30 , 31 , 32 The specific source and nature of these infections have not yet been fully investigated; however, there is evidence indicating that multidrug‐resistant bacteria are among those microbes responsible for the development of these secondary infections. In one study, five cases (5.1%) with bacterial co‐infections including Acinetobacter baumannii and Klebsiella pneumoniae were found among 99 patients, 146 while in another study, four cases (9.8%) with secondary bacterial infections were reported among 41 patients. 147 In a study performed by Zhang et al., 148 221 patients with SARS‐CoV‐2 pneumonia were admitted to Zhongnan Hospital, Wuhan, China. Among them, 25.8% (57/221) patients were afflicted with co‐infections, and among these patients with co‐infections, 29.8% (17/57) were co‐infected with bacteria. In a study conducted by Blasco et al., 149 they detected one patient who was positive for M. pneumoniae coinfection among patients with COVID‐19 pneumonia. Also, Claire et al. 150 reported a fatal case of necrotizing pneumonia induced by Panton‐Valentine leukocidin–secreting S. aureus in a patient who was affected by COVID‐19.
Some patients infected with SARS‐CoV‐2 showed the increased levels of biomarkers and inflammatory cytokines related to co‐infection by bacteria, caused by dysregulation in the immune system. 33 The management of the severe form of SARS‐CoV‐2 is similar to most viral pneumonia‐causing respiratory failure. In a study carried out by Bordi et al., 151 they detected M. pneumoniae in five patients (4.0%), while only one patient was infected with L. pneumophila and S. pneumoniae (0.8%), and mixed infections were also observed in a small number of cases. They found the importance of using a broad‐spectrum molecular diagnostic panel for rapid detection of the most common respiratory pathogens to improve evaluation and clinical management of patients with a respiratory syndrome consistent with COVID‐19. 151 A list of bacterial co‐infection with COVID‐19 is depicted in Table 3.
TABLE 3.
List of bacterial co‐infection with COVID‐19
| Bacterium | Infection | References |
|---|---|---|
| Staphylococcus aureus | Necrotizing pneumonia | 152 |
| Mycoplasma pneumoniae | Exacerbate clinical symptoms, increase morbidity and prolonged intensive care unit stay | 153 |
| Legionella pneumophila | Pneumonia | 154 |
|
Enterobacter cloacae |
Pneumonia | 155 |
| Acinetobacter baumannii | Pneumonia | 146, 155 |
| Klebsiella pneumoniae | Pneumonia | 146 |
| Mycoplasma pneumoniae | Interstitial pneumonia | 149 |
| Mycoplasma pneumoniae | Not reported | 151 |
| Legionella pneumophila | Not reported | 151 |
| Streptococcus pneumoniae | Not reported | 151 |
| Prevotella | Not reported | 156, 157, 158 |
| Haemophilus | Not reported | 158, 159 |
| Lautropia | Not reported | 159 |
| Cutibacterium | Not reported | 159 |
One reason for this bacterial co‐infection is due to many hospital‐associated bacteria being adapted to develop an infection in individuals with a weakened immune system. It has been noted that the SARS‐CoV‐2 infection can damage the cells and the lung infrastructure. 160 Subsequently, the changed condition enables bacteria to increase adherence and invasion (Figure 1). It has been found that the mortality rate of viral pandemics is heavily impacted by secondary bacterial infections with myriad numbers of people in the 1918 influenza pandemic as well as the 2009 pandemic, who died from secondary bacterial infections rather than the virus alone. 26 , 27 , 161
FIGURE 1.
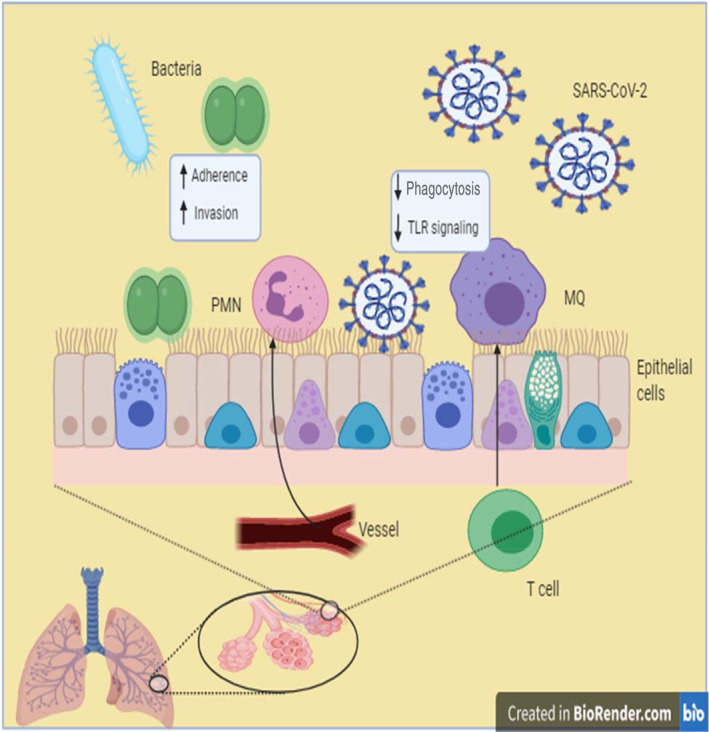
Postulated schematic of bacterial coinfection with SARS‐CoV‐2 infection. It has been proposed when SARS‐ CoV‐2 infects lung cells can damage the cells and the lung infrastructure. 160 This situation attracts neutrophil and macrophages to the site of infection and promoting the inflammation. 160 Finally, the changed situation and epithelial damage can cause bacteria to adhere to and invasion of the cells and proliferation. MQ, macrophage; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
Despite the proven significance of co‐infections in the severity of respiratory diseases, these kinds of infections during large outbreaks of respiratory infections are underdetermined. 162 For the precise diagnosis and evaluation of co‐infections during the COVID‐19 pandemic, samples must be taken longitudinally throughout the disease course via techniques independent of the culture for identifying mixed infections such as whole‐genome metagenomics (WGMs). 162 Such works offer valuable monitoring findings on co‐infection pathogens and drug‐resistance infections resulting in improved antibiotic prescribing options. 162 Zhou et al. 163 found that in the current COVID‐19 pandemic, 50% of patients who died from Covid‐19 had bacterial co‐infections. Correspondingly, Chen et al. 146 have reported fungal and bacterial co‐infections in patients with COVID‐19. COVID‐19 patients are hospitalized on invasive mechanical ventilation for a long time, leading to higher chances of using a ventilator and hospital‐acquired infections. 162 Thus, the rapid diagnosis of a broad range of potential pathogens and antimicrobial resistances for subsequent monitoring of co‐infection would be crucial. The metagenome of COVID‐19 patients has shown that Prevotella is a key player in immune response in a Chinese study, 156 while other opportunistic pathogens were found in a study conducted in the USA. 157 , 158 , 159 , 164 There is a wide range of bacterial pathogens including Haemophilus, Lautropia, Prevotella have been detected as co‐infection in Brazil, China, and the USA. 157 , 158 , 159 , 164
However, a further concern with the rapid expansion of critical care capacity to manage SARS‐CoV‐2 can potentially increase the rate of nosocomial infection within the hospital environment. 165 To date, although the role of viral or bacterial co‐infection in SARS‐CoV‐2 remains elusive, only a few SARS‐CoV‐2 patients worldwide have had documented evidence of co‐infection; however, there is still a concern on this issue as many reports claim that a significant proportion of COVID‐19 patients developed bacterial co‐infections.
