Abstract
SARS‐CoV‐2 or COVID‐19 pandemic global outbreak created the most unstable situation of human health–economy. In the past two decades different parts of the word experienced smaller or bigger outbreak related to human coronaviruses. The spike glycoproteins of the COVID‐19 (similar to SARS‐CoV) attach to the angiotensin‐converting enzyme (ACE2) and transit over a stabilized open state for the viral internalization to the host cells and propagate with great efficacy. Higher rate of mutability makes this virus unpredictable/less sensitive to the protein/nucleic acid based drugs. In this emergent situation, drug‐induced destabilization of spike binding to RBD could be a good strategy. In the current study we demonstrated by bioinformatics (CASTp: computed atlas of surface topography of protein, PyMol: molecular visualization) and molecular docking (PatchDock and Autodock) experiments that tea flavonoids catechin products mainly epigallocatechin gallate or other like theaflavin gallate demonstrated higher atomic contact energy (ACE) value, binding energy, Ki value, ligand efficiency, surface area and more amino acid interactions than hydroxychloroquine (HCQ) during binding in the central channel of the spike protein. Moreover, out of three distinct binding sites (I, II and III) of spike core when HCQ binds only with site III (farthest from the nCoV‐RBD of ACE2 contact), epigallocatechin gallate and theaflavin gallate bind all three sites. As sites I and II are in closer contact with open state location and viral–host contact area, these drugs might have significant effects. Taking into account the toxicity/side effects by chloroquine/HCQ, present drugs may be important. Our laboratory is working on tea flavonoids and other phytochemicals in the protection from toxicity, DNA/mitochondrial damage, inflammation and so on. The present data might be helpful for further analysis of flavonoids in this emergent pandemic situation.
Keywords: ACE2, hydroxychloroquine, pandemic global outbreak, PatchDock, SARS‐CoV‐2 or COVID‐19, spike glycoprotein, tea flavonoids
1. INTRODUCTION
SARS‐CoV‐2 (COVID‐19) has created a global health crisis. This pandemic threatening closely resembles the SARS‐CoV outbreak occurred in 2003 (Walls et al., 2020). However, the present one is highly spreading with extremely higher degree of virulence. As of today (April 1, 2020), it killed 5,11,250 people from a total 10,523,410 infected people in 213 countries (https://www.worldometers.info/coronavirus/). Global economy has been in a jerk for 4 months because of more period of lockdown all over the globe. Thus the therapeutic/preventive intervention is an immediate requirement and challenging act against this highly stable and frequently mutable viral strain. There are no established preventive/therapeutic measures against this infection. Some of the old drugs are used on the basis of previous experiences from similar kind of infections. Some tests have been performed in some in vitro model with inclusive results. For example, one recent report suggests that remdesivir and chloroquine effectively inhibit this infection in an in vitro experimental model (Cortegiani, Ingoglia, Ippolito, Giarratano, & Einav, 2020; Gao, Tian, & Yang, 2020; Wang et al., 2020). However, it is now considered that chloroquine has a high level of toxicity and hydroxychloroquine (HCQ) is predicted to be less toxic (Colson, Rolain, Lagier, Brouqui, & Raoult, 2020). In a trial and survey‐type experiment with a very small sample size, it is shown that HCQ treatment is significantly associated with viral load reduction/disappearance in COVID‐19 patients and its effect is reinforced by azithromycin (Gautret et al., 2020). Out of few combination of medication, presently HCQ is being used in COVID‐19 cases (Wang et al., 2020). However, this drug also has a varied range of toxicity (Cortegiani et al., 2020). Apart from that, this drug is also used in systemic lupus erythematosus (SLE) and rheumatoid arthritis (RA), so strategies should be taken to resolve the issue of sufficient supply of this medicine for these patients also (Yazdany & Kim, 2020). When there is almost nothing to do, chloroquine group of drugs are of use, but alarming call has been raised by scientists and clinicians for the wide range of side effects of this drug (Yazdany & Kim, 2020). Besides lung, liver and the kidney tissues also face a significant degree of injuries and in this situation use of drugs needs critical analysis and careful approach (Jiang, 2020; Rismanbaf & Zarei, 2020). Other drugs like remdesivir, lopinavir, ribavirin and ritonavir have shown efficacy to inhibit coronavirus in vitro. Teicoplanin, an antibiotic used to treat staphylococcal infections, has been shown to inhibit MERS‐CoV in human cells. So, this drug may be rechecked in the present situation also (Baron, Devaux, Colson, Raoult, & Rolain, 2020). Molecular docking (MD) results suggest that few drugs, namely sofosbuvir, galidesivir and tenofovir, can be used against SARS‐CoV‐2 RNA‐dependent RNA polymerase inactivation (Elfiky, 2020).
Our laboratory has been working for last several years on the tea (Camelia sinensis) flavonoids for their different therapeutic and disease protective roles. Tea flavonoids have been demonstrated as strong antitoxicant, antioxidant and antiinflammatory agents. Antitumerigenic role of catechin derivatives especially Epigallocatechin gallate (EGCG) has been shown decisively by several laboratories (Acharyya, Ali, Deb, Chattopadhyay, & Maiti, 2015; Acharyya, Chattopadhyay, & Maiti, 2014; Maiti et al., 2017). Regarding the antiviral role, tea flavonoids have been shown to be strong agent (Li et al., 2020). Report demonstrates that catechin can prevent influenza A (H1N1) virus infection and gallic acid can inhibit influenza virus infection (You et al., 2018). In the past three decades several studies suggested that the regular consumption of green tea decreases influenza infection and some cold symptoms. The gargling with tea extracts and/or flavonoids may protect from influenza virus (Furushima, Ide, & Yamada, 2018). EGCG‐fatty acid derivative is important because fatty acid on the phenolic hydroxyl group increases viral and cellular membrane permeability which protects from viral infection (Kaihatsu, Yamabe, & Ebara, 2018). Preliminary attachment to cellular glycans is a critical step for entry of several human viruses. Some viruses, such as herpes simplex virus type 1 (HSV‐1) and hepatitis C virus (HCV), bind to heparan sulfate, whereas others, such as influenza A virus (IAV), bind to sialic acid (Colpitts & Schang, 2014). The SARS‐CoV‐2 is also covered by a large number of NAG molecules.
