Abstract
Background
To determine the utility of admission laboratory markers in the assessment and prognostication of coronavirus disease‐2019 (COVID‐19), a systematic review and meta‐analysis were conducted on the association between admission laboratory values in hospitalized COVID‐19 patients and subsequent disease severity and mortality.
Material and Methods
Searches were conducted in MEDLINE, Pubmed, Embase, and the WHO Global Research Database from December 1,2019 to May 1, 2020 for relevant articles. A random effects meta‐analysis was used to calculate the weighted mean difference (WMD) and 95% confidence interval (95% CI) for each of 27 laboratory markers. The impact of age and sex on WMDs was estimated using meta‐regression techniques for 11 markers.
Results
In total, 64 studies met the inclusion criteria. The most marked WMDs were for neutrophils (ANC) at 3.82 × 109/L (2.76, 4.87), lymphocytes (ALC) at −0.34 × 109/L (−0.45, −0.23), interleukin‐6 (IL‐6) at 32.59 pg/mL (23.99, 41.19), ferritin at 814.14 ng/mL (551.48, 1076.81), C‐reactive protein (CRP) at 66.11 mg/L (52.16, 80.06), D‐dimer at 5.74 mg/L (3.91, 7.58), LDH at 232.41 U/L (178.31, 286.52), and high sensitivity troponin I at 90.47 pg/mL (47.79, 133.14) when comparing fatal to nonfatal cases. Similar trends were observed comparing severe to non‐severe groups. There were no statistically significant associations between age or sex and WMD for any of the markers included in the meta‐regression.
Conclusion
The results highlight that hyper inflammation, blunted adaptive immune response, and intravascular coagulation play key roles in the pathogenesis of COVID‐19. Markers of these processes are good candidates to identify patients for early intervention and, importantly, are likely reliable regardless of age or sex in adult patients.
Keywords: COVID‐19, laboratory values, meta‐analysis, meta‐regression, systematic review
1. INTRODUCTION
The disease caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV2), known as coronavirus disease‐2019 (COVID‐19), is primarily a respiratory condition that can range from being asymptomatic to causing respiratory failure and other potentially fatal complications. 1 Approximately 20% of cases develop severe dyspnea due to an often‐bilateral viral pneumonia that requires hospitalization. 2 The virus has caused a global pandemic with growing case numbers, but early studies of seroprevalence estimate that the proportion of infected individuals does not exceed 20% even in regions with large case burdens, leaving most of the population susceptible. 3 , 4 , 5 As such, hospitals across the world remain at risk of spikes in patient load that may stretch or exceed their capacity, thereby contributing to worsening COVID‐19 morbidity and mortality.
As has been demonstrated with other health conditions, clinical tools incorporating laboratory parameters have been invaluable to the efficient use of health care resources, and improvement of patient outcomes. 6 Such tools are often based on an understanding of disease pathophysiology, and in the case of COVID‐19, cytokine storm syndrome and thromboinflammation have surfaced as central and interconnected factors in the development of severe and fatal illness. 7 , 8 , 9 These disease processes can be monitored using various biochemical and hematologic markers that are routinely measured at the time of hospitalization, potentially contributing to the accurate prediction of severity and mortality among patients hospitalized for COVID‐19 and allowing for early intervention. 10 , 11 However, the development of useful predictive tools incorporating laboratory parameters will require studies with large sample sizes covering broad population groups to be accurate and generalizable. Usually, individual studies are small and hence meta‐analyses could provide critical evidence needed for clinical and policy decisions.
To date, meta‐analyses assessing the impact of COVID‐19 on laboratory markers have suffered from various methodological and reporting issues, including a lack of consideration for potentially overlapping datasets, incorrect estimation of study means and error, and not including data on mortality. 10 , 11 In addition, given the importance of age and sex as predictors of disease severity, the contribution of these factors to any associations between laboratory parameters and COVID‐19 severity has not received adequate attention. 2 , 10 , 11 To resolve these limitations, we first conducted a systematic review and meta‐analysis on the association between admission laboratory values and disease severity and mortality in patients hospitalized for COVID‐19. We then assessed the contributions of age and sex to the observed associations using meta‐regression techniques.
2. METHODS
2.1. Search strategy
A broad initial search was conducted on March 17, 2020 in PubMed, Ovid Embase, and Ovid Medline, limited to publications from 2019 onwards. In addition, the WHO Global Research Database 12 was manually searched up to March 17, 2020 for relevant publications. Shortly after, a search strategy was created by Wolters Kluwer, 13 which was used to supplement the search in Medline.
An updated search was run on May 1, 2020 to capture additional publications entered into PubMed and Embase since March 16, 2020. The PubMed strategy included the new MeSH supplementary concepts, “severe acute respiratory syndrome coronavirus 2” and “COVID‐19.” We combined the COVID‐19 PubMed strategy provided by the Stephen B. Thacker CDC Library 14 with the newly released Embase strategy by Wolters Kluwer 15 and translated them for the appropriate databases. To expand the search while maintaining relevance to the topic, we searched keywords in multi‐purpose or.mp fields. Conference abstracts were excluded from Embase, and additional limiters, including English language and entry dates, were applied in both databases. The search strategies are presented in Supplement 1. Backward chaining was employed to identify potentially relevant references in the included studies and published reviews.
2.2. Screening and data extraction
Articles were imported to Covidence, a systematic review manager. 16 The majority of duplicate records were automatically excluded at the article importing stage. Two reviewers (JK and MD) independently screened the titles, abstracts, and full texts based on eligibility criteria. Studies were eligible for inclusion if they provided hospital‐based data and reported summary values (eg, mean) with precision estimates (eg, standard deviation (SD)). Studies were excluded at full‐text assessment stage if they were not relevant to the review topic, were exact duplicates, were not in the English language, did not report results by disease severity or mortality (wrong outcomes), were not original research articles (wrong study design), were not available in full‐text, or were focused on ineligible patient populations (eg pregnant women and children).
Reviewers extracted data on study characteristics (ie, authors, date of publication, study location, sample size, and study design), characteristics of the participants (ie, mean age and sex), disease status (ie, severe vs non‐severe, deceased vs survived), laboratory markers at admission (ie, mean and SD, median and interquartile range (IQR), or range of minimum and maximum observations), and inclusion and exclusion criteria.
2.3. Disease status and laboratory markers
Across all studies, COVID‐19 cases were diagnosed based on clinical suspicion and all cases were ultimately confirmed via reverse transcription‐polymerase chain reaction (RT‐PCR). Disease status was evaluated as COVID‐19‐related disease severity and mortality, which were compared to non‐severe and surviving categories, respectively. COVID‐19 severity was classified using various criteria across studies. Most studies used the Chinese National Commission of Health Guidelines, which provide the following categories for COVID‐19 severity 17 : (a) Mild—May include fever, respiratory symptoms, and signs of pneumonia on radiological imaging; (b) Severe—Respiratory distress with the respiratory rate (RR) ≥ 30, oxygen (O2) saturation ≤ 93% at rest on room air, or PaO2/FiO2 ≤ 300 mm Hg; (c) Critical – Respiratory failure, the requirement for mechanical ventilation, shock, or ICU admission. Some studies used the American Thoracic Society (ATS) Guidelines for Community‐Acquired Pneumonia (CAP) which define severe pneumonia as either the development of septic shock with vasopressor requirement, respiratory failure requiring mechanical ventilation, or at least three of the following: RR > 30, PaO2/FiO2 < 250 mm Hg, multilobar infiltrates, confusion, uremia (BUN ≥ 20 mg/dL), leukopenia (<4.0 × 109 cells/L), thrombocytopenia (<100 × 109 cells/L), hypothermia (core temperature < 36 C), and hypotension requiring aggressive fluid resuscitation. 18 Other studies defined severity based on one of the following: admission to Intensive Care Unit (ICU), development of acute respiratory distress syndrome (ARDS), oxygen saturation at rest, the requirement of invasive mechanical ventilation (IMV), development of acute cardiac injury, or custom composite endpoints. A table reflecting the major classification schemes is presented in Supplement 2.
The following laboratory markers were considered in this study: Hemoglobin (Hb), white blood cell count (WBC), absolute neutrophil count (ANC), absolute lymphocyte count (ALC), platelet count (PLT), C‐reactive protein (CRP), erythrocyte sedimentation rate (ESR), ferritin, interleukin‐6 (IL‐6), interleukin‐10 (IL‐10), procalcitonin (PCT), albumin, total bilirubin, prothrombin time (PT), creatinine (Cr), blood urea nitrogen (BUN), activated partial thromboplastin time (APTT), Ddimer, lactate dehydrogenase (LDH), creatine kinase (CK), high sensitivity troponin I (hsTropI), troponin I (TropI), creatine kinase myocardial band (CKMB), alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP), and γ‐glutamyl transferase (GGT).
2.4. Risk of bias
This systematic review and meta‐analysis follow the Preferred Reporting Items for Systematic Review and Meta‐Analysis (PRISMA) guidelines. 19 The quality of included studies was assessed by using the Institute of Health Economics (IHE) quality appraisal of the case series studies checklist. 20 Although the checklist includes twenty critical appraisal items for quality assessment, only fifteen criteria were relevant to those included in this review, of which both reviewers agreed that seven were especially important to the risk of bias. These seven items were: Consecutive recruitment, reporting of patient characteristics, clear eligibility criteria, similar disease point at study entry, appropriate outcome measurement, sufficient follow‐up, and estimates of random variability. 20 Based on reviewers’ judgment, the risk of bias was categorized as low, medium, or high, if 0, 1, or ≥ 2 checklist items were marked as no or unclear, respectively.
2.5. Risk of duplicate data
To determine the potential of duplicate data from the studies selected for inclusion, we compared studies based on their research teams (eg, authors list), study location (ie, city and hospital), and reported study period. If two or more studies shared study sites and had overlapping study periods, one study was designated as a reference study and assigned a low duplicate risk and the others were considered a high duplicate risk. Reference studies were selected based on the length of the study period, the number of patients in the sample, and number of laboratory markers reported and had to be agreed upon by both reviewers. Furthermore, the risk of duplication was separately considered for each laboratory marker in those studies considered to be at high risk of duplicate reporting. Within a high duplicate risk study, laboratory markers not reported in the associated reference study were considered as low risk of duplication.
2.6. Statistical analyses
Reported means and SDs for laboratory parameters in each included study were used to estimate weighted mean differences (WMD) and 95% confidence intervals (95% CI) for severe versus non‐severe and deceased versus surviving patients. In the absence of mean and SD values, sample sizes, medians, and measures of precision (ie IQR or range) were used to calculate mean and SD (Supplement 3 and 4). 21 These data were then pooled to provide overall WMDs and their 95% CIs using the DerSimonian and Laird random‐effects model. 22 To quantify heterogeneity, the I 2 (%) statistic was calculated as a measure of inconsistency. 23 I 2 thresholds of 25%, 50%, and 75%, indicated low, medium, and high levels of heterogeneity, respectively. 23
To assess the potential impact of duplicate data, as a sensitivity analysis, we excluded studies categorized as high risk of duplication. Additional sensitivity analyses were performed by excluding studies with a high risk of bias and studies with confidence intervals not overlapping with the 95% CIs of the pooled estimates (ie, “outlier studies”). 11
To account for Type I error rate for multiple hypothesis testing (ie, 27 and 23 laboratory markers for disease severity and mortality, respectively), Bonferroni correction was used to declare the significance levels of P values. 24 The Bonferroni corrected P value for the disease severity tests including overall estimates, sensitivity analysis by the risk of duplicates, and sensitivity analysis by the risk of bias was .002 (ie, corrected P = .05/27) and for sensitivity analysis of outliers, where only 15 laboratory markers were tested, was .003 (ie, corrected P = .05/15). The Bonferroni corrected p‐value for the mortality analyses including overall estimates, sensitivity analysis by the risk of duplicates, and sensitivity analysis by the risk of bias was .002 (ie, corrected P = .05/23), and for sensitivity analysis of outliers, where only 8 laboratory markers were tested, was .006 (ie, corrected P = .05/8).
Finally, to assess the potential impact of age and sex on WMD variation when comparing severe to non‐severe or fatal to nonfatal cases, univariate random‐effects meta‐regression, using the method of moments, was conducted on 11 laboratory markers, including ALC, ANC, WBC, Ddimer, PT, ferritin, IL‐6, IL‐10, CRP, ESR, and albumin. These markers were selected because they reflect the key pathogenetic mechanisms involved and may vary by age and sex. We calculated tests for covariates using a minimum of 10 000 Monte Carlo random permutations. 25 All statistical analyses were performed using STATA software (version 16.1; StataCorp, College Station, TX). 26
3. RESULTS
In total 15 314 studies were identified through systematic search and backward chaining (Figure 1). Following the removal of duplicates and title/abstract screening, 527 studies remained for full‐text review. 64 studies remained for data extraction and analysis after excluding ineligible studies for the following reasons: irrelevant to study objectives (N = 292), wrong study outcomes (N = 68), ineligible study design (N = 30), wrong patient population (N = 28), not available in full‐text (N = 23), duplicate of included study (N = 13), and not in English (N = 9). All included studies were case series.
Figure 1.
