Abstract
Coronavirus disease 2019 (COVID‐19) is caused by severe acute respiratory syndrome coronavirus 2 infection. This study aims to examine the changes in peripheral blood parameters during the early stages of COVID‐19 and influenza. We analyzed the peripheral blood parameters of 169 COVID‐19 patients and 131 influenza patients during the early‐onset stage. Results from the patients with COVID‐19 were compared with those from healthy controls and influenza patients. In addition, results from patients with common and severe COVID‐19 were further compared. There were significant differences between COVID‐19 and influenza patients in terms of age, white blood cell count, platelet count, percentage of neutrophils, percentage of lymphocytes, percentage of monocytes, percentage of eosinophils, percentage of basophils, neutrophil, count and monocyte count. Two parameters (monocyte count and percentage of basophils) were combined to clarify the diagnostic efficacy of COVID‐19 and influenza and the area under the curve was found to be 0.772. Comparison of peripheral blood parameters from common COVID‐19, severe COVID‐19, and influenza patients revealed many differences during the early disease stages. The diagnostic formula developed by this study will be of benefit for physicians in the differentiation of COVID‐19 and influenza.
Keywords: biochemical parameters, blood routine parameters, COVID‐19, influenza, SARS‐CoV‐2
Abbreviations
- ALB
albumin
- ALT
alanine aminotransferase
- AST
aspartate aminotransferase
- BA#
basophil count
- BA%
percentage of basophils
- CK
creatine kinase
- COVID‐19
coronavirus disease 2019
- CRP
C‐reactive protein
- EO#
eosinophil count
- EO%
percentage of eosinophils
- GGT
gamma‐glutamyl transferase
- LDH
lactate dehydrogenase
- LY#
lymphocyte count
- LY%
percentage of lymphocytes
- MO#
monocyte count
- MO%
percentage of monocytes
- NE#
neutrophil count
- NE%
percentage of neutrophils
- PLT
platelet
- RT‐PCR
real‐time reverse transcriptase‐polymerase chain reaction
- SARS‐CoV‐2
severe acute respiratory syndrome coronavirus 2
- TP
total protein
- WBC
white blood cell
1. INTRODUCTION
The severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) that caused the coronavirus disease 2019 (COVID‐19) was discovered due to a viral pneumonia case in Wuhan in December 2019. With the spread of the epidemic disease, more and more countries and regions have successively discovered similar cases. As of 8 July 2020, the number of COVID‐19 infections worldwide has exceeded 11 million and the cumulative death toll has exceeded 530 000. Nucleic acid detection is the most direct means of diagnosing COVID‐19; however, the positive rate of real‐time reverse transcriptase‐polymerase chain reaction (RT‐PCR) detection of viral nucleic acid is 38% to 59%. 1 , 2 SARS‐CoV‐2‐specific immunoglobulin M appeared 1 week after the onset of COVID‐19 and its positive rate was 52.68% to 69%. 3 , 4 However, many countries and regions possess inadequate detection capability or the detection costs are prohibitive. COVID‐19 and influenza have similar symptoms during the early stages of the illness, resulting in many COVID‐19 cases being missed or misdiagnosed. Our study explored changes in peripheral blood parameters of patients during the early stages of COVID‐19 and influenza, and provides a reliable reference for better understanding the changes in laboratory test indicators of these patients as well as potential diagnostic markers for the COVID‐19 disease.
2. METHODS
2.1. Study population
A total of 169 patients diagnosed with COVID‐19 who were admitted to the Affiliated Hospital of Shaoxing University, Wenzhou Central Hospital, and Shaoxing People's Hospital from December 2019 to March 2020 were included in the present study. All COVID‐19 cases were confirmed by RT‐PCR assay of nasal and pharyngeal swab specimens. The patients with COVID‐19 were diagnosed according to the Novel Coronavirus and Pneumonia Diagnosis and Treatment Interim Guidance Report by the National Health Commission of the People's Republic of China. 5 Common cases were those who had a fever, respiratory tract symptoms, and pneumonia on imaging. Severe cases were those who had one of the following three clinical manifestations: (a) shortness of breath with a respiratory rate greater than 30 breaths/min; (b) mean oxygen saturation ≤93% in the resting state; and (c) partial pressure of arterial oxygen/oxygen concentration ≤300 mm Hg (1 mm Hg = 0.133 kPa). Severe cases also included the progressed of lesions by more than 50% within 24 to 48 hours, as detected by pulmonary imaging.
A total of 131 patients with influenza were also included in the present study. Of these, 78 patients had influenza A and 53 patients had influenza B. All influenza cases were confirmed by RT‐PCR assay of nasal and pharyngeal swab specimens. The control group was subjected to tests including clinical examination, computed tomography, hepatitis B virus‐DNA, anti‐hepatitis C virus antibody, human immunodeficiency virus antigen and antibody tests, and RT‐PCR for SARS‐CoV‐2, and the results of all tests were negative. The control group excluded respiratory diseases.
Peripheral blood from COVID‐19 and influenza patients was collected at the hospitals as part of the first examination, and the parameters measured. The study was approved by the Ethics Committee of the Affiliated Hospital of Shaoxing University (IRB‐AF‐016‐1.0).
