Abstract
An outbreak of pneumonia, caused by a novel coronavirus (SARS-CoV-2), was identified in China in December 2019. This virus expanded worldwide, causing global concern. Although clinical, laboratory, and imaging features of COVID-19 are characterized in some observational studies, we undertook a systematic review and meta-analysis to assess the frequency of these features. We did a systematic review and meta-analysis using three databases to identify clinical, laboratory, and computerized tomography (CT) scanning features of rRT-PCR confirmed cases of COVID-19. Data for 3420 patients from 30 observational studies were included. Overall, the results showed that fever (84.2%, 95% CI 82.6-85.7), cough (62%, 95% CI 60-64), and fatigue (39.4%, 95% CI 37.2-41.6%) are the most prevalent symptoms in COVID-19 patients. Increased CRP level, decreased lymphocyte count, and increased D-dimer level were the most common laboratory findings. Among COVID-19 patients, 92% had a positive CT finding, most prevalently ground-glass opacification (GGO) (60%, 95% CI 58-62) and peripheral distribution opacification (64%, 95% CI 60-69). These results demonstrate the clinical, paraclinical, and imaging features of COVID-19.
1. Background
In December 2019, the first case of unknown origin pneumonia was identified in Wuhan, the capital city of Hubei Province. By January 7, 2020, Chinese scientists had isolated a novel virus belongs to coronaviruses family and classified it as a type of RNA virus [1].
Although the outbreak has been started from a primary zoonotic virus transmission in a large seafood market (Huanan Seafood Market), person-to-person transmission of the virus started a pandemic involving 197 countries [2, 3].
The clinical outcomes of SARS-CoV-2 infection are various, including asymptomatic infection, mild upper respiratory tract illness, severe viral pneumonia, and even death. Patients are presented with various clinical manifestations such as fever, dyspnea, and cough [4].
Initially, some studies have observed particular imaging patterns on chest radiography and computed tomography in COVID-19 patients [5]. As our knowledge increased, recent studies claimed that the sensitivity of CT scan is higher compared to rRT-PCR in the diagnosis of COVID-19 [6].
Laboratory findings are essential in order to evaluate patients' complications and triaging them [7]. Complete blood count as an easy and affordable test detects disorders such as leukopenia, anemia, and thrombocytopenia that are contributed to patients' prognosis [8]. In response to inflammation induced by COVID-19, acute-phase reactants may increase or decrease [9]. These factors may contribute to the patients' outcomes. This meta-analysis is aimed at measuring the most common clinical, laboratory, and imaging findings among COVID-19 patients.
2. Methods
2.1. Protocol
In this study, we used a protocol based on the transparent reporting of systematic reviews and meta-analysis (PRISMA) (Figures 1–3).
Figure 1.

PRISMA flow chart of search, inclusion and exclusion screening, and accepted studies of the review on the clinical characteristics.
Figure 2.
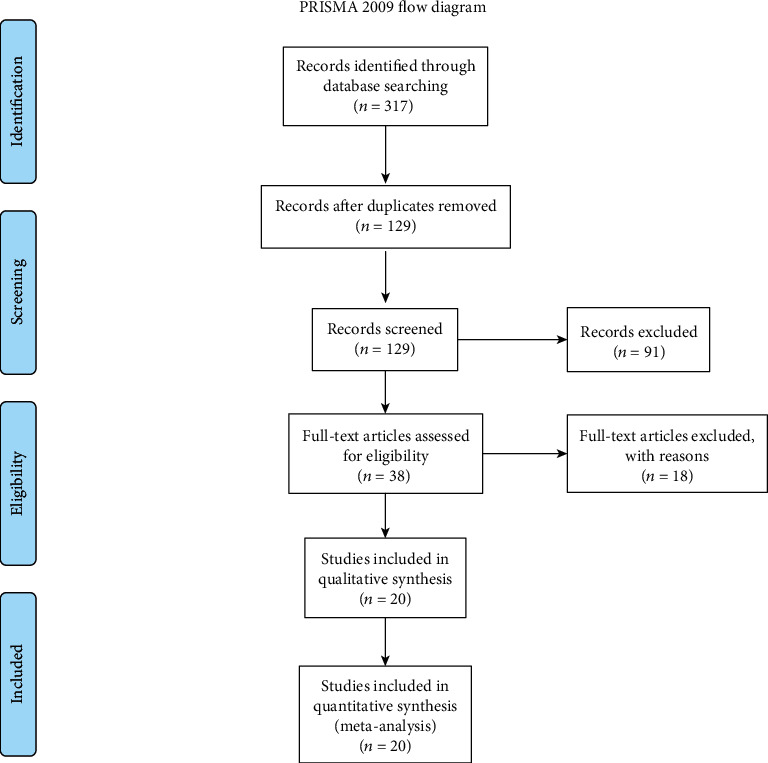
PRISMA flow chart of search, inclusion and exclusion screening, and accepted studies of the review on the imaging studies.
Figure 3.
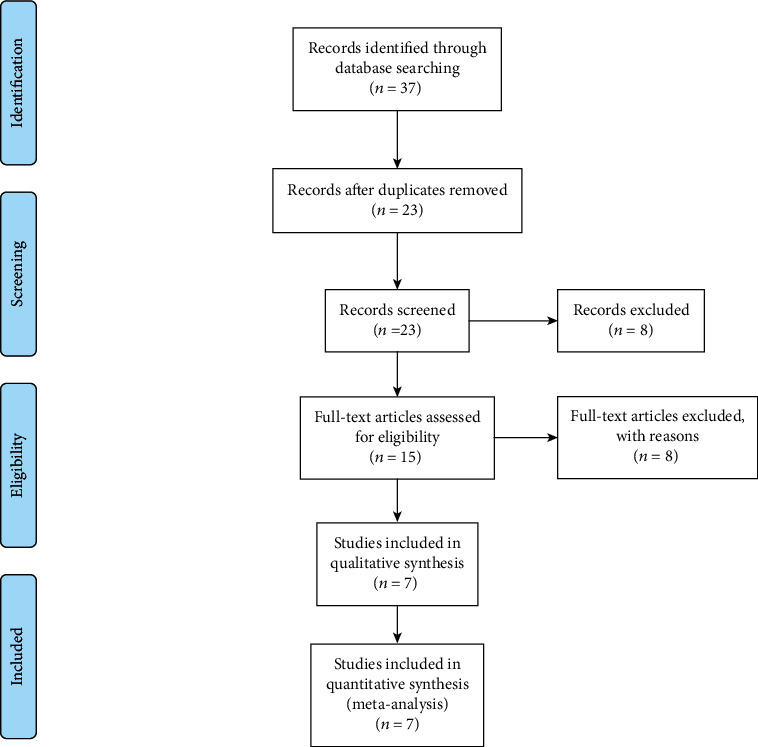
PRISMA flow chart of search, inclusion and exclusion screening, and accepted studies of the review on the laboratory findings.
2.2. Eligibility Criteria
In this study, all included patients were confirmed using real-time reverse transcriptase-polymerase chain reaction (rRT-PCR). All searched articles were cross-sectional studies, reporting descriptive data, and no language restrictions were conducted. All articles published before drafting the manuscript have been included. Review articles, opinion articles, and letters not presenting original data were excluded from the analysis.
2.3. Information Sources and Search Strategy
Three systematic searches were performed using Medline/PubMed, Scopus, and Web of Science. Systematic search was conducted prior to March 25, 2020, and three independent researchers evaluated all papers. The search was conducted based on the following keywords, and all studies were divided in three groups: (1) For clinical characteristics group: (“clinical manifestation” AND COVID-19) or (“clinical manifestation” AND 2019-nCoV) or (“clinical manifestation” AND COVID) or (“clinical manifestation” AND Corona) or (“clinical characteristics” AND COVID-19) or (“clinical characteristics” AND 2019-nCoV) or (“clinical characteristics” AND COVID) or (“clinical characteristics” AND corona). (2) For laboratory findings group: (liver AND COVID-19) or (liver AND 2019-nCoV) or (liver AND COVID) or (liver AND Corona) or (“blood gas” AND COVID-19) or (“blood gas” AND 2019-nCoV) or (“blood gas” AND COVID) or (“blood gas” AND corona). (3) For imaging studies group: (COVID-19 AND radiography) or (2019-nCoV AND radiography) or (Corona AND radiography) or (COVID AND radiography) or (COVID-19 AND radiographic) or (2019-nCoV AND radiographic) or (Corona AND radiographic) or (COVID AND radiographic) or (COVID-19 AND CT) or (2019-nCoV AND CT) or (Corona AND CT) or (COVID AND CT) or (COVID-19 AND “computed tomography”) or (2019-nCoV AND “computed tomography”) or (Corona AND “computed tomography”) or (COVID AND “computed tomography”) or (CBC AND corona) or (CBC AND COVID) or (CBC AND COVID-19) or (CBC AND 2019-nCoV).