1.5. The era of post‐COVID‐19 and antimicrobial resistance bacteria
It has been noted during the current pandemic, the antibiotic administration has been frequently used for COVID‐19 patients who were admitted to the intensive care unit. 165 While scientists attempt to understand and control the COVID‐19 pandemic, it would be also critical to prepare for the effect of the current and future viral pandemics on secondary bacterial infections, resulting in antimicrobial resistance in the near future. In combination with using an antimalarial drug, hydroxyl‐chloroquine, azithromycin has become a popular therapeutic option for COVID‐19 patients. Reports demonstrated that a combination of hydroxyl‐chloroquine and azithromycin was effective for a large proportion of Covid‐19 patients. 166 , 167 It is hard to estimate how often this combination is prescribed, but such a rate would be high enough to cause a shortage of azithromycin. However, 30–40% of common types of bacterial agents are already resistant to azithromycin, and overuse could render this or other antibiotics even less effective. 168 The findings could help experts' advice on using the antibiotics in COVID‐19 patients and help them to better understand the spread of co‐infections in hospitals and the mechanism of bacterial‐viral coinfection. One factor that involves in the antibiotic resistance in bacterial co‐infection is the widespread use of antibiotics in COVID‐19 patients. Emerging data show that more than 90% of COVID‐19 patients receive antibacterial drugs. 169 , 170 This rapid increase in antibiotic administration can cause a strong selective pressure on bacterial pathogens to evolve resistance leading to the increased incidence of drug‐resistant bacterial infections in the years subsequent to the COVID‐19 pandemic. It was estimated that 10 million people could die from an antibiotic‐resistant bacterial infection in the year of 2050, 171 but such prediction may be altered and shortened because of the devastating impact of the COVID‐19 pandemic on the usage of antibiotics, so this timeline will almost have to be modified. Nevertheless, concerted efforts must be made to better understand antibiotic administration in COVID‐19 patients. Antibiotics do not directly act on viral infections but viral respiratory infections often lead to bacterial co‐infections. 30 , 31 , 32 The current pandemic highlights the necessity for understanding the complex relationship between viral and bacterial infections. Of note, in patients who have treated with high‐dose antibiotics may have more co‐infections with drug‐resistant bacteria.
Additionally, a recent clinical trial conducted by Hagan et al 172 demonstrated that the use of broad‐spectrum antibiotics (which led to depleting gut microbiota) decreased and impaired the immune system's ability to generate antibodies. Also, the current study shows that the use of antibiotics perturbed bile acid metabolism and induced inflammatory responses. 173 Hence, improved functional therapies, including antibiotics and alternative therapies as well as the prediction of bacterial respiratory infections using vaccines, should be regarded as potential therapeutic approaches. Besides, standard guidelines should also be established for the administration of the antibiotics. In addition to the direct effect on drug‐resistant bacteria as a result of enhanced antibiotic administration, the transmission of drug‐resistant bacteria through the medical system should be taken into account. The COVID‐19 pandemic has highlighted the importance of vaccination, the need for functional antimicrobials, as well as the necessity for supporting research into the understanding and control of co‐infections. Rapid characterization of co‐infection is essential in the treatment of the most COVID‐19 patients, and could help to save lives, and will improve antimicrobial stewardship during the pandemic. Additionally, mixed bacterial‐viral infections can result in antibiotic treatment failure. These observations show that a better understanding of the underlying mechanisms will enable researchers to design effective preventive and therapeutic options. Several functional suggestions for management and control of bacterial co‐infection with COVID‐19 are offered in Table 4.
TABLE 4.
Several functional suggestions for management and control of bacterial co‐infection with COVID‐19
| Suggestion | Description |
|---|---|
| Using a broad‐spectrum diagnostic panel | Improves diagnosis, evaluation, and clinical management of patients with other respiratory viral infection concurrent with COVID‐19 |
| Developing novel treatment and prevention strategies | Increases our knowledge about the underlying molecular mechanisms accounting for viral‐bacterial co‐infection to promote novel therapeutic and prevention approaches |
|
Performing antibacterial susceptibility tests and potential therapy |
Prevents reduced antimicrobial susceptibility and treatment failure due to co‐infections 174 |
| Considering the biofilm‐associated bacterial infections | Facilitates treatment management as biofilm formation on artificial devices has been observed previously, thereby affecting infection outcomes, especially in COVID‐19 patients under mechanical ventilation 175 |
| Classifying mechanisms of pathogen interactions | Increases the ability of infection control as the extension of chemotherapy‐resistant pathogens is a severe global obstacle 174 |
2. CONCLUSION
Respiratory viruses such as SARS‐CoV‐2 are well‐characterized to cause severe disorders and pneumonia, particularly in individuals with serious medical comorbidities and aged populations. Additionally, respiratory virus infection could usually lead to enhanced susceptibility to secondary bacterial infections. However, the mechanisms responsible for bacterial‐SARS‐CoV‐2 co‐infection require further study. It has been noted that an elicited adaptive immune reaction toward viral infection fails the reaction of the host innate immunity against bacterial infection. This situation can explain why bacterial co‐infections occur when the virus starts to be eradicated from the lungs of patients with COVID‐19. This is accompanied by a shift in phagocytic activity of lung cells that mediate basal levels of innate protection via phagocytosis and pro‐inflammatory cytokines formation to cells better attuned to antigen presentation and stimulation of adaptive immune reactions. Additionally, recently it has been found the microbiome diversity shapes our immune system. In line with this, the depletion of the gut microbiome hinders the immune system's ability to create a humoral response against viruses like the flu virus. However, this novel paradigm ultimately allows the development of new immune intervention approaches for the prevention and management of viral‐bacterial co‐infections in COVID‐19 patients. The COVID‐19 pandemic reinforces the importance of preventative measures such as vaccination and antimicrobial treatments in maintaining human health.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
Mirzaei R, Goodarzi P, Asadi M, et al. Bacterial co‐infections with SARS‐CoV‐2. IUBMB Life. 2020;72:2097–2111. 10.1002/iub.2356
Contributor Information
Rasoul Mirzaei, Email: rasul.micro92@gmail.com.