2.2. Statistical analyses
SPSS 19.0 was used for statistical analyses. Continuous variables were expressed as mean (±standard deviation) or median (P25, P75), and were compared using an unpaired Student t test or the nonparametric Mann‐Whitney test. Categorical variables were presented as counts and percentages, which were compared using χ 2 statistics or Fisher's exact test. Single‐factor parameters (P < .05) were included in the multivariate logistic regression analysis and the regression equation was constructed based on multiple factors. The receiver operating characteristic (ROC) curve was used to evaluate the efficiency of diagnosing the disease stage of the patients. Statistical significance was defined as P < .05.
3. RESULTS
3.1. Analysis of peripheral blood cell parameters from COVID‐19 and influenza patients
Table 1 lists the parameters measured in the peripheral blood of the COVID‐19 and influenza patients. Of these, there were significant differences between COVID‐19 and influenza patients in terms of age, the white blood cell (WBC) count, platelet (PLT) count, percentage of neutrophils (NE%), percentage of lymphocytes (LY%), percentage of monocytes (MO%), percentage of eosinophils (EO%), percentage of basophils (BA%), neutrophil count (NE#), and monocyte count (MO#).
Table 1.
Various parameters from the blood samples of the COVID‐19, control, common COVID‐19, severe COVID‐19, and influenza groups
| COVID‐19 (N = 169) | Control (N = 80) | Common group (N = 145) | Severe group (N = 24) | Influenza (N = 131) | P a | P b | P c | P d | P e | |
|---|---|---|---|---|---|---|---|---|---|---|
| Age, y | 45.81 ± 14.84 | 46.0 ± 14.16 | 44.75 ± 14.69 | 52.21 ± 14.46 | 37.59 ± 21.19 | .934 | .022 | .000 | .001 | .000 |
| Male, N(%) | 87 (51.5) | 44 (55.0) | 70 (48.3) | 17 (70.8) | 60 (45.8) | .603 | .041 | .329 | .681 | .024 |
| WBC (×109/L) | 4.92 ± 1.75 | 6.43 ± 1.42 | 4.95 ± 1.80 | 4.72 ± 1.46 | 6.33 ± 2.35 | .000 | .543 | .000 | .000 | .000 |
| RBC (×1012/L) | 4.63 ± 0.50 | 4.60 ± 0.52 | 4.63 ± 0.51 | 4.62 ± 0.50 | 4.70 ± 0.65 | .689 | .918 | .265 | .288 | .537 |
| Hb, g/L | 135.44 ± 15.58 | 135.79 ± 15.07 | 135.27 ± 15.68 | 136.50 ± 15.24 | 136.69 ± 17.59 | .993 | .721 | .515 | .477 | .960 |
| PLT (×109/L) | 187.79 ± 63.93 | 256.88 ± 56.48 | 191.39 ± 63.46 | 166.04 ± 63.73 | 205.12 ± 69.96 | .000 | .072 | .026 | .088 | .012 |
| NE% | 64.50 ± 11.64 | 59.18 ± 9.24 | 63.26 ± 10.97 | 72.04 ± 12.94 | 68.42 ± 14.69 | .000 | .001 | .011 | .001 | .260 |
| LY% | 26.30 ± 10.52 | 31.20 ± 8.58 | 27.38 ± 10.07 | 19.75 ± 11.04 | 21.07 ± 12.85 | .000 | .001 | .000 | .000 | .638 |
| MO% | 7.60 (6.20‐9.95) | 7.20 (6.23‐8.48) | 7.70 (6.35‐10.00) | 6.70 (5.58‐9.55) | 9.0 (7.20‐11.40) | .059 | .224 | .000 | .000 | .007 |
| EO% | 0.60 (0.30‐1.15) | 1.50 (0.80‐2.18) | 0.60 (0.40‐1.20) | 0.30 (0.05‐0.50) | 0.40 (0.10‐1.10) | .000 | .000 | .038 | .000 | .163 |
| BA% | 0.20 (0.10‐0.30) | 0.40 (0.30‐0.70) | 0.20 (0.10‐0.30) | 0.10 (0.00‐0.20) | 0.10 (0.10‐0.30) | .000 | .000 | .001 | .000 | .118 |
| NE# (×109/L) | 2.93 (2.26‐3.79) | 3.63 (3.01‐4.50) | 2.89 (2.20‐3.79) | 3.03 (2.78‐4.02) | 4.26 (3.00‐5.74) | .000 | .229 | .000 | .000 | .018 |
| LY# (×109/L) | 1.12 (0.81‐1.54) | 1.92 (1.55‐2.29) | 1.17 (0.86‐1.63) | 0.73 (0.58‐1.01) | 1.08 (0.76‐1.54) | .000 | .000 | .364 | .074 | .012 |
| MO# (×109/L) | 0.36 (0.28‐0.48) | 0.45 (0.37‐0.55) | 0.36 (0.29‐0.48) | 0.33 (0.22‐0.49) | 0.55 (0.4‐0.71) | .000 | .299 | .000 | .000 | .000 |
| EO# (×109/L) | 0.03 (0.01‐0.05) | 0.10 (0.05‐0.14) | 0.03 (0.02‐0.06) | 0.01 (0.00‐0.02) | 0.02 (0.01‐0.06) | .000 | .000 | .418 | .107 | .021 |
| BA# (×109/L) | 0.01 (0.01‐0.01) | 0.03 (0.02‐0.04) | 0.01 (0.01‐0.02) | 0.01 (0.00‐0.01) | 0.01 (0.00‐0.01) | .000 | .000 | .359 | .047 | .002 |
Note: Data were expressed as mean (SD), medians (P25, P75), or N (%).