2.4. Study Selection
In the initial search, we assessed the title and abstract, followed by a full-text evaluation based on previously described inclusion and exclusion criteria. When two articles reported one patient's characteristics, we merged all reported data and assumed as a single individual. Descriptive studies reporting clinical symptoms, laboratory, and radiological findings were used to perform a meta-analysis. The characteristics of the included studies are shown in Table 1. The modified appraisal tool for cross-sectional studies (AXIS) was used to determine the methodological quality of the research designs of the included studies (Table 2). AXIS is used to assess research papers systematically and to judge the reliability of the study being presented in the paper. It also helps in assessing the worth and relevance of the study. Studies with total scores of ten or less were excluded.
Table 1.
Characteristics of the included studies.
| Row | Author | Journal | Type | Date | Country | Sample size | Reference |
|---|---|---|---|---|---|---|---|
| Imaging | |||||||
| 1 | Fang et al. | Radiology | Cross-sectional | Feb 19 | China | 51 | [6] |
| 2 | Zhao et al. | American Journal of Roentgenology | Cross-sectional | Feb 18 | China | 101 | [38] |
| 3 | Shi et al. | The Lancet | Cross-sectional | Feb 24 | China | 81 | [40] |
| 4 | Pan et al. | European Radiology | Cross-sectional | Feb 13 | China | 63 | [41] |
| 5 | Xu et al. | European Journal of Nuclear Medicine and Molecular Imaging | Cross-sectional | Feb 28 | China | 90 | [42] |
| 6 | Zhang et al. | European Respiratory Journal | Cross-sectional | Mar 25 | China | 17 | [43] |
| 7 | Chen et al. | The Lancet | Cross-sectional | Jan 30 | China | 99 | [44] |
| 8 | Huang | The Lancet | Cross-sectional | Feb 21 | China | 41 | [18] |
| 9 | Wu et al. | Clinical Infectious Diseases | Cross-sectional | Feb 29 | China | 80 | [19] |
| 10 | Guan et al. | The New England Journal of Medicine | Cross-sectional | Feb 28 | China | 1099 | [45] |
| 11 | Wang et al. | Clinical Infectious Diseases | Cross-sectional | Feb 29 | China | 138 | [46] |
| 12 | Yang et al. | Journal of Infection | Cross-sectional | Feb 26 | China | 149 | [47] |
| 13 | Xu et al. | Journal of Infection | Cross-sectional | Feb 25 | China | 50 | [48] |
| 14 | Wu et al. | Investigative Radiology | Cross-sectional | Feb 29 | China | 80 | [49] |
| 15 | Li et al. | Investigative Radiology | Cross-sectional | Feb 29 | China | 83 | [50] |
| 16 | Xia et al. | Pediatric Pulmonology | Cross-sectional | Mar 05 | China | 20 | [51] |
| 17 | Zhang et al. | Allergy | Cross-sectional | Feb 19 | China | 140 | [52] |
| 18 | Zhou et al. | American Journal of Roentgenology | Cross-sectional | Feb 16 | China | 62 | [53] |
| 19 | Wang et al. | Journal of Zhejiang University | Cross-sectional | Feb 24 | China | 52 | [54] |
| 20 | Yoon et al. | Korean J Radiol | Cross-sectional | Apr 21 | China | 9 | [55] |
| Laboratory | |||||||
| 21 | Qian et al. | An International Journal of Medicine | Cross-sectional | Feb 21 | China | 91 | [56] |
| 22 | Liu et al. | medRxiv | Cross-sectional | Feb 21 | China | 109 | [57] |
| 23 | Chen et al. | medRxiv | Cross-sectional | Feb 14 | China | 21 | [58] |
| 24 | Young et al. | Jama | Cross-sectional | Feb 24 | China | 18 | [59] |
| 25 | Fan et al. | The Lancet | Cross-sectional | Mar 05 | China | 148 | [30] |
| 26 | Wu et al. | Clinical Infectious Diseases | Cross-sectional | Feb 29 | China | 80 | [19] |
| 27 | Guan et al. | New England Journal of Medicine | Cross-sectional | Feb 28 | China | 1099 | [45] |
| Clinical characteristics | |||||||
| 28 | Chen et al. | The Lancet | Cross-sectional | Feb 21 | China | 99 | [44] |
| 29 | Deng and Peng | Journal Clinical Medicine | Cross-sectional | Feb 14 | China | 41 | [60] |
| 30 | Huang | Clinical Gastroenterology and Hepatology | Cross-sectional | Feb 21 | China | 41 | [18] |
| 31 | Guan et al. | The New England Journal of Medicine | Cross-sectional | Feb 19 | China | 1099 | [45] |
| 32 | Huang et al. | Travel Medicine and Infectious Disease | Cross-sectional | Feb 24 | China | 34 | [61] |
| 33 | Kui et al. | Chinese Medical Journal | Cross-sectional | Feb 07 | China | 137 | [62] |
| 34 | Tian et al. | Journal of Infection | Cross-sectional | Feb 27 | China | 262 | [63] |
| 35 | Wang et al. | Jama | Cross-sectional | Feb 21 | China | 138 | [46] |
| 36 | Wu et al. | Clinical Infectious Diseases | Cross-sectional | Feb 29 | China | 80 | [19] |
| 37 | Xu et al. | European Journal of Nuclear Medicine and Molecular Imaging | Cross-sectional | Feb 28 | China | 90 | [42] |
| 38 | Xiao-Wei et al. | BMJ: British Medical Journal | Cross-sectional | Feb 19 | China | 62 | [64] |
| 39 | Xu et al. | Journal of Infection | Cross-sectional | Feb 25 | China | 50 | [48] |
| 40 | Yang et al. | Journal of Infection | Cross-sectional | Feb 26 | China | 149 | [47] |
| 41 | Zhang et al. | Allergy | Cross-sectional | Feb 19 | China | 140 | [52] |
Table 2.
Appraisal tool for cross-sectional studies (AXIS) scores of included studies.