Hossein Keyvani, Email: H.keyvani@iums.ac.ir.
Sajad Karampoor, Email: sajadkarampour1987@gmail.com.
REFERENCES
- 1. Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Organization T.W.H . Coronavirus disease (COVID‐19) pandemic. 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
- 3. Zimmermann P, Curtis N. Coronavirus infections in children including COVID‐19: An overview of the epidemiology, clinical features, diagnosis, treatment and prevention options in children. Pediatr Infect Dis J. 2020;39:355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McArdle AJ, Turkova A, Cunnington AJ. When do co‐infections matter? Curr Opin Infect Dis. 2018;31:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Paget C, Trottein F. Mechanisms of bacterial superinfection post‐influenza: A role for unconventional T cells. Front Immunol. 2019;10:336–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beadling C, Slifka MK. How do viral infections predispose patients to bacterial infections? Curr Opin Infect Dis. 2004;17:185–191. [DOI] [PubMed] [Google Scholar]
- 7. Metzger DW, Sun K. Immune dysfunction and bacterial coinfections following influenza. J Immunol (Baltimore, MD: 1950). 2013;191:2047–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jia L, Xie J, Zhao J, et al. Mechanisms of severe mortality‐associated bacterial co‐infections following influenza virus infection. Front Cell Infect Microbiol. 2017;7:338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Katsurada N, Suzuki M, Aoshima M, Yaegashi M, Ishifuji T, et al. The impact of virus infections on pneumonia mortality is complex in adults: A prospective multicentre observational study. BMC Infect Dis. 2017;17:755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Quah J, Jiang B, Tan PC, Siau C, Tan TY. Impact of microbial Aetiology on mortality in severe community‐acquired pneumonia. BMC Infect Dis. 2018;18:451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Smith AM, McCullers JA. Secondary bacterial infections in influenza virus infection pathogenesis. Curr Top Microbiol Immunol. 2014;385:327–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abbasi S, Pendergrass LB, Leggiadro RJ. Influenza complicated by Moraxella catarrhalis bacteremia. Pediatr Infect Dis J. 1994;13:937–938. [DOI] [PubMed] [Google Scholar]
- 13. Jacobs JH, Viboud C, Tchetgen ET, Schwartz J, Steiner C, et al. The association of meningococcal disease with influenza in the United States, 1989‐2009. PloS One. 2014;9:e107486‐e107486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mulcahy ME, McLoughlin RM. Staphylococcus aureus and influenza A virus: Partners in coinfection. mBio. 2016;7:e02068–e02016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ochi F, Tauchi H, Jogamoto T, et al. Sepsis and pleural empyema caused by Streptococcus pyogenes after influenza a virus infection. Case Rep Pediatr. 2018;2018:4509847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rowe HM, Meliopoulos VA, Iverson A, et al. Direct interactions with influenza promote bacterial adherence during respiratory infections. Nat Microbiol. 2019;4:1328–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Su I‐C, Lee K‐L, Liu H‐Y, Chuang H‐C, Chen L‐Y, Lee YJ. Severe community‐acquired pneumonia due to Pseudomonas aeruginosa coinfection in an influenza a (H1N1) pdm09 patient. J Microbiol Immunol Infect. 2019;52:365–366. [DOI] [PubMed] [Google Scholar]
- 18. Sun K, Metzger DW. Influenza and Staphylococcus aureus coinfection: TLR9 at play. Trends Microbiol. 2019;27:383–384. [DOI] [PubMed] [Google Scholar]
- 19. Dasaraju PV, Liu C. Chapter 93: Infections of the respiratory system. Medical microbiology. 4th ed. Galveston: University of Texas Medical Branch at Galveston, 1996. [PubMed] [Google Scholar]
- 20. MacIntyre CR, Bui CM. Pandemics, public health emergencies and antimicrobial resistance ‐ putting the threat in an epidemiologic and risk analysis context. Arch Public Health. 2017;75:54–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bengoechea JA, Bamford CG. SARS‐CoV‐2, bacterial co‐infections, and AMR: The deadly trio in COVID‐19? EMBO Mol Med. 2020;12(7):e12560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hendaus MA, Jomha FA. Covid‐19 induced superimposed bacterial infection. J Biomol Struct Dyn. 2020;1–10. [DOI] [PubMed] [Google Scholar]
- 23. Rawson TM, Moore LS, Zhu N, et al. Bacterial and fungal co‐infection in individuals with coronavirus: A rapid review to support COVID‐19 antimicrobial prescribing. Clin Infect Dis. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cauley LS, Vella AT. Why is coinfection with influenza virus and bacteria so difficult to control. Discov Med. 2015;19:33–40. [PMC free article] [PubMed] [Google Scholar]
- 25. Sun K, Yajjala VK. Nox2‐derived oxidative stress results in inefficacy of antibiotics against post‐influenza S. aureus pneumonia. J Exp Med. 2016;213:1851–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. A tool for the potential fall 2009 wave of pandemic H1N1 to guide public health decision‐making: An overview of the Public Health Agency of Canada's planning considerations, September 2009. Can Commun Dis Rep. 2010;36:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Morris DE, Cleary DW, Clarke SC. Secondary bacterial infections associated with influenza pandemics. Front Microbiol. 2017;8:1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Smith H, Sweet C. Cooperation between viral and bacterial pathogens in causing human respiratory disease. Polymicrobial Diseases. ASM Press, 2002. [Google Scholar]