Abbreviations: BA#, basophil count; BA%, percentage of basophil; COVID‐19, coronavirus disease 2019; EO#, eosinophil count; EO%, percentage of eosinophil; Hb, haemoglobin; LY#, lymphocyte count; LY%, percentage of lymphocyte; MO#, monocyte count; MO%, percentage of monocytes; NE#, neutrophil count; NE%, percentage of neutrophils; PLT, platelet; RBC, red blood cell; WBC, white blood cell.
Comparison between COVID‐19 and control groups.
Comparison between common and severe groups.
Comparison between COVID‐19 and influenza groups.
Comparison between common and influenza groups.
Comparison between severe and influenza groups.
A multivariate analysis was performed to obtain the regression formula. Using stepwise forward logistic regression analysis, we found that two variables (MO# and BA%) were independently related to COVID‐19 (Table 2). Then, the logistic regression equation was used to calculate the following formula:
Table 2.
Multivariate logistic regression analysis of blood routine parameters of the COVID‐19 and influenza patients
| β Coefficient | Odds ratio (95% CI) | P | |
|---|---|---|---|
| MO# | −5.182 | 0.006 (0.001‐0.026) | .000 |
| BA% | 2.388 | 10.895 (2.093‐56.700) | .005 |
Note: The values of the three variables were found to be independently associated with COVID‐19 based on the results from the logistic regression analysis.
Abbreviations: BA%, percentage of basophil; CI, confidence interval; COVID‐19, coronavirus disease 2019; MO#, monocyte count.
On the basis of ROC curve (Figure 1), the best cutoff point for the joint probability was found to be 0.45, the diagnostic sensitivity was 71.6%, and the specificity was 74.8%. Therefore, COVID‐19 should be considered as the diagnosis when the joint probability is greater than 0.45, while influenza should be considered when the joint probability is less than 0.45.
Figure 1.
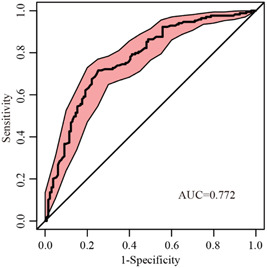
Receiver operating characteristic curve for the joint probability derived from the logistic regression model. AUC, area under the curve
The average age and the proportion of men in the severe COVID‐19 group were older and greater, respectively, than in the common COVID‐19 and influenza groups. Lymphocyte count (LY#) and eosinophil count (EO#) of the severe COVID‐19 group were significantly lower than in the common COVID‐19 and influenza groups. The PLT count of the severe COVID‐19 group was significantly lower than that of the common COVID‐19 group. MO%, MO#, and NE# of the influenza group were significantly higher than in the severe and common COVID‐19 groups. LY%, EO%, and BA% of influenza and severe COVID‐19 groups were significantly lower than in the common COVID‐19 group. NE% of influenza and severe COVID‐19 groups were significantly higher than in the common COVID‐19 group.
During the early stages of the disease, male sex, higher age, and lower lymphocyte, eosinophil, and basophil levels predicted that the COVID‐19 disease was more likely to be a severe case. Table 1 shows that WBC count, PLT count, LY%, EO%, BA%, LY#, MO#, EO#, and basophil count (BA#) during the early stage of COVID‐19 were significantly lower than in the control group (P < .001). NE% of COVID‐19 was significantly higher than that of the control group (P < .001).
3.2. Analysis of the abnormal rate of COVID‐19 and influenza
The WBC count in the influenza group was higher than in the severe and common COVID‐19 groups, and the proportion of WBC count in the influenza group that was above the upper limit of the reference interval (9.2%) was higher than that of the common (0.7%) and severe COVID‐19 (0%) groups (Table 3). The abnormal rates of NE%, LY%, and EO% in influenza and severe COVID‐19 groups were significantly higher than in the common COVID‐19 group. The percentages of LY% in influenza and severe COVID‐19 groups that were below the lower limit of the reference interval were 55.7% and 54.2, respectively, which were higher than in the common COVID‐19 group (24.1%). The proportion of EO% in the severe COVID‐19 group (62.5%) that was lower than the lower limit of the reference interval was higher than in influenza (45.8%) and common COVID‐19 (21.4%) groups. The proportion of EO# in the severe COVID‐19 group (58.3%) that was lower than the lower limit of the reference interval was higher than in influenza (40.5%) and common COVID‐19 (22.1%) groups. The proportion of LY# in the severe COVID‐19 group (79.2%) that was lower than the lower limit of the reference interval was higher than in influenza (52.7%) and common COVID‐19 (40%) groups. The proportion of MO# in the influenza group (36.6%) that was higher than the upper limit of the reference interval was significantly higher than in the severe (4.2%) and common COVID‐19 (6.2%) groups.
Table 3.
