Editor
SARS-CoV-2 is the causative agent for the COVID-19 pandemic. COVID-19 has necessitated rapid changes in surgical practice and organisation through both the initial peak and ongoing recovery period1. SARS-CoV-2 infects cells by interacting with the host cell surface protein ACE2 and utilises TMPRSS2 in viral spike protein priming to facilitate cell entry (Fig. 1a)2. Whilst COVID-19 is predominantly a respiratory disease approximately 15% of patients have concurrent gastrointestinal symptoms3. SARS-CoV-2 RNA and live virus have been identified in stool from COVID-19 patients and SARS-CoV-2 readily infects intestinal organoids4–6. Despite these circumstantial data, gastrointestinal transmission has not yet been formally confirmed. Cancers commonly express different genes from the tissue of origin and it is largely unexplored whether tumours can be infected with SARS-CoV-2. We sought to explore the expression of ACE2 and TMPRSS2 in large publicly available normal tissue and pan-cancer expression data sets to understand whether levels of these genes identify susceptible tissues.
Fig. 1.
ACE2 and TMPRSS2 are expressed heterogeneously in the normal intestine and colorectal cancer
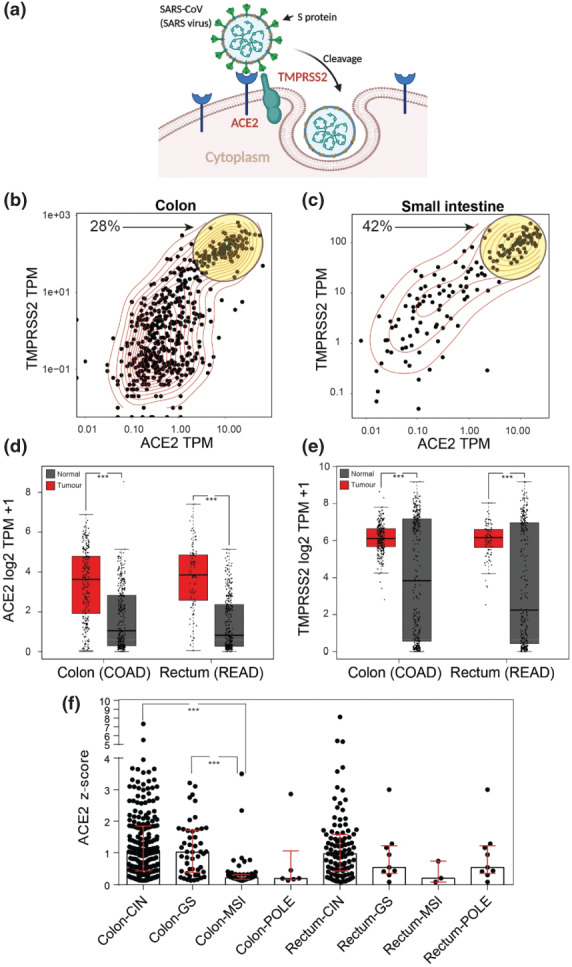
(a) Schematic demonstrating the mode of entry of SARS-CoV-2 into cells via interactions with the cell surface proteins ACE2 and TMPRSS2. (b) Scatter plot of the expression of ACE2 and TMPRSS2 across all colon GTEx samples. Yellow circle highlights high co-expressing samples. (c) Scatter plot of the expression of ACE2 and TMPRSS2 across all small intestine GTEx samples. Yellow circle highlights high co-expressing samples. (d) Box and whisker plot of expression of ACE2 between normal large intestine (Grey, TCGA and GTEx) and colon and rectal cancer (Red, TCGA) samples. Median +/- IQR. One-way ANOVA. ***, P < 0.001. TPM = Transcripts per million. (e) Box and whisker plot of expression of TMPRSS2 between normal large intestine (Grey, TCGA and GTEx) and colon and rectal cancer (Red, TCGA) samples. Median +/- IQR. One-way ANOVA. ***, P < 0.001. TPM = Transcripts per million. (f) Box and scatter plot of ACE2 expression levels from TCGA COAD/READ data sets between tumour subtypes CIN = chromosomal instability, MSI = microsatellite instability, GS = genome stable and POLE = DNA polymerase epsilon. Median +/- IQR. Kruskal-Wallis. ***, p < 0.001.