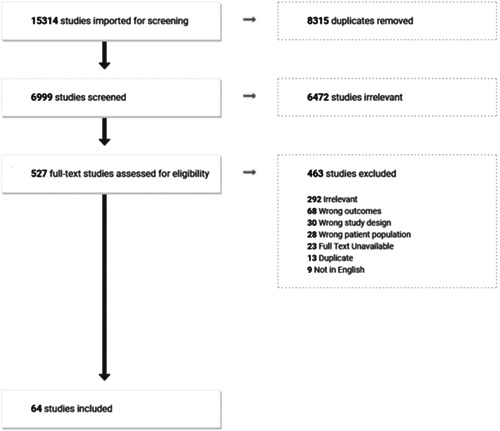
PRISMA flow diagram outlining study selection
The key characteristics of included studies are presented in Table 1. Forty‐nine studies reported severity outcomes, 27 , 28 , 29 , 30 , 31 , 32 , 33 , 34 , 35 , 36 , 37 , 38 , 39 , 40 , 41 , 42 , 43 , 44 , 45 , 46 , 47 , 48 , 49 , 50 , 51 , 52 , 53 , 54 , 55 , 56 , 57 , 58 , 59 , 60 , 61 , 62 , 63 , 64 , 65 , 66 , 67 , 68 , 69 , 70 , 71 , 72 , 73 , 74 , 75 fourteen reported mortality outcomes 27 , 28 , 29 , 76 , 77 , 78 , 79 , 80 , 81 , 82 , 83 , 84 , 85 , 86 and one reported both mortality and severity. 87 COVID‐19 severity was classified using Chinese National Commission of Health Guidelines in 31 studies, American Thoracic Society Guidelines for Community‐Acquired Pneumonia in three studies, oxygen saturation at rest in two studies, ICU admission in seven studies, ARDS in one study, cardiac injury in one study, IMV in two studies, and custom composite endpoints in three studies.
Table 1.
Main characteristics of included studies
| Study (ID) | Outcomes | Study characteristics | Patient characteristics | Risk of bias | Duplicate risk |
|---|---|---|---|---|---|
| Xu et al (54) 76 | ALC, ALT, ANC, AST, BUN, CK, Cr, CRP, Ddimer, IL‐6, IL‐10, PCT, PT, WBC | Setting: Hubei Provincial Hospital of Chinese and Western Medicine, Wuhan, China | Survivors: 117 | Low | Low |
| Male, n: 59 | |||||
| Study period: Dec 26‐Mar 1 | Age, y: 72.75 ± 7.26 | ||||
| Follow‐up: Unclear | Deceased: 28 | ||||
| Sample size: 145 | Male, n: 17 | ||||
| Exposure: Mortality | Age, y: 55 ± 17.25 | ||||
| Zhou et al (13) 77 | Alb, ALC, ALT, Ddimer, Ferr, Hb, IL‐6, LDH, PLT, PCT, PT, hsTropI, WBC | Setting: Jin Yintan Hospital, Wuhan Pulmonary Hospital, Wuhan, China | Survivors: 137 | Low | Low (reference study) |
| Male, n: 81 | |||||
| Study period: Dec 29‐Jan 31 | Age, y: 51.7 ± 9.7 | ||||
| Follow‐up: Jan 31 (complete) | Deceased: 54 | ||||
| Sample size: 191 | Male, n: 38 | ||||
| Exposure: Mortality (death or discharge by study end) | Age, y: 69.3 ± 9.9 | ||||
| Yang et al (15) 78 | ALC, Bili, Cr, Hb, PLT | Setting: Jin Yintan Hospital, Wuhan, China | Survivors: 20 | Low | High |
| Reasons: | |||||
| Study period: Dec 24‐Jan 26 | Male, n: 14 | Setting and period shared with Zhou (13), Wu (6) | |||
| Follow‐up: Feb 9 (28 d for mortality) | Age, y: 51.9 ± 12.9 | ||||
| Sample size: 52 | Deceased: 32 | ||||
| Exposure: Mortality (28‐d mortality) | Male, n: 21 | ||||
| Age, y: 64.6 ± 11.2 | |||||
| Ruan et al (11) 79 | Alb, ALC, ALT, AST, Bili, Cr, CRP, Ferritin, Hb, IL‐6, LDH, PLT, hsTropI, WBC | Setting: Jin Yintan Hospital, Tongji Hospital, Wuhan, China | Survivors: 82 | High | HighReasons: Setting – Zhou (13), Wu (6), Yang (15)Study period – Unclear |
| Study period: Unclear (before Mar 3) | Male, n: 53 | ||||
| Follow‐up: Unclear | Age, y: 58.33 ± 27.9 | ||||
| Sample size: 150 | Deceased: 68 | ||||
| Exposure: Mortality | Male, n: 49 | ||||
| Age, y: 54.33 ± 50 | |||||
| Wu et al (6) 87 | Alb, ALC, ALT, ANC, AST, Bili, BUN, Cr, CRP, Ddimer, ESR, Ferritin, IL‐6, LDH, PLT, PT, WBC | Setting: Jin Yintan Hospital, Wuhan, China | Survivors: 40 | Low | High |
| Reasons: | |||||
| Study period: Dec 25‐Jan 26 | Male, n: 31 | Setting and period shared with Zhou (13), Yang (15) | |||
| Follow‐up: Feb 13 (incomplete) | Age, y: 49.03 ± 12.7 | ||||
| Sample size: 84 | Deceased: 44 | ||||
| Exposure: Mortality (Acute Respiratory Distress Syndrome (ARDS) patients only) | Male, n: 29 | ||||
| Age, y: 67.6 ± 12.1 | |||||
| Zhang et al (59) 80 | Alb, CRP, Ddimer | Setting: Liyuan Hospital, Wuhan, China | Survivors: 11 | Med | Low |
| Study period: Jan 16‐20 Feb | Male, n: 6 | ||||
| Follow‐up: Unclear | Age, y: 64.33 ± 42.8 | ||||
| Sample size: 19 | Deceased: 8 | ||||
| Exposure: Mortality (ICU patients only) | Male, n: 5 | ||||
| Age, y: 78 ± 25.3 | |||||
| Wang et al (47) 81 | ALC, ALT, ANC, APTT, AST, BUN, CK, Cr, CRP, Ddimer, Hb, IL‐6, LDH, PLT, PCT, PT, hsTropI, WBC | Setting: Renmin Hospital, Wuhan, China | Survivors: 274 | Low | Low |
| Male, n: 127 | |||||
| Study period: Jan 1‐Feb 6 | Age, y: 68.67 ± 7.6 | ||||
| Follow‐up: 5 Mar (complete) | Deceased: 65 | ||||
| Sample size: 339 | Male, n: 39 | ||||
| Exposure: Mortality (Patients over 60 only) | Age, y: 76.33 ± 9.6 | ||||
| Wang et al (49) 82 | Alb, ALC, ALT, ANC, AST, Bili, BUN, CK, Cr, CRP, Ddimer, IL‐6, IL‐10, LDH, PLT, PCT, PT, hsTropI, WBC | Setting: Tongji Hospital, Wuhan, China | Survivors: 211 | Low | Low (reference study) |
| Study period: Jan 25‐Feb 25 | Male, n: 105 | ||||
| Follow‐up: Mar 24 (complete) | Age, y: 57.7 ± 16.3 | ||||
| Sample size: 344 | |||||
| Exposure: Mortality (ICU patients only) | Deceased: 133 | ||||
| Male, n: 74 | |||||
| Age, y: 69.7 ± 11.2 | |||||
| Chen et al (24) 83 | Alb, ALC, ALT, ANC, APTT, AST, Bili, BUN, CK, Cr, CRP, Ddimer, ESR, Ferritin, Hb, IL‐6, IL‐10, LDH, PLT, PCT, PT, hsTropI, WBC | Setting: Tongji Hospital, Wuhan, China | Survivors: 161 | Low | High |
| Reasons: | |||||
| Study period: Jan 13‐Feb 12 | Male, n: 88 | Setting and period shared with Wang (49) | |||
| Follow‐up: Feb 18 (complete) | Age, y: 51.33 ± 21.7 | ||||
| Sample size: 339 | Deceased: 113 | ||||
| Exposure: Mortality | Male, n: 83 | ||||
| Age, y: 69 ± 11.3 | |||||
| Tang et al (12) 84 | Ddimer, PT | Setting: Tongji Hospital, Wuhan, China | Survivors: 162 | Med | High |
| Reasons: | |||||
| Study period: Jan 1‐Feb 3 | Male, n: 82 | Setting and period shared with Tang (43), Fibrinogen OK | |||
| Follow‐up: Feb 13 (incomplete) | Age, y: 52.4 ± 15.6 | ||||
| Sample size: 183 | |||||
| Exposure: Mortality | Deceased: 21 | ||||
| Male, n: 16 | |||||
| Age, y: 64 ± 20.7 | |||||
| Tang et al (43) 85 | Ddimer, PLT, PT | Setting: Tongji Hospital, Wuhan, China | Survivors: 315 | Low | Low |
| Study period: Jan 1‐Feb 13 | Male, n: 178 | ||||
| Follow‐up: Mar 13 (complete) | Age, y: 63.7 ± 12.2 | ||||
| Sample size: 449 | |||||
| Exposure: Mortality (Severely ill patients only) | Deceased: 134 | ||||
| Male, n: 90 | |||||
| Age, y: 68.7 ± 11.4 | |||||
| Yan et al (55) 86 | Alb, ALC, ALT, ANC, APTT, AST, Bili, BUN, CK, Cr, CRP, Ddimer, ESR, Ferritin, Hb, IL‐6, LDH, PLT, PCT, PT, hsTropI, WBC | Setting: Tongji Hospital, Wuhan, China | Survivors: 9 | Med | High |
| Male, n: 3 | Reasons: | ||||
| Study period: Jan 10‐Feb 24 | Age, y: 64.7 ± 7.3 | Setting and period shared with Wang (49), Ferritin, Hemoglobin, Thromboplastin time OK | |||
| Follow‐up: Unclear | Deceased: 39 | ||||
| Sample size: 48 | Male, n: 30 | ||||
| Exposure: Mortality (Diabetic patients only) | Age, y: 70.5 ± 10.1 | ||||
| Du et al (27) 27 | Alb, ALC, ALT, ANC, APTT, AST, Bili, Cr, CRP, Ddimer, PCT, PT, WBC | Setting: Wuhan Pulmonary Hospital, Wuhan, China | Survivors: 158 | Low | Low (reference study) |
| Study period: Dec 25‐Feb 7 | Male, n: 87 | ||||
| Follow‐up: Complete | Age, y: 56 ± 13.5 | ||||
| Sample size: 179 | |||||
| Exposure: Mortality | Deceased: 21 | ||||
| Male, n: 10 | |||||
| Age, y: 70.2 ± 7.7 | |||||
| Gao et al (14) 28 | ALC, ALT, AST, Bili, Cr, Ddimer, Hb, PLT, PCT, PT, hsTropI, WBC | Setting: Wuhan Pulmonary Hospital, Wuhan, China | Survivors: 8 | Med | High |
| Reasons: | |||||
| Male, n: 6 | |||||
| Study period: Jan 1‐Jan 29 | Age, y: 56.3 ± 10 | Setting and period shared with Du (27) | |||
| Follow‐up: Feb 9 (incomplete) | |||||
| Sample size: 15 | Deceased: 7 | ||||
| Exposure: Mortality | Male, n: 4 | ||||
| Age, y: 68 ± 3.3 | |||||
| Chen et al (23) 29 | Alb, ALC, ALT, ANC, AST, CK, Cr, CRP, Ddimer, ESR, IL‐6, LDH, PLT, PCT, WBC | Setting: Zhongnan Hospital, Wuhan, China | Survivors: 36 | High | Low |
| Study period: Jan 1‐ Feb 10 | Male, n: 18 | ||||
| Follow‐up: Feb 20 (incomplete) | Age, y: 72 ± ? | ||||
| Sample size: 55 | Deceased: 19 | ||||
| Exposure: Mortality (Patients over 65 only) | Male, n: 16 | ||||
| Age, y: 77 ± ? | |||||
| Myers et al (37) 30 | ALC, ALT, ANC, AST, Bili, Cr, LDH, WBC | Setting: Multicenter, Northern California, USA | Non‐severe: 264 | Med | Low |
| Study period: Mar 1‐Mar 31 | Male, n: 138 | ||||
| Follow‐up: Apr 9 (incomplete) | Age, y: 60.33 ± 17.04 | ||||
| Sample size: 277 | Severe: 113 | ||||
| Exposure: Severity (ICU or non‐ICU) | Male, n: 74 | ||||
| Age, y: 63 ± 15.02 | |||||
| Lu et al (35) 31 | BUN, Cr | Setting: Multicenter, 42 hospitals in Hubei, Sichuan, and Chongqing, China | Non‐severe: 196 | Low | High |
| Male, n: 116 | Reasons: | ||||
| Study period: Jan 13‐Feb 18 | Age, y: 39.7 ± 13.4 | Multicenter study across 3 provinces in China | |||
| Follow‐up: Complete | Severe: 108 | ||||
| Sample size: 304 | Male, n: 66 | ||||
| Exposure: Severity (severe or mild, Chinese classification) | Age, y: 60.6 ± 19.7 | ||||
| Guan et al (5) 32 | ALC, Hb, PLT, WBC | Setting: Multicenter, 522 hospitals in China | Non‐severe: 1032 | High | High |
| Reasons: | |||||
| Study period: Dec 11‐Jan 29 | Male, n: 595 | Multicenter study across many hospitals in China | |||
| Follow‐up: Jan 31 (incomplete) | Age, y: 46 ± 16.3 | ||||
| Sample size: 1099 | Severe: 67 | ||||
| Exposure: Severity (ICU/IMV/Death or not) | Male, n: 45 | ||||
| Age, y: 62.3 ± 13.6 | |||||
| Zheng et al (63) 33 | ALC, ALT, ANC, AST, CRP, Ddimer, PT, WBC | Setting: Chengdu Public Health Clinical Medical Center, Chengdu, China | Non‐severe: 67 | Med | Low |