Abnormal parameter rates from the blood samples in the common COVID‐19, severe COVID‐19, and influenza groups
| Common group (N = 145) | Severe group (N = 24) | Influenza (N = 131) | P a | P b | P c | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Reference interval | Overall (N = 169) N (%) | Overall N (%) | Below the lower reference limit N (%) | Above the higher reference limit N (%) | Overall N (%) | Below the lower reference limit N (%) | Above the higher reference limit N (%) | Overall N (%) | Below the lower reference limit N (%) | Above the higher reference limit N (%) | ||||
| WBC (×109/L) | 3.5‐9.5 | 33 (19.5) | 30 (20.7) | 29 (20.0) | 1 (0.7) | 3 (12.5) | 3 (12.5) | 0 (0) | 22 (16.8) | 10 (7.6) | 12 (9.2) | .510 | .409 | .768 |
| RBC (×1012/L) | Male: 4.30‐5.80 Female: 3.80‐5.10 | 20 (11.8) | 18 (12.4) | 9 (6.2) | 9 (6.2) | 2 (8.3) | 2 (8.3) | 0 (0) | 28 (21.3) | 18 (13.7) | 10 (7.6) | .816 | .046 | .0169 |
| Hb, g/L | Male: 130‐175 Female: 115‐150 | 23 (13.6) | 19 (13.1) | 18 (12.4) | 1 (0.7) | 4 (16.7) | 4 (16.7) | 0 (0) | 22 (16.8) | 20 (15.3) | 2 (1.5) | .881 | .389 | 1.000 |
| PLT (×109/L) | 125‐350 | 24 (14.2) | 17 (11.7) | 14 (9.7) | 3 (2.1) | 7 (29.2) | 6 (25.0) | 1 (4.2) | 17 (13.0) | 13 (9.9) | 4 (3.1) | .051 | .752 | .062 |
| NE% | 40.0‐75.0 | 35 (20.7) | 22 (15.2) | 2 (1.4) | 20 (13.8) | 13 (54.2) | 1 (4.2) | 12 (50.0) | 51 (38.9) | 8 (6.1) | 43 (32.8) | .000 | .000 | .163 |
| LY% | 20.0‐50.0 | 50 (29.6) | 36 (24.8) | 35 (24.1) | 1 (0.7) | 14 (58.3) | 13 (54.2) | 1 (4.2) | 79 (60.3) | 73 (55.7) | 6 (4.6) | .001 | .000 | .856 |
| MO% | 3.0‐10.0 | 41 (24.3) | 35 (24.1) | 0 (0) | 35 (24.1) | 6 (25.0) | 1 (4.2) | 5 (20.8) | 51 (38.9) | 0 (0) | 51 (38.9) | .927 | .008 | .193 |
| EO% | 0.4‐8.0 | 47 (27.8) | 32 (22.1) | 31 (21.4) | 1 (0.7) | 15 (62.5) | 15 (62.5) | 0 (0) | 60 (45.8) | 60 (45.8) | 0 (0) | .000 | .000 | .132 |
| BA% | 0‐1.0 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | … | … | … |
| NE# (×109/L) | 1.8‐6.3 | 26 (15.4) | 21 (14.5) | 18 (12.4) | 3 (2.1) | 5 (20.8) | 3 (12.5) | 2 (8.3) | 38 (29.0) | 13 (9.9) | 25 (19.1) | .622 | .003 | .411 |
| LY# (×109/L) | 1.1‐3.2 | 80 (47.3) | 61 (42.1) | 58 (40.0) | 3 (2.1) | 19 (79.2) | 19 (79.2) | 0 (0) | 71 (54.2) | 69 (52.7) | 2 (1.5) | .001 | .044 | .023 |
| MO# (×109/L) | 0.1‐0.6 | 13 (7.7) | 11 (7.6) | 2 (1.4) | 9 (6.2) | 2 (8.3) | 1 (4.2) | 1 (4.2) | 48 (36.6) | 0 (0) | 48 (36.6) | 1.000 | .000 | .006 |
| EO# (×109/L) | 0.02‐0.52 | 46 (27.2) | 32 (22.1) | 32 (22.1) | 0 (0) | 14 (58.3) | 14 (58.3) | 0 (0) | 53 (40.5) | 53 (40.5) | 0 (0) | .000 | .001 | .104 |
| BA# (×109/L) | 0.00‐0.06 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | … | … | … |
Abbreviations: BA#, basophil count; BA%, percentage of basophil; COVID‐19, coronavirus disease 2019; EO#, eosinophil count; EO%, percentage of eosinophil; Hb, haemoglobin; LY#, lymphocyte count; LY%, percentage of lymphocyte; MO#, monocyte count; MO%, percentage of monocytes; NE#, neutrophil count; NE%, percentage of neutrophils; PLT, platelet; RBC, red blood cell; WBC, white blood cell.
Comparison of total abnormal rate between common and severe groups.
Comparison of total abnormal rate between common and influenza groups.
Comparison of total abnormal rate between severe and influenza groups.
3.3. Diagnostic efficacy of peripheral blood cell parameters between severe COVID‐19 and common COVID‐19 groups
BA%, BA#, EO#, EO%, LY%, and LY# had diagnostic efficacy in the severe COVID‐19 group. The severe COVID‐19 group was set as the positive group and the common COVID‐19 group was set as the negative group. The area under the curve (AUC) of BA% was 0.765 (95% confidence interval, CI [0.672‐0.858], P < .001); AUC of BA# was 0.756 (95% CI [0.661‐0.851], P < .001); AUC of EO# was 0.746 (95% CI [0.640‐0.852], P < .001); AUC of EO% was 0.729 (95% CI [0.610‐0.848], P < .001); AUC of LY% was 0.726 (95% CI [0.613‐0.838], P < .001); and AUC of LY# was 0.726 (95% CI [0.611‐0.841], P < .001) (Figure 2).
Figure 2.