In this background, we have been intended to test different flavonoid effects in the process of SARS‐CoV‐2 spike glycoproteins binding to its host cell receptor ACE2. In the present bioinformatics, cheminformatics and molecular modeling studies, we initially characterized the nature of nCoV and ACE2 binding and surface contact areas. The role of specific amino acids and their binding energy have been evaluated. Furthermore, different flavonoids have been docked on two targets; nCoV spike proteins and host cell receptors and all the bindings were carefully characterized. The current study will help to screen some suitable drug that can block the viral binding and further host cell attachment/entry.
2. MATERIALS AND METHODS
2.1. Protein structure retrieval
The electron microscopic structure of human coronavirus spike glycoprotein (PDB ID: 6vsb) and X‐ray diffraction structure of human angiotensin‐converting enzyme 2 (ACE2; PDB ID: 4aph) were retrieved from RCSB Protein Data Bank (https://www.rcsb.org/) in PDB format. The structure 6vsb was selected because it was found with number of N‐acetyl glucosamine (NAG) attached in different locations of the protein. On the other hand ACE2 (PDB ID: 4aph) was selected as it was a complete tertiary structures with no such viral spike protein contaminations. Other two viral spike protein structures like 6VXX and 6VYB and one spike‐ACE2 (receptor) complex structure 6M0J were also retrieved for analysis.
2.2. Ligand structure retrieval
The three‐dimensional (3D) structures of different selected tea flavonoid molecules like, catechin, catechin gallate, epicatechin 3‐O‐gallate, epigallocatechin, epigallocatechin 3‐gallate, gallocatechin, gallocatechin gallate, theaflavin monogallate (TFMG) and theaflavin digallate (TFDG) were retrieved in .sdf format from world's largest chemical information database, PubChem (https://pubchem.ncbi.nlm.nih.gov/). As control or for comparative study, 3D structure of HCQ was also retrieved from PubChem in .sdf format.
2.3. Preparation of both receptor and ligand molecules
Receptor molecules like human coronavirus (COVID‐19) spike glycoprotein (PDB ID: 6vsb) and human ACE2 protein (PDB ID: 4aph) were found with numerous water molecules with their structures. The structures were individually dehydrated using PyMol molecular visualization system and edited structures were saved in .pdb format. Additionally, angiotensin II molecule was removed from human ACE2 protein also. Ligand molecules were retrieved in .sdf format and then they were converted to protein data bank format and saved as .pdb format using PyMol and used for further analysis. The chemical structures, compound PubChem Compound ID (CID), molecular formula (MF) and molecular weight (MW) of selected ligands were listed in Table 1.
TABLE 1.
Chemical structure of selected ligands used for MD study with human ACE2 and coronavirus spike protein
|
|
|
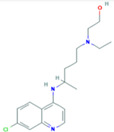
| HCQ (compound CID: 3652; MF: C18H26ClN3O; MW: 335.9 g/mol) | Catechin (compound CID: 9064; MF: C15H14O6; MW: 290.27 g/mol) | Catechin gallate (compound CID: 6419835; MF: C22H18O10; MW: 442.4 g/mol) |
|
|
|
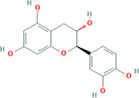
| Epicatechin 3‐O‐gallate (compound CID: 442678; MF: C37H30O17; MW: 746.6 g/mol) | Epigallocatechin (compound CID: 72277; MF: C15H14O7; MW: 306.27 g/mol) | Epigallocatechin 3‐gallate (compound CID: 65064; MF: C22H18O11; MW: 458.4 g/mol) |
|
|
|
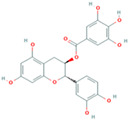
| Gallocatechin (compound CID: 65084; MF: C15H14O7; MW: 306.27 g/mol) | Gallocatechin gallate (compound CID: 199472; MF: C22H18O11; MW: 458.4 g/mol) | TFMG |
|
|
|
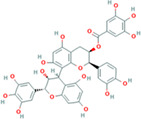
| Chloroquine (compound CID: 2719, MF: C18H26ClN3, MW: 319.87 g/mol) | TFDG (compound CID: 135403795; MF: C43H32O20; MW: 868.7 g/mol) | Number of galloyl group favors greater ACE value, surface‐area (Table 2), more contacting amino acids (Figure 5) |
2.4. Surface topology calculation of selected protein molecule
Basically two different terminologies generally used to define the solvent accessible and nonaccessible area of a protein. Pocket represents the accessible area and consists of a mouth opening for solvent to get in, whereas cavity or void are the closed and solvent nonaccessible areas of protein. To identify the pockets within the selected nCoV2 spike glycoprotein, computed atlas of surface topography of protein (CASTp; http://sts.bioe.uic.edu/castp/index.html?j_5e8c7bec25090) was used. According to Tian, Chen, Lei, Zhao, and Liang (2018), protein pockets are those, on infinite cross section at least one will show the larger value then the mouth opening of that pocket.
2.5. Molecular docking
The MD between human coronavirus spike protein selected ligands and human ACE2 protein selected ligands were performed using PatchDock web server and offline Autodock software for interactive MD study. PatchDock is developed as geometry‐based MD algorithm. It calculates the docking transformation between two molecules to get the best molecular interface complementarity which finds out the ligand posture in receptor with maximum interface area covered and minimum steric hindrance (Schneidman‐Duhovny, Inbar, Nussinov, & Wolfson, 2005). It also calculates the atomic contact energy (ACE) value of each docking positions to indicate the amount of required desolvation free energies to transfer the ligand molecule from water to protein (receptor) interior. Autodock is actually an authentic suite to determine the small molecule or drug binding to a known 3D structure of protein. Autodock consists of two different software generations: AutoDock4 and AutoDock Vina. AutoDock Vina is an automated grid calculating software, whereas AutoDock4 is essential for gradual optimization‐based grid calculating software. Initially, it runs autogrid to detect the best possible grids for ligand docking; hence, it generates automatic affinity grid to find out the better binding posture of ligand (Morris et al., 2009).