| Male, n: 32 | |||||
| Study period: Jan 11‐Feb 20 | Age, y: 42.5 ± 15.1 | ||||
| Follow‐up: Feb 23 (incomplete) | Severe: 32 | ||||
| Sample size: 99 | Male, n: 19 | ||||
| Exposure: Severity (Critical or noncritical, Chinese classification) | Age, y: 63.8 ± 16.5 | ||||
| Yao et al (57) 34 | Alb, ALC, ALT, ANC, Cr, CRP, Ddimer, Hb, PCT, WBC | Setting: Dabieshan Medical Center, Huanggang City, China | Non‐severe: 83 | Low | Low |
| Study period: Jan 30‐Feb 11 | Male, n: 30 | ||||
| Follow‐up: Mar 3 (complete) | Age, y: 46.7 ± 16.6 | ||||
| Sample size: 108 | Severe: 25 | ||||
| Exposure: Severity (Severe or non‐severe, ATS classification) | Male, n: 13 | ||||
| Age, y: 60 ± 16.5 | |||||
| Liu et al (34) 35 | ALC, APTT, CK, CKMB, Ddimer, IL‐6, IL‐10, LDH, PT | Setting: First Affiliated Hospital of Nanchang University, China | Non‐severe: 30 | High | Low |
| Male, n: NR | |||||
| Study period: Jan 22‐Feb 15 | Age, y: NR | ||||
| Follow‐up: Unclear | Severe: 46 | ||||
| Sample size: 76 | Male, n: NR | ||||
| Exposure: Severity (Severe or mild, Chinese classification) | Age, y: NR | ||||
| Wu et al (52) 36 | Alb, ALC, ALT, ANC, APTT, AST, Bili, BUN, CK, CKMB, Cr, CRP, Ddimer, ESR, Hb, LDH, PLT, PCT, PT, WBC | Setting: First People's Hospital of Yancheng City, Second People's Hospital of Yancheng City, Second People's Hospital of Fuyang City, Fifth People's Hospital of Wuxi, China | Non‐severe: 197 | High | Low |
| Male, n: 106 | |||||
| Age, y: 37.6 ± 17.1 | |||||
| Study period: Jan 20‐Feb 19 | Severe: 83 | ||||
| Follow‐up: Feb 19 (incomplete) | Male, n: 45 | ||||
| Sample size: 280 | Age, y: 63 ± 10.2 | ||||
| Exposure: Severity (Critical/severe or mild, Chinese classification) | |||||
| Qian et al (39) 37 | Alb, ALC, ALT, ANC, AST, BUN, Cr, CRP, Ddimer, Hb, PLT, PCT, WBC | Setting: Five hospitals in Zheijang, China | Non‐severe: 82 | High | Low (reference study) |
| Male, n: NR | |||||
| Study period: Jan 20‐Feb 11 | Age, y: 46.8 ± 15.6 | ||||
| Follow‐up: Feb 16 (incomplete) | Severe: 9 | ||||
| Sample size: 91 | Male, n: NR | ||||
| Exposure: Severity (Severe or mild, Chinese classification) | Age, y: 66.7 ± 22.7 | ||||
| Sun et al (42) 38 | ALC, ANC, Hb, PLT, WBC | Setting: Hospitals in Wenzhou, Zheijang, China | Non‐severe: 89 | Med | High |
| Reasons: | |||||
| Study period: Jan 19‐Feb 20 | Male, n: 42 | Setting and period shared with Qian (39) | |||
| Follow‐up: Unclear | Age, y: 57.7 ± 11.5 | ||||
| Sample size: 116 | Severe: 18 | ||||
| Exposure: Severity (Severe or mild, Chinese classification) | Male, n: 66 | ||||
| Age, y: 69.2 ± 10.2 | |||||
| Gao et al (30) 39 | ALC, ALT, ANC, APTT, AST, BUN, Cr, CRP, Ddimer, IL‐6, PCT, PT, WBC | Setting: Second People's Hospital of Fuyang, China | Non‐severe: 28 | High | High |
| Male, n: 17 | Reasons: | ||||
| Study period: Jan 23‐Feb 2 | Age, y: 43 ± 14 | Setting and period shared with Wu (52), Interleukin‐6 OK | |||
| Follow‐up: Unclear | Severe: 15 | ||||
| Sample size: 43 | Male, n: 9 | ||||
| Exposure: Severity (ARDS/ICU or not) | Age, y: 45.2 ± 7.7 | ||||
| Wang et al (48) 40 | ALC, ALT, ANC, AST, Bili, BUN, Cr, CRP, Hb, IL‐6, PLT, PCT, WBC | Setting: Second People's Hospital of Fuyang, China | Non‐severe: 100 | Med | High |
| Reasons: | |||||
| Study period: Jan 20‐Feb 9 | Male, n: 55 | Setting and period shared with Wu (52), Gao (30) | |||
| Follow‐up: Feb 18 (incomplete) | Age, y: 39.5 ± 14.8 | ||||
| Sample size: 125 | |||||
| Exposure: Severity (Critical or noncritical, Chinese classification) | Severe: 16 | ||||
| Male, n: 19 | |||||
| Age, y: 49.4 ± 13.6 | |||||
| Fan et al (2) 41 | ALC, ANC, Hb, LDH, PLT, WBC | Setting: National Center for Infectious Diseases, Singapore | Non‐severe: 58 | Med | Low (reference study) |
| Study period: Jan 23‐Feb 28 | Male, n: 31 | ||||
| Follow‐up: Unclear (incomplete) | Age, y: 42 ± 16 | ||||
| Sample size: 67 | Severe: 9 | ||||
| Exposure: Severity (ICU or non‐ICU) | Male, n: 6 | ||||
| Age, y: 54.3 ± 13.1 | |||||
| Young et al (9) 42 | ALC, ANC, CRP, Hb, LDH, PLT, WBC | Setting: Four hospitals in Singapore | Non‐severe: 12 | Low | High |
| Male, n: 7 | Reasons: | ||||
| Study period: Jan 23‐Feb 3 | Age, y: 41.3 ± 21.9 | Setting and period shared with Fan (2) | |||
| Follow‐up: Unclear (incomplete) | |||||
| Sample size: 18 | Severe: 2 | ||||
| Exposure: Severity (O2 saturation < 92% on room air or not) | Male, n: 6 | ||||
| Age, y: 55.3 ± 16.1 | |||||
| Zhou et al (65) 43 | Alb, ALC, ALP, ALT, ANC, APTT, AST, Bili, BUN, Cr, CRP, Ddimer, GGT, Hb, IL‐6, PLT, PCT, PT, WBC | Setting: Huangshi Central Hospital, Huangshi, Hubei, China | Non‐severe: 8 | Med | Low |
| Male, n: 3 | |||||
| Study period: Jan 28‐Mar 2 | Age, y: 64 ± 15.5 | ||||
| Follow‐up: Unclear | Severe: 13 | ||||
| Sample size: 21 | Male, n: 10 | ||||
| Exposure: Severity (Critical or severe, Chinese classification) | Age, y: 67.4 ± 13.4 | ||||
| Qu et al (40) 44 | ALC, ALT, AST, LDH, PLT | Setting: Huizhou Municipal Central Hospital, Guangdong, China | Non‐severe: 27 | Med | Low |
| Male, n: NR | |||||
| Study period: Jan‐Feb | Age, y: 49.4 ± 14.9 | ||||
| Follow‐up: Feb 21 (complete) | Severe: 3 | ||||
| Sample size: 30 | |||||
| Exposure: Severity (Severe or mild, Chinese classification) | Male, n: NR | ||||
| Age, y: 60 ± 5.3 | |||||
| Zhu et al (66) 45 | ALC, ANC, CRP, Ddimer, ESR, IL‐6, IL‐10, PLT, TropI, WBC | Setting: Hwa Mei Hospital, Ningbo, Zheijang, China | Non‐severe: 111 | Med | Low |
| Male, n: 73 | |||||
| Study period: Jan 23‐Feb 20 | Age, y: 50 ± 15.5 | ||||
| Follow‐up: Unclear | |||||
| Sample size: 127 | Severe: 16 | ||||
| Exposure: Severity (Severe or mild, Chinese classification) | Male, n: 9 | ||||
| Age, y: 57.5 ± 11.7 | |||||
| Huang et al (3) 46 | Alb, ALC, ALT, ANC, AST, Bili, Cr, Ddimer, Hb, LDH, PLT, PT, hsTropI, WBC | Setting: Jin Yintan Hospital, Wuhan, Hubei, China | Non‐severe: 28 | Med | Low |
| Male, n: 19 | |||||
| Study period: Dec 16‐Jan 2 | Age, y: 49.2 ± 13 | ||||
| Follow‐up: Unclear | |||||
| Sample size: 41 | Severe: 13 | ||||
| Exposure: Severity (ICU or non‐ICU) | Male, n: 11 | ||||
| Age, y: 50.3 ± 16.6 | |||||
| Xie et al (53) 47 | ALC, ALP, ALT, ANC, AST, Bili, Cr, CRP, Ddimer, ESR, GGT, WBC | Setting: Jin Yintan Hospital, Wuhan, Hubei, China | Non‐severe: 51 | Med | Low |
| Male, n: 26 | |||||
| Study period: Feb 2‐Feb 23 | Age, y: 57 ± 15.3 | ||||
| Follow‐up: Unclear | Severe: 28 | ||||
| Sample size: 79 | Male, n: 18 | ||||
| Exposure: Severity (Severe or mild, Chinese classification) | Age, y: 60.3 ± 13.6 | ||||
| Feng et al (29) 48 | Alb, ALC, ANC, Bili, BUN, CK, CKMB, Cr, CRP, Ddimer, ESR, Hb, LDH, PLT, PCT, WBC | Setting: Jin Yintan Hospital, Wuhan, Hubei, Shanghai Public Health Center, Shanghai, Tongling People's Hospital, China | Non‐severe: 352 | High | Low (reference study) |
| Male, n: 190 | |||||
| Study period: Jan 1‐Feb 15 | Age, y: 50.3 ± 19.3 | ||||
| Follow‐up: Mar 21 | Severe: 124 | ||||
| Sample size: 476 | Male, n: 81 | ||||
| Exposure: Severity (Critical/severe or mild, Chinese classification) | Age, y: 58.6 ± 14.4 | ||||
| Zou et al (67) 49 | APTT, Ddimer, PT | Setting: Shanghai Public Health Center, Shanghai, China | Non‐severe: 277 | High | High |
| Male, n: 138 | Reasons: | ||||
| Study period: Jan 20‐Feb 24 | Age, y: 49.7 ± 20 | Setting and period shared with Feng (29), Thromboplastin time and fibrinogen OK | |||
| Follow‐up: Unclear (incomplete) | |||||
| Sample size: 303 | Severe: 26 | ||||
| Exposure: Severity (Severe or mild, Chinese classification) | Male, n: 20 | ||||
| Age, y: 68 ± 10.2 | |||||
| Wu et al (6) 87 | Alb, ALC, ALT, ANC, AST, Bili, BUN, Cr, CRP, Ddimer, ESR, Ferritin, IL‐6, LDH, PLT, PT, WBC | Setting: Jin Yintan Hospital, Wuhan, Hubei, China | Non‐severe: 117 | Low | High |
| Male, n: 68 | Reasons: | ||||
| Study period: Dec 25‐Jan 26 | Age, y: 49 ± 12.7 | Setting and period shared with Feng (29), ALT, AST, Interleukin‐6, Ferritin, Prothrombin Time OK | |||
| Follow‐up: Feb 13 | Severe: 84 | ||||
| Sample size: 201 | Male, n: 60 | ||||
| Exposure: Severity (ARDS or not) | Age, y: 67.6 ± 12 | ||||
| Zhang et al (7) 50 | ALC, CRP, Ddimer, PCT, WBC | Setting: No. 7 Hospital, Wuhan, Hubei, China | Non‐severe: 82 | Med | Low |
| Male, n: 38 | |||||
| Study period: Jan 16‐Feb 3 | Age, y: 51.8 ± 39.2 | ||||
| Follow‐up: Unclear | Severe: 58 | ||||
| Sample size: 140 | Male, n: 33 | ||||
| Exposure: Severity (Severe or mild, Chinese classification) | Age, y: 58.7 ± 47.1 | ||||
| Zheng et al (62) 51 | ALC, ALT, AST, Bili, CK, Cr, CRP, Hb, LDH, PLT, WBC | Setting: North Hospital of Changsha First Hospital, Changsha, Hunan, China | Non‐severe: 131 | Med | Low |
| Male, n: 66 | |||||
| Study period: Jan 17‐Feb 7 | Age, y: 40.7 ± 15 | ||||
| Follow‐up: Unclear | Severe: 30 | ||||
| Sample size: 161 | Male, n: 14 | ||||
| Exposure: Severity (Severe or mild, Chinese classification) | Age, y: 56.5 ± 15.2 | ||||
| Petrilli et al (69) 52 | ALC, ALT, AST, Cr, CRP, Ddimer, Ferritin, PCT, TropI | Setting: New York University Langone Health, New York, USA | Non‐severe: 1739 | Low | Low |
| Male, n: 1016 | |||||
| Study period: Mar 1‐Apr 8 | Age, y: 59.7 ± 17.0 | ||||
| Follow‐up: May 5 (complete) | |||||
| Sample size: 2729 | Severe: 990 | ||||