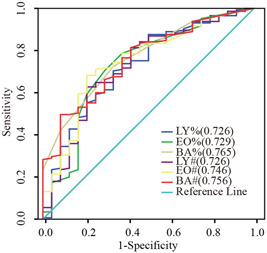
Receiver operating characteristic curve of the parameters from the blood samples in the severe common coronavirus disease 2019 (COVID‐19) and common COVID‐19 groups. BA#, basophil count; BA%, percentage of basophil; EO#, eosinophil count; EO%, percentage of eosinophil; LY#, lymphocyte count; LY%, percentage of lymphocyte
3.4. Analysis of the biochemical parameters of COVID‐19
During the early stage of COVID‐19 disease, lower albumin (ALB) and higher C‐reactive protein (CRP), lactate dehydrogenase (LDH), gamma‐glutamyl transferase (GGT), aspartate aminotransferase (AST), and creatine kinase (CK) predicted that it was more likely to develop into severe COVID‐19 disease. Table 4 shows that the concentrations of total protein (TP), ALB, globulin, alkaline phosphatase, total bilirubin, blood urea nitrogen, and total cholesterol of patients in the early stages of COVID‐19 were significantly lower than those of the control group, whereas the concentrations of AST, serum creatinine, CK, CK‐myocardial band, LDH, and CRP were significantly higher than in the control group (P < .05). The ratio of ALB in the COVID‐19 group above the upper reference limit was 0%. TP and ALB concentrations in the severe COVID‐19 group were significantly lower than in the common group, and the proportion of the ALB in the severe COVID‐19 group that was lower than the reference lower limit was 58.3% compared to 29.0% for the common COVID‐19 group. Alanine aminotransferase (ALT), AST, GGT, CK, LDH, and CRP concentrations in the severe COVID‐19 group were significantly higher than in the common COVID‐19 group. The ratios of CRP, LDH, GGT, and AST in the severe COVID‐19 group that were above the reference upper limit were 87.5%, 62.5%, 50.0%, and 45.8%, respectively, which were significantly higher than in the common COVID‐19 group, being 45.5%, 21.4%, 20.7%, and 15.2%, respectively (Table 5).
Table 4.
Biochemical parameters of the COVID‐19, control, common COVID‐19, and severe COVID‐19 groups
| COVID‐19 (N = 169) | Controls (N = 80) | Statistics | P a | Common group (N = 145) | Severe group (N = 24) | Statistics | P b | |
|---|---|---|---|---|---|---|---|---|
| TP | 70.62 ± 5.94 | 76.15 ± 4.27 | t = −8.369 | .000 | 71.08 ± 5.94 | 67.82 ± 5.23 | t = 2.529 | .012 |
| ALB | 41.39 ± 4.25 | 45.06 ± 2.25 | t = −8.908 | .000 | 41.80 ± 4.12 | 38.87 ± 4.21 | t = 3.222 | .002 |
| GLB | 29.23 ± 3.84 | 31.09 ± 3.30 | t = −3.723 | .000 | 29.28 ± 3.87 | 28.95 ± 3.69 | t = 0.403 | .687 |
| ALT | 21.1 (13.0‐32.0) | 19.0 (14.0‐28.8) | Z = −0.641 | .522 | 18.0 (13.0‐29.0) | 31.2 (24.3‐61.6) | Z = −3.864 | .000 |
| AST | 24.0 (19.0‐34.1) | 19.5 (17.0‐23.0) | Z = −4.953 | .000 | 23.0 (19.0‐31.0) | 35.1 (26.3‐46.0) | Z = −3.648 | .000 |
| GGT | 27.0 (16.0‐56.5) | 20.0 (15.3‐32.8) | Z = −1.499 | .134 | 25.0 (15.0‐48.5) | 55.5 (25.0‐157.8) | Z = −3.929 | .000 |
| ALP | 54.0 (41.0‐64.0) | 71.0 (62.0‐86.3) | Z = −6.792 | .000 | 54.0 (41.0‐63.5) | 55.5 (40.6‐70.0) | Z = −0.419 | .675 |
| TBIL | 11.3 (8.6‐15.2) | 15.2 (13.1‐19.8) | Z = −5.912 | .000 | 11.0 (8.5‐15.1) | 12.3 (8.8‐15.4) | Z = −0.669 | .504 |
| SCr | 65.0 (55.0‐79.5) | 58.5 (51.3‐72.8) | Z = −2.244 | .025 | 64.7 (54.0‐79.5) | 71.3 (62.5‐80.8) | Z = −1.552 | .121 |
| BUN | 3.7 (3.1‐4.4) | 4.7 (3.6‐5.4) | Z = −5.436 | .000 | 3.6 (3.1‐4.3) | 4.2 (3.2‐4.7) | Z = −1.586 | .113 |
| CK | 70.7 (48.2‐113.5) | 95.8 (73.0‐115.0) | Z = −3.569 | .000 | 70.2 (47.0‐106.0) | 90.5 (66.8‐210.2) | Z = −2.567 | .010 |
| CK‐MB | 12.3 (9.4‐15.7) | 12.5 (10.0‐15.0) | Z = −0.134 | .894 | 12.3 (9.4‐15.1) | 12.9 (9.0‐16.4) | Z = −0.401 | .689 |
| LDH | 203.0 (164.5‐256.3) | 189.5 (165.8‐210.0) | Z = −2.127 | .033 | 198.0 (159.0‐235.5) | 272.0 (221.2‐340.8) | Z = −4.195 | .000 |
| TC | 3.78 ± 0.87 | 4.96 ± 0.83 | t = −10.000 | .000 | 3.82 ± 0.87 | 3.58 ± 0.87 | t = 1.259 | .210 |
| TG | 1.1 (0.9‐1.7) | 1.2 (0.9‐1.9) | Z = −0.664 | .507 | 1.1 (0.9‐1.7) | 1.1 (0.8‐1.6) | Z = −0.432 | .665 |
| CRP | 10.8 (3.0‐25.8) | 0.3 (0.2‐1.3) | Z = −9.701 | .000 | 7.9 (2.3‐21.1) | 37.2 (22.3‐61.8) | Z = −4.855 | .000 |
Note: Data were expressed as mean (SD) and medians (P25, P75).