- 29. Almand EA, Moore MD, Jaykus L‐A. Virus‐bacteria interactions: An emerging topic in human infection. Viruses. 2017;9:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rasmussen SA, Smulian JC, Lednicky JA, Wen TS, Jamieson DJ. Coronavirus disease 2019 (COVID‐19) and pregnancy: What obstetricians need to know. Am J Obstet Gynecol. 2020;222(5):415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ritchie AI, Singanayagam A. Immunosuppression for hyperinflammation in COVID‐19: A double‐edged sword? Lancet. 2020;395:1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tetro JA. Is COVID‐19 receiving ADE from other coronaviruses? Microbes Infect. 2020;22:72–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang B, Liu S, Tan T, et al. Treatment with convalescent plasma for critically ill patients with SARS‐CoV‐2 infection. Chest. 2020;158(1):e9–e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Crotty MP, Meyers S, Hampton N, et al. Epidemiology, co‐infections, and outcomes of viral pneumonia in adults: An observational cohort study. Medicine (Baltimore). 2015;94:e2332–e2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Morens DM, Taubenberger JK, Fauci AS. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: Implications for pandemic influenza preparedness. J Infect Dis. 2008;198:962–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Investigators AI. Critical care services and 2009 H1N1 influenza in Australia and New Zealand. N Engl J Med. 2009;361:1925–1934. [DOI] [PubMed] [Google Scholar]
- 37. Cillóniz C, Ewig S, Menéndez R, et al. Bacterial co‐infection with H1N1 infection in patients admitted with community acquired pneumonia. J Infect. 2012;65:223–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rice TW, Rubinson L, Uyeki TM, et al. Critical illness from 2009 pandemic influenza a (H1N1) virus and bacterial co‐infection in the United States. Crit Care Med. 2012;40:1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McCullers JA, Bartmess KC. Role of neuraminidase in lethal synergism between influenza virus and Streptococcus pneumoniae . J Infect Dis. 2003;187:1000–1009. [DOI] [PubMed] [Google Scholar]
- 40. Peltola VT, Murti KG, McCullers JA. Influenza virus neuraminidase contributes to secondary bacterial pneumonia. J Infect Dis. 2005;192:249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rothberg MB, Haessler SD, Brown RB. Complications of viral influenza. Am J Med. 2008;121:258–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bakaletz LO. Viral‐bacterial co‐infections in the respiratory tract. Curr Opin Microbiol. 2017;35:30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Korppi M, Don M, Valent F, Canciani M. The value of clinical features in differentiating between viral, pneumococcal and atypical bacterial pneumonia in children. Acta Paediatr. 2008;97:943–947. [DOI] [PubMed] [Google Scholar]
- 44. Allander T, Tammi MT, Eriksson M, et al. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci. 2005;102:12891–12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gatherer D. The 2009 H1N1 influenza outbreak in its historical context. J Clin Virol. 2009;45:174–178. [DOI] [PubMed] [Google Scholar]
- 46. van der Hoek L, Ihorst G, Sure K, et al. Burden of disease due to human coronavirus NL63 infections and periodicity of infection. J Clin Virol. 2010;48:104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Abdel‐Moneim AS. Middle East respiratory syndrome coronavirus (MERS‐CoV): Evidence and speculations. Arch Virol. 2014;159:1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. El ZME, Järhult JD. From SARS to COVID‐19: A previously unknown SARS‐CoV‐2 virus of pandemic potential infecting humans–call for a one health approach. One Health. 2020;9:100124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Basarab M, Macrae MB, Curtis CM. Atypical pneumonia. Curr Opin Pulm Med. 2014;20:247–251. [DOI] [PubMed] [Google Scholar]
- 50. Baroudy NRE, Refay ASE, Hamid TAA, et al. Respiratory viruses and atypical bacteria co‐infection in children with acute respiratory infection. Open Access Maced J Med Sci. 2018;6:1588–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. McCullers JA. Insights into the interaction between influenza virus and pneumococcus. Clin Microbiol Rev. 2006;19:571–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Iverson AR, Boyd KL, McAuley JL, Plano LR, Hart ME, McCullers JA. Influenza virus primes mice for pneumonia from Staphylococcus aureus . J Infect Dis. 2011;203:880–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Robertson L, Caley J, Moore J. Importance of Staphylococcus aureus in pneumonia in the 1957, epidemic of influenza a. Lancet. 1958;272:233–236. [DOI] [PubMed] [Google Scholar]
- 54. Severe methicillin‐resistant Staphylococcus aureus community‐acquired pneumonia associated with influenza–Louisiana and Georgia, December 2006–January 2007. MMWR Morb Mortal Wkly Rep. 2007;56:325–329. [PubMed] [Google Scholar]
- 55. Finelli L, Fiore A, Dhara R, et al. Influenza‐associated pediatric mortality in the United States: Increase of Staphylococcus aureus coinfection. Pediatrics. 2008;122:805–811. [DOI] [PubMed] [Google Scholar]
- 56. Reed C, Kallen AJ, Patton M, et al. Infection with community‐onset Staphylococcus aureus and influenza virus in hospitalized children. Pediatr Infect Dis J. 2009;28:572–576. [DOI] [PubMed] [Google Scholar]
- 57. Gao H‐N, Lu H‐Z, Cao B, et al. Clinical findings in 111 cases of influenza a (H7N9) virus infection. N Engl J Med. 2013;368:2277–2285. [DOI] [PubMed] [Google Scholar]
- 58. Palacios G, Hornig M, Cisterna D, et al. Streptococcus pneumoniae coinfection is correlated with the severity of H1N1 pandemic influenza. PloS One. 2009;4:e8540–e8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang X‐Y, Kilgore PE, Lim KA, Wang S‐M, Lee J, et al. Influenza and bacterial pathogen coinfections in the 20th century. Interdisc Perspect Infect Dis. 2011;2011:146376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Doern GV, Jones RN, Pfaller MA, Kugler K, Group S.P . Haemophilus influenzae and Moraxella catarrhalis from patients with community‐acquired respiratory tract infections: Antimicrobial susceptibility patterns from the SENTRY antimicrobial surveillance program (United States and Canada, 1997). Antimicrob Agents Chemother. 1999;43:385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Simmons WP. Moraxella catarrhalis pneumonia and bacteremia in an otherwise healthy child. Clin Pediatr. 1999;38:560–561. [DOI] [PubMed] [Google Scholar]
- 62. Young LS, LaForce FM, Head JJ, Feeley JC, Bennett JV. A simultaneous outbreak of meningococcal and influenza infections. N Engl J Med. 1972;287:5–9. [DOI] [PubMed] [Google Scholar]