3. RESULTS
A large body of evidences has clarified the health benefit of tea components catchin and theaflavin. Tea is a regular beverage that is consumed worldwide and specifically may be more in India, China and other parts of Southeast Asian countries. Moreover, daily food/nutrients intake in these places is based on various vegetable combinations, fruits and green leaves. These are highly enriched with phytochemicals, polyphenols like flavonoids. Earlier our laboratory has demonstrated the anticarcinogenic role of green tea extract (Acharyya, Ali, et al., 2015; Maiti et al., 2017). It decisively helps in DNA protections by scavenging free radicals and that efficacy was found to be even greater than the known specific radical scavengers (Acharyya, Ali, et al., 2015; Acharyya, Chattopadhyay, et al., 2014). According to the relationships between structure and antiviral activity of catechin derivatives, the 3‐galloyl and 5′‐OH group of catechin derivatives appear critical to antiviral activities (Kaihatsu et al., 2018). In vitro activity of QR‐435 green‐tea extract against Influenza A Virus (IAV) demonstrated its strong virucide role by prevention of viral transmission. Masks impregnated with QR‐435 (a flavonoid component) were highly effective in blocking the passage of live H3N2 virus (Oxford et al., 2007a). in vivo prophylactic activity of QR‐435 was demonstrated against H3N2 influenza virus infection (Oxford et al., 2007b).
3.1. Structure selection
The human coronavirus spike glycoprotein (PDB ID: 6vsb) and X‐ray diffraction structure of human ACE2 (PDB ID: 4aph) were retrieved to analyze the attachment pattern and find out the types of amino acid involved in proper attachment, prefusion and postfusion posture of coronavirus spike glycoprotein and finally potential spike protein blocker. Here we have selected green tea components like catechin (CID: 9064), catechin gallate (CID: 6419835), epicatechin 3‐O‐gallate (CID: 442678), epigallocatechin (CID: 72277), epigallocatechin 3‐gallate (CID: 65064), gallocatechin (CID: 65084), gallocatechin gallate (CID: 199472), TFMG (CID: 135458102) and TFDG (CID: 135403795) along with HCQ (CID: 3652) as molecular docker (Table 1). The surfaces of all selected ligand were also reported in Table 1. All the selected ligands have minimum one benzene ring. The ligands were retrieved in 2D as well as 3D forms. Two‐dimensional forms were used in Table 1.
3.2. Mechanism of COVID‐19 spike glycoprotein attachment with human receptor ACE2
According to literature, human SERS‐COV‐2 or COVID‐19 have 14–15 different conserved regions in their spike glycoproteins (Wrapp et al., 2020). Among them, S1 region is responsible for surface attachment and bond formation with human cell membrane receptor ACE2. During this attachment a significant conformational change was observed within the spike glycoprotein (Figure 1).
FIGURE 1.
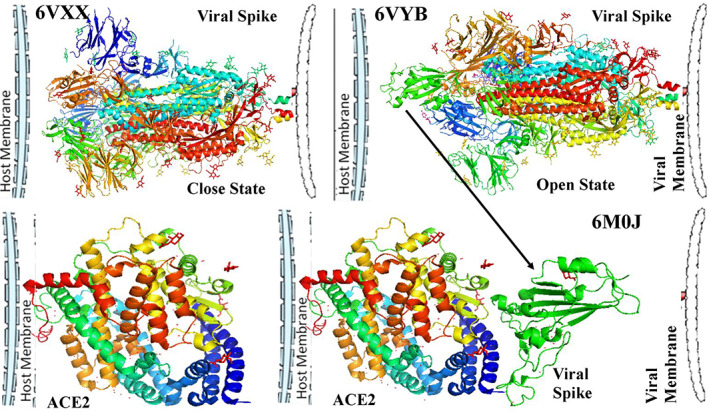
Two different stages of coronavirus spike protein; closed state of receptor attachment site (a), open state of receptor attachment site (b). Structure of membrane bound viral receptor protein ACE2 (c). Attachment of viral spike protein with membrane bound ACE2 protein (d)
PDB structure 6VXX showed a closed state structure unable to bind with host membrane, whereas in PDB structure 6VYB a raised structure was observed that was an open state conformation of spike glycoprotein (Figure 1b). Viruses are host‐specific particulate. This specificity depends upon the presence of a particular receptor protein molecule with proper interactive site exposed out of the cell. This attachments lead to internalization of virus within the cell (Ciesek et al., 2011; Yamada, Takuma, Daimon, & Hara, 2006). For COVID‐19, ACE2 plays that role through proper protein–protein interaction (Figure 1c,d). The spike protein interaction is a result of internal hydrogen bond cleavage and formation of a hinge like structure (Figure 2a). With the cleavage of internal bonds, ACE2 binding site is exposed at the top of the open conformation. Comparatively, in closed condition the interactive site remains embedded (Figure 2b). At the surface of ACE2 and spike glycoprotein interaction, number of rigid as well as H‐bonds forms (Figure 2c). So this attachment site demands a proper posture which is become ready after hinge region up conformation (Figure 2d–f). However, specific bonds between ARG983–GLY3821 and ARG983–SER383 were observed in closed conformation (Figure 2g–i).
FIGURE 2.