Abbreviations: ALB, albumin; ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; BUN, blood urea nitrogen; CK, creatine kinase; CK‐MB, creatine kinase‐myocardial band; COVID‐19, coronavirus disease 2019; CRP, C‐reactive protein; GGT, gamma‐glutamyl transferase; GLB, globulin; LDH, lactate dehydrogenase; SCr, serum creatinine concentration; TBil, total bilirubin; TC, total cholesterol; TG, triglyceride; TP, total protein.
Comparison between the COVID‐19 and control groups.
Comparison between common COVID‐19 and severe COVID‐19 groups.
Table 5.
Biochemical parameters of the common COVID‐19 and severe COVID‐19 groups
| Common group (N = 145) | Severe group (N = 24) | χ 2 | P a | ||||||
|---|---|---|---|---|---|---|---|---|---|
| All patients N (%) | Overall N (%) | Below the lower reference limit N (%) | Above the higher reference limit N (%) | Overall N(%) | Below the lower reference limit N (%) | Above the higher reference limit N (%) | |||
| TP | 27 (16.0) | 20 (13.8) | 19 (13.1) | 1 (0.7) | 7 (29.2) | 7 (29.2) | 0 (0) | 2.571 | .109 |
| ALB | 56 (33.1) | 42 (29.0) | 42 (29.0) | 0 (0) | 14 (58.3) | 14 (58.3) | 0 (0) | 8.016 | .005 |
| GLB | 2 (1.2) | 2 (1.4) | 0 (0) | 2 (1.4) | 0 (0) | 0 (0) | 0 (0) | 0.000 | 1.000 |
| ALT | 32 (18.9) | 25 (17.2) | 5 (3.5) | 20 (13.8) | 7 (29.2) | 0 (0) | 7 (29.2) | 1.210 | .271 |
| AST | 36 (21.3) | 24 (16.6) | 2 (1.4) | 22 (15.2) | 12 (50.0) | 1 (4.2) | 11 (45.8) | 13.742 | .000 |
| GGT | 44 (26.0) | 32 (22.1) | 2 (1.4) | 30 (20.7) | 12 (50.0) | 0 (0) | 12 (50.0) | 8.342 | .004 |
| ALP | 35 (20.7) | 27 (18.6) | 24 (16.6) | 3 (2.1) | 8 (33.3) | 6 (25.0) | 2 (8.3) | 2.714 | .099 |
| TBIL | 22 (13.0) | 19 (13.1) | 8 (5.5) | 11 (7.6) | 3 (12.5) | 1 (4.2) | 2 (8.3) | 0.000 | 1.000 |
| SCr | 9 (5.3) | 6 (4.1) | 4 (2.8) | 2 (1.4) | 3 (12.5) | 2 (8.3) | 1 (4.2) | 1.438 | .230 |
| BUN | 78 (46.2) | 69 (47.6) | 68 (46.9) | 1 (0.7) | 9 (37.5) | 9 (37.5) | 0 (0) | 0.843 | .359 |
| CK | 26 (15.4) | 20 (13.8) | 16 (11.0) | 4 (2.8) | 6 (25.0) | 0 (0) | 6 (25.0) | 1.219 | .270 |
| CK‐MB | 10 (5.9) | 8 (5.5) | 0 (0) | 8 (5.5) | 2 (8.3) | 0 (0) | 2 (8.3) | 0.006 | .941 |
| LDH | 50 (29.6) | 35 (24.1) | 4 (2.8) | 31 (21.4) | 15 (62.5) | 0 (0) | 15 (62.5) | 14.546 | .000 |
| TC | 40 (23.7) | 32 (22.1) | 25 (17.2) | 7 (4.8) | 8 (33.3) | 7 (29.2) | 1 (4.2) | 1.446 | .229 |
| TG | 39 (23.1) | 35 (24.1) | 2 (1.4) | 33 (22.8) | 4 (16.7) | 0 (0) | 4 (16.7) | 0.648 | .421 |
| CRP | 87 (51.5) | 66 (45.5) | 0 (0) | 66 (45.5) | 21 (87.5) | 0 (0) | 21 (87.5) | 14.530 | .000 |
Abbreviations: ALB, albumin; ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; BUN, blood urea nitrogen; CK, creatine kinase; CK‐MB, creatine kinase‐myocardial band; COVID‐19, coronavirus disease 2019; CRP, C‐reactive protein; GGT, gamma‐glutamyl transferase; GLB, globulin; LDH, lactate dehydrogenase; SCr, serum creatinine concentration; TBil, total bilirubin; TC, total cholesterol; TG, triglyceride; TP, total protein.
Comparison of total abnormal rate between common and severe groups.