| Row | Author | Date | AXIS score | Reference | |||||
|---|---|---|---|---|---|---|---|---|---|
| Introduction | Methods | Results | Discussion | Others | Total | ||||
| 1 | Fang et al. | Feb 19 | 1 | 6 | 4 | 2 | 2 | 15 | [6] |
| 2 | Zhao et al. | Feb 18 | 1 | 8 | 3 | 2 | 2 | 16 | [38] |
| 3 | Shi et al. | Feb 24 | 1 | 9 | 3 | 2 | 2 | 17 | [40] |
| 4 | Pan et al. | Feb 13 | 1 | 7 | 2 | 2 | 2 | 14 | [41] |
| 5 | Xu et al. | Feb 28 | 1 | 6 | 4 | 2 | 2 | 15 | [42] |
| 6 | Zhang et al. | Mar 25 | 1 | 7 | 2 | 2 | 2 | 14 | [43] |
| 7 | Chen et al. | Jan 30 | 1 | 8 | 5 | 2 | 2 | 18 | [44] |
| 8 | Huang | Feb 21 | 1 | 8 | 5 | 2 | 2 | 18 | [18] |
| 9 | Wu et al. | Feb 29 | 1 | 6 | 2 | 2 | 1 | 12 | [19] |
| 10 | Guan et al. | Feb 28 | 1 | 9 | 5 | 2 | 2 | 19 | [45] |
| 11 | Wang et al. | Feb 29 | 1 | 9 | 5 | 2 | 2 | 19 | [46] |
| 12 | Yang et al. | Feb 26 | 1 | 5 | 3 | 2 | 2 | 13 | [47] |
| 13 | Xu et al. | Feb 25 | 1 | 6 | 4 | 2 | 2 | 15 | [48] |
| 14 | Wu et al. | Feb 29 | 1 | 9 | 3 | 2 | 2 | 17 | [49] |
| 15 | Li et al. | Feb 29 | 1 | 8 | 5 | 2 | 2 | 18 | [50] |
| 16 | Xia et al. | Mar 05 | 1 | 5 | 2 | 2 | 2 | 12 | [51] |
| 17 | Zhang et al. | Feb 19 | 1 | 6 | 2 | 1 | 2 | 12 | [52] |
| 18 | Zhou et al. | Feb 16 | 1 | 5 | 4 | 2 | 2 | 14 | [53] |
| 19 | Wang et al. | Feb 24 | 1 | 7 | 3 | 2 | 2 | 15 | [54] |
| 20 | Yoon et al. | Apr 21 | 1 | 4 | 3 | 2 | 2 | 12 | [55] |
| 21 | Qian et al. | Feb 21 | 1 | 7 | 4 | 2 | 2 | 16 | [56] |
| 22 | Liu et al. | 1 | 6 | 4 | 2 | 2 | 15 | [57] | |
| 23 | Chen et al. | 1 | 5 | 2 | 2 | 2 | 12 | [58] | |
| 24 | Young et al. | Feb 24 | 1 | 8 | 4 | 2 | 2 | 17 | [59] |
| 25 | Fan et al. | Mar 05 | 1 | 8 | 3 | 2 | 2 | 16 | [30] |
| 29 | Deng and Peng | Feb 14 | 1 | 5 | 5 | 2 | 2 | 15 | [60] |
| 32 | Huang et al. | Feb 24 | 1 | 5 | 3 | 2 | 2 | 13 | [61] |
| 33 | Kui et al. | Feb 07 | 1 | 5 | 3 | 2 | 2 | 13 | [62] |
| 34 | Tian et al. | Feb 27 | 1 | 5 | 3 | 2 | 2 | 13 | [63] |
| 38 | Xiao-Wei et al. | Feb 19 | 1 | 7 | 2 | 2 | 2 | 14 | [64] |
2.5. Data Collection Process and Data Items
Three independent researchers filled data extraction forms containing study type, journal, publication date, sample size, age, gender, clinical characteristics, laboratory, and radiological findings. Conflicts were resolved by another researcher.
2.6. Assessment of Methodological Quality and Risk of Bias
Publication bias was assessed with a funnel plot for the standard error and considering that the interpretation of the plot is subjective (Figures 4–6). Also, bias was quantified by using the Egger regression test.
Figure 4.

Funnel plot for clinical characteristics group studies.
Figure 5.
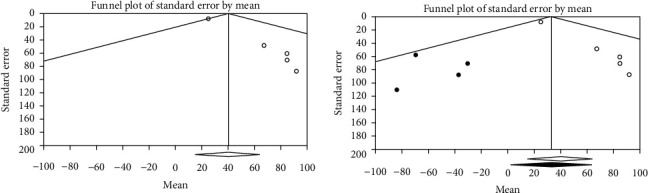
Funnel plots for observed studies and imputed studies (trim and fill test) of laboratory findings. The observed studies are shown as open circles, and the four imputed studies are shown as filled circles. The observed point estimate is shown as an open diamond, and the imputed point estimate is shown as a filled diamond.
Figure 6.

Funnel plots for observed studies and imputed studies (trim and fill test) of imaging studies. The observed studies are shown as open circles, and the four imputed studies are shown as filled circles. The observed point estimate is shown as an open diamond, and the imputed point estimate is shown as a filled diamond.
Sensitivity analysis and adjusting for risk bias were performed by the attractive test (trim and fill method). We initially identified and trimmed the asymmetric (missing) studies, followed by estimating the unbiased summary effect. Sensitivity analysis was also performed using the “Remove-One” analysis by running the analysis with each of the studies removed. The result of the impact of each study on the pooled estimate is shown in the forest plot (Figure 7).
Figure 7.

Sensitivity analysis of the meta-analysis for each of the included studies.
The percentage of total variation across studies (heterogeneity) was measured by the inconsistency index tool (I squared). The I squared index measured for each of the clinical characteristics, imaging studies, and laboratory findings groups. I squared index value in the ranges of <25%, 25–50%, 50–75%, and >75% was interpreted as low, moderate, high, and very high heterogeneity, respectively [10].
We conducted a random-effects analysis because it was assumed that some of the included studies did not share a common effect size (heterogeneity). Findings in each group are summarized as forest plots in Figures 8–10.
Figure 8.

Pool prevalence forest plots of clinical manifestations.
Figure 9.
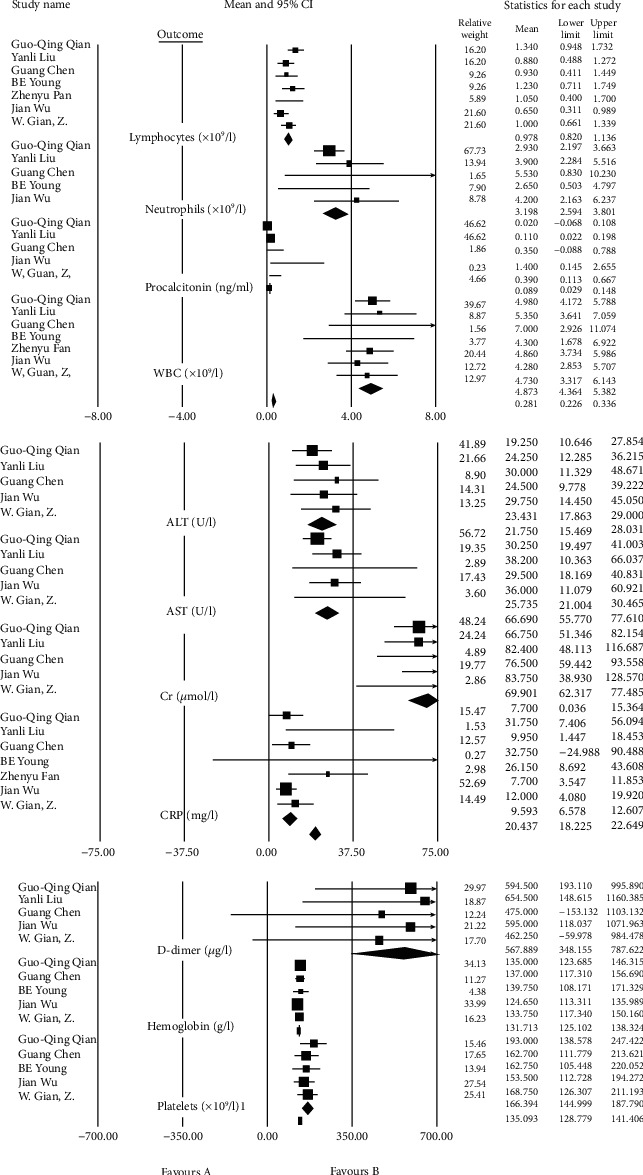
Pool prevalence forest plots of laboratory findings.
Figure 10.

Pool prevalence forest plots of CT scan findings.
2.7. Statistical Approach
Effect size pooled estimate for imaging and clinical data was measured based on event rate, logit event rate, and standard error.
Considering that the computational index of laboratory data was median in order to meta-analyze them in CMA v.2. software, we use the following formula:
Estimating the mean and variance from the median:
| (1) |
where m is the median, a is the smallest value (minimum), b is the largest value (maximum), and n is the size of the sample [11].
The meta-analysis was performed using STATA, the software OpenMeta[Analyst], and Comprehensive Meta-Analysis Software (CMA) ve.2. Pooled estimate and their 95% confidence intervals (95% CIs) were used to summarize the weighted effect size for each study grouping variable.