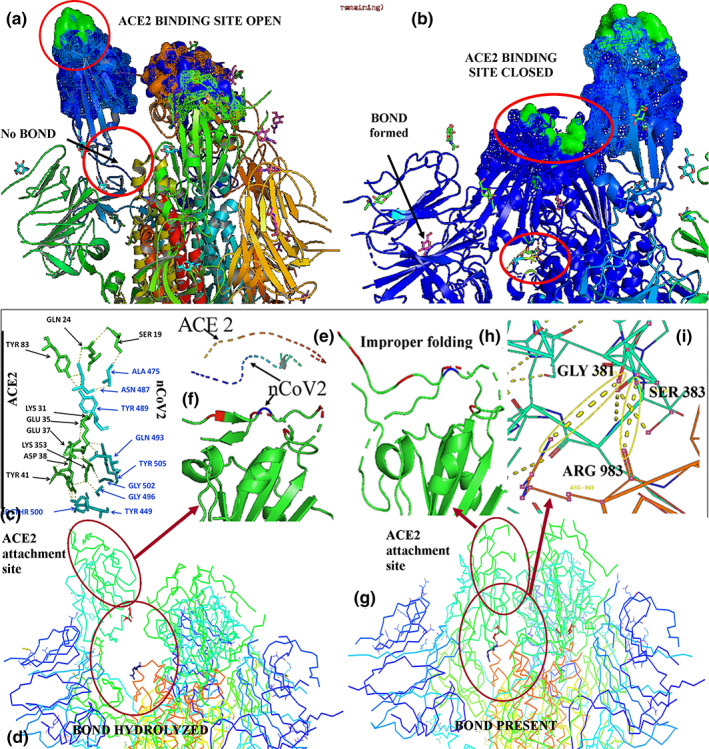
Complete exposure of coronavirus spike protein (nCoV2) attachment surface during opened state (a). Partial exposure of attachment surface during closed state (b). Attachment between ACE2 and nCoV2 stabilized by different stable and H‐bond formation (c). Attachment site opened due to specific internal bond hydrolysis (d) which gives proper conformation to viral attachment site (e, f). Specific internal bonds stabilizes closed state (g) leading to improper folding of attachment site (h). The closed state is maintained by the H‐bond formation among GLY381, SER383 and ARG983 (i)
3.3. Surface topology calculation of selected protein molecules
To understand the solvent accessible area of the selected proteins molecules nCoV2 and ACE2, CASTp server was used, where the internal channel of ACE2 was observed to have protein pocket with a surface area of 2674.824 Å2 and surface area volume was 2,291.847 Å3. In spike glycoprotein, the solvent accessibility was observed starting from the flexible ACE2 binding region to most rigid central channel with a surface area value of 33706.386 Å2 and surface area volume of 63378.539 Å3 (Fig. 3).
FIGURE 3.
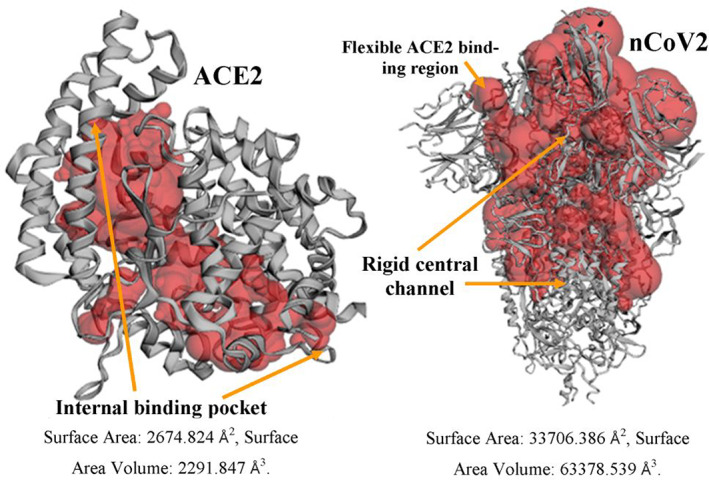
Surface topology calculation of COVID‐19 spike glycoprotein (nCoV2) and its human receptor ACE2. Surface area and volume is much higher in nCoV2 then ACE2 and the ligand binding pockets were exposed in nCoV2 protein
3.4. MD study
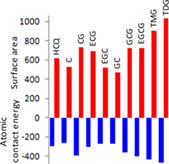
To find out potent viral spike protein destabilizing drug, MD study was performed using PatchDock and Autodock. In Patchdock, best 20 docking results for each interaction were considered for position‐specific docking posture analysis and selection of best one with lowest ACE value calculation. All the selected drug molecules showed higher affinity to viral spike protein in comparison with ACE2 receptor proteins (Table 2). Here HCQ, the drug recently used for the treatment of COVID‐19 on the basis of some previous inconsistent report, showed ACE value of −154.06 with receptor molecule. However, the highest affinity was observed for TFMG–ACE2 interaction (−287.14). Epicatechin 3‐O‐gallate also showed affinity to receptor molecule (−265.13). Overall, the affinity of our several selected drugs toward the viral spike protein was observed to be very high. The highest value was observed for TFDG (−465.17) followed by TFMG (−434.42) and epigallocatechin 3‐gallate (−407.58). In contrast, HCQ was observed with ACE value of −293.32. Moderate ACE value were found for catechin gallate (−393.05), gallocatechin gallate (−364.16) and epicatechin 3‐O‐gallate (−308.25). The corresponding surface area occupancy on nCoV2 spike glycoprotein of all the selected drugs have also been listed in Table 2.
TABLE 2.
Comparative analysis of HCQ, catechin, catechin gallate, epicatechin 3‐O‐gallate, epigallocatechin, epigallocatechin 3‐gallate, gallocatechin, gallocatechin gallate, TFMG and TFDG binding with ACE2 and nCoV2 through MD using Patchdock and Autodock
| S. No. | Compound | Patchdock ACE value with nCoV2 (area in Å2) | Autodock | |
|---|---|---|---|---|
| Binding energy (Ki value in μmol) | Ligand efficiency | |||
| 1 | HCQ | −293.32 (616.90) | −3.64 (2.14) | −0.16 |
| 2 | Catechin | −266.41 (525.20) | −4.23(799.34) | −0.2 |
| 3 | Catechin gallate | −393.05 (732.90) | −4.16 (885.65) | −0.13 |
| 4 | Epicatechin 3‐O‐gallate | −308.25 (689.60) | −3.55 (2.51) | −0.11 |
| 5 | Epigallocatechin | −270.01 (523.40) | −3.94 (1.29) | −0.18 |
| 6 | Epigallocatechin 3‐gallate | −407.58 (723.90) | −2.98 (6.54) | −0.09 |
| 7 | Gallocatechin | −274.72 (471.40) | −4.76 (322.09) | −0.22 |
| 8 | Gallocatechin gallate | −364.16 (722.70) | −3.79 (1.67) | −0.11 |
| 9 | TFMG | −434.42 (906.20) | −6.72 (11.9) | −0.13 |
| 10 | TFDG | −465.17 (1034.60) | −1.85 (44.19) | −0.06 |
Note: Bold values are highly significant rather than low significant value in HCQ (−293.32).