| Exposure: Severity (ICU/IMV/Hospice/Death or not) | Male, n: 656 | ||||
| Age, y: 68 ± 14.8 | |||||
| He et al (32) 53 | ALC, ALT, ANC, AST, BUN, CK, Cr, CRP, Ddimer, IL‐6, IL‐10, LDH, PLT, PCT, PT, hsTropI, WBC | Setting: Renmin Hospital, Wuhan, Hubei, China | Non‐severe: 135 | Med | Low (reference study) |
| Male, n: 42 | |||||
| Study period: Jan 10‐Feb 13 | Age, y: 42.3 ± 16.5 | ||||
| Follow‐up: Feb 13 (incomplete) | |||||
| Sample size: 204 | Severe: 69 | ||||
| Exposure: Severity (Severe or mild, Chinese classification) | Male, n: 37 | ||||
| Age, y: 62.3 ± 16.6 | |||||
| Deng et al (26) 54 | CK, CKMB, CRP, Ddimer, Hb, LDH, PCT, TropI | Setting: Renmin Hospital, Wuhan, Hubei, China | Non‐severe: 45 | Med | High |
| Male, n: 19 | Reasons: | ||||
| Study period: Jan 6‐Feb 20 | Age, y: 67.3 ± 15.2 | Setting and period shared with He (32), Shi (41) | |||
| Follow‐up: Mar 11 (incomplete) | |||||
| Sample size: 112 | Severe: 67 | ||||
| Exposure: Severity (Severe or mild, Chinese classification) | Male, n: 38 | ||||
| Age, y: 54 ± 21.4 | |||||
| Han et al (31) 55 | hsTropI | Setting: Renmin Hospital, Wuhan, Hubei, China | Non‐severe: 198 | High | High |
| Reasons: | |||||
| Study period: Jan 1‐Feb 18 | Male, n: 71 | Setting and period shared with He (32), Shi (41), Deng (26) | |||
| Follow‐up: Unclear | Age, y: 59 ± 10.8 | ||||
| Sample size: 173 | |||||
| Exposure: Severity (Critical/severe or mild, Chinese classification) | Severe: 75 | ||||
| Male, n: 26 | |||||
| Age, y: 58.6 ± 14.9 | |||||
| Shi et al (41) 56 | Alb, ALC, ALT, AST, Cr, CRP, Hb, PLT, PCT, WBC | Setting: Renmin Hospital, Wuhan, Hubei, China | Non‐severe: 334 | Med | High |
| Reasons: | |||||
| Study period: Jan 20‐Feb 10 | Male, n: 161 | Setting and period shared with He (32) | |||
| Follow‐up: Feb 15 (incomplete) | Age, y: 57.7 ± 11.5 | ||||
| Sample size: 416 | Severe: 82 | ||||
| Exposure: Severity (Cardiac injury or not) | Male, n: 44 | ||||
| Age, y: 69.2 ± 10.2 | |||||
| Lei et al (33) 57 | ALC, ALT, ANC, APTT, AST, Bili, BUN, CK, Cr, CRP, Ddimer, LDH, PLT, PT, WBC | Setting: Renmin Hospital, Zhongnan Hospital, Tongji Hospital, Central Hospital, Wuhan, Hubei, China | Non‐severe: 19 | Low | High |
| Male, n: 9 | Reasons: | ||||
| Study period: Jan 1‐Feb 5 | Age, y: 44.7 ± 23.4 | Multicenter from three major COVID‐19 Hospitals in China | |||
| Follow‐up: Mar 10 (complete) | Severe: 5 | ||||
| Sample size: 34 | Male, n: 44 | ||||
| Exposure: Severity (ICU or non‐ICU) | Age, y: 57.7 ± 24.9 | ||||
| Li et al (1) 58 | ALC, ANC, CRP, PCT, WBC | Setting: Second Affiliated Hospital of Chongqing Medical University, Chongqing Three Gorges Central Hospital, Yanzhuang Central Hospital of Gancheng District, China | Non‐severe: 58 | Med | High |
| Male, n: 29 | Reasons: | ||||
| Age, y: 41.9 ± 10.6 | Multicenter study from China | ||||
| Study period: Jan‐Feb | Severe: 25 | ||||
| Follow‐up: Unclear (incomplete) | Male, n: 15 | ||||
| Sample size: 83 | Age, y: 53.7 ± 12.3 | ||||
| Exposure: Severity (Critical/severe or mild, Chinese classification) | |||||
| Chen et al (22) 59 | Alb, ALC, ALP, ALT, ANC, APTT, AST, Bili, BUN, Cr, CRP, Ddimer, ESR, GGT, Hb, LDH, PLT, PCT, PT, TropI, WBC | Setting: Taizhou Public Health Center, Enze Hospital, Zheijang, China | Non‐severe: 102 | Med | Low |
| Male, n: 56 | |||||
| Study Period: Jan 1‐Mar 11 | Age, y: 45.3 ± 13.6 | ||||
| Follow‐up: Mar 11 | |||||
| Sample size: 145 | Severe: 43 | ||||
| Male, n: 23 | |||||
| Exposure: Severity (Severe or mild, Chinese classification) | Age, y: 52.8 ± 15.5 | ||||
| Cai et al (18) 60 | ALC, ALP, ALT, ANC, AST, Bili, BUN, CK, CKMB, Cr, CRP, Ddimer, ESR, GGT, IL‐6, LDH, PCT, WBC | Setting: Third People's Hospital of Shenzhen, Shenzhen, China | Non‐severe: 240 | Low | Low |
| Male, n: 106 | |||||
| Study Period: Jan 11‐Feb 6 | Age, y: 42.7 ± 18.5 | ||||
| Follow‐up: Mar 6 (complete) | |||||
| Sample size: 298 | Severe: 58 | ||||
| Exposure: Severity (Severe or non‐severe, ATS classification) | Male, n: 39 | ||||
| Age, y: 61.5 ± 7.6 | |||||
| Wan et al (44) 61 | Alb, ALC, ALT, ANC, APTT, AST, Bili, CK, Cr, CRP, Ddimer, Hb, LDH, PLT, PCT, PT, WBC | Setting: Three Gorges Hospital, Chongqing, China | Non‐severe: 95 | High | Low (reference study) |
| Male, n: 52 | |||||
| Study period: Jan 23‐Feb 8 | Age, y: 42 ± 12 | ||||
| Follow‐up: Feb 8 (incomplete) | |||||
| Sample size: 135 | Severe: 40 | ||||
| Exposure: Severity (Severe or mild, Chinese classification) | Male, n: 21 | ||||
| Age, y: 60.3 ± 16.2 | |||||
| Wan et al (45) 62 | ALC, ANC, IL‐6, IL‐10, WBC | Setting: Three Gorges Hospital, Chongqing, China | Non‐severe: 102 | High | High |
| Male, n: NR | Reasons: | ||||
| Study period: Jan 26‐Feb 4 | Age, y: 43 ± 13.1 | Setting and period shared with Wan (44), Interleukin‐6 and 10 OK | |||
| Follow‐up: Unclear | |||||
| Sample size: 123 | Severe: 21 | ||||
| Exposure: Severity (Severe or mild, Chinese classification) | Male, n: NR | ||||
| Age, y: 61.3 ± 15.6 | |||||
| Chuan et al (8) 63 | ALC, ANC, CRP, ESR, Ferritin, IL‐6, PCT, WBC | Setting: Tongji Hospital, Wuhan, Hubei China | Non‐severe: 166 | Med | Low (reference study) |
| Study period: Jan 10‐Feb 12 | Male, n: 80 | ||||
| Follow‐up: Unclear | Age, y: 52 ± 15.5 | ||||
| Sample size: 452 | Severe: 286 | ||||
| Exposure: Severity (Severe or mild, Chinese classification) | Male, n: 155 | ||||
| Age, y, y: 60.3 ± 13.3 | |||||
| Pei et al (38) 64 | Alb, ALC, ALT, ANC, AST, BUN, Cr, CRP, Ddimer, ESR, IL‐6, IL‐10, PT, hsTropI | Setting: Tongji Hospital, Wuhan, Hubei China | Non‐severe: 144 | High | High |
| Male, n: 67 | Reasons: | ||||
| Study period: Jan 28‐Feb 9 | Age, y: 50.9 ± 12.5 | Setting and period shared with Chuan (8), ALT, AST, Trop, Alb, BUN, Cr, PT, Ddimer, IL‐10 OK | |||
| Follow‐up: Feb 23 (incomplete) | |||||
| Sample size: 333 | Severe: 189 | ||||
| Exposure: Severity (Critical/severe or mild, Chinese classification) | Male, n: 115 | ||||
| Age, y: 59.8 ± 12.1 | |||||
| Wang et al (46) 65 | Alb, ALC, ALP, ANC, Bili, BUN, Cr, CRP, Ddimer, Ferritin, GGT, LDH, PCT, WBC | Setting: Tongji Hospital, Wuhan, Hubei China | Non‐severe: 30 | High | High |
| Male, n: NR | Reasons: | ||||
| Study period: Jan | Age, y: 55.2 ± 12.4 | Setting and period shared with Chuan (8), Chen (21) | |||
| Follow‐up: Unclear | |||||
| Sample size: 65 | Severe: 35 | ||||
| Exposure: Severity (Critical/severe or mild, Chinese classification) | Male, n: NR | ||||
| Age, y: 61.3 ± 12.2 | |||||
| Chen et al (21) 66 | Alb, ALC, ALT, ANC, APTT, AST, Bili, BUN, CK, Cr, CRP, Ddimer, Ferritin, Hb, IL‐6, IL‐10, LDH, PLT, PCT, PT, WBC | Setting: Tongji Hospital, Wuhan, Hubei China | Non‐severe: 10 | Med | High |
| Reasons: | |||||
| Study period: Dec‐Jan 27 | Male, n: 7 | Setting and period shared with Chuan (8), Wang (46), IL‐10 OK | |||
| Follow‐up: Unclear | Age, y: 55.2 ± 12.4 | ||||
| Sample size: 21 | Severe: 11 | ||||
| Exposure: Severity (Severe or mild, Chinese classification) | Male, n: 10 | ||||
| Age, y: 61.3 ± 12.2 | |||||
| Goyal et al (68) 67 | Alb, BUN, ESR, Hb | Setting: Two Hospitals in Manhattan, USA | Non‐severe: 263 | Med | Low |
| Study period: Mar 3‐Mar 27 | Male, n: 146 | ||||
| Follow‐up: Apr 10 (incomplete) | Age, y: 61.2 ± 20.7 | ||||
| Sample size: 393 | Severe: 130 | ||||
| Exposure: Severity (Invasive mechanical ventilation or not) | Male, n: 92 | ||||
| Age, y: 63.3 ± 16.4 | |||||
| Wang et al (10) 68 | ALC, ALT, ANC, AST, Cr, CRP, ESR, Hb, IL‐6, LDH, PLT, PCT, WBC | Setting: Union Hospital Tongji Medical College, Wuhan, Hubei, China | Non‐severe: 55 | Low | Low |
| Male, n: 25 | |||||
| Study period: Jan 16‐Jan 29 | Age, y: 40 ± 14.5 | ||||
| Follow‐up: Feb 4 (incomplete) | Severe: 14 | ||||
| Sample size: 69 | Male, n: 7 | ||||
| Exposure: Severity (O2 saturation < 90% on room air or not) | Age, y: 69.8 ± 12.4 | ||||
| Mao et al (36) 69 | ALC, ALT, ANC, AST, BUN, CK, Cr, CRP, Ddimer, LDH, PLT, WBC | Setting: Union Hospital Tongji Medical College, Wuhan, Hubei, China | Non‐severe: 88 | Med | High |
| Male, n: 43 | Reasons: | ||||
| Study period: Jan 16‐Feb 19 | Age, y: 48.9 ± 14.7 | Setting and period shared with Wang (10) | |||
| Follow‐up: Unclear | Severe: 126 | ||||
| Sample size: 214 | Male, n: 44 | ||||
| Exposure: Severity (Severe or non‐severe, ATS classification) | Age, y: 58.2 ± 15 | ||||
| Wei et al (50) 70 | Alb, ALC, Bili, BUN, CK, Cr, IL‐6, IL‐10, LDH, PCT, hsTropI, WBC | Setting: Union Hospital Tongji Medical College, Wuhan, Hubei, China | Non‐severe: 131 | High | Low |
| Male, n: 59 | |||||
| Study period: Feb 13‐Mar 3 | Age, y: 60.1 ± 12.4 | ||||
| Follow‐up: Unclear | Severe: 121 | ||||
| Sample size: 252 | Male, n: 71 | ||||
| Exposure: Severity (Critical/severe or mild, Chinese classification) | Age, y: 69.8 ± 12.4 | ||||
| Zhou et al (64) 71 | ALC, ALT, ANC, AST, CK, Cr, CRP, LDH, hsTropI, WBC | Setting: Union Hospital Tongji Medical College, Wuhan, Hubei, China | Non‐severe: 26 | Med | High |
| Male, n: 12 | Reasons: | ||||
| Study period: Feb 5‐Feb 13 | Age, y: 63.3 ± 8.6 | Setting and period shared with Wang (10), Trop, CK, CKMB OK | |||
| Follow‐up: Unclear | Severe: 8 | ||||
| Sample size: 34 | Male, n: 5 | ||||
| Exposure: Severity (Critical or severe, Chinese classification) | Age, y: 69.3 ± 9 | ||||