3. Results
3.1. Study Selection and Characteristics
Two hundred seven articles were included based on a search strategy, which are previously described (Table 1). The full text of 65 articles was evaluated after the title and abstract assessment. Twenty-four articles were excluded due to inadequate data. Finally, the meta-analysis was performed on 30 articles (three different subjects). The article's data summary is reported in Table 2. Also, demographic characteristics and comorbidities of patients participated in the included studies are demonstrated in Table 3.
Table 3.
Demographical characteristics and comorbidities of patients in the included studies.
| Row | Author | Date | Sample size | Mean age (y. old) | Age range | Sex (male) | Diabetes | Hypertension | Cardiovascular disease | COPD | Malignancies | Digestive system disease |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Fang et al. | Feb 19 | 51 | 45 | 39-55 | 29 | — | — | — | — | — | — |
| 2 | Zhao et al. | Feb 18 | 101 | 44/44 | 17-75 | 56 | — | — | 15/8 | 4/9 | — | — |
| 3 | Shi et al. | Feb 24 | 81 | 49/5 | 39-61 | 42 | 12 | 15 | 10 | 11 | 5 | 9 |
| 4 | Pan et al. | Feb 13 | 63 | 44/9 | 31-62 | 33 | — | — | — | — | — | — |
| 5 | Xu et al. | Feb 28 | 90 | 50 | 18-86 | 39 | 6 | 19 | 3 | 1 | 2 | 2 |
| 6 | Zhang et al. | Mar 25 | 17 | 48/6 | 23-74 | 8 | — | 11/7 | 5 | 11 | — | 11/7 |
| 7 | Chen et al. | Jan 30 | 99 | 55/5 | 21-82 | 67 | — | — | 40 | 1 | 1 | 11 |
| 8 | Huang | Feb 21 | 41 | 49 | 41-58 | 30 | 20 | 15 | 15 | 2 | 2 | 2 |
| 9 | Wu et al. | Feb 29 | 80 | 46 | 18-65 | 39 | — | — | 31/25 | 1/25 | 1/25 | 3/75 |
| 10 | Guan et al. | Feb 28 | 1096 | 49 | 35-58 | 637 | 7/4 | 15 | 3/9 | 1/1 | 0/9 | 2/1 |
| 11 | Wang et al. | Feb 29 | 138 | 56 | 42-68 | 75 | 10/1 | 31/2 | 14/5 | 2/9 | 7/2 | 2/9 |
| 12 | Yang et al. | Feb 26 | 149 | 45/1 | 30-68 | 81 | — | — | 18/79 | 0/67 | 1/34 | 5/37 |
| 13 | Xu et al. | Feb 25 | 50 | 43 | 3-85 | 29 | — | — | — | — | — | — |
| 14 | Wu et al. | Feb 29 | 80 | 44 | 30-52 | 42 | 5 | 5 | 1 | 4 | — | — |
| 15 | Li et al. | Feb 29 | 83 | 45/5 | 25-64 | 44 | 7/8 | 6 | 1/2 | 6 | — | — |
| 16 | Xia et al. | Mar 05 | 20 | 1 | 0-7 | 13 | — | — | — | — | — | — |
| 17 | Zhang et al. | Feb 19 | 135 | 57 | 25-87 | 71 | 12 | 30 | 12/1 | 2/8 | — | 10/7 |
| 18 | Zhou et al. | Feb 16 | 62 | 52/8 | 30-77 | 39 | 6 | 6 | 1 | — | — | — |
| 19 | Wang et al. | Feb 24 | 52 | — | — | — | — | — | — | — | — | — |
| 20 | Yoon et al. | Apr 21 | 9 | 54 | — | — | — | — | — | — | — | — |
| 21 | Qian et al. | Feb 21 | 91 | 50 | 36-57 | 37 | 8/79 | 16/48 | 3/3 | — | — | — |
| 22 | Liu et al. | 109 | 55 | 43-66 | 59 | 11 | 33 | 6/4 | 3/7 | — | — | |
| 23 | Chen et al. | 21 | 56 | — | 17 | 14/3 | 23/8 | — | — | — | — | |
| 24 | Young et al. | Feb 24 | 18 | 47 | 31-73 | 9 | — | — | — | — | — | — |
| 25 | Fan et al. | Mar 05 | 148 | 50 | 36-64 | 73 | — | — | — | — | — | — |
| 26 | Xu et al. | Feb 19 | 62 | 41 | 32-52 | 36 | 2 | 8 | 2 | 2 | — | 11 |
| 27 | Deng and Peng | Feb 14 | 41 | 55/5 | 25-89 | 42/3 | 53/8 | 19/2 | 19/2 | — | — | |
| 28 | Huang et al. | Feb 24 | 34 | 56/24 | 26-88 | 14 | 11/8 | 23/5 | 17/6 | 2/9 | 8/8 | 2/9 |
| 29 | Kui et al. | Feb 07 | 137 | 57 | 20-83 | 61 | 9/5 | 10/2 | 7/3 | 1/5 | 1/5 | — |
| 30 | Tian et al. | Feb 27 | 262 | 47/5 | 1-94 | 127 | — | — | — | — | — | — |
In this study, we evaluate 30 articles. All papers were from China, and 3420 individual's data were evaluated. All studies were cross-sectional, and 27 variables were included.
3.2. Heterogeneity
Evaluating the heterogeneity of the studies indicated that in the clinical characteristics and imaging studies groups, the combined effect of the I squared index is high (68.43, 68.53). While the laboratory findings groups combined effect of the I squared index is considered low (6.12). I squared index for each of the outcomes is shown in Tables 4–7.
Table 4.
A meta-analysis of clinical characteristics group.
| Clinical group | Effect size and 95% confidence interval | Test of null (2-tail) | Heterogeneity | ||||
|---|---|---|---|---|---|---|---|
| Outcome | Number of studies | Point estimate | Lower limit | Upper limit | Z value | P value | I squared |
| Cough | 14 | 0.601 | 0.535 | 0.664 | 2.975 | 0.003 | 87.304 |
| Fever | 14 | 0.843 | 0.786 | 0.887 | 8.650 | 0.000 | 87.006 |
| Headache | 12 | 0.091 | 0.070 | 0.118 | -15.933 | 0.000 | 57.760 |
| Diarrhea | 11 | 0.064 | 0.043 | 0.095 | -12.258 | 0.000 | 71.281 |
| Fatigue | 11 | 0.394 | 0.291 | 0.508 | -1.827 | 0.068 | 94.073 |
| Dyspnea | 8 | 0.171 | 0.091 | 0.298 | -4.290 | 0.000 | 92.769 |
| Expectoration | 5 | 0.239 | 0.164 | 0.334 | -4.840 | 0.000 | 77.659 |
| Hemoptysis | 5 | 0.023 | 0.009 | 0.057 | -7.652 | 0.000 | 70.741 |
| Shortness of breath | 5 | 0.241 | 0.151 | 0.361 | -3.898 | 0.000 | 87.758 |
| Sore throat | 5 | 0.130 | 0.085 | 0.193 | -7.866 | 0.000 | 78.329 |
| Muscle ache | 4 | 0.114 | 0.052 | 0.231 | -4.749 | 0.000 | 83.406 |
| Nausea | 2 | 0.109 | 0.038 | 0.275 | -3.633 | 0.000 | 81.783 |
Table 5.