The basic tendency of each 20 docking results for one combination was plotted in same receptor molecules. In this situation, HCQ was observed at the interior core of ACE2 receptor molecule. No influence was observed at or near spike protein binding site. Best docking site of HCQ within coronavirus spike protein central core region with ACE value −293.32 (Figure 4b–d), whereas best docking site of TFDG at the hinge region of virus–receptor attachment site opening with ACE value −465.17 observed (Figure 4g,h).
FIGURE 4.
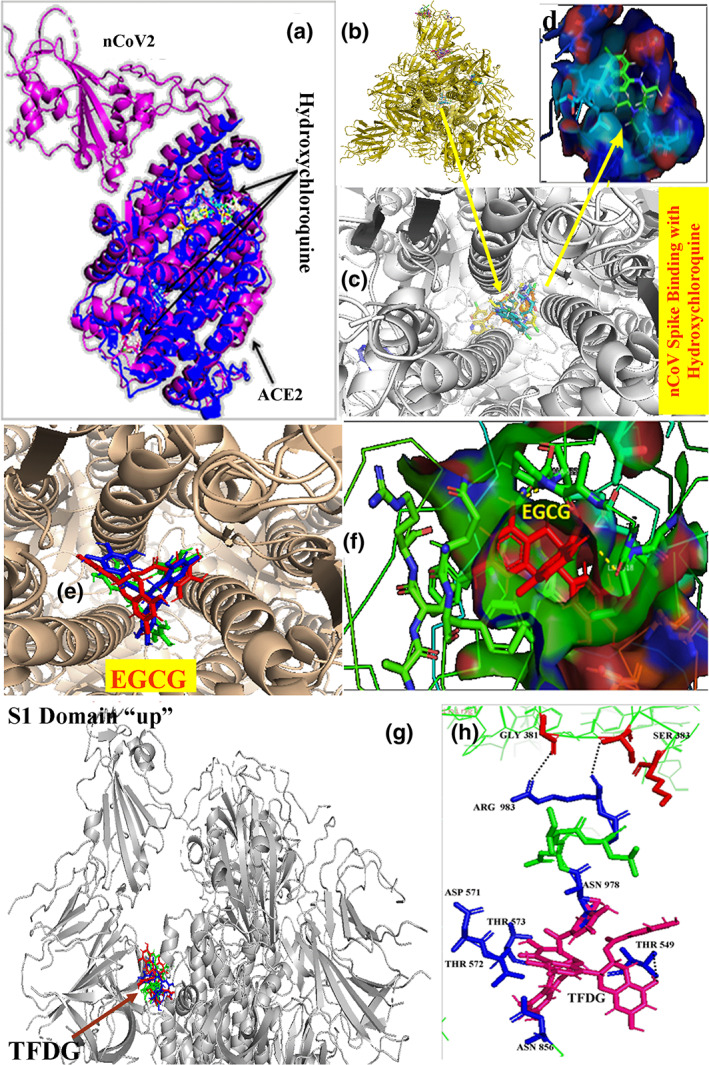
Interaction of HCQ with ACE2 viral receptor protein (a). Best docking site of HCQ within coronavirus spike protein central core region with ACE value −293.32 (b–d). Best docking site of TFDG at the hinge region of virus–receptor attachment site opening with ACE value −465.17 (g). TFDG forms rigid bonds with ASN978, THR549, ASN856, THR572, THR573 and ASP578 near the ARG983 (h)
According to Autodock result, highest binding energy was found for TFMG (−6.72) with a Ki value of 11.9 μmol and the ligand efficiency value of −0.13 (Table 2). The second highest binding energy value was observed for gallocatechin (−4.76) with a ligand efficiency value of −0.22. The Ki value was very high (322.09 μmol), whereas the HCQ showed Ki value of 2.14 μmol with a ligand efficiency value of −0.16. However, its binding energy value was very low in comparison to TFMG.
Finally, the attachment location of different selected drug molecules within the central core region of spike glycoprotein was analyzed. Different ligand binding sites were denoted in Figure 5a. Among them the central core region was focused elaborately, where, in general, the core region is being stabilized by interchain and intrachain hydrogen bonding. Three locations were found which mainly stabilizes the channel‐like structure through intrachain H‐bonding (Figure 5b). In the central channel site, A, B and C are the locations where intrachain hydrogen bonding observed which normally stabilizes the quaternary structure of nCoV2 spike glycoprotein. At site A, ASP994 and ARG995 are the key molecules at each unit which formed H‐bond at alternating pattern. Selected drug molecule TFDG and TFMG both were found to interact with ASP994 and ARG995 at site A. EGCG formed H‐bond with ARG995. No such binding was observed for HCQ. At site B, GLN1002, GLN1005 and THR1006 of each unit formed alternate H‐bond pattern. Here only TFMG formed bond with all abovementioned amino acids. Drug binding sites I and II were the exact location of intramolecular H‐bond forming sites A and B, respectively. However, the site C did not match with drug binding site III. Site C was stabilized by alternating amino acids GLU1031 and ARG1039. At this location all four drug molecules were found. HCQ attached with only two amino acids ARG1019 and ALA1020. Highest affinity was shown by TFDG (−417.62). So, efficiency of these three molecules could have been better than HCQ to destabilize the coronavirus spike proteins.
FIGURE 5.