- 63. Jansen AGSC, Sanders EAM, Van Der Ende A, et al. Invasive pneumococcal and meningococcal disease: Association with influenza virus and respiratory syncytial virus activity? Epidemiol Infect. 2008;136:1448–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mina M, Burke R, Klugman K. Estimating the prevalence of coinfection with influenza virus and the atypical bacteria Bordetella pertussis, Chlamydophila pneumoniae, and Mycoplasma pneumoniae . Eur J Clin Microbiol Infect Dis. 2014;33:1585–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wolf DG, Greenberg D, Shemer‐Avni Y, Givon‐Lavi N, Bar‐Ziv J, Dagan R. Association of human metapneumovirus with radiologically diagnosed community‐acquired alveolar pneumonia in young children. J Pediatr. 2010;156:115–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Van Ewijk BE, Wolfs TF, Aerts PC, et al. RSV mediates Pseudomonas aeruginosa binding to cystic fibrosis and normal epithelial cells. Pediatr Res. 2007;61:398–403. [DOI] [PubMed] [Google Scholar]
- 67. Petersen NT, Høiby N, Mordhorst CH, Lind K, Flensborg EW, BRUUN B. Respiratory infections in cystic fibrosis patients caused by virus, chlamydia and mycoplasma–possible synergism with Pseudomonas aeruginosa . Acta Paediatr. 1981;70:623–628. [DOI] [PubMed] [Google Scholar]
- 68. Korppi M, Leinonen M, Mäkelä PH, Launiala K. Mixed infection is common in children with respiratory adenovirus infection. Acta Paediatr. 1991;80:413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Korppi M, Leinonen M, Mäkelä PH, Launiala K. Bacterial involvement in parainfluenza virus infection in children. Scand J Infect Dis. 1990;22:307–312. [DOI] [PubMed] [Google Scholar]
- 70. Tong J, Fu Y, Meng F, et al. The sialic acid binding activity of human parainfluenza virus 3 and mumps virus glycoproteins enhances the adherence of group B streptococci to HEp‐2 cells. Front Cell Infect Microbiol. 2018;8:280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Katiyar R, Agarwal V, Chowdhary S, et al. Incidence of human rhinovirus coinfection with Staphylococcus aureus among HIV patients suffering from flu like illness. Int J Pharm Sci Res. 2017;8:4441–4446. [Google Scholar]
- 72. Zahariadis G, Gooley TA, Ryall P, et al. Risk of ruling out severe acute respiratory syndrome by ruling in another diagnosis: Variable incidence of atypical bacteria coinfection based on diagnostic assays. Can Respir J. 2006;13:17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yap FH, Gomersall CD, Fung KS, et al. Increase in methicillin‐resistant Staphylococcus aureus acquisition rate and change in pathogen pattern associated with an outbreak of severe acute respiratory syndrome. Clin Infect Dis. 2004;39:511–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Bassetti S, Bischoff WE, Sherertz RJ. Outbreak of methicillin‐resistant Staphylococcus aureus infection associated with an outbreak of severe acute respiratory syndrome. Clin Infect Dis. 2005;40:633–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Alfaraj SH, Al‐Tawfiq JA, Altuwaijri TA, Memish ZA. Middle East respiratory syndrome coronavirus and pulmonary tuberculosis coinfection: Implications for infection control. Intervirology. 2017;60:53–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Arabi YM, Deeb AM, Al‐Hameed F, et al. Macrolides in critically ill patients with Middle East respiratory syndrome. Int J Infect Dis. 2019;81:184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. McCullers JA. The co‐pathogenesis of influenza viruses with bacteria in the lung. Nat Rev Microbiol. 2014;12:252–262. [DOI] [PubMed] [Google Scholar]
- 78. Peltola VT, Boyd KL, McAuley JL, Rehg JE, McCullers JA. Bacterial sinusitis and otitis media following influenza virus infection in ferrets. Infect Immun. 2006;74:2562–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ballinger MN, Standiford TJ. Postinfluenza bacterial pneumonia: Host defenses gone awry. J Interferon Cytokine Res. 2010;30:643–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Li N, Ren A, Wang X, et al. Influenza viral neuraminidase primes bacterial coinfection through TGF‐β–mediated expression of host cell receptors. Proc Natl Acad Sci. 2015;112:238–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Nakamura S, Davis KM, Weiser JN. Synergistic stimulation of type I interferons during influenza virus coinfection promotes Streptococcus pneumoniae colonization in mice. J Clin Invest. 2011;121:3657–3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Smith AM, Adler FR, Ribeiro RM, et al. Kinetics of coinfection with influenza a virus and Streptococcus pneumoniae . PLoS Pathog. 2013;9:e1003238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Avadhanula V, Rodriguez CA, DeVincenzo JP, et al. Respiratory viruses augment the adhesion of bacterial pathogens to respiratory epithelium in a viral species‐and cell type‐dependent manner. J Virol. 2006;80:1629–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Novotny LA, Bakaletz LO. Intercellular adhesion molecule 1 serves as a primary cognate receptor for the type IV pilus of nontypeable Haemophilus influenzae . Cell Microbiol. 2016;18:1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hament J‐M, Aerts PC, Fleer A, et al. Enhanced adherence of Streptococcus pneumoniae to human epithelial cells infected with respiratory syncytial virus. Pediatr Res. 2004;55:972–978. [DOI] [PubMed] [Google Scholar]
- 86. Esposito S, Zampiero A, Terranova L, et al. Pneumococcal bacterial load colonization as a marker of mixed infection in children with alveolar community‐acquired pneumonia and respiratory syncytial virus or rhinovirus infection. Pediatr Infect Dis J. 2013;32:1199–1204. [DOI] [PubMed] [Google Scholar]
- 87. Wolter N, Tempia S, Cohen C, et al. High nasopharyngeal pneumococcal density, increased by viral coinfection, is associated with invasive pneumococcal pneumonia. J Infect Dis. 2014;210:1649–1657. [DOI] [PubMed] [Google Scholar]