Analysis of the normal tissue Genotype Tissue Expression project (GTEx) dataset showed high ACE2 expression in the testis, small intestine, kidney, heart, thyroid and adipose tissue (Fig. S1a, supporting information). TMPRSS2 levels were highest in the prostate, stomach, small intestine, pancreas, lung, salivary gland, kidney, thyroid and liver (Fig. S1b, supporting information). Whilst initial analysis suggested only kidney and thyroid co-expressed high levels of ACE2 and TMPRSS2 closer inspection of small intestinal and colonic samples revealed heterogeneity with a sub-population (30-40%) also co-expressing high levels that was confirmed by K-means clustering (Fig. 1b,c, Fig. S1c,d, supporting information). In colonic tissue high levels of both genes were found in younger patients and greater TMPRSS2 expression in females, although neither factor defined the sub-population observed (Fig. S2a-j, supporting information). Biopsy location and tissue compartment from colon samples were explored and higher levels of both genes were found in the mucosa and proximal colon although these factors also failed to fully define the high co-expressing sub-population (Fig. S2k-m, supporting information). These normal expression data appear to identify a proportion of individuals who, in the gastrointestinal tract, express high levels of both genes known to be involved in cell entry of SARS-CoV-2.
The Cancer Genome Atlas program (TCGA) is the largest pan-tumour collection of genomic and transcriptomic sequencing data. Having identified the heterogeneous gene expression within the normal gastrointestinal tract we interrogated TCGA and GTEx gene expression data to identify relative expression of ACE2 and TMPRSS2 in tumours compared to their tissue of origin. Generally, there was no correlation between high expression of ACE2 and TPMRSS2 in normal tissue and high tumour expression (Fig. S3a-d, supporting information). However, we identified colorectal cancer as unique amongst human malignancies by co-expressing higher levels of both ACE2 and TMPRSS2 relative to normal (Fig. 1d,e). We explored TCGA data to identify if molecular subgroups of colorectal cancers, specific mutations or other commonly collected clinical variables could define tumours with varying expression of ACE2 and TMPRSS2. There was a subtle yet significant trend for higher ACE2 expression with younger age but no association with sex (Fig. S4a, supporting information). Possession of a BRAF mutation was found to predict lower levels of tumour ACE2 however TMPRSS2 expression was unchanged (Fig. S4b,c, supporting information). Strikingly, ACE2 levels were very low in tumours with microsatellite instability (MSI) (Fig. 1f).
Cumulatively these data identify a proportion of healthy individuals as susceptible to putative SARS-CoV-2 intestinal infection and that patients with colorectal cancer may be at even greater risk of infection. Further clinical studies are urgently required to explore this mode of transmission of COVID-19.
Conflicts of Interest and Sources of Funding
There are no conflicts of interests.
SJAB is supported by an Advanced Clinician Scientist Fellowship grant from Cancer Research UK C14094/A27178; and core funding from Wellcome and MRC to the Wellcome-MRC Cambridge Stem Cell Institute.
Supplementary Material
Appendix S1. Supporting Information
References
- 1. Søreide K, Hallet J, Matthews JB, Schnitzbauer AA, Line PD, Lai PBSet al. . Immediate and long-term impact of the COVID-19 pandemic on delivery of surgical services. Br J Surg 2020; 10.1002/bjs.11670 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen Set al. . SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020; 181: 271–280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yang L, Tu L. Implications of gastrointestinal manifestations of COVID-19. Lancet Gastroenterol Hepatol 2020; 5: 629–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wu Y, Guo C, Tang L, Hong Z, Zhou J, Dong Xet al. . Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol 2020; 5: 434–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang W, Xu Y, Gao R, Lu R, Han K, Wu Get al. . Detection of SARS-CoV-2 in Different Types of Clinical Specimens. Jama 2020; 323: 1843–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lamers MM, Beumer J, van der Vaart J, Knoops K, Puschhof J, Breugem TIet al. . SARS-CoV-2 productively infects human gut enterocytes. Science 2020; 10.1126/science.abc1669 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Supporting Information