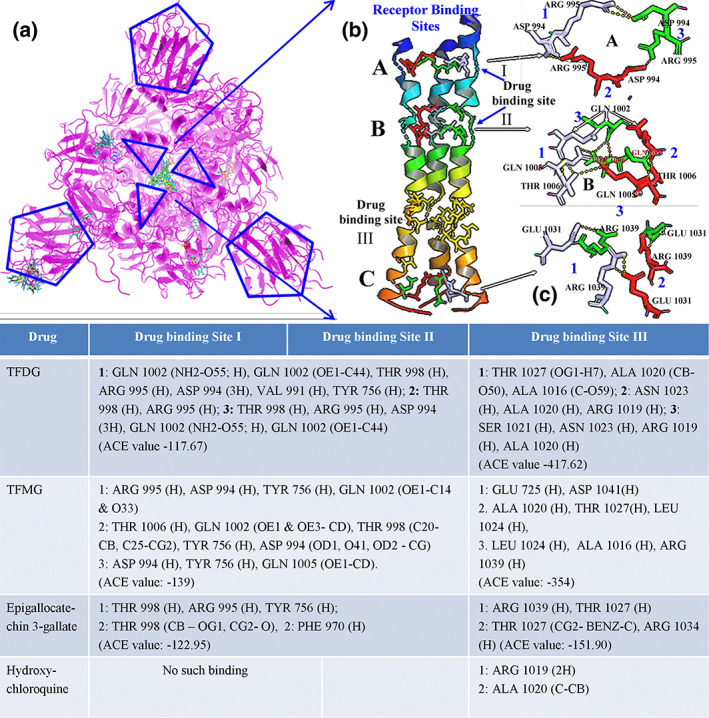
Homotrimer coronavirus spike protein and its probable ligand binding site (a). Hydrogen bond stabilizing the central core of coronavirus spike protein (b). Different ligand‐binding efficiencies in different drug binding sites with ACE value. When ECGC and TG interact three different drug binding sites involving with more number of amino acids the HCQ interacts only on site III which is far from RBD involving with less number of amino acids
4. DISCUSSION
Present pandemic situation demands an obvious getaway through development of potent drug against COVID‐19. In our study we have selected most of the common ingredients present in tea leaves for potent drug selection. Inhibitory effects of catechin derivatives on the activities of HIV RT and cellular polymerases have been demonstrated (Nakane & Ono, 1990). Especially, EGCG significantly inhibits the HIV reverse transcription step (Li, Hattori, & Kodama, 2011). Along with this, those components also have different therapeutic and disease protective roles as strong antitoxicant, antioxidant and antiinflammatory agents. Antitumerigenic role of EGCG has also been shown by several laboratories (Acharyya, Ali, et al., 2015; Acharyya, Chattopadhyay, et al., 2014; Maiti et al., 2017).
According to the nature of viral spike protein attachment with its human receptor protein ACE2, there are several steps behind it. According to Duquerroy, Vigouroux, Rottier, Rey, and Bosch (2005), SARS coronavirus spike glycoprotein possesses a central channel stabilized by several H‐bonds among asparagines/glutamine zipper which ultimately helps to form postfusion hairpin conformation during receptor protein attachment. COVID‐19 have similar genetic makeup as SARS coronavirus. This nCoV2 spike protein is composed of similar three homologous protein units like SARS coronavirus. As in Figure 5, those units have been stabilized by intermolecular and intramolecular hydrogen bonds. This unit functionally has two parts; one is the central core region or channel which provides structural rigidity to the protein, and another was the flexible receptor binding region (Figure 1). The flexibility depends upon the destabilization of H‐bond between GLY381–ARG983–SER383 amino acids. An attachment of TFDG molecule was observed at the hinge region of S1 up domain (Figure 4g) just beneath the ARG983 binding site (Figure 4h) which maintains the close conformation of spike glycoprotein. In future TFDG could be used for the chemically fused therapeutic molecule synthesis for more strengthening the closed S1 conformation to avoid proper attachment of viral spike protein with receptor ACE2.
On the other hand, docking results revealed different ACE value for different drug molecules. ACE value indicated the energy required for the transfer of a molecule from water dissolve condition to desired protein's interior core (Schneidman‐Duhovny et al., 2005). From our result, lowest energy required for TFDG (−465.17) then gradually TFMG (−434.42) and epigallocatechin 3‐gallate (−407.58), whereas HCQ showed energy value of −293.32 for the binding to CoV spike. This indicated that transfer of HCQ to viral spike protein from dissolved state requires more contact energy value.
Structural chemistry as well as the molecular charge distribution directly influences the molecular interaction of a drug. Here in our study catechin gallate showed higher ACE value of −393.05 with a surface area of 732.90 Å2 (Table 2), whereas only catechin showed lower ACE value of −266.41 with a less surface area occupancy of 525.20 Å2. Same result was also observed for epigallocatechin (ACE value: −270.01, area: 523.40 Å2), epicatechin 3‐O‐gallate (ACE value: −308.25, area: 689.60) and epigallocatechin 3‐gallate (ACE value: −407.58, area: 723.90 Å2), in between TFMG (ACE value: −434.42, area: 906.20 Å2) and TFDG (ACE value: −465.17, area: 1034.60 Å2). So, from this result it is being evaluated that gallate group increases the drug accessibility to the receptor protein may be due to the presence of extra –OH groups. This comparative study was represented in Table 1. On the other hand, molecular plane arrangement also influences ligand affinity to receptor protein. During Autodock analysis, EGCG the third highest ACE value containing ligand (−407.58) showed the lowest Ki value of 6.54 μmol in comparison with TFDG and TFMG which indicated that less amount of drug can inhibit the protein efficiently. A versatile role of EGCG has been demonstrated in the last decade regarding the antiviral effects. EGCG prevented neurotoxicity mediated by HIV‐1 proteins gp120 and immune inflammatory responses via IFN‐gamma where role JAK/STAT1 signaling has been found to be involved (Giunta et al., 2006). This suggests that beside the disfavoring role to the viral entry and propagation, EGCG also significantly restricted the cellular inflammatory burst. Role of overproduction of cytokines like IL1, IL‐6 and TNF‐α and cytokine storm may also be prevented by EGCG. The EGCG can play as inhibitor of hepatitis C and hepatitis B virus entry into the host (Calland et al., 2012; Huang et al., 2014). Ability to block the several viral entries might indicate the interferences in the viral spike protein destabilization in the preinfusion and postinfusion events. These finding may be validated by the synthetic EGCG‐palmitate efficiency against porcine reproductive/respiratory syndrome virus (Zhao, Liu, Li, Yang, & Zu, 2014). The green tea catechin, EGCG has also been shown to inhibit chikungunya virus infection (Weber, Sliva, Von, Kummerer, & Schnierle, 2015).