- 88. Braciale TJ, Sun J, Kim TS. Regulating the adaptive immune response to respiratory virus infection. Nat Rev Immunol. 2012;12:295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Lijek RS, Weiser JN. Co‐infection subverts mucosal immunity in the upper respiratory tract. Curr Opin Immunol. 2012;24:417–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ghoneim HE, Thomas PG, McCullers JA. Depletion of alveolar macrophages during influenza infection facilitates bacterial superinfections. J Immunol. 2013;191:1250–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Sun K, Metzger DW. Influenza infection suppresses NADPH oxidase–dependent phagocytic bacterial clearance and enhances susceptibility to secondary methicillin‐resistant Staphylococcus aureus infection. J Immunol. 2014;192:3301–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Davidson S, Maini MK, Wack A. Disease‐promoting effects of type I interferons in viral, bacterial, and coinfections. J Interferon Cytokine Res. 2015;35:252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Mehta D, Petes C, Gee K, Basta S. The role of virus infection in deregulating the cytokine response to secondary bacterial infection. J Interferon Cytokine Res. 2015;35:925–934. [DOI] [PubMed] [Google Scholar]
- 94. Robinson KM, Kolls JK, Alcorn JF. The immunology of influenza virus‐associated bacterial pneumonia. Curr Opin Immunol. 2015;34:59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. McGillivary G, Mason KM, Jurcisek JA, Peeples ME, Bakaletz LO. Respiratory syncytial virus‐induced dysregulation of expression of a mucosal β‐defensin augments colonization of the upper airway by non‐typeable Haemophilus influenzae . Cell Microbiol. 2009;11:1399–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Sanford BA, Shelokov A, Ramsay MA. Bacterial adherence to virus‐infected cells: A cell culture model of bacterial superinfection. J Infect Dis. 1978;137:176–181. [DOI] [PubMed] [Google Scholar]
- 97. Ramphal R, Small P, Shands J, Fischlschweiger W, Small P. Adherence of Pseudomonas aeruginosa to tracheal cells injured by influenza infection or by endotracheal intubation. Infect Immun. 1980;27:614–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Galina L. Possible mechanisms of viral‐bacterial interaction in swine. Swine Health Prod. 1995;3:9–14. [Google Scholar]
- 99. Babiuk LA, Lawman M, Ohmann HB. Viral‐bacterial synergistic interaction in respiratory disease. Advances in virus research. Volume 35. Elsevier, 1988; p. 219–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Iglesias G, Pijoan C. Effect of swine fever live vaccine on the mucociliary apparatus of swine, and its interaction with Pasteurella multocida . Rev Lat Microbiol. 1980;22:52. [Google Scholar]
- 101. Kleinerman ES, Daniels C, Polisson R, Snyderman R. Effect of virus infection on the inflammatory response. Depression of macrophage accumulation in influenza‐infected mice. Am J Pathol. 1976;85:373. [PMC free article] [PubMed] [Google Scholar]
- 102. Warr G, Jakab G, Hearst J. Alterations in lung macrophage immune receptor (s) activity associated with viral pneumonia. RES J Reticuloendothel Soc. 1979;26:357–366. [PubMed] [Google Scholar]
- 103. Warr GA, Jakab GJ. Alterations in lung macrophage antimicrobial activity associated with viral pneumonia. Infect Immun. 1979;26:492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Taneja R, Sharma A, Hallett M, Findlay G, Morris M. Immature circulating neutrophils in sepsis have impaired phagocytosis and calcium signaling. Shock (Augusta, GA). 2008;30:618–622. [DOI] [PubMed] [Google Scholar]
- 105. Grudzinska FS, Brodlie M, Scholefield BR, et al. Neutrophils in community‐acquired pneumonia: Parallels in dysfunction at the extremes of age. Thorax. 2020;75:164–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Ciencewicki J, Gowdy K, Krantz QT, et al. Diesel exhaust enhanced susceptibility to influenza infection is associated with decreased surfactant protein expression. Inhal Toxicol. 2007;19:1121–1133. [DOI] [PubMed] [Google Scholar]
- 107. Kongchanagul A, Suptawiwat O, Boonarkart C, et al. Decreased expression of surfactant protein D mRNA in human lungs in fatal cases of H5N1 avian influenza. J Med Virol. 2011;83:1410–1417. [DOI] [PubMed] [Google Scholar]
- 108. Hanada S, Pirzadeh M, Carver KY, Deng JC. Respiratory viral infection‐induced microbiome alterations and secondary bacterial pneumonia. Front Immunol. 2018;9:2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Qin Z, Yang Y, Wang H, et al. Role of autophagy and apoptosis in the Postinfluenza bacterial pneumonia. Biomed Res Int. 2016;2016:3801026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Sadler AJ, Williams BR. Interferon‐inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Shahangian A, Chow EK, Tian X, et al. Type I IFNs mediate development of postinfluenza bacterial pneumonia in mice. J Clin Invest. 2009;119:1910–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Robinson KM, McHugh KJ, Mandalapu S, et al. Influenza a virus exacerbates Staphylococcus aureus pneumonia in mice by attenuating antimicrobial peptide production. J Infect Dis. 2014;209:865–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Lee B, Robinson KM, McHugh KJ, et al. Influenza‐induced type I interferon enhances susceptibility to gram‐negative and gram‐positive bacterial pneumonia in mice. Am J Physiol Lung Cell Mol Physiol. 2015;309:L158–L167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Rynda‐Apple A, Robinson KM, Alcorn JF. Influenza and bacterial superinfection: Illuminating the immunologic mechanisms of disease. Infect Immun. 2015;83:3764–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Melvin JA, Bomberger JM. Compromised defenses: Exploitation of epithelial responses during viral‐bacterial co‐infection of the respiratory tract. PLoS Pathog. 2016;12:e1005797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Siegel SJ, Roche AM, Weiser JN. Influenza promotes pneumococcal growth during coinfection by providing host sialylated substrates as a nutrient source. Cell Host Microbe. 2014;16:55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Hendricks MR, Lashua LP, Fischer DK, et al. Respiratory syncytial virus infection enhances Pseudomonas aeruginosa biofilm growth through dysregulation of nutritional immunity. Proc Natl Acad Sci. 2016;113:1642–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Hennigar SR, McClung JP. Nutritional immunity: Starving pathogens of trace minerals. Am J Lifestyle Med. 2016;10:170–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Pawlowski A, Jansson M, Sköld M, Rottenberg ME, Källenius G. Tuberculosis and HIV co‐infection. PLoS Pathog. 2012;8:e1002464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Murray JF. Epidemiology of human immunodeficiency virus–associated pulmonary disease. Clin Chest Med. 2013;34:165–179. [DOI] [PubMed] [Google Scholar]