| Herold et al (70) 72 | ALC, Bili, Cr, CRP, Ddimer, Ferritin, IL‐6, LDH, PLT, PCT, WBC | Setting: University Hospital, Munich, Germany | Non‐severe: 27 | Med | Low |
| Male, n: 16 | |||||
| Study period: Feb 29‐Mar 27 | Age, y: 51.7 ± 15.3 | ||||
| Follow‐up: Unclear | |||||
| Sample size: 34 | Severe: 13 | ||||
| Exposure: Severity (invasive mechanical ventilation or not) | Male, n: 13 | ||||
| Age, y: 63.5 ± 10.8 | |||||
| Wei et al (51) 73 | Alb, ALC, ALT, ANC, AST, BUN, CK, CKMB, CRP, Ddimer, IL‐6, LDH, PCT, WBC | Setting: Unknown Hospital(s) in Anhui, China | Non‐severe: 137 | High | High |
| Male, n: 75 | Reasons: | ||||
| Study period: Unclear | Age, y: 40.8 ± 15.5 | Unknown hospital(s), Unclear study period | |||
| Follow‐up: Unclear | Severe: 30 | ||||
| Sample size: 167 | Male, n: 20 | ||||
| Exposure: Severity (Severe or mild, Chinese classification) | Age, y: 49 ± 12.6 | ||||
| Aggarwal et al (16) 74 | ALC, ALT, ANC, AST, Cr, CRP, Hb, PLT, WBC | Setting: Unspecified hospital in Midwestern USA | Non‐severe: 8 | Med | Low |
| Study period: Through Apr 4 | Male, n: 7 | ||||
| Follow‐up: Unclear | Age, y: 68.2 ± 19 | ||||
| Sample size: 16 | Severe: 8 | ||||
| Exposure: Severity (ICU/IMV/Death/Inotropes or not) | Male, n: 5 | ||||
| Age, y: 59.7 ± 11.2 | |||||
| Du et al (28) 75 | Alb, ALC, ALT, ANC, APTT, AST, BUN, CKMB, Cr, CRP, Ddimer, Hb, PLT, PCT, PT, TropI, WBC | Setting: Wuhan Pulmonary Hospital, Tianyou Hospital, Shanghai, Central Hospital of Wuhan, China | Non‐severe: 58 | High | Low |
| Male, n: 38 | |||||
| Study period: Dec 25‐Feb 24 | Age, y: 72.7 ± 11.6 | ||||
| Follow‐up: Complete | Severe: 51 | ||||
| Sample size: 109 | Male, n: 36 | ||||
| Exposure: Severity (ICU or non‐ICU) | Age, y: 68.4 ± 9.7 | ||||
| Zhang et al (58) 88 | ALC, ALT, ANC, APTT, AST, Bili, BUN, CK, CKMB, Cr, Ddimer, LDH, PLT, PT, hsTropI, WBC | Setting: Zhongnan Hospital, Wuhan, Hubei, China | Non‐severe: 166 | Med | Low (reference study) |
| Male, n: 73 | |||||
| Study period: Jan 2‐Feb 10 | Age, y: 50.4 ± 21.2 | ||||
| Follow‐up: Feb 15 (incomplete) | |||||
| Sample size: 221 | Severe: 55 | ||||
| Exposure: Severity (Severe or mild, Chinese classification) | Male, n: 35 | ||||
| Age, y: 62.7 ± 16.8 | |||||
| Wang et al (4) 89 | ALC, ALT, ANC, AST, Bili, Cr, Ddimer, LDH, PLT, PT, hsTropI, WBC | Setting: Zhongnan Hospital, Wuhan, Hubei, China | Non‐severe: 102 | Med | High |
| Male, n: 53 | Reasons: | ||||
| Study period: Jan 1‐Jan 28 | Age, y: 50 ± 18.8 | Setting and period shared with Zhang (58), Trop, Procalcitonin OK | |||
| Follow‐up: Feb 3 (incomplete) | |||||
| Sample size: 138 | Severe: 36 | ||||
| Exposure: Severity (ICU or non‐ICU) | Male, n: 22 | ||||
| Age, y: 67 ± 16.3 | |||||
| Zhang et al (60) 115 | Alb, ALP, ALT, AST, Bili, CRP, GGT, LDH | Setting: Zhongnan Hospital, Wuhan, Hubei, China | Non‐severe: 84 | Med | Low |
| Male, n: 29 | |||||
| Study period: Jan 22‐Feb 22 | Age, y: 44 ± 14.8 | ||||
| Follow‐up: Mar 9 (incomplete) | |||||
| Sample size: 115 | Severe: 20 | ||||
| Exposure: Severity (Severe or mild, Chinese classification) | Male, n: 35 | ||||
| Age, y: 64.6 ± 13.3 |
The 50 severity studies contributed a total of 11 173 patients, of which 7845 were from at least 522 hospitals across China, 85 were from at least 4 hospitals in Singapore, 3315 were from at least 7 hospitals in the United States, and 40 were from one hospital in Germany. The 15 mortality studies contributed a total of 2525 patients from 6 hospitals in Wuhan, Hubei, China. Blood samples were collected within 2 days of hospital admission in all but two studies. 48 , 81 15 severity studies 34 , 40 , 45 , 47 , 48 , 49 , 50 , 58 , 59 , 60 , 64 , 66 , 67 , 88 , 89 and two mortality studies 29 , 83 reported median time from symptom onset to hospital admission, which ranged from 3.5 40 to 12 83 days. There were significant differences in time to admission for three severity studies 48 , 58 , 89 and one mortality study (Table 1). 29
3.1. Meta‐analysis of laboratory tests and severity
A summary of meta‐analysis results for severity is presented in Table 2. In total, 27 laboratory markers were analyzed for associations with disease severity. Only Hb, PLT, Cr, APTT, TropI, CKMB, ALP, and GGT did not achieve statistical significance. Among markers involved in a complete blood count (CBC), the WMDs of WBC and ANC in patients with severe vs those with the non‐severe disease were 1.23 × 109 (0.85, 1.60) and 1.49 × 109 (0.96, 2.01) cells/L, respectively. ALC and PLT showed associations in the opposite direction, with WMDs of −0.30 × 109 (−0.37, −0.24) and −16.69 × 109 (−35.35, 1.96) cells/L, respectively. Among inflammatory markers, the most pronounced difference was for ferritin at 423.13 ng/mL (281.41, 582.85). Other inflammatory markers including IL‐6 and IL‐10 also showed statistically significant associations. Among markers for tissue damage the most marked differences were for LDH, CK, and hsTropI at 120.31 U/L (93.50, 147.12), 45.33 U/L (18.60, 72.07), and 11.07 pg/mL (3.64, 18.50), respectively. TropI did not reach statistical significance at a WMD of 0.04 ng/mL (−0.01, 0.09), but was only reported in three studies (Table 2).
Table 2.
Weighted mean differences in admission laboratory values comparing patients with severe disease to those with non‐severe disease
| Laboratory marker | Studies, n | Patients, n | Weighted mean difference (95% CI) | I 2 (%) | P value |
|---|---|---|---|---|---|
| Complete blood count | |||||
| Hemoglobin (Hb, g/L) | 21 | 3819 | −3.91 (−6.47, −1.35) | 62.4 | .003NS |
| White blood cell count (WBC, 109/L) | 40 | 6705 | 1.23 (0.85, 1.60) | 77.7 | .000 |
| Absolute neutrophil count (ANC, 109/L) | 35 | 4930 | 1.49 (0.96, 2.01) | 92.3 | .000 |
| Absolute lymphocyte count (ALC, 109/L) | 44 | 9873 | −0.30 (−0.37, −0.24) | 89 | .000 |
| Platelet count (PLT, 109/L) | 27 | 4515 | −16.69 (−35.35, 1.96) | 92.6 | .080NS |
| Inflammatory markers | |||||
| C‐reactive protein (CRP, mg/L) | 35 | 7660 | 39.91 (33.17, 46.64) | 84.5 | .000 |
| Erythrocyte sedimentation rate (ESR, mm/h) | 11 | 2653 | 6.84 (3.37, 10.31) | 69.4 | .000 |
| Ferritin (ng/mL) | 6 | 3508 | 432.13 (281.41, 582.85) | 72.0 | .000 |
| Interleukin‐6 (IL‐6, pg/mL) | 16 | 2526 | 12.25 (7.00, 17.50) | 95.8 | .000 |
| Interleukin‐10 (IL‐10, pg/mL) | 7 | 1136 | 1.86 (1.07, 2.64) | 91.3 | .000 |
| Procalcitonin (ng/mL) | 22 | 6481 | 0.08 (0.05, 0.11) | 94.2 | .000 |
| Liver function tests | |||||
| Albumin (g/L) | 18 | 3169 | −4.36 (−5.08, −3.64) | 75.1 | .000 |
| Bilirubin (Bili, µM) | 20 | 3025 | 1.93 (1.28, 2.57) | 42.9 | .000 |
| Prothrombin time (PT, s) | 17 | 2304 | 0.44 (0.24, 0.64) | 78.0 | .000 |
| Renal function tests | |||||
| Creatinine (Cr, µM) | 32 | 7580 | 7.34 (2.59, 12.10) | 91.7 | .002NS |
| Blood urea nitrogen (BUN, mM) | 21 | 3797 | 1.28 (0.82, 1.74) | 88.2 | .000 |
| Coagulation markers | |||||
| Activated partial thromboplastin time (APTT, s) | 11 | 1288 | 0.59 (−1.22, 2.39) | 83.9 | .523NS |
| D‐dimer (mg/L) | 30 | 6950 | 0.67 (0.52, 0.82) | 92.7 | .000 |
| Markers of tissue damage | |||||
| Lactate dehydrogenase (LDH, U/L) | 27 | 3791 | 120.31 (93.50, 147.12) | 89.3 | .000 |
| Creatine kinase (CK, U/L) | 15 | 2585 | 45.33 (18.60, 72.07) | 93.5 | .001 |
| High sensitivity troponin I (hsTropI, pg/mL) | 6 | 1223 | 11.07 (3.64, 18.50) | 73.7 | .004NS |
| Troponin I (TropI, ng/mL) | 3 | 3222 | 0.04 (−0.01, 0.09) | 87.8 | .134NS |
| Creatine kinase myocardial band (CKMB, U/L) | 8 | 1639 | 2.44 (0.78, 4.11) | 90.9 | .004NS |
| Alanine aminotransferase (ALT, U/L) | 31 | 6854 | 6.25 (3.09, 9.42) | 86.6 | .000 |
| Aspartate aminotransferase (AST, U/L) | 30 | 6746 | 8.52 (4.98, 12.06) | 91.1 | .000 |
| Alkaline phosphatase (ALP, U/L) | 6 | 723 | −1.50 (−6.41, 3.41) | 28.7 | .549NS |
| Gamma glutamyl transferase (GGT, U/L) | 6 | 723 | 5.01 (−3.16, 13.17) | 48.0 | .230NS |
I 2 statistic – Describes the percentage of variation across studies estimated to be due to heterogeneity.
Denotes nonsignificant P values. Bonferroni corrected significance levels is .002.
3.2. Meta‐analysis of laboratory tests and mortality
A summary of meta‐analysis results for mortality is presented in Table 3. In general, the trends in WMDs were the same as for severity, but with larger absolute differences. Of the 22 laboratory markers that were analyzed for associations with mortality, only Hb, ESR, and APTT did not achieve statistical significance. Among the CBC markers, the WMDs of WBC, ANC, and PLT in patients who died vs those that survived were 3.49 × 109 (2.71, 4.27), 3.82 × 109 (2.76, 4.87), and −43.41 × 109 (−54.55, −32.27) cells/L, respectively. The WMD for ALC was −0.34 × 109 (−0.45, −0.23) cells/L, which is similar to the value seen for severity. Among the inflammatory markers and acute phase reactants, ferritin showed the most marked elevation at 814.14 ng/mL (551.48, 1076.81). CRP, ESR, IL‐6, IL‐10, and PCT also showed positive associations. Of the liver, coagulation, and renal function tests, D‐dimer was the most markedly elevated at 5.74 mg/L (3.91, 7.58). Among markers of tissue damage, LDH, CK, and hsTropI all showed marked elevations at 232.41 U/L (178.31, 286.52), 97.18 U/L (60.01, 134.25), and 90.47 pg/mL (47.79, 133.14), respectively.
Table 3.