Meta-analysis of laboratory findings before removing Fan et al. study.
| Group LAB test | Effect size and 95% confidence interval | Test of null (2-tail) | Heterogeneity | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Outcome | Number of studies | Point estimate | Standard error | Variance | Lower limit | Upper limit | Z value | P value | I squared |
| CRP (mg/l) | 7 | 10.78 | 2.21 | 4.89 | 6.44 | 15.11 | 4.87 | 0.0000 | 30.63 |
| Lymphocytes (×109/l) | 7 | 1.00 | 0.13 | 0.02 | 0.73 | 1.26 | 7.48 | 0.0000 | 45.87 |
| WBC (×109/l) | 7 | 4.87 | 0.26 | 0.07 | 4.36 | 5.38 | 18.76 | 0.0000 | 0.00 |
| ALT (U/l) | 5 | 23.43 | 2.84 | 8.07 | 17.86 | 29.00 | 8.25 | 0.0000 | 0.00 |
| AST (U/l) | 5 | 25.84 | 2.47 | 6.08 | 21.01 | 30.68 | 10.48 | 0.0000 | 1.71 |
| Cr (μmol/l) | 5 | 69.90 | 3.87 | 14.97 | 62.32 | 77.48 | 18.06 | 0.0000 | 0.00 |
| D-dimer (μg/l) | 5 | 567.89 | 112.11 | 12568.89 | 348.15 | 787.62 | 5.07 | 0.0000 | 0.00 |
| Hemoglobin (g/l) | 5 | 131.71 | 3.37 | 11.38 | 125.10 | 138.32 | 39.05 | 0.0000 | 0.00 |
| LDH (U/l) | 5 | 258.56 | 26.39 | 696.51 | 206.84 | 310.29 | 9.80 | 0.0000 | 11.06 |
| Neutrophils (×109/l) | 5 | 3.20 | 0.31 | 0.09 | 2.59 | 3.80 | 10.38 | 0.0000 | 0.00 |
| Platelets (×109/l) | 5 | 166.39 | 10.92 | 119.17 | 145.00 | 187.79 | 15.24 | 0.0000 | 0.00 |
| Procalcitonin (ng/ml) | 5 | 0.17 | 0.08 | 0.01 | 0.01 | 0.32 | 2.15 | 0.0312 | 68.47 |
| Creatine kinase (U/l) | 4 | 110.18 | 19.92 | 396.72 | 71.14 | 149.22 | 5.53 | 0.0000 | 0.00 |
| Total bilirubin (mmol/l) | 4 | 8.36 | 1.08 | 1.16 | 6.25 | 10.48 | 7.76 | 0.0000 | 0.00 |
| Albumin (g/l) | 3 | 38.61 | 1.46 | 2.14 | 35.75 | 41.48 | 26.41 | 0.0000 | 44.52 |
| BUN (mmol/l) | 3 | 4.83 | 0.55 | 0.30 | 3.75 | 5.91 | 8.77 | 0.0000 | 0.00 |
| ESR | 2 | 40.79 | 24.93 | 621.75 | -8.08 | 89.67 | 1.64 | 0.1018 | 91.15 |
| Fibrinogen (g/l) | 2 | 3.21 | 0.22 | 0.05 | 2.78 | 3.64 | 14.71 | 0.0000 | 0.00 |
Table 6.
Meta-analysis of laboratory findings after removing Fan et al. study.
| Laboratory findings group (one study removed) | Effect size and 95% confidence interval | Test of null (2-tail) | Heterogeneity | ||||
|---|---|---|---|---|---|---|---|
| Outcome | Number of studies | Point estimate | Lower limit | Upper limit | Z value | P value | I squared |
| CRP (mg/l) | 6 | 9.13 | 6.00 | 12.27 | 5.71 | 0.00 | 1.73 |
| Lymphocytes (×109/l) | 6 | 0.99 | 0.78 | 1.20 | 9.20 | 0.00 | 37.93 |
| WBC (×109/l) | 6 | 4.88 | 4.31 | 5.45 | 16.75 | 0.00 | 0.00 |
| ALT (U/l) | 5 | 23.43 | 17.86 | 29.00 | 8.25 | 0.00 | 0.00 |
| AST (U/l) | 5 | 25.84 | 21.01 | 30.68 | 10.48 | 0.00 | 1.71 |
| Cr (μmol/l) | 5 | 69.90 | 62.32 | 77.48 | 18.06 | 0.00 | 0.00 |
| D-dimer (μg/l) | 5 | 567.89 | 348.15 | 787.62 | 5.07 | 0.00 | 0.00 |
| Hemoglobin (g/l) | 5 | 131.71 | 125.10 | 138.32 | 39.05 | 0.00 | 0.00 |
| LDH (U/l) | 5 | 258.56 | 206.84 | 310.29 | 9.80 | 0.00 | 11.06 |
| Neutrophils (×109/l) | 5 | 3.20 | 2.59 | 3.80 | 10.38 | 0.00 | 0.00 |
| Platelets (×109/l) | 5 | 166.39 | 145.00 | 187.79 | 15.24 | 0.00 | 0.00 |
| Procalcitonin (ng/ml) | 5 | 0.17 | 0.01 | 0.32 | 2.15 | 0.03 | 68.47 |
| Creatine kinase (U/l) | 4 | 110.18 | 71.14 | 149.22 | 5.53 | 0.00 | 0.00 |
| Total bilirubin (mmol/l) | 4 | 8.36 | 6.25 | 10.48 | 7.76 | 0.00 | 0.00 |
| Albumin (g/l) | 3 | 38.61 | 35.75 | 41.48 | 26.41 | 0.00 | 44.52 |
| BUN (mmol/l) | 3 | 4.83 | 3.75 | 5.91 | 8.77 | 0.00 | 0.00 |
| Fibrinogen (g/l) | 2 | 3.21 | 2.78 | 3.64 | 14.71 | 0.00 | 0.00 |
Table 7.
A meta-analysis of imaging study outcomes.
| CT group | Number of studies | Effect size and 95% confidence interval | Test of null (2-tail) | Heterogeneity | |||
|---|---|---|---|---|---|---|---|
| Outcome | Point estimate | Lower limit | Upper limit | Z value | P value | I squared | |
| Total (CT+) | 20 | 0.923 | 0.877 | 0.953 | 9.290 | 0.000 | 81.398 |
| (1) Ground-glass opacification (GGO) | 14 | 0.698 | 0.603 | 0.779 | 3.890 | 0.000 | 90.700 |
| (4) Bilateral involvement | 11 | 0.794 | 0.669 | 0.881 | 4.081 | 0.000 | 93.032 |
| (5) Consolidation | 11 | 0.378 | 0.264 | 0.508 | -1.845 | 0.065 | 90.564 |
| (3) Peripheral distribution | 6 | 0.668 | 0.500 | 0.802 | 1.955 | 0.051 | 88.951 |
| (10) Unilateral involvement | 5 | 0.156 | 0.082 | 0.278 | -4.510 | 0.000 | 71.765 |
| (∗∗) Mixed opacity | 4 | 0.328 | 0.121 | 0.635 | -1.106 | 0.269 | 97.487 |
| (11) Air bronchogram | 4 | 0.426 | 0.225 | 0.656 | -0.618 | 0.537 | 87.500 |
| (6) Pleural effusion | 4 | 0.066 | 0.036 | 0.120 | -7.950 | 0.000 | 14.830 |
| (7) Adjacent pleura thickening | 4 | 0.409 | 0.211 | 0.641 | -0.764 | 0.445 | 92.842 |
| (12) Fibrous stripes | 3 | 0.229 | 0.056 | 0.596 | -1.486 | 0.137 | 93.828 |
| (2) Lower lung predominant | 3 | 0.624 | 0.467 | 0.758 | 1.558 | 0.119 | 64.196 |
| (8) Interlobular septal thickening | 3 | 0.535 | 0.359 | 0.703 | 0.379 | 0.705 | 85.123 |
| 1 lobe affected | 2 | 0.207 | 0.087 | 0.417 | -2.608 | 0.009 | 83.897 |
| 2 lobes affected | 2 | 0.061 | 0.032 | 0.113 | -7.927 | 0.000 | 0.000 |
| 3 lobes affected | 2 | 0.105 | 0.047 | 0.220 | -4.796 | 0.000 | 57.131 |
| 4 lobes affected | 2 | 0.099 | 0.060 | 0.157 | -8.135 | 0.000 | 0.000 |
| 5 lobes affected | 2 | 0.394 | 0.312 | 0.483 | -2.331 | 0.020 | 18.313 |
| (9) Cavitation | 2 | 0.012 | 0.002 | 0.082 | -4.350 | 0.000 | 0.000 |
| Crazy-paving pattern | 2 | 0.222 | 0.067 | 0.530 | -1.786 | 0.074 | 92.080 |
| Linear opacities | 2 | 0.630 | 0.555 | 0.699 | 3.372 | 0.001 | 0.000 |
3.3. Publication Bias and Sensitivity Analysis
The Funnel plot for clinical characteristics group studies is almost symmetric confirmed by the Egger regression test (intercept = −0.28, P value = 0.20). By using the random-effects model, the summary estimate and 95% confidence interval for the combined studies is 0.25 (0.22, 0.29). These findings indicated no publication bias in the clinical characteristics group (Figure 4).