- 121. Vittor AY, Garland JM, Gilman RH. Molecular diagnosis of TB in the HIV positive population. Ann Glob Health. 2014;80:476–485. [DOI] [PubMed] [Google Scholar]
- 122. Tashiro M, Ciborowski P, Klenk H‐D, Pulverer G, Rott R. Role of Staphylococcus protease in the development of influenza pneumonia. Nature. 1987;325:536–537. [DOI] [PubMed] [Google Scholar]
- 123. Conenello GM, Zamarin D, Perrone LA, Tumpey T, Palese P. A single mutation in the PB1‐F2 of H5N1 (HK/97) and 1918 influenza a viruses contributes to increased virulence. PLoS Pathog. 2007;3:1414–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Chattoraj SS, Ganesan S, Jones AM, et al. Rhinovirus infection liberates planktonic bacteria from biofilm and increases chemokine responses in cystic fibrosis airway epithelial cells. Thorax. 2011;66:333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Pettigrew MM, Marks LR, Kong Y, Gent JF, Roche‐Hakansson H, Hakansson AP. Dynamic changes in the Streptococcus pneumoniae transcriptome during transition from biofilm formation to invasive disease upon influenza a virus infection. Infect Immun. 2014;82:4607–4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Chao Y, Marks LR, Pettigrew MM, Hakansson AP. Streptococcus pneumoniae biofilm formation and dispersion during colonization and disease. Front Cell Infect Microbiol. 2015;4:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. di Mauro G, Cristina S, Concetta R, Francesco R, Annalisa C. SARS‐Cov‐2 infection: Response of human immune system and possible implications for the rapid test and treatment. Int Immunopharmacol. 2020;84:106519–106519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Nikolich‐Zugich J, Knox KS, Rios CT, et al. SARS‐CoV‐2 and COVID‐19 in older adults: What we may expect regarding pathogenesis, immune responses, and outcomes. GeroScience. 2020;42(2):505–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Li CK‐f, Xu X. Host immune responses to SARS coronavirus in humans. Molecular biology of the SARS‐coronavirus, 2009; p. 259–278. [Google Scholar]
- 130. Frieman M, Baric R. Mechanisms of severe acute respiratory syndrome pathogenesis and innate immunomodulation. Microbiology and molecular biology reviews: MMBR 72, 672–685, Table of Contents, 2008. [DOI] [PMC free article] [PubMed]
- 131. Cinatl J, Morgenstern B, Bauer G, et al. Treatment of SARS with human interferons. Lancet (London, England). 2003;362:293–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Spiegel M, Pichlmair A, Martinez‐Sobrido L, et al. Inhibition of Beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two‐step model for activation of interferon regulatory factor 3. J Virol. 2005;79:2079–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Haagmans BL, Kuiken T, Martina BE, Fouchier RA, Rimmelzwaan GF, et al. Pegylated interferon‐alpha protects type 1 pneumocytes against SARS coronavirus infection in macaques. Nat Med. 2004;10:290–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Freundt EC, Yu L, Park E, Lenardo MJ, Xu XN. Molecular determinants for subcellular localization of the severe acute respiratory syndrome coronavirus open reading frame 3b protein. J Virol. 2009;83:6631–6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Narayanan K, Huang C, Lokugamage K, et al. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J Virol. 2008;82:4471–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Devaraj SG, Wang N, Chen Z, et al. Regulation of IRF‐3‐dependent innate immunity by the papain‐like protease domain of the severe acute respiratory syndrome coronavirus. J Biol Chem. 2007;282:32208–32221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Krahling V, Stein DA, Spiegel M, Weber F, Muhlberger E. Severe acute respiratory syndrome coronavirus triggers apoptosis via protein kinase R but is resistant to its antiviral activity. J Virol. 2009;83:2298–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Bezerra PG, Britto MC, Correia JB, Duarte MdCM, Fonceca AM, et al. Viral and atypical bacterial detection in acute respiratory infection in children under five years. PloS One. 2011;6:e18928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Binks MJ, Cheng AC, Smith‐Vaughan H, et al. Viral‐bacterial co‐infection in Australian indigenous children with acute otitis media. BMC Infect Dis. 2011;11:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Johansson N, Kalin M, Hedlund J. Clinical impact of combined viral and bacterial infection in patients with community‐acquired pneumonia. Scand J Infect Dis. 2011;43:609–615. [DOI] [PubMed] [Google Scholar]
- 141. Edrada EM, Lopez EB, Villarama JB, Villarama EPS, Dagoc BF, et al. First COVID‐19 infections in the Philippines: A case report. Trop Med Health. 2020;48:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Kaiser L, Regamey N, Roiha H, Deffernez C, Frey U. Human coronavirus NL63 associated with lower respiratory tract symptoms in early life. Pediatr Infect Dis J. 2005;24:1015–1017. [DOI] [PubMed] [Google Scholar]
- 143. Golda A, Malek N, Dudek B, et al. Infection with human coronavirus NL63 enhances streptococcal adherence to epithelial cells. J Gen Virol. 2011;92:1358–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Dijkman R, Jebbink MF, El Idrissi NB, et al. Human coronavirus NL63 and 229E seroconversion in children. J Clin Microbiol. 2008;46:2368–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Wang J‐b, Xu N, Shi H‐z, Huang X‐z, Lin L. Organism distribution and drug resistance in 7 cases of severe acute respiratory syndrome death patients with secondary bacteria infection. Chin Crit Care Med. 2003;15:523–525. [PubMed] [Google Scholar]