Weighted mean differences in admission laboratory values comparing patients that died to those that survived
| Laboratory marker | Studies (n) | Patients (n) | Weighted mean difference (95% CI) | I 2 (%) | P value |
|---|---|---|---|---|---|
| Complete blood count | |||||
| Hemoglobin (Hb, g/L) | 7 | 1069 | −0.15 (−2.49, 2.19) | 0.0 | .901NS |
| White blood cell count (WBC, 109/L) | 10 | 1824 | 3.49 (2.71, 4.27) | 59.8 | .000 |
| Absolute neutrophil count (ANC, 109/L) | 8 | 1468 | 3.82 (2.76, 4.87) | 72.4 | .000 |
| Absolute lymphocyte count (ALC, 109/L) | 12 | 1876 | −0.34 (−0.45, −0.23) | 81.7 | .000 |
| Platelet count (PLT, 109/L) | 11 | 2001 | −43.41 (−54.55, −32.27) | 40.5 | .000 |
| Inflammatory markers | |||||
| C‐reactive protein (CRP, mg/L) | 10 | 1635 | 66.11 (52.16, 80.06) | 68.1 | .000 |
| Erythrocyte sedimentation rate (ESR, mm/h) | 4 | 461 | 8.73 (3.23, 14.24) | 0.0 | .002NS |
| Ferritin (ng/mL) | 5 | 747 | 814.14 (551.48, 1076.81) | 70.1 | .000 |
| Interleukin‐6 (IL‐6, pg/mL) | 9 | 1630 | 32.59 (23.99, 41.19) | 97.4 | .000 |
| Interleukin‐10 (IL‐10, pg/mL) | 3 | 763 | 7.55 (6.44, 8.65) | 0.0 | .000 |
| Procalcitonin (ng/mL) | 8 | 1399 | 0.29 (0.20, 0.38) | 69.1 | .000 |
| Liver function tests | |||||
| Albumin (g/L) | 9 | 1342 | −3.98 (−5.23, −2.72) | 81.0 | .000 |
| Bilirubin (bili, µM) | 8 | 1146 | 4.49 (3.56, 5.43) | 20.5 | .000 |
| Prothrombin time (PT, s) | 11 | 2251 | 1.21 (0.77, 1.64) | 81.0 | .000 |
| Renal function tests | |||||
| Creatinine (Cr, µM) | 11 | 1630 | 16.50 (10.74, 22.25) | 61.2 | .000 |
| Blood urea nitrogen (BUN, mM) | 6 | 1234 | 3.99 (2.93, 5.04) | 81.5 | .000 |
| Coagulation markers | |||||
| Activated partial thromboplastin time (APTT, s) | 4 | 840 | 0.97 (0.07, 1.86) | 0.0 | .034NS |
| D‐dimer (mg/L) | 12 | 2323 | 5.74 (3.91, 7.58) | 74.6 | .000 |
| Markers of tissue damage | |||||
| Lactate dehydrogenase (LDH, U/L) | 8 | 1435 | 232.41 (178.31, 286.52) | 82.5 | .000 |
| Creatine kinase (CK, U/L) | 6 | 1205 | 97.18 (60.01, 134.35) | 80.9 | .000 |
| High sensitivity troponin I (hsTropI, pg/mL) | 6 | 1346 | 90.47 (47.79, 133.14) | 91.9 | .000 |
| Alanine aminotransferase (ALT, U/L) | 11 | 1842 | 5.45 (2.64, 8.26) | 28.4 | .000 |
| Aspartate aminotransferase (AST, U/L) | 10 | 1633 | 13.89 (8.16, 19.63) | 69.3 | .000 |
I 2 statistic—Describes the percentage of variation across studies estimated to be due to heterogeneity.
Denotes nonsignificant P values. Bonferroni corrected significance level is .002.
3.3. Meta‐regression analyses for age and sex
Meta‐regressions were conducted for age and sex as described in the Methods section. All associations were nonsignificant when compared to Bonferroni corrected significance levels (data not provided).
3.4. Sensitivity analyses for bias risk, duplicate risk, and outlier studies
Of the 64 included studies, 17, 31, and 16 were assigned high, medium, and low risks of bias, respectively (Table 1). Insufficient or unclear follow‐up durations and nonconsecutive recruitment were the most common shortcomings among high‐risk studies. When studies at high risk of bias were excluded for sensitivity analysis, final estimates and statistical significance were unchanged, except for IL‐6 in association with disease severity; the WMD dropped from 12.25 pg/mL (7.00, 17.50) to 2.58 pg/mL (−1.53, 6.69) (Supplement 5). A total of 27 studies were assessed to be at high risk of duplication (Table 1). Excluding these studies resulted in loss of statistical significance for IL‐6 (WMD = 8.37 pg/mL; 2.76, 13.99) and CK (WMD = 27.65 U/L; 10.19, 45.11) in association with severity, and for IL‐6 (WMD = 46.34 pg/mL; 4.35, 88.33) and ALT (WMD = 4.60 U/L; 1.03, 8.17) in association with mortality (Supplement 5 and 6). Excluding outlier studies (8 among studies of mortality and 15 among studies of severity) reduced heterogeneity but had minimal impact on overall estimates, except for Cr and CKMB in association with disease severity, which achieved statistical significance at 6.69uM (3.31, 10.06) and 2.31 U/L (0.61, 4.02), respectively (Supplement 5).
4. DISCUSSION
In this systematic review and meta‐analysis, we observed significant differences in many admission laboratory values between severe and non‐severe, and fatal and nonfatal cases of COVID‐19. Although the WMDs for most parameters were statistically significant, those that were most pronounced, and thus those would be most clinically useful, are markers of overactive inflammatory response, blunted adaptive response, intravascular coagulation, and cell death, reflecting the pathophysiology of severe and fatal COVID‐19. 7 , 8 , 9
In support of the role that hyper inflammation plays in COVID‐19, the levels of all included acute phase reactants were significantly altered at admission when comparing severe to non‐severe and fatal to nonfatal cases. Among the acute phase proteins, the largest difference was observed for ferritin, which is driven by IL‐18. 90 Although IL‐18 was not reported in any of the included studies, we expect elevated levels. CRP was also markedly elevated, and albumin was decreased, both of which are acute phase reactions driven by IL‐6, a major pro‐inflammatory cytokine. 91 As expected by the derangements in acute phase reactants, IL‐6 levels were increased and were more prominent for fatal than for severe cases.
Inflammation is a major component of innate immunity and is typically a transient initial response to any pathogen or injury, eventually subsiding and being replaced by a focused immune response when the trigger is infectious. 92 The inflammatory response is driven and sustained by numerous pro‐inflammatory cytokines and chemokines including IL‐6, TNF‐α, and CXCL10, which are not only elevated in COVID‐19 but have also been implicated in the pathogenesis of disease caused by the related respiratory coronaviruses SARS‐CoV and MERS‐CoV. 9 , 92 As such, a prolonged hyper inflammatory state caused by dysregulated release of pro‐inflammatory cytokines, known as cytokine storm syndrome, is thought to be central to the pathogenesis of severe and fatal COVID‐19. 93
Although T‐lymphocytes are usually the major producers of many cytokines including IL‐6, 94 ALC was decreased for both severe and fatal disease, consistent with hypotheses proposing alternate major sources of cytokines in COVID‐19. 95 In fact, even in patients with relatively mild illness, lymphopenia is a common and characteristic feature of COVID‐19, suggesting that the adaptive immune response is blunted and may be delayed or insufficient. 27 , 29 , 30 , 32 , 43 , 44 , 52 , 87 , 88 One possible explanation is that, like a number of other viruses, 96 SARS‐CoV2 may directly infect lymphocytes. SARS‐CoV2 relies on angiotensin‐converting enzyme 2 (ACE2) for cellular entry, 97 and it has been reported that a small proportion of lymphocytes are ACE2 positive. 98 Another possibility is that inflammatory cytokines such as IL‐6 induce chemotaxis of lymphocytes to lymphoid organs, thus reducing circulating concentrations. 99 Functional exhaustion of lymphocytes due to SARS‐CoV2‐induced inhibitory cytokines such as IL‐10, which was significantly elevated in our analysis, has also been suggested. 95 , 100 However, IL‐10 is an important anti‐inflammatory cytokine that may in fact not be elevated enough to combat inflammation in fatal COVID‐19. 101
In contrast to ALC, ANC was elevated with a more pronounced difference observed for fatal than for severe illness. Neutrophils play a major role in inflammation and are not typically elevated in viral infections. However, in COVID‐19, not only are their concentrations increased, but they have been suggested to be major producers of pro‐inflammatory cytokines 102 , 103 and to contribute to the development of acute respiratory distress syndrome (ARDS) through the formation of neutrophil extracellular traps (NETs) 102 , 104 and direct tissue infiltration causing vascular leakage. 105 On the other hand, neutrophilia is classically a marker of bacterial infection; thus, it is possible that the observed elevations in ANC seen in severe/fatal COVID‐19 reflect bacterial super‐infection contributing to severe illness. However, procalcitonin, which is a more specific marker of bacterial infection, 106 has been reported to fall within normal reference ranges even in patients with fatal illness, 27 , 28 , 29 , 76 , 77 , 81 , 82 , 83 , 86 suggesting that this explanation may not be sufficient. In SARS‐CoV, neutrophilia is an independent predictor of severe illness and is associated with hypersensitivity pneumonitis. 107 Hence, a similar mechanism might be plausible for the neutrophilia seen in severe/fatal COVID‐19.
In addition to causing localized damage at sites of inflammation, prolonged activation of neutrophils may also contribute to systemic damage in other ways. There were significant abnormalities in Ddimer, PLT, and PT, three of the analyzed coagulation parameters. The largest difference was for Ddimer, which is a fibrin degradation product indicative of intravascular thrombosis. The significant elevation in PT and decrease in PLT is likely due to the development of consumptive coagulopathy, as clotting factors and platelets are used up in forming microthrombi. Elevated Ddimer in severe and fatal COVID‐19 may be explained by NETs, which can play a major role in the formation of intravascular thrombi. 102 , 108 In addition, inflammatory cytokines such as IL‐6 have procoagulant effects that contribute to an inflammation‐induced hypercoagulable state known as thromboinflammation, 7 reinforcing the connection between the innate immune system and thrombosis. Once ARDS has developed and a patient becomes hypoxemic, thrombosis may also be promoted via a hypoxia‐inducible factor‐mediated pathway. 85 , 109
Tissue damage is an inevitable and unsurprising result of the disease processes described, as evidenced by significant increases in most markers of tissue damage that were analyzed. LDH is a ubiquitous intracellular enzyme that was markedly elevated in both severe and fatal cases. CK, which is highly expressed in skeletal muscle and the aminotransferases, which are expressed in hepatocytes, were also significantly elevated, but to a lesser degree than LDH. Cardiac troponin I, a marker of heart muscle damage, was also markedly increased, especially in fatal illness. Although hypoxemia and shock are the most likely causes of myocardial damage, it is possible that direct infection of cardiomyocytes by SARS‐CoV2 110 plays a role in some cases. This hypothesis is given plausibility by the presence of ACE2 on cardiomyocytes 111 and case reports of COVID‐19 associated myocarditis. 112
Meta‐regression analyses conducted for age and sex on key markers involved in inflammation, poor adaptive response and intravascular coagulation did not show any significant associations between age or sex and observed marker levels. This is an important result reinforcing the utility of these markers for predicting disease severity among all adults, as males and the elderly have been overrepresented among severe cases. 2
4.1. Study strengths and limitations
To our knowledge, this is the first meta‐analysis that assessed COVID‐19 disease severity and mortality in association with laboratory markers and included a meta‐regression for age and sex. In addition, the potential impacts of duplicate reporting and important sources of bias were considered. Rather than simply exclude duplicate studies, as was done in a previous systematic review, 11 we conducted a sensitivity analysis showing that exclusion had little impact on overall results. Despite these strengths, there are multiple limitations to our study. When not available, we estimated means and standard deviations from reported medians, IQRs, and ranges. Estimates from studies with small sample sizes can be imprecise, contributing to greater heterogeneity. Furthermore, we did not assess the risk of publication bias in this study. Due to the pandemic nature of COVID‐19, most published studies on clinical outcomes, especially during the first months, were small case‐reports and case‐series. Hence, it is unlikely that small studies reporting on COVID‐19 disease severity and mortality remained unpublished because of null and/or nonsignificant results. There are also limitations to the data set. For example, our entire mortality data set and 70% of the severity data set is from China, potentially limiting the generalizability of the results. Additionally, 42 of 64 studies had unreported or insufficient follow‐up, which could bias the results by incorrectly classifying a patient as non‐severe or living, only to develop the more severe disease after the follow‐up period. This requires updating analyses once more data becomes available outside of China. Diverse classification schemes for disease severity among different studies potentially contributed to high levels of the observed heterogeneity. Another potential contributor to heterogeneity is that time to hospitalization was unreported in all but 17 studies, however, we assume most patients were hospitalized shortly after developing severe respiratory symptoms such as dyspnea.