The funnel plots in the imaging studies group and findings studies seem asymmetric and skewed (Figures 5 and 6). Also, the Egger regression test has indicated an intercept of 1.55, and P value of 0.01 for the laboratory findings group and an intercept of 0.94 and P value of 0.04 for the imaging studies group.
In the laboratory findings group, using the random-effects model, the point estimate and 95% confidence interval for the combined studies is 3.01 (2.22, 3.80). Using trim and fill (four trimmed studies), the imputed summary estimate is 3.30 (2.30, 3.38) (Figure 5).
In the imaging studies group, the summary estimate and 95% confidence interval for the combined studies is 0.51 (0.48, 0.54). Using trim and fill, the imputed (four trimmed studies) summary estimate is 0.50 (0.47, 0.53) (Figure 6).
In conclusion, the finding of trim and fill analysis has indicated only minimal changes, which do not seem to be a threat to the validity of the effect size estimates.
Sensitivity analysis by using the “Remove-One” analysis did not show any change in the combined effect of the clinical characteristics and imaging studies groups after removing any of the studies. However, in the laboratory findings group, removing only one of the studies (Fan et al.) changed the combined effect significantly (Figure 7). The combined effect before and after removing Fan et al. study is presented in Tables 5 and 6.
3.4. Clinical Characteristics Group
According to clinical manifestations, fever (84.3%, 95% CI 78.6-88.7), cough (60.1%, 95% CI 53.5-66.4), and fatigue (39.4%, 95% CI 29.1-50.8) are the most prevalent clinical symptoms among patients (Table 4) (Figure 8).
3.5. Laboratory Findings Group
Laboratory studies show increased level in following tests: CRP (10.78 mg/l, 95% CI 6.44-15.11) with the normal range of 0-3.0 mg/l, D-dimer (567.89 ng/ml, 95% CI 348.15-787.62) with the normal range of 0-500 ng/ml, LDH (258.56 U/l, 95% CI 206.84-310.29) with the normal range of 135-250 U/l, and procalcitonin (0.17 ng/ml, 95% CI 0.01-0.32) which is normally less than 0.05 ng/ml in a healthy individual.
Also, the level of some laboratory factors is lower than normal, such as lymphocyte (1.00∗109/l, 95% CI 0.73-1.26) and albumin (38.61 g/l, 95% CI 35.75-41.48) concerning the normal range of 1‐4∗109/l and 40-55 g/l, respectively (Tables 5 and 6) (Figure 9).
3.6. Imaging Studies Group
Among all patients infected by SARS-CoV-2 (confirmed by RT-PCR), 85% had abnormalities in CT scans. In most of them, the bilateral pneumonia was dominant (79.4%, 95% CI 66.9-88.1). Ground-glass opacification (GGO) (69.8%, 95% CI 60.3-77.9), peripheral distribution (66.8%, 95% CI 50.0-80.2), and consolidation (37.8%, 95% CI 26.4-50.8) in those with CT scan results are presented (Table 7) (Figure 10).
4. Discussion
From December 2019, more than 500,000 cases of new unknown origin pneumonia have been confirmed all over the world [12]. It was primarily known as 2019-nCov; then, WHO decided to name this novel coronavirus “SARS-CoV-2” [13]. COVID-19 is a severe condition that is compromising the health condition of people in all countries worldwide [14]. Identifying the various characteristics of this infection is vital for controlling the outbreak in different countries [15]. Clinical, laboratory, and imaging findings are essential to evaluate the different aspects of infection [16]. Different outcomes of COVID-19 (from an asymptomatic infection to death) and contagiousness of this virus, even in its incubation period [17], emphasize why discovering different characteristics are crucial in controlling this pandemic.
In this systematic review and meta-analysis, we describe the most common clinical data on COVID-19 confirmed cases that were published during the first months of the outbreak. We analyzed 2422, rRT-PCR confirmed patients, for different clinical manifestations. Our findings are robust due to the pooled results after combining all the studies' data.
As expected from initial studies in China, COVID-19 patients presented predominantly with cough and fever, as well as headache, diarrhea, and fatigue, among other clinical features [18]. This was consistently found in many of the included studies [19, 20]. Fever frequency is similar in other β-CoV-associated infections such as SARS and MERS, but studies showed that the cough frequency is higher in SARS and COVID-19 than MERS (<50%) [21, 22]. In SARS and MERS, diarrhea is reported in about a quarter of patients, but our data shows that only 6 percent of COVID-19 patients present with diarrhea (Table 4). Our data also suggest that about 11 percent of patients are presented with headache as a symptom. Unlike SARS, which is well characterized in the two-stage clinical course of the disease [23], COVID-19 still needs further definition to identify the disease process.
Studies on epidemiological features of COVID-19 showed that about 80 percent of patients are asymptomatic or are presented with mild manifestations [24, 25], but almost all of the patients included in our study had moderate-to-severe characteristics. It seems that fever and cough are the most common clinical features among moderate-to-severe patients (Table 4).
Studies show different laboratory abnormalities in COVID-19 patients, such as hypoalbuminemia or elevated inflammatory markers [26]. However, our data suggest that C-reactive protein is the most elevated factor among infected cases (Tables 5 and 6). D-dimer, LDH, and procalcitonin are also elevated in patients, which confirmed that measuring inflammatory markers are essential to investigate new cases [27]. Also, seven studies showed lymphopenia and albuminuria as other common laboratory findings. Data from new studies suggest lymphocytopenia or an increase in WBC as prognostic factors in COVID-19 patients. Studies on the SARS outbreak in 2003 indicate that lymphopenia, leukopenia, and thrombocytopenia, elevated levels of LDH, alanine transaminase (ALT), AST, and creatine-kinase are the most affected laboratory findings [28].
Nevertheless, not significantly seen in COVID-19, the novel corona virus can affect the liver and other organs [29]. AST and ALT are normal in most cases, but impaired liver function tests are associated with poor prognosis and higher mortality rates [30, 31]. Coagulation function tests (such as INR) are affected in the prognosis of this infection [32]. Lymphopenia in COVID-19 patients suggests that this virus might act on lymphocytes (mainly T cells), but some studies suggest that B cells are also affected [26, 33].
CT scan is one of the most useful methods to diagnose the respiratory tract diseases diagnosis. CT scan high sensitivity and availability makes it one of the most common tests for lung disease screening [34, 35]. In COVID-19 patients, different results could present in the early stages of infection [36, 37]; even some studies demonstrate that CT scan sensitivity is higher than rRT-PCR [6]. Our data shows that 92% of rRT-PCR confirmed cases had abnormal CT scan results, which suggest CT scan as a reliable method. As seen in Table 7, CT scan meta-analysis outcomes are performed in random-effects analyses. Our meta-analysis on 15 studies showed that ground-glass opacification (GGO) and peripheral distribution are seen on 69.8% and 66.8% of patients, respectively. 79.4% of the patients had bilateral involvements, which is contributed to poor prognosis. CT scan is useful in monitoring the treatment, and it is crucial in classifying patients and identifying who should be treated with aggressive treatments [38]. Other findings such as consolidation or reverse halo or atoll sign are reported in some studies [39], which were not included in our analysis.
5. Limitations
This review has several limitations. Few studies are available on COVID-19, and most of them are from China. Many countries such as Italy, the United States, and Iran reported several new COVID-19 patients, but data about clinical characteristics or laboratory findings are limited. By publishing more studies worldwide, researchers are going to get a more comprehensive understanding of COVID-19. Patients' detailed information, especially in clinical outcomes, was unavailable in most studies at the time of analysis. In this study, we used random-effects model for analysis in all three groups. In comparison to the fixed model, random model findings have wider confidence intervals and less accurate results, although heterogeneity in included studies could not be considered in the fixed model.