According to our study, EGCG, TFDG and TFMG were the most potent three molecules, which could be used for the COVID‐19 treatment as such or after some chemical modification by making it more efficient in interfering the transition between open and closed state of the viral spike. It was proposed in a previous study that gargling with tea catechin extracts may be practiced for the prevention of influenza infection in suspect/sensitive individuals. For the better bioavailability and interaction with different viral components, spike‐glycoprotein nanoparticulated catechin derivatives may be used alone or in combination of other bioactive peptides/chitosan/lectins (Hu et al., 2012). To serve promptly this bioinformatics, MD and virtual binding/inhibition study was finished with great pace. We believe that our present work will add some new findings in the present emergent situation developed pandemic COVID‐19 outbreak.
CONFLICT OF INTEREST
The authors declare no potential conflict of interest.
ACKNOWLEDGEMENT
This work was partially funded by Department of Science and Technology, Government of West Bengal, India.
Maiti S, Banerjee A. Epigallocatechin gallate and theaflavin gallate interaction in SARS‐CoV‐2 spike‐protein central channel with reference to the hydroxychloroquine interaction: Bioinformatics and molecular docking study. Drug Dev Res. 2021;82:86–96. 10.1002/ddr.21730
Funding information Partially by WB State DST, Grant/Award Number: 05/2016
REFERENCES
- Acharyya, N. , Ali, S.‐S. , Deb, B. , Chattopadhyay, S. , & Maiti, S. (2015). Green tea (Camellia sinensis) alleviates arsenic‐induced damages to DNA and intestinal tissues in rat and in situ intestinal loop by reinforcing antioxidant system. Environmental Toxicology, 30(9), 1033–1044. 10.1002/tox.21977 [DOI] [PubMed] [Google Scholar]
- Acharyya, N. , Chattopadhyay, S. , & Maiti, S. (2014). Chemoprevention against arsenic‐induced mutagenic DNA breakage and apoptotic liver damage in rat via antioxidant and SOD1 upregulation by green tea (Camellia sinensis) which recovers broken DNA resulted from arsenic‐H2O2 related in vitro oxidant stress. Journal of Environmental Science Health, Part C: Environmental Carcinogenesis & Ecotoxicology Reviews, 32(4), 338–361. 10.1080/10590501.2014.967061 [DOI] [PubMed] [Google Scholar]
- Baron, S.‐A. , Devaux, C. , Colson, P. , Raoult, D. , & Rolain, J.‐M. (2020). Teicoplanin: An alternative drug for the treatment of COVID‐19? International Journal of Antimicrobial Agents, 55, 105944. 10.1016/j.ijantimicag.2020.105944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calland, N. , Albecka, A. , Belouzard, S. , Wychowski, C. , Duverlie, G. , Descamps, V. , … Seron, K. (2012). (−)‐Epigallocatechin‐3‐gallate is a new inhibitor of hepatitis C virus entry. Hepatology, 55, 720–729. [DOI] [PubMed] [Google Scholar]
- Ciesek, S. , Von, H.‐T. , Colpitts, C.‐C. , Schang, L.‐M. , Friesland, M. , Steinmann, J. , … Steinmann, E. (2011). The green tea polyphenol, epigallocatechin‐3‐gallate, inhibits hepatitis C virus entry. Hepatology, 54, 1947–1955. [DOI] [PubMed] [Google Scholar]
- Colpitts, C.‐C. , & Schang, L.‐M. (2014). A small molecule inhibits virion attachment to heparan sulfate‐ or sialic acid‐containing glycans. Journal of Virology, 88(14), 7806–7817. 10.1128/JVI.00896-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson, P. , Rolain, J.‐M. , Lagier, J.‐C. , Brouqui, P. , & Raoult, D. (2020). Chloroquine and hydroxychloroquine as available weapons to fight COVID‐19. International Journal of Antimicrobial Agents., 55, 105932. 10.1016/j.ijantimicag.2020.105932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortegiani, A. , Ingoglia, G. , Ippolito, M. , Giarratano, A. , & Einav, S. (2020). A systematic review on the efficacy and safety of chloroquine for the treatment of COVID‐19. Journal of Critical Care, 57, 279–283. 10.1016/j.jcrc.2020.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duquerroy, S. , Vigouroux, A. , Rottier, P.‐J.‐M. , Rey, F.‐A. , & Bosch, B.‐J. (2005). Central ions and lateral asparagine/glutamine zippers stabilize the post‐fusion hairpin conformation of the SARS coronavirus spike glycoprotein. Virology, 335, 276–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky, A.‐A. (2020). Ribavirin, remdesivir, sofosbuvir, galidesivir, and tenofovir against SARS‐CoV‐2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Science, 253, 117592. 10.1016/j.lfs.2020.117592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furushima, D. , Ide, K. , & Yamada, H. (2018). Effect of tea catechins on influenza infection and the common cold with a focus on epidemiological/clinical studies. Molecules, 23(7), 1795. 10.3390/molecules23071795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, J. , Tian, Z. , & Yang, X. (2020). Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID‐19 associated pneumonia in clinical studies. Bioscience Trends, 14(1), 72–73. 10.5582/bst.2020.01047 [DOI] [PubMed] [Google Scholar]
- Gautret, P. , Lagier, J.‐C. , Parola, P. , Hoang, V. T. , Meddeb, L. , Mailhe, M. , … Raoult, D. (2020). Hydroxychloroquine and azithromycin as a treatment of COVID‐19: Results of an open‐label non‐randomized clinical trial. International Journal of Antimicrobial Agents, 56(1), 105949. 10.1016/j.ijantimicag.2020.105949 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Giunta, B. , Obregon, D. , Hou, H. , Zeng, J. , Sun, N. , Nikolic, V. , … Tan, J. (2006). EGCG mitigates neurotoxicity mediated by HIV‐1 proteins gp120 and Tat in the presence of IFN‐gamma: Role of JAK/STAT1 signaling and implications for HIV‐associated dementia. Brain Research, 1123, 216–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, B. , Ting, Y. , Yang, X. , Tang, W. , Zeng, X. , & Huang, Q. (2012). Nanochemoprevention by encapsulation of (−)‐epigallocatechin‐3‐gallate with bioactive peptides/chitosan nanoparticles for enhancement of its bioavailability. Chemical Communication, 48, 2421–2423. [DOI] [PubMed] [Google Scholar]