5. CONCLUSIONS
The associations between markers of inflammation (ANC, IL‐6, ferritin, CRP, albumin), poor adaptive immune response (ALC), intravascular coagulation (Ddimer), and tissue damage (LDH, hsTropI) observed with a severe and fatal disease in this meta‐analysis not only support the key roles of these processes in COVID‐19 but also provide evidence that there are identifiable biochemical and hematologic differences that exist between severe and non‐severe, and fatal and nonfatal cases before the development of potentially lethal complications such as ARDS. Although these disease processes are certainly not unique to COVID‐19, they appear to be key pathways involved in the development of severe/fatal disease and can all be connected to hyper inflammation and cytokine storm. Importantly, the results of the meta‐regression suggest that these markers are likely reliable regardless of age or sex in adult patients. Assessment of these markers at admission contributes both to an understanding of the disease mechanisms involved, as well as guiding attempts at predicting severe illness, thus allowing for identification of patients likely to benefit from early interventions. There are no widely accepted disease prediction models yet for COVID‐19, 113 but accurate tools will likely need to incorporate markers of the main pathogenetic pathways involved: inflammation, blunted adaptive response, and thrombosis. These pathways are likely also ideal targets for therapy, such as the IL‐6 inhibitor tocilizumab to target inflammation, 114 or heparin to target coagulation. 85 The results also indicate that further research is warranted in the utility of different immunomodulators and anticoagulants in the treatment of COVID‐19.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
MD and JK conceived and designed the study. MD and ERH analyzed the data. JK, NZJ, SC, ERH, PB, and MD interpreted the data. JK drafted the manuscript. N.Z.J, SC, ERH, PB, and MD critically revised the manuscript. MD supervised the study. All authors approved the final version of the manuscript for publication.
Supporting information
Supporting information
Khinda J, Janjua NZ, Cheng S, van den Heuvel ER, Bhatti P, Darvishian M. Association between markers of immune response at hospital admission and COVID‐19 disease severity and mortality: A meta‐analysis and meta‐regression. J Med Virol. 2021;93:1078–1098. 10.1002/jmv.26411
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Rodriguez‐Morales AJ, Cardona‐Ospina JA, Gutiérrez‐Ocampo E, et al. Clinical, laboratory and imaging features of COVID‐19: A systematic review and meta‐analysis. Travel Med Infect Dis. 2020;34:101623. 10.1016/j.tmaid.2020.101623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID‐19) outbreak in China: summary of a report of 72 314 cases from the Chinese center for disease control and prevention. JAMA. 2020;323(13):1239‐1242. 10.1001/jama.2020.2648 [DOI] [PubMed] [Google Scholar]
- 3. Worldometers.info. Coronavirus Update (Live) : 6,150,483 Cases and 370,506 Deaths from COVID‐19 virus pandemic—Worldometer . https://www.worldometers.info/coronavirus/. Accessed 30 May 2020.
- 4. Center for Systems Science and Engineering at the Whiting School of Engineering . COVID‐19 Map. Johns Hopkins Coronavirus Resource Center . https://coronavirus.jhu.edu/map.html. Accessed 30 May 2020.
- 5. Levesque J, Maybury DW. A note on COVID‐19 seroprevalence studies: a meta‐analysis using hierarchical modelling [published online ahead of print January 1, 2020]. medRxiv. 2020. 10.1101/2020.05.03.20089201 [DOI] [Google Scholar]
- 6. Chong K, Chan S, Jia KM. Clinical scores and risk factors to predict patient outcomes: how useful are they? Hong Kong Med J. 2018:552‐553. 10.12809/hkmj185085 [DOI] [Google Scholar]
- 7. Connors JM, Levy JH. COVID‐19 and its implications for thrombosis and anticoagulation. Blood. 2020;135(23):2033‐2040. 10.1182/blood.2020006000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jose RJ, Manuel A. COVID‐19 cytokine storm: the interplay between inflammation and coagulation. Lancet Respir Med. 2020;8(6):e46‐e47. 10.1016/S2213-2600(20)30216-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Coperchini F, Chiovato L, Croce L, Magri F, Rotondi M. The cytokine storm in COVID‐19: An overview of the involvement of the chemokine/chemokine‐receptor system. Cytokin Growth Factor Rev. 2020;53:25‐32. 10.1016/j.cytogfr.2020.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Henry BM, Oliveira MHS, , de Benoit S , Plebani M, Lippi G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID‐19): a meta‐analysis. Clin Chem Lab Med. 2020. 10.1515/cclm-2020-0369 [DOI] [PubMed] [Google Scholar]
- 11. Moutchia J, Pokharel P, Kerri A, et al. Clinical laboratory parameters associated with severe or critical novel coronavirus disease 2019 (COVID‐19): a systematic review and meta‐analysis [published online ahead of print Jaunary 1, 2020]. medRxiv. 2020. 10.1101/2020.04.24.20078782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. World Health Organization (WHO) Global Research Database. 2019. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/global-research-on-novel-coronavirus-2019-ncov
- 13. COVID‐19 Tools & Resources for Clinicians . Wolters Kluwer. http://tools.ovid.com/coronavirus/Covid-19%20search%20notes.pdf. Accessed 17 March 2020.
- 14. COVID‐19 PubMed Search Alert . Stephen B Thacker CDC Library Published April 17, 2020. https://www.cdc.gov/library/researchguides/2019novelcoronavirus/pubmedsearchalert.html. Accessed 1 May 2020.
- 15. COVID‐19 Tools & Resources for Clinicians . Wolters Kluwer. http://tools.ovid.com/coronavirus/Covid-19%20search%20notes%20Embase.pdf. Accessed 1 May, 2020.
- 16. Covidence systematic review software, Veritas Health Innovation, Melbourne, Australia. www.covidence.org [Google Scholar]
- 17. New coronavirus pneumonia prevention and control program (6th ed). 2020. (in Chinese). http://www.nhc.gov.cn/yzygj/s7653p/202002/8334a8326dd94d329df351d7da8aefc2/files/b218cfeb1bc54639af227f922bf6b817.pdf
- 18. Metlay JP, Waterer GW, Long AC, et al. Diagnosis and treatment of adults with community‐acquired pneumonia. An official clinical practice guideline of the American Thoracic Society and Infectious Diseases Society of America. Am J Respir Crit Care Med. 2019;200(7):e45‐e67. 10.1164/rccm.201908-1581ST [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moher D, Liberati A, Tetzlaff J, Altman DG. The PG. Preferred reporting items for systematic reviews and meta‐analyses: The PRISMA Statement. PLOS Med. 2009;6(7):e1000097. 10.1371/journal.pmed.1000097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Institute of Health Economics (IHE) . Quality Appraisal of Case Series Studies Checklist. Edmonton (AB): Institute of Health Economics; 2014. http://www.ihe.ca/research-programs/rmd/cssqac/cssqac-about [Google Scholar]
- 21. Wan X, Wang W, Liu J, Tong T. Estimating the sample mean and standard deviation from the sample size, median, range and/or interquartile range. BMC Med Res Methodol. 2014;14:135. 10.1186/1471-2288-14-135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. DerSimonian R, Laird N. Meta‐analysis in clinical trials. Controlled Clin Trials. 1986;7:177e88‐88. [DOI] [PubMed] [Google Scholar]
- 23. Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta‐analyses. BMJ. 2003;327:557‐560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cooper HM, Hedges LV, Valentine JC. Handbook of Research Synthesis and Meta‐analysis. New York: Russell Sage Foundation; 2019. [Google Scholar]
- 25. Higgins JP, Thompson SG. Controlling the risk of spurious findings from meta‐regression. Stat Med. 2004;23:1663‐82. [DOI] [PubMed] [Google Scholar]
- 26. StataCorp . Stata Statistical Software: Release 16. College Station, TX: StataCorp LLC; 2019. [Google Scholar]
- 27. Du R‐H, Liang L‐R, Yang C‐Q, et al. Predictors of mortality for patients with COVID‐19 pneumonia caused by SARS‐CoV‐2: a prospective cohort study. Eur Respir J. 2020;55(5):2000524. 10.1183/13993003.00524-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou W, Liu Y, Tian D, et al. Potential benefits of precise corticosteroids therapy for severe 2019‐nCoV pneumonia. Sig Transduct Target Ther. 2020;5(1):18. 10.1038/s41392-020-0127-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen T, Dai Z, Mo P, et al. Clinical characteristics and outcomes of older patients with coronavirus disease 2019 (COVID‐19) in Wuhan, China: a single‐centered, retrospective study. J Gerontol Ser A. 2020:glaa089. 10.1093/gerona/glaa089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Myers LC, Parodi SM, Escobar GJ, Liu VX. Characteristics of hospitalized adults with COVID‐19 in an integrated health care system in California. JAMA. 2020;323(21):2195‐2198. 10.1001/jama.2020.7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lu L, Xiong W, Liu D, et al. New onset acute symptomatic seizure and risk factors in coronavirus disease 2019: a retrospective multicenter study. Epilepsia. 2020;61(6):e49‐e53. 10.1111/epi.16524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708‐1720. 10.1056/NEJMoa2002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zheng Y, Xu H, Yang M, et al. Epidemiological characteristics and clinical features of 32 critical and 67 noncritical cases of COVID‐19 in Chengdu. J Clin Virol. 2020;127:104366. 10.1016/j.jcv.2020.104366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yao Q, Wang P, Wang X, et al. Retrospective study of risk factors for severe SARS‐Cov‐2 infections in hospitalized adult patients. Polish Arch Int Med. 2020;130:390‐399. 10.20452/pamw.15312 [DOI] [PubMed] [Google Scholar]
- 35. Liu Y, Liao W, Wan L, Xiang T, Zhang W. Correlation between relative nasopharyngeal virus RNA load and lymphocyte count disease severity in patients with COVID‐19. Viral Immunol. 2020;2020:0062. 10.1089/vim.2020.0062 [DOI] [PubMed] [Google Scholar]
- 36. Wu J, Li W, Shi X, et al. Early antiviral treatment contributes to alleviate the severity and improve the prognosis of patients with novel coronavirus disease (COVID‐19). J Int Med. 2020;288(1):128‐138. 10.1111/joim.13063 [DOI] [PubMed] [Google Scholar]
- 37. Qian G‐Q, Yang N‐B, Ding F, et al. Epidemiologic and clinical characteristics of 91 hospitalized patients with COVID‐19 in Zhejiang, China: a retrospective, multi‐centre case series. QJM. 2020;113:474‐481. 10.1093/qjmed/hcaa089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sun S, Cai X, Wang H, et al. Abnormalities of peripheral blood system in patients with COVID‐19 in Wenzhou, China. Clin Chim Acta. 2020;507:174‐180. 10.1016/j.cca.2020.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gao Y, Li T, Han M, et al. Diagnostic utility of clinical laboratory data determinations for patients with the severe COVID‐19. J Med Virol. 2020;92:791‐796. 10.1002/jmv.25770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang R, Pan M, Zhang X, et al. Epidemiological and clinical features of 125 hospitalized patients with COVID‐19 in Fuyang, Anhui, China. Int J Infect Dis. 2020;95:421‐428. 10.1016/j.ijid.2020.03.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fan BE, Chong VCL, Chan SSW, et al. Hematologic parameters in patients with COVID‐19 infection. Am J Hematol. 2020;95(6):E131‐E134. 10.1002/ajh.25774 [DOI] [PubMed] [Google Scholar]
- 42. Young BE, Ong SWX, Kalimuddin S, et al. Epidemiologic features and clinical course of patients infected with SARS‐CoV‐2 in Singapore. JAMA. 2020;323(15):1488‐1494. 10.1001/jama.2020.3204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhou Y, Han T, Chen J, et al. Clinical and autoimmune characteristics of severe and critical cases of COVID‐19. Clin Trans Sci. 10.1111/cts.12805 [DOI] [PMC free article] [PubMed]
- 44. Qu R, Ling Y, Zhang YH, et al. Platelet‐to‐lymphocyte ratio is associated with prognosis in patients with coronavirus disease‐19 [published online ahead of print March 26, 2020]. J Med Virol. 2020:1‐9. 10.1002/jmv.25767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhu Z, Cai T, Fan L, et al. Clinical value of immune‐inflammatory parameters to assess the severity of coronavirus disease 2019. Int J Infect Dis. 2020;95:332‐339. 10.1016/j.ijid.2020.04.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497‐506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xie H, Zhao J, Lian N, Lin S, Xie Q, Zhuo H. Clinical characteristics of non‐ICU hospitalized patients with coronavirus disease 2019 and liver injury: a retrospective study. Liver Int. 2020;40:1321‐1326. 10.1111/liv.14449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Feng Y, Ling Y, Bai T, et al. COVID‐19 with different severities: a multicenter study of clinical features. Am J Respir Crit Care Med. 2020;201(11):1380‐1388. 10.1164/rccm.202002-0445OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zou Y, Guo H, Zhang Y, et al. Analysis of coagulation parameters in patients with COVID‐19 in Shanghai, China. BioScience Trends. 2020. 10.5582/bst.2020.03086 [DOI] [PubMed] [Google Scholar]
- 50. Zhang J, Dong X, Cao Y, et al. Clinical characteristics of 140 patients infected with SARS‐CoV‐2 in Wuhan, China. Allergy , 2020;75(7):1734‐1741 10.1111/all.14238 [DOI] [PubMed] [Google Scholar]
- 51. Zheng F, Tang W, Li H, Huang Y‐X, Xie Y‐L, Zhou Z‐G. Clinical characteristics of 161 cases of corona virus disease 2019 (COVID‐19) in Changsha. Eur Rev Med Pharmacol Sci. 2020;24(6):3404‐3410. 10.26355/eurrev_202003_20711 [DOI] [PubMed] [Google Scholar]