- 146. Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet. 2020;395:507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Zhang G, Hu C, Luo L, et al. Clinical features and outcomes of 221 patients with COVID‐19 in Wuhan, China. J Clin Virol. 2020;127:104364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Blasco ML, Buesa J, Colomina J, et al. Co‐detection of respiratory pathogens in patients hospitalized with coronavirus viral disease‐2019 pneumonia. J Med Virol. 2020. [DOI] [PubMed] [Google Scholar]
- 150. Claire D, Rémi Le G, Claire T, et al. Panton‐valentine Leukocidin–secreting Staphylococcus aureus pneumonia complicating COVID‐19. Emerg Infect Dis J. 2020;26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Bordi L, Nicastri E, Scorzolini L, et al. Differential diagnosis of illness in patients under investigation for the novel coronavirus (SARS‐CoV‐2), Italy, February 2020. Eurosurveillance. 2020;25:2000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Duployez C, Le Guern R, Tinez C, et al. Panton‐valentine Leukocidin‐secreting Staphylococcus aureus pneumonia complicating COVID‐19. Emerg Infect Dis. 2020;26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Fan BE, Lim KGE, Chong VCL, Chan SSW, Ong KH, Kuperan P. COVID‐19 and Mycoplasma pneumoniae coinfection. Am J Hematol. 2020;95:723–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Yu N, Li W, Kang Q, et al. Clinical features and obstetric and neonatal outcomes of pregnant patients with COVID‐19 in Wuhan, China: A retrospective, single‐Centre, descriptive study. Lancet Infect Dis. 2020;20:559–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Wang Z, Yang B, Li Q, Wen L, Zhang R. Clinical features of 69 cases with coronavirus disease 2019 in Wuhan, China. Clin Infect Dis. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Chakraborty S. Metagenome of SARS‐Cov2 patients in Shenzhen with travel to Wuhan shows a wide range of species‐Lautropia, Cutibacterium, Haemophilus being most abundant‐and Campylobacter explaining diarrhea, 2020.
- 157. Chakraborty S. The 2019 Wuhan outbreak could be caused by the bacteria Prevotella, which is aided by the coronavirus, possibly to adhere to epithelial cells‐Prevotella is present in huge amounts in patients from both China and Hong Kong, 2020.
- 158. Chakraborty S. San Diego county Nanopore SARS‐Cov2 sequencing data shows metagenomic Prevotella, Haemophilus parainfluenzae, a lot of unknown species and chimeric reads, 2020.
- 159. Chakraborty S. Metagenome of SARS‐Cov2 from a patient in Brazil shows a wide range of bacterial species‐Lautropia, Prevotella, Haemophilus‐overshadowing viral reads, which does not even add up to a full genome, explaining false negatives, 2020.
- 160. Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LF. The trinity of COVID‐19: Immunity, inflammation and intervention. Nat Rev Immunol. 2020;20:363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. MacIntyre CR, Chughtai AA, Barnes M, et al. The role of pneumonia and secondary bacterial infection in fatal and serious outcomes of pandemic influenza a(H1N1)pdm09. BMC Infect Dis. 2018;18:637–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Cox MJ, Loman N, Bogaert D, O'Grady J. Co‐infections: Potentially lethal and unexplored in COVID‐19. Lancet Microbe. 2020;1(1):e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID‐19 in Wuhan, China: A retrospective cohort study. Lancet. 2020;395:1054–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Wu F, Zhao S, Yu B, et al. Complete genome characterisation of a novel coronavirus associated with severe human respiratory disease in Wuhan, China. bioRxiv. 2020. [Google Scholar]
- 165. Phua J, Weng L, Ling L, et al. Intensive care management of coronavirus disease 2019 (COVID‐19): Challenges and recommendations. Lancet Respir Med. 2020;8:506–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Gautret P, Lagier J‐C, Parola P, et al. Hydroxychloroquine and azithromycin as a treatment of COVID‐19: Results of an open‐label non‐randomized clinical trial. Int J Antimicrob Agents. 2020;105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 167. Singh AK, Singh A, Shaikh A, Singh R, Misra A. Chloroquine and hydroxychloroquine in the treatment of COVID‐19 with or without diabetes: A systematic search and a narrative review with a special reference to India and other developing countries. Diabetes Metab Syndr. 2020;14:241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Serisier DJ. Risks of population antimicrobial resistance associated with chronic macrolide use for inflammatory airway diseases. Lancet Respir Med. 2013;1:262–274. [DOI] [PubMed] [Google Scholar]
- 169. Devaux, C , Colson P, Raoult D, Rolain J‐M. Teicoplanin: An alternative drug for the treatment of coronavirus COVID‐19. Int J Antimicrob Agents. 2020;(4):105944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Du Y, Tu L, Zhu P, et al. Clinical features of 85 fatal cases of COVID‐19 from Wuhan: A retrospective observational study. Am J Respir Crit Care Med. 2020;201:1372–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Willyard C. The drug‐resistant bacteria that pose the greatest health threats. Nat News. 2017;543:15. [DOI] [PubMed] [Google Scholar]
- 172. Hagan T, Cortese M, Rouphael N, et al. Antibiotics‐driven gut microbiome perturbation alters immunity to vaccines in humans. Cell. 2019;178:1313–1328.e1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Kelly PN. Antibiotics blunt flu immunity. Science. 2019;366:197–198. [Google Scholar]
- 174. Birger RB, Kouyos RD, Cohen T, et al. The potential impact of coinfection on antimicrobial chemotherapy and drug resistance. Trends Microbiol. 2015;23:537–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Römling U, Balsalobre C. Biofilm infections, their resilience to therapy and innovative treatment strategies. J Intern Med. 2012;272:541–561. [DOI] [PubMed] [Google Scholar]