3.5. Diagnostic efficacy of biochemical parameters between severe COVID‐19 and common COVID‐19 groups
CRP, LDH, GGT, ALT, and AST had diagnostic efficacy for the severe COVID‐19 group. The severe COVID‐19 group was set as the positive group and the common COVID‐19 group was set as the negative group. The AUC of CRP was 0.805 (95% CI [0.717‐0.893], P < .001); AUC of LDH was 0.770 (95% CI [0.665‐0.874], P < .001); AUC of GGT was 0.748 (95% CI [0.654‐0.842], P < .001); AUC of ALT was 0.746 (95% CI [0.659‐0.833], P < .001); and AUC of AST was 0.733 (95% CI [0.628‐0.838], P < .001) (Figure 3).
Figure 3.
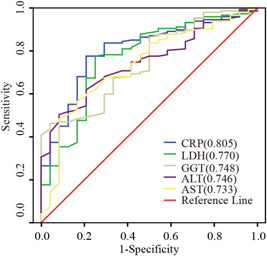
Receiver operating characteristic curves of biochemical parameters for the diagnosis of the common coronavirus disease 2019 (COVID‐19) and severe COVID‐19 groups. ALT, alanine aminotransferase; AST, aspartate aminotransferase; CRP, C‐reactive protein; GGT, gamma‐glutamyl transferase; LDH, lactate dehydrogenase
4. DISCUSSION
Coronaviruses are a large family of viruses known to cause serious diseases including the Middle East respiratory syndrome (MERS) and severe acute respiratory syndrome (SARS). SARS‐CoV‐2 is a new strain of coronavirus that has never been found in humans before. At present, the whole‐genome sequencing of the virus has been completed. 6 However, changes in the disease, diagnosis, treatment, and prognosis are not well understood. Influenza viruses (including influenza A and B) are another cause of contagious respiratory disease. The clinical manifestations of COVID‐19 and influenza are very similar; both can lead to increased mortality, with COVID‐19 having a higher mortality rate than influenza. 7 , 8 The mortality rate of severe COVID‐19 is significantly increased. 9 Therefore, it is very important to distinguish between COVID‐19 and influenza early and to carry out appropriate treatment. The present study analyzed data from COVID‐19 and influenza patients in an attempt to find patterns regarding the development of these diseases to provide useful information for clinical diagnosis and treatment.
Analysis of peripheral blood parameters revealed significant differences between COVID‐19 and influenza patients in many indicators. These included age, WBC count, PLT count, NE%, LY%, MO%, EO%, BA%, NE#, and MO#. In addition, many indicators in the severe COVID‐19 and influenza groups were significantly different from the common COVID‐19 group, such as NE%, LY%, EO%, BA%, NE% abnormal rate, LY% abnormal rate, and EO% abnormal rate. Many indicators in the influenza group were intermediate to the severe COVID‐19 and common COVID‐19 groups, such as NE%, LY%, EO%, BA%, LY#, EO#, and BA#.
Monocytes and macrophages play central roles in the immune response of humans and in protecting the body from influenza infection. They are necessary for the influenza virus to infect lymphocytes and regulate lymphocyte apoptosis by synthesizing and expressing viral neuraminidase. 10 Increased numbers of peripheral blood monocytes have been found in patients with influenza. 11 The decrease in eosinophils and basophils may be due to the stress response in the case of acute lung injury caused by a viral infection, wherein glucocorticoid secretion in the bone marrow would suppress the release of eosinophils and basophils. During the later stages of COVID‐19, eosinophils continue to increase in number and this is synchronous with improvements in radiology and symptoms. 12
This study showed that the peripheral blood MO# of the patients with influenza was significantly higher than that of the patients with COVID‐19. The BA# in the COVID‐19 group was lower than that in the control group, whereas the BA# in the patients with influenza was lower than that in the patients with COVID‐19. Two parameters, MO# and BA%, were combined to derive an equation using logistic regression analysis. The AUC was found to be 0.772 according to the ROC curve. This diagnostic formula could help physicians differentiate between COVID‐19 and influenza.
Many viral infections can cause thrombocytopenia. Both MERS and SARS can cause peripheral blood PLT reduction. 13 , 14 In this study, patients with COVID‐19 also showed significantly lower PLT counts, and the PLT count of severe COVID‐19 patients was significantly lower than that of common COVID‐19 and influenza patients. PLT count is an independent risk factor for COVID‐19. 15 This study also found that the severe COVID‐19 group was older and had a higher percentage of men than that of the common COVID‐19 and influenza groups. The age of the patient can be related to the prognosis of the disease. 16 Previous studies have shown that men are more susceptible to SARS‐CoV‐2 and have a higher mortality rate, which is related to endogenous testosterone. 17
Studies have shown that liver injury is common in patients with COVID‐19 18 and autopsy results of patients with COVID‐19 have shown hepatocyte degeneration, neutrophil infiltrating focal necrosis, lymphocytes and mononuclear cells in the hepatic lumen area, cell infiltration, and microthrombosis. Liver injury is associated with longer hospital stays and may be related to the prognosis of patients with COVID‐19. Some COVID‐19 patients without a history of liver disease are found with a liver injury before using any medication. 19 This study showed that during the early stage of the disease, the TP and ALB in the severe COVID‐19 group were lower than in the common COVID‐19 group, whereas the CRP, GGT, ALT, and AST were significantly higher. These indicators are significant in their ability to predict, in the early stage of the disease, patients who will develop severe COVID‐19. During the early stage of COVID‐19, LDH and CK in the severe COVID‐19 group were significantly higher than in the common COVID‐19 group. The decreases of LDH and CK in serum are related to the elimination of viral messenger RNA (mRNA), with the COVID‐19 viral mRNA clearance time positively correlated with a hospital stay. Decreases in LDH and CK may indicate a good prognosis for COVID‐19. 20
In summary, COVID‐19 and influenza can cause different changes in peripheral blood parameters, which should be considered in the early stages of COVID‐19 and influenza. The diagnostic formula developed in this study should help us to enable differentiation of COVID‐19 and influenza during their early stages.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Project administration: JZ. Data curation: JC, YP, SY, and YX. Methodology: WX and LZ. Resources: GL, PL, and YP. Writing and original draft: JC. Writing, review, and editing: YP and JZ.