- 52. Petrilli CM, Jones SA, Yang J, et al. Factors associated with hospital admission and critical illness among 5279 people with coronavirus disease 2019 in New York City: prospective cohort study. BMJ. 2020;369:369. 10.1136/bmj.m1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. He R, Lu Z, Zhang L, et al. The clinical course and its correlated immune status in COVID‐19 pneumonia. J Clin Virol. 2020;127:104361. 10.1016/j.jcv.2020.104361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Deng Q, Hu B, Zhang Y, et al. Suspected myocardial injury in patients with COVID‐19: Evidence from front‐line clinical observation in Wuhan, China. Int J Cardiol. 2020;311:116‐121. 10.1016/j.ijcard.2020.03.087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Han H, Xie L, Liu R, et al. Analysis of heart injury laboratory parameters in 273 COVID‐19 patients in one hospital in Wuhan, China. J Med Virol. 2020;92:819‐823. 10.1002/jmv.25809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shi S, Qin M, Shen B, et al. Association of cardiac injury with mortality in hospitalized patients with COVID‐19 in Wuhan, China. JAMA Cardiol. 2020;5(7):802–810. 10.1001/jamacardio.2020.0950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lei S, Jiang F, Su W, et al. Clinical characteristics and outcomes of patients undergoing surgeries during the incubation period of COVID‐19 infection. E Clin Med. 2020;21:100331. 10.1016/j.eclinm.2020.100331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li K, Wu J, Wu F, et al. The clinical and chest CT features associated with severe and critical COVID‐19 pneumonia. Invest Radiol. 2020;55:327‐331. 10.1097/RLI.0000000000000672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chen Q, Zheng Z, Zhang C, et al. Clinical characteristics of 145 patients with corona virus disease 2019 (COVID‐19) in Taizhou, Zhejiang, China. Infection. 2020;48(4):543‐551. 10.1007/s15010-020-01432-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cai Q, Huang D, Ou P, et al. COVID‐19 in a designated infectious diseases hospital outside Hubei Province, China. Allergy. 2020;75(7):1742‐1752. 10.1111/all.14309 [DOI] [PubMed] [Google Scholar]
- 61. Wan S, Xiang Y, Fang W, et al. Clinical features and treatment of COVID‐19 patients in northeast Chongqing. Journal of Medical Virology. 2020;92(7):797‐806. 10.1002/jmv.25783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wan S, Yi Q, Fan S, et al. Relationships among lymphocyte subsets, cytokines, and the pulmonary inflammation index in coronavirus (COVID‐19) infected patients. Br J Haematol. 2020;189(3):428‐437. 10.1111/bjh.16659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Qin C, Zhou L, Hu Z, et al. Dysregulation of immune response in patients with COVID‐19 in Wuhan, China. Clin Infect Dis. 2020;71:762‐768. 10.1093/cid/ciaa248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pei G, Zhang Z, Peng J, et al. Renal involvement and early prognosis in patients with COVID‐19 Pneumonia. J Am Soc Nephrol. 2020;31(6):1157‐1165. 10.1681/ASN.2020030276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wang F, Hou H, Luo Y, et al. The laboratory tests and host immunity of COVID‐19 patients with different severity of illness. JCI Insight. 2020;5(10):e137799 10.1172/jci.insight.137799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chen G, Wu D, Guo W, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130(5):2620‐2629. 10.1172/JCI137244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Goyal P, Choi JJ, Pinheiro LC, et al. Clinical characteristics of covid‐19 in New York City. N Engl J Med. 2020;382:2372‐2374. 10.1056/NEJMc2010419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang Z, Yang B, Li Q, Wen L, Zhang R. Clinical features of 69 cases with coronavirus disease 2019 in Wuhan, China. Clin Infect Dis. 2020;71(15):769‐777. 10.1093/cid/ciaa272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mao L, Jin H, Wang M, et al. Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurol. 2020;77(6):683–690. 10.1001/jamaneurol.2020.1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wei X, Su J, Yang K, et al. Elevations of serum cancer biomarkers correlate with severity of COVID‐19. J Med Virol. 2020. 10.1002/jmv.25957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhou B, She J, Wang Y, Ma X. The clinical characteristics of myocardial injury in severe and very severe patients with 2019 novel coronavirus disease. J Infect. 2020;81:147‐178. 10.1016/j.jinf.2020.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Herold T, Jurinovic V, Arnreich C, et al. Elevated levels of IL‐6 and CRP predict the need for mechanical ventilation in COVID‐19. Journal of Allergy and Clinical Immunology. 2020;146(1):128–136.e4. 10.1101/2020.04.01.20047381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wei Y‐Y, Wang R‐R, Zhang D‐W, et al. Risk factors for severe COVID‐19: Evidence from 167 hospitalized patients in Anhui, China. J Infect. 2020;81(1):e89‐e92. 10.1016/j.jinf.2020.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Aggarwal S, Garcia‐Telles N, Aggarwal G, Lavie C, Lippi G, Henry BM. Clinical features, laboratory characteristics, and outcomes of patients hospitalized with coronavirus disease 2019 (COVID‐19): Early report from the United States. Diagnosis. 2020;7(2):91‐96. 10.1515/dx-2020-0046 [DOI] [PubMed] [Google Scholar]
- 75. Du R‐H, Liu L‐M, Yin W, et al. Hospitalization and critical care of 109 decedents with COVID‐19 pneumonia in Wuhan, China. Ann Am Thorac Soc. 2020;17(7):839‐846. 10.1513/AnnalsATS.202003-225OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Xu B, Fan CY, Wang AL, et al. Suppressed T cell‐mediated immunity in patients with COVID‐19: a clinical retrospective study in Wuhan, China. J Infect. 2020;81:51. 10.1016/j.jinf.2020.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID‐19 in Wuhan, China: a retrospective cohort study. Lancet 2020;395(10229):1054‐1062. 10.1016/S0140-6736(20)30566-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Yang X, Yu Y, Xu J, et al. Clinical course and outcomes of critically ill patients with SARS‐CoV‐2 pneumonia in Wuhan, China: a single‐centered, retrospective, observational study. Lancet Respir Med. 2020;8(5):475‐481. 10.1016/S2213-2600(20)30079-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ruan Q, Yang K, Wang W, Jiang L, Song J. Clinical predictors of mortality due to COVID‐19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020;46(5):846‐848. 10.1007/s00134-020-05991-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zhang J, Liu P, Wang M, et al. The clinical data from 19 critically ill patients with coronavirus disease 2019: a single‐centered, retrospective, observational study [published online ahead of print April 21, 2020]. Z Gesundh Wiss. 2020:1‐4. 10.1007/s10389-020-01291-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wang L, He W, Yu X, et al. Coronavirus disease 2019 in elderly patients: characteristics and prognostic factors based on 4‐week follow‐up. J Infect. 2020;80(6):639‐645. 10.1016/j.jinf.2020.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wang Y, Lu X, Li Y, et al. Clinical course and outcomes of 344 intensive care patients with COVID‐19. Am J Respir Crit Care Med. 2020;201(11):1430‐1434. 10.1164/rccm.202003-0736LE [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chen T, Wu D, Chen H, et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. 2020;368:368. 10.1136/bmj.m1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tang N, Li D, Wang X, Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemostasis. 2020;18(4):844‐847. 10.1111/jth.14768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tang N, Bai H, Chen X, Gong J, Li D, Sun Z. Anticoagulant treatment is associated with decreased mortality in severe coronavirus disease 2019 patients with coagulopathy. J Thromb Haemostasis. 2020;18(4):1094‐1099. 10.1111/jth.14817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Yan Y, Yang Y, Wang F, et al. Clinical characteristics and outcomes of patients with severe covid‐19 with diabetes. BMJ Open Diab Res Care. 2020;8(1):e001343. 10.1136/bmjdrc-2020-001343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wu C, Chen X, Cai Y, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. 2020;180(7):934–943. 10.1001/jamainternmed.2020.0994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zhang G, Hu C, Luo L, et al. Clinical features and short‐term outcomes of 221 patients with COVID‐19 in Wuhan, China. J Clin Virol. 2020;127:104364. 10.1016/j.jcv.2020.104364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus‐infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061‐1069. 10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Slaats J, ten Oever J, van de Veerdonk FL, Netea MG. IL‐1β/IL‐6/CRP and IL‐18/ferritin: distinct inflammatory programs in infections. PLOS Pathog. 2016;12(12):1005973. 10.1371/journal.ppat.1005973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Gauldie J, Richards C, Harnish D, Lansdorp P, Baumann H. Interferon beta 2/B‐cell stimulatory factor type 2 shares identity with monocyte‐derived hepatocyte‐stimulating factor and regulates the major acute phase protein response in liver cells. Proc Natl Acad Sci USA. 1987;84(20):7251‐7255. 10.1073/pnas.84.20.7251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Chen L, Deng H, Cui H, et al. Inflammatory responses and inflammation‐associated diseases in organs. Oncotarget. 2017;9(6):7204‐7218. 10.18632/oncotarget.23208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ. COVID‐19: consider cytokine storm syndromes and immunosuppression. Lancet 2020;395(10229):1033‐1034. 10.1016/S0140-6736(20)30628-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Kany S, Vollrath JT, Relja B. Cytokines in inflammatory disease. Int J Mol Sci. 2019;20(23):6008. 10.3390/ijms20236008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Diao B, Wang C, Tan Y, et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID‐19). Front Immunol. 2020;11:11. 10.3389/fimmu.2020.00827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Denman AM. Lymphocyte function and virus infections. J Clin Pathol Suppl (R Coll Pathol). 1979;13:39‐47. 10.1136/jcp.s3-13.1.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Hoffmann M, Kleine‐Weber H, Schroeder S, et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271‐280. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Xu H, Zhong L, Deng J, et al. High expression of ACE2 receptor of 2019‐nCoV on the epithelial cells of oral mucosa. Int J Oral Sci. 2020;12(1):8. 10.1038/s41368-020-0074-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Evans SS, Repasky EA, Fisher DT. Fever and the thermal regulation of immunity: the immune system feels the heat. Nat Rev Immunol. 2015;15(6):335‐49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Zheng M, Gao Y, Wang G, et al. Functional exhaustion of antiviral lymphocytes in COVID‐19 patients. Cell Mol Immunol. 2020;17(5):533‐535. 10.1038/s41423-020-0402-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Couper KN, Blount DG, Riley EM. IL‐10: the master regulator of immunity to infection. J Immunol. 2008;180(9):5771‐5777. 10.4049/jimmunol.180.9.5771 [DOI] [PubMed] [Google Scholar]
- 102. Barnes BJ, Adrover JM, Baxter‐Stoltzfus A, et al. Targeting potential drivers of COVID‐19: Neutrophil extracellular traps. J Exp Med. 2020;217(6):e20200652 10.1084/jem.20200652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Tecchio C, Micheletti A, Cassatella MA. Neutrophil‐derived cytokines: facts beyond expression. Front Immunol 2014;5:5. 10.3389/fimmu.2014.00508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Caudrillier A, Kessenbrock K, Gilliss BM, et al. Platelets induce neutrophil extracellular traps in transfusion‐related acute lung injury. J Clin Invest. 2012;122(7):2661‐2671. 10.1172/JCI61303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Drescher B, Bai F. Neutrophil in viral infections, friend or foe? Virus Res. 2013;171(1):1‐7. 10.1016/j.virusres.2012.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Schuetz P, Albrich W, Mueller B. Procalcitonin for diagnosis of infection and guide to antibiotic decisions: past, present and future. BMC Med. 2011;9:107. 10.1186/1741-7015-9-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Tsui PT, Kwok ML, Yuen H, Lai ST. Severe acute respiratory syndrome: clinical outcome and prognostic correlates. Emerg Infect Dis. 2003;9(9):1064‐1069. 10.3201/eid0909.030362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Fuchs TA, Brill A, Wagner DD. NET impact on deep vein thrombosis. Arterioscler Thromb Vasc Biol. 2012;32(8):1777‐1783. 10.1161/ATVBAHA.111.242859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Gupta N, Zhao YY, Evans CE. The stimulation of thrombosis by hypoxia. Thromb Res. 2019;181:77‐83. 10.1016/j.thromres.2019.07.013 [DOI] [PubMed] [Google Scholar]
- 110. Siripanthong B, Nazarian S, Muser D, et al. Recognizing COVID‐19–related myocarditis: the possible pathophysiology and proposed guideline for diagnosis and management. Heart Rhythm. 2020;S1547‐5271(20):30422–30427. 10.1016/j.hrthm.2020.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Li M‐Y, Li L, Zhang Y, Wang X‐S. Expression of the SARS‐CoV‐2 cell receptor gene ACE2 in a wide variety of human tissues. Infect Dis Poverty. 2020;9(1):45. 10.1186/s40249-020-00662-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Doyen D, Moceri P, Ducreux D, Dellamonica J. Myocarditis in a patient with COVID‐19: a cause of raised troponin and ECG changes. Lancet. 2020;395(10235):1516. 10.1016/S0140-6736(20)30912-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Wynants L, Van Calster B, Collins GS, et al. Prediction models for diagnosis and prognosis of covid‐19 infection: systematic review and critical appraisal. BMJ. 2020;369:m1328. 10.1136/bmj.m1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Hassoun A, Thottacherry ED, Muklewicz J, Aziz QU, Edwards J. Utilizing tocilizumab for the treatment of cytokine release syndrome in COVID‐19. J Clin Virol. 2020;128:104443. 10.1016/j.jcv.2020.104443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Zhang Y, Zheng L, Liu L, Zhao M, Xiao J, Zhao Q. Liver impairment in COVID‐19 patients: a retrospective analysis of 115 cases from a single centre in Wuhan city, China. Liver Int. 2020;40(9):2095–2103. 10.1111/liv.14455 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