6. Conclusion
COVID-19 presents in the majority of cases with fever and cough. Laboratory findings such as elevated inflammatory markers can assist the diagnosis. Other laboratory indices, such as AST, ALT, or INR, are also affected in these patients. 92% of the RT-PCR confirmed that patients have abnormalities in CT scan most frequently bilateral involvement. Additional research with higher sample sizes is needed in order to describe the patients' characteristics more precisely.
Abbreviations
- COVID-19:
Corona virus disease of 2019
- rRT-PCR:
Real-time reverse transcription-polymerase chain reaction
- CT scan:
Computed tomography scan
- CI:
Confidence intervals
- CRP:
C-reactive protein
- RNA:
Ribonucleic acid
- SARS-CoV-2:
Severe acute respiratory syndrome corona virus #2
- PRISMA:
Protocol based on the transparent reporting of systematic reviews and meta-analysis
- CBC:
Complete blood count
- CMA:
Comprehensive meta-analysis
- GGO:
Ground-glass opacification
- β-CoV:
Beta coronavirus
- MERS:
Middle East respiratory syndrome
- SARS:
Severe acute respiratory syndrome
- LDH:
Lactate dehydrogenase
- WBC:
White blood cell
- ALT:
Alanine transaminase
- AST:
Aspartate aminotransferase
- INR:
International normalized ratio.
Conflicts of Interest
The authors declare that they have no competing interest regarding the publication of this paper.
Authors' Contributions
HH managed the group and participated in preparing the manuscript. SS participated in meta-analysis. SM and NT participated in the systematic search and substantially drafted the manuscript. PM revised the manuscript critically. All authors read and approved the final version of the manuscript.
References
- 1.Zhu N., Zhang D., Wang W., et al. A novel coronavirus from patients with pneumonia in China, 2019. New England Journal of Medicine. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang C., Horby P. W., Hayden F. G., Gao G. F. A novel coronavirus outbreak of global health concern. The Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee P.-I., Hsueh P.-R. Emerging threats from zoonotic coronaviruses-from SARS and MERS to 2019-nCoV. Journal of Microbiology, Immunology and Infection. 2020;53(3):365–367. doi: 10.1016/j.jmii.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen H., Guo J., Wang C., et al. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: a retrospective review of medical records. The Lancet. 2020;395(10226):809–815. doi: 10.1016/S0140-6736(20)30360-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernheim A., Mei X., Huang M., et al. Chest CT findings in coronavirus disease-19 (COVID-19): relationship to duration of infection. Radiology. 2020;295(3, article 200463) doi: 10.1148/radiol.2020200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang Y., Zhang H., Xie J., et al. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;296(2, article 200432):E115–E117. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.World Health Organization. Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases: interim guidance, 2 March 2020. 2020.
- 8.Li Y. X., Wu W., Yang T., et al. Characteristics of peripheral blood leukocyte differential counts in patients with COVID-19. Zhonghua Nei Ke Za Zhi. 2020;59, article E003 [PubMed] [Google Scholar]
- 9.Luo X., Zhou W., Yan X., et al. Prognostic value of C-reactive protein in patients with COVID-19. 2020, https://www.medrxiv.org/content/10.1101/2020.03.21.20040360v1.
- 10.Higgins J. P., Thompson S. G., Deeks J. J., Altman D. G. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hozo S. P., Djulbegovic B., Hozo I. Estimating the mean and variance from the median, range, and the size of a sample. BMC Medical Research Methodology. 2005;5(1) doi: 10.1186/1471-2288-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.WHO. Coronavirus disease 2019 (COVID-19) situation report – 69. 2020, https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200329-sitrep-69-covid-19.pdf?sfvrsn=8d6620fa_2.
- 13.Gorbalenya A. E., Baker S. C., Baric R. S., et al. Severe acute respiratory syndrome-related coronavirus–the species and its viruses, a statement of the Coronavirus Study Group. 2020, https://www.biorxiv.org/content/10.1101/2020.02.07.937862v1.
- 14.Heymann D. L., Shindo N., WHO Scientific and Technical Advisory Group for Infectious Hazards COVID-19: what is next for public health? The Lancet. 2020;395(10224):542–545. doi: 10.1016/S0140-6736(20)30374-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahase E. Coronavirus: covid-19 has killed more people than SARS and MERS combined, despite lower case fatality rate. BMJ. 2020;368 doi: 10.1136/bmj.m641. [DOI] [PubMed] [Google Scholar]
- 16.Wu Z., McGoogan J. M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China. JAMA. 2020;323(13) doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 17.Bai Y., Yao L., Wei T., et al. Presumed asymptomatic carrier transmission of COVID-19. JAMA. 2020;323(14) doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang C., Wang Y., Li X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu J., Liu J., Zhao X., et al. Clinical characteristics of imported cases of COVID-19 in Jiangsu Province: a multicenter descriptive study. Clinical Infectious Diseases. 2020;10 doi: 10.1093/cid/ciaa199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang F., Deng L., Zhang L., Cai Y., Cheung C. W., Xia Z. Review of the clinical characteristics of coronavirus disease 2019 (COVID-19) Journal of General Internal Medicine. 2020;35(5):1545–1549. doi: 10.1007/s11606-020-05762-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meo S., Alhowikan A. M., al-Khlaiwi T., et al. Novel coronavirus 2019-nCoV: prevalence, biological and clinical characteristics comparison with SARS-CoV and MERS-CoV. European Review for Medical and Pharmacological Sciences. 2020;24(4):2012–2019. doi: 10.26355/eurrev_202002_20379. [DOI] [PubMed] [Google Scholar]
- 22.Guarner J. Three emerging coronaviruses in two Decades. American Journal of Clinical Pathology. 2020;153(4):420–421. doi: 10.1093/ajcp/aqaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J.-T., Sheng W. H., Fang C. T., et al. Clinical manifestations, laboratory findings, and treatment outcomes of SARS patients. Emerging Infectious Diseases. 2004;10(5):818–824. doi: 10.3201/eid1005.030640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baud D., Qi X., Nielsen-Saines K., Musso D., Pomar L., Favre G. Real estimates of mortality following COVID-19 infection. The Lancet Infectious Diseases. 2020;20(7):p. 773. doi: 10.1016/S1473-3099(20)30195-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu H., Shao J., Guo Y., et al. Data-driven discovery of clinical routes for severity detection in COVID-19 pediatric cases. 2020, https://www.medrxiv.org/content/medrxiv/early/2020/03/10/2020.03.09.20032219.full.pdf.