ACKNOWLEDGMENTS
This study was supported by the Science Technology Bureau of Shaoxing (Grant No. 2018C30017). None of the funders had any role in the study design or in the collection, analysis, and interpretation of data, or in the writing of the article and the decision to submit it for publication. The researchers confirm their independence from funders and sponsors.
Chen J, Pan Y, Li G, et al. Distinguishing between COVID‐19 and influenza during the early stages by measurement of peripheral blood parameters. J Med Virol. 2021;93:1029–1037. 10.1002/jmv.26384
DATA AVAILABILITY STATEMENT
Research data are not shared.
REFERENCES
- 1. Liu R, Han H, Liu F, et al. Positive rate of RT‐PCR detection of SARS‐CoV‐2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clin Chim Acta. 2020;505:172‐175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ai T, Yang Z, Hou H, et al. Correlation of chest CT and RT‐PCR testing in coronavirus disease 2019 (COVID‐19) in China: a report of 1014 cases. Radiology. 2020;296:E32‐E40. 10.1148/radiol.2020200642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang G, Nie S, Zhang Z, Zhang Z. Longitudinal change of SARS‐CoV2 antibodies in patients with COVID‐19. J Infect Dis. 2020;222(2):183‐188. 10.1093/infdis/jiaa229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hoffman T, Nissen K, Krambrich J, et al. Evaluation of a COVID‐19 IgM and IgG rapid test; an efficient tool for assessment of past exposure to SARS‐CoV‐2. Infect Ecol Epidemiol. 2020;10(1):1754538. 10.1080/20008686.2020.1754538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. National Health Commission of the People's Republic of China . Diagnosis and treatment protocols of the novel coronavirus pneumonia (trial version 7). Chin Med J. 2020;133:1087‐1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in china, 2019. N Engl J Med. 2020;382(8):727‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deng M, Jiang L, Ren. Y , Ren Y, Liao J. Can we reduce mortality of COVID‐19 if we do better in glucose control? Med Drug Discov. 2020;7:100048. 10.1016/j.medidd.2020.100048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. 2019‐2020 U.S. Flu Season: Preliminary Burden Estimates | CDC. https://www.cdc.gov/flu/about/burden/preliminary-in-season-estimates.htm. Accessed 17 May 2020.
- 9. Yang X, Yu Y, Xu. J , et al. Clinical course and outcomes of critically ill patients with SARS‐CoV‐2 pneumonia in Wuhan, China: a single‐centered, retrospective, observational study. Lancet Respir Med. 2020;8:475‐481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Roberts NJ Jr. Diverse and unexpected roles of human monocytes/macrophages in the immune response to influenza virus. Viruses. 2020;12(4):E379. 10.3390/v12040379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Turner JS, Lei T, Schmitz AJ, et al. Impaired cellular immune responses during the first week of severe acute influenza infection. J Infect Dis. 2020. 10.1093/infdis/jiaa226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu F, Xu A, Zhang Y, et al. Patients of COVID‐19 may benefit from sustained Lopinavir‐combined regimen and the increase of Eosinophil may predict the outcome of COVID‐19 progression. Int J Infect Dis. 2020;95:183‐191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Joob B, Wiwanitkit V. Magnitude to thrombocytopenia among the patients with novel Middle East respiratory syndrome. Platelets. 2015;26(6):612. 10.3109/09537104.2014.934796. [DOI] [PubMed] [Google Scholar]
- 14. Yang M, Li CK, Li K, et al. Hematological findings in SARS patients and possible mechanisms (review). Int J Mol Med. 2004;14(2):311‐315. [PubMed] [Google Scholar]
- 15. Bi X, Su Z, Yan. H , et al. Prediction of severe illness due to COVID‐19 based on an analysis of initial fibrinogen to albumin ratio and platelet count. Platelets. 2020;31(5):674‐679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu J, Liu S. The management of coronavirus disease 2019 (COVID‐19). J Med Virol. 2020. 10.1002/jmv.25965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Salonia A, Corona G, Giwercman A, et al. SARS‐CoV‐2, testosterone and frailty in males (PROTEGGIMI): a multidimensional research project. Andrology. 2020. 10.1111/andr.12811 [DOI] [PubMed] [Google Scholar]
- 18. Xu L, Liu J, Lu M, Yang D, Zheng X. Liver injury during highly pathogenic human coronavirus infections. Liver Int. 2020;40(5):998‐1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xie H, Zhao J, Lian N, Lin S, Xie Q, Zhuo H. Clinical characteristics of non‐ICU hospitalized patients with coronavirus disease 2019 and liver injury: a retrospective study. Liver Int. 2020;40:1321‐1326. 10.1111/liv.14449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yuan J, Zou R, Zeng L, et al. The correlation between viral clearance and biochemical outcomes of 94 COVID‐19 infected discharged patients. Inflamm Res. 2020;69(6):599‐606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Research data are not shared.