- Huang, H.‐C. , Tao, M.‐H. , Hung, T.‐M. , Chen, J.‐C. , Lin, Z.‐J. , & Huang, C. (2014). (−)‐Epigallocatechin‐3‐gallate inhibits entry of hepatitis B virus into hepatocytes. Antiviral Research, 111, 100–111. [DOI] [PubMed] [Google Scholar]
- Jiang, S. (2020). Don't rush to deploy COVID‐19 vaccines and drugs without sufficient safety guarantees. Nature, 579(7799), 321. 10.1038/d41586-020-00751-9 [DOI] [PubMed] [Google Scholar]
- Kaihatsu, K. , Yamabe, M. , & Ebara, Y. (2018). Antiviral mechanism of action of epigallocatechin‐3‐O‐gallate and its fatty acid esters. Molecules, 23(10), 2475. 10.3390/molecules23102475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Song, D. , Wang, S. , Dai, Y. , Zhou, J. , & Gu, J. (2020). Antiviral effect of epigallocatechin gallate via impairing porcine circovirus type 2 attachment to host cell receptor. Viruses, 12(2), 176. 10.3390/v12020176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. , Hattori, T. , & Kodama, E. N. (2011). Epigallocatechin gallate inhibits the HIV reverse transcription step. Antiviral Chemistry and Chemotherapy, 21, 239–243. [DOI] [PubMed] [Google Scholar]
- Maiti, S. , Acharyya, N. , Ghosh, T.‐K. , Ali, S.‐S. , Manna, E. , Nazmeen, A. , & Sinha, N.‐K. (2017). Green tea (Camellia sinensis) protects against arsenic neurotoxicity via antioxidative mechanism and activation of superoxide dismutase activity. Central Nervous System Agents in Medical Chemistry, 17(3), 187–195. 10.2174/187152491766617020114510 [DOI] [PubMed] [Google Scholar]
- Morris, G.‐M. , Huey, R. , Lindstrom, W. , Sanner, M. F. , Belew, R. K. , Goodsell, D. S. , & Olson, A.‐J. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry, 30(16), 2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakane, H. , & Ono, K. (1990). Differential inhibitory effects of some catechin derivatives on the activities ofhuman immunodeficiency virus reverse transcriptase and cellular deoxyribonucleic and ribonucleic acid polymerases. Biochemistry, 29, 2841–2845. [DOI] [PubMed] [Google Scholar]
- Oxford, J. S. , Lambkin, R. , Guralnik, M. , Rosenbloom, R.‐A. , Petteruti, M.‐P. , Digian, K. , & LeFante, C. (2007b). In vivo prophylactic activity of QR‐435 against H3N2 influenza virus infection. American Journal of Therapeutics, 14, 462–468. [DOI] [PubMed] [Google Scholar]
- Oxford, J.‐S. , Lambkin, R. , Guralnik, M. , Rosenbloom, R.‐A. , Petteruti, M.‐P. , Digian, K. , & Lefante, C. (2007a). Preclinical in vitro activity of QR‐435 against influenza A virus as a virucide and in paper masks for prevention of viral transmission. American Journal of Therapeutics, 14, 455–461. [DOI] [PubMed] [Google Scholar]
- Rismanbaf, A. , & Zarei, S. (2020). Liver and kidney injuries in COVID‐19 and their effects on drug therapy; a letter to editor. Archives of Academic Emergency Medicine, 8(1), e17. [PMC free article] [PubMed] [Google Scholar]
- Schneidman‐Duhovny, D. , Inbar, Y. , Nussinov, R. , & Wolfson, H.‐J. (2005). PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucleic Acids Research, 33, W363–W367. 10.1093/nar/gki481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, W. , Chen, C. , Lei, X. , Zhao, J. , & Liang, J. (2018). CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Research, 46(W1), W363–W367. 10.1093/nar/gky473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls, A.‐C. , Park, Y.‐J. , Tortorici, M.‐A. , Wall, A. , McGuire, A.‐T. , & Veesler, D. (2020). Structure, function, and antigenicity of the SARS‐CoV‐2 spike glycoprotein. Cell, 181, 281–292. 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. , Cao, R. , Zhang, L. , Yang, X. , Liu, J. , … Xiao, G. (2020). Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019‐nCoV) in vitro. Cell Research, 30(3), 269–271. 10.1038/s41422-020-0282-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, C. , Sliva, K. , Von, R.‐C. , Kummerer, B.‐M. , & Schnierle, B.‐S. (2015). The green tea catechin, epigallocatechin gallate inhibits chikungunya virus infection. Antiviral Research, 113(1–3), 1–3. [DOI] [PubMed] [Google Scholar]
- Wrapp, D. , Wang, N. , Corbett, K.‐S. , Goldsmith, J.‐A. , Hsieh, C.‐L. , … McLellan, J.‐S. (2020). Cryo‐EM structure of the 2019‐nCoV spike in the prefusion conformation. Science, 367(6483), 1260–1263. 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, H. , Takuma, N. , Daimon, T. , & Hara, Y. (2006). Gargling with tea catechin extracts for the prevention ofinfluenza infection in elderly nursing home residents: A prospective clinical study. Journal of Alternative and Complementary Medicine, 12, 669–672. [DOI] [PubMed] [Google Scholar]
- Yazdany, J. , & Kim, A.‐H. (2020). Use of hydroxychloroquine and chloroquine during the COVID‐19 pandemic: What every clinician should know. Annals of Internal Medicine, 172, 754–755. 10.7326/M20-1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You, H.‐L. , Huang, C.‐C. , Chen, C.‐J. , Chang, C.‐C. , Liao, P.‐L. , & Huang, S.‐T. (2018). Anti‐pandemic influenza A (H1N1) virus potential of catechin and gallic acid. Journal of Chinese Medical Association, 81(5), 458–468. 10.1016/j.jcma.2017.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, C. , Liu, S. , Li, C. , Yang, L. , & Zu, Y. (2014). In vitro evaluation of the antiviral activity of the synthetic epigallocatechin gallate analog‐epigallocatechin gallate (EGCG) palmitate against porcine reproductive andrespiratory syndrome virus. Viruses, 6, 938–950. [DOI] [PMC free article] [PubMed] [Google Scholar]