- 26.Tan L., Wang Q., Zhang D., et al. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. 2020, https://www.medrxiv.org/content/10.1101/2020.03.01.20029074v1. [DOI] [PMC free article] [PubMed]
- 27.Gong J., Dong H., Xia S. Q., et al. Correlation analysis between disease severity and inflammation-related parameters in patients with COVID-19 pneumonia. 2020, https://www.medrxiv.org/content/10.1101/2020.02.25.20025643v1. [DOI] [PMC free article] [PubMed]
- 28.Liang G., Chen Q., Xu J., et al. Laboratory diagnosis of four recent sporadic cases of community-acquired SARS, Guangdong Province, China. Emerging Infectious Diseases. 2004;10(10):1774–1781. doi: 10.3201/eid1010.040445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang C., Shi L., Wang F.-S. Liver injury in COVID-19: management and challenges. The Lancet Gastroenterology & Hepatology. 2020;5(5):428–430. doi: 10.1016/s2468-1253(20)30057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan Z., Chen L., Li J., et al. Clinical features of COVID-19-related liver damage. SSRN Electronic Journal. 2020 doi: 10.2139/ssrn.3546077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao N., Wang S. N., Lian J. Q., et al. Clinical characteristics and influencing factors of patients with novel coronavirus pneumonia combined with liver injury in Shaanxi region. Zhonghua gan zang bing za zhi= Zhonghua ganzangbing zazhi= Chinese Journal of Hepatology. 2020;28 doi: 10.3760/cma.j.cn501113-20200226-00070. [DOI] [PubMed] [Google Scholar]
- 32.Li T., Lu H., Zhang W. Clinical observation and management of COVID-19 patients. Emerging Microbes & Infections. 2020;9(1):687–690. doi: 10.1080/22221751.2020.1741327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qin C., Zhou L., Hu Z., et al. Dysregulation of immune response in patients with Coronavirus 2019 (COVID-19) in Wuhan, China. Clinical Infectious Diseases. 2020;71(15):762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathieson J. R., Mayo J. R., Staples C. A., Müller N. L. Chronic diffuse infiltrative lung disease: comparison of diagnostic accuracy of CT and chest radiography. Radiology. 1989;171(1):111–116. doi: 10.1148/radiology.171.1.2928513. [DOI] [PubMed] [Google Scholar]
- 35.Tan R. T., Kuzo R., Goodman L. R., Siegel R., Haasler G. R., Presberg K. W. Utility of CT scan evaluation for predicting pulmonary hypertension in patients with parenchymal lung disease. Chest. 1998;113(5):1250–1256. doi: 10.1378/chest.113.5.1250. [DOI] [PubMed] [Google Scholar]
- 36.Li M., Lei P., Zeng B., et al. Coronavirus disease (COVID-19): spectrum of CT findings and temporal progression of the disease. Academic Radiology. 2020;27(5):603–608. doi: 10.1016/j.acra.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye Z., Zhang Y., Wang Y., Huang Z., Song B. Chest CT manifestations of new coronavirus disease 2019 (COVID-19): a pictorial review. European Radiology. 2020;30(8):4381–4389. doi: 10.1007/s00330-020-06801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao W., Zhong Z., Xie X., Yu Q., Liu J. Relation between chest CT findings and clinical conditions of coronavirus disease (COVID-19) pneumonia: a multicenter study. American Journal of Roentgenology. 2020;214(5):1072–1077. doi: 10.2214/AJR.20.22976. [DOI] [PubMed] [Google Scholar]
- 39.Li X., Zeng X., Liu B., Yu Y. COVID-19 infection presenting with CT halo sign. Radiology: Cardiothoracic Imaging. 2020;2(1, article e200026) doi: 10.1148/ryct.2020200026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi H., Han X., Jiang N., et al. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. The Lancet Infectious Diseases. 2020;20(4):425–434. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pan Y., Guan H., Zhou S., et al. Initial CT findings and temporal changes in patients with the novel coronavirus pneumonia (2019-nCoV): a study of 63 patients in Wuhan, China. European Radiology. 2020;30(6):3306–3309. doi: 10.1007/s00330-020-06731-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu X., Yu C., Qu J., et al. Imaging and clinical features of patients with 2019 novel coronavirus SARS-CoV-2. European Journal of Nuclear Medicine and Molecular Imaging. 2020;47(5):1275–1280. doi: 10.1007/s00259-020-04735-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang S., Li H., Huang S., You W., Sun H. High-resolution computed tomography features of 17 cases of coronavirus disease 2019 in Sichuan province, China. European Respiratory Journal. 2020;55(4, article 2000334) doi: 10.1183/13993003.00334-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen N., Zhou M., Dong X., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. The Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guan W.-j., Ni Z. Y., Hu Y., et al. Clinical characteristics of coronavirus disease 2019 in China. New England Journal of Medicine. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang D., Hu B., Hu C., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020;323(11):p. 1061. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang W., Cao Q., Qin L., et al. Clinical characteristics and imaging manifestations of the 2019 novel coronavirus disease (COVID-19):A multi-center study in Wenzhou city, Zhejiang, China. Journal of Infection. 2020;80(4):388–393. doi: 10.1016/j.jinf.2020.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu Y.-H., Dong J. H., An W. M., et al. Clinical and computed tomographic imaging features of novel coronavirus pneumonia caused by SARS-CoV-2. Journal of Infection. 2020;80(4):394–400. doi: 10.1016/j.jinf.2020.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu J., Wu X., Zeng W., et al. Chest CT findings in patients with Coronavirus disease 2019 and its relationship with clinical features. Investigative Radiology. 2020;55(5):257–261. doi: 10.1097/rli.0000000000000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li K., Wu J., Wu F., et al. The clinical and chest CT features associated with severe and critical COVID-19 pneumonia. Investigative Radiology. 2020;55(6):327–331. doi: 10.1097/RLI.0000000000000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xia W., Shao J., Guo Y., Peng X., Li Z., Hu D. Clinical and CT features in pediatric patients with COVID-19 infection: different points from adults. Pediatric Pulmonology. 2020;55(5):1169–1174. doi: 10.1002/ppul.24718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang J.-j., Dong X., Cao Y.-y., et al. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy. 2020;75(7):1730–1741. doi: 10.1111/all.14238. [DOI] [PubMed] [Google Scholar]
- 53.Zhou S., Wang Y., Zhu T., Xia L. CT features of coronavirus disease 2019 (COVID-19) pneumonia in 62 patients in Wuhan, China. American Journal of Roentgenology. 2020;214(6):1287–1294. doi: 10.2214/ajr.20.22975. [DOI] [PubMed] [Google Scholar]
- 54.Wang J., Liu J., Wang Y., et al. Dynamic changes of chest CT imaging in patients with corona virus disease-19 (COVID-19) Journal of Zhejiang University (Medical Science) 2020;49(1) doi: 10.3785/j.issn.1008-9292.2020.02.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoon S. H., Lee K. H., Kim J. Y., et al. Chest radiographic and CT findings of the 2019 novel coronavirus disease (COVID-19): analysis of nine patients treated in Korea. Korean Journal of Radiology. 2020;21(4):494–500. doi: 10.3348/kjr.2020.0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qian G.-Q., Yang N.-B., Ding F., et al. Epidemiologic and clinical characteristics of 91 hospitalized patients with COVID-19 in Zhejiang, China: a retrospective, multi-centre case series. 2020, https://www.medrxiv.org/content/10.1101/2020.02.23.20026856v2. [DOI] [PMC free article] [PubMed]
- 57.Liu Y., Sun W., Chen L., Wang Y., Zhang L., Yu L. Clinical characteristics and progression of 2019 novel coronavirus-infected patients concurrent acute respiratory distress syndrome. 2020, https://www.medrxiv.org/content/10.1101/2020.02.17.20024166v1.article-info?versioned=true.
- 58.Chen G., Di Wu W. G., Cao Y., et al. Clinical and immunologic features in severe and moderate forms of coronavirus disease 2019. 2020, https://www.medrxiv.org/content/10.1101/2020.02.16.20023903v1. [DOI] [PMC free article] [PubMed]
- 59.Young B. E., Ong S. W. X., Kalimuddin S., et al. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA. 2020;323(15) doi: 10.1001/jama.2020.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deng S.-Q., Peng H.-J. Characteristics of and public health responses to the coronavirus disease 2019 outbreak in China. Journal of Clinical Medicine. 2020;9(2):p. 575. doi: 10.3390/jcm9020575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang Y., Tu M., Wang S., et al. Clinical characteristics of laboratory confirmed positive cases of SARS-CoV-2 infection in Wuhan, China: A retrospective single center analysis. Travel medicine and infectious disease. 2020;4, article 101606 doi: 10.1016/j.tmaid.2020.101606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu K., Fang Y.-Y., Deng Y., et al. Clinical characteristics of novel coronavirus cases in tertiary hospitals in Hubei Province. Chinese Medical Journal. 2020;133(9):1025–1031. doi: 10.1097/cm9.0000000000000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tian S., Hu N., Lou J., et al. Characteristics of COVID-19 infection in Beijing. Journal of Infection. 2020;80(4):401–406. doi: 10.1016/j.jinf.2020.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu X.-W., Wu X.-X., Jiang X.-G., et al. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-Cov-2) outside of Wuhan, China: retrospective case series. BMJ. 2020;368 doi: 10.1136/bmj.m606. [DOI] [PMC free article] [PubMed] [Google Scholar]


