Abstract
Coronavirus (CoV) pandemics have become a huge threat to the public health worldwide in the recent decades. Typically, severe acute respiratory syndrome CoV (SARS‐CoV) caused SARS pandemic in 2003 and SARS‐CoV‐2 caused the ongoing COVID‐19 pandemic. Both viruses are most likely originated from bats. Thus, direct or indirect inter‐species transmission from bats to humans is required for the viruses to cause pandemics. Receptor utilization is a key factor determining the host range of viruses which is critical to the inter‐species transmission. Angiotensin‐converting enzyme 2 (ACE2) is the receptor of both SARS‐CoV and SARS‐CoV‐2, but only ACE2s of certain animals can be utilized by the viruses. Here, we employed pseudovirus cell‐entry assay to evaluate the receptor‐utilizing capability of ACE2s of 20 animals by the two viruses and found that SARS‐CoV‐2 utilized less ACE2s than SARS‐CoV, indicating a narrower host range of SARS‐CoV‐2. Especially, SARS‐CoV‐2 tended not to use murine or non‐mammal ACE2s. Meanwhile, pangolin‐CoV, another SARS‐related coronavirus highly homologous to SARS‐CoV‐2 in its genome, yet showed similar ACE2 utilization profile with SARS‐CoV rather than SARS‐CoV‐2. Nevertheless, the actual susceptibility of these animals to the coronaviruses should be further verified by in vivo studies. To clarify the mechanism underlying the receptor utilization, we compared the amino acid sequences of the 20 ACE2s and found 5 amino acid residues potentially critical for ACE2 utilization, including the N‐terminal 20th and 42nd amino acid residues that might determine the different receptor utilization of SARS‐CoV, SARS‐CoV‐2 and pangolin‐CoV. Our studies enhance the understanding of receptor utilization of pandemic coronaviruses, potentially contributing to the virus tracing, intermediate host screening and epidemic prevention for pathogenic coronaviruses.
Keywords: angiotensin‐converting enzyme 2 (ACE2), coronavirus, host range, inter‐species transmission, receptor utilization, SARS‐CoV, SARS‐CoV‐2
1. INTRODUCTION
Coronaviruses are enveloped non‐segmented positive‐sense RNA viruses. In the recent two decades, human coronaviruses (HCoVs) have caused at least three major pandemics and posed a huge threat to the public health worldwide (Zhou et al., 2020). Severe acute respiratory syndrome coronavirus (SARS‐CoV) and SARS‐CoV‐2 are the most pathogenic HCoVs (Meo et al., 2020). SARS‐CoV caused the SARS pandemic in years of 2002–2003, resulting in more than 8,000 clinical cases with a mortality of 10% (Parry, 2003; Stadler et al., 2003). After the SARS pandemic, plenty of research and measurements have been done to prevent the re‐emergence of coronavirus epidemics (de Wit, van Doremalen, Falzarano, & Munster, 2016). Nevertheless, 17 years later, SARS‐CoV‐2 brought a much more severe and widespread pandemic of coronavirus disease 2019 (COVID‐19) to the world (Wu et al., 2020; Zhu et al., 2020). Up to 17 June 2020, about 6 months after the first reported case of COVID‐19, SARS‐CoV‐2 has caused 8,179,520 confirmed infections and 441,491 deaths worldwide, far surpassing the SARS pandemic. SARS‐CoV‐2 emerges a high transmissibility whose R0 value is currently estimated as 2.3 but could be as high as 5.7 when more infection cases are identified (Bulut & Kato, 2020). The high transmissibility of SARS‐CoV‐2 is probably a major reason for the rapid development of COVID‐19 epidemic and more studies on the transmission of pathogenic HCoV is urgently required.
Inter‐species transmission from wide animals to humans is a major cause of the epidemics of highly pathogenic coronaviruses. Previous studies have shown that Chinese horseshoe bats are natural reservoirs of SARS‐CoV (Ge et al., 2013; Hon et al., 2008), and a recent phylogenetic analysis has revealed that SARS‐CoV‐2 might also be originated from bat‐SARSr‐CoV (Lu et al., 2020). Some small mammals, such as civets and raccoon dogs, can serve as the intermediate hosts of SARS‐CoV and might be the direct sources of the SARS epidemic in early 2003 (Guan et al., 2003). Similarly, SARS‐CoV‐2‐like CoVs were detected in Malayan pangolins, indicating pangolins might serve as an intermediate host for SARS‐CoV‐2 (Lam et al., 2020; Zhang, Wu, & Zhang, 2020). The host range of a virus is an essential factor determining its intermediate hosts. Thus, research on the host range of viruses is of great importance for virus tracing and epidemic control.
A main factor determining the host range of viruses is the recognition and binding between viral particles and their receptors on the host cells. Angiotensin‐converting enzyme 2 (ACE2) is utilized by SARS‐CoV and SARS‐CoV‐2 as their cellular receptor (Xiao, Chakraborti, Chakraborti, Dimitrov, Gramatikoff, & Dimitrov, 2003; Zhou et al., 2020). Discovered in the year 2000, ACE2 was initially identified as an exopeptidase that catalyses the conversion of angiotensins (Donoghue et al., 2000; Ferrario, Trask, Trask, & Jessup, 2005). ACE2 is ubiquitously expressed in most vertebrates, but not all ACE2s can serve as the receptor for SARS‐CoV and SARS‐CoV‐2. For instance, SARS‐CoV can use mouse ACE2 as its receptor but SARS‐CoV‐2 cannot, indicating that mouse is a potential host for SARS‐CoV but not for SARS‐CoV‐2 (Zhou et al., 2020). Our previous study predicted the ACE2 utilization of SARS‐CoV‐2 and 9 amino acid (aa) residues in ACE2 critical for SARS‐CoV‐2 utilization (Qiu et al., 2020). However, this study was mainly based on the aa sequence analysis and lacked experimental evidence, which could be hardly referred to clarify the host range of SARS‐CoV‐2.
In this study, we ectopically expressed ACE2 of 20 different animals in HeLa cells, a cell line lacking ACE2 expression naturally, and then infected the cells with HIV‐based pseudoviral particles carrying coronavirus spike proteins to test their utilization of these ACE2s. The result showed that both SARS‐CoV and SARS‐CoV‐2 could use most mammalian ACE2s as their receptors but not fish or reptilian ACE2s. Interestingly, similar to mouse ACE2, SARS‐CoV but not SARS‐CoV‐2 was capable of using chicken ACE2, indicating a narrower host range of SARS‐CoV‐2, especially in murine and birds. By alignment of the aa sequence of the 20 ACE2 orthologs, we further confirmed several aa residues critical for SARS‐CoV‐2 utilization, including T20, K31, Q42 and Y83. Especially, T20 of ACE2 probably played critical roles in spike–ACE2 binding by interacting with S477 and T478 within the receptor‐binding motif (RBM) of SARS‐CoV‐2 spike protein. These aa residues might partially determine the unique receptor utilization and host range of SARS‐CoV‐2. In summary, our study provides a more clear view of ACE2 utilization by SARS‐CoV‐2, which may contribute to a better understanding about the virus–receptor interaction and the host range of SARS‐CoV‐2.
2. METHODS AND MATERIALS
2.1. Cell lines and plasmids
HEK293T and HeLa cells were obtained from the American Tissue Culture Collection and cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (FBS) in a humidified 5% CO2 incubator at 37°C.
Full‐length SARS‐CoV‐2 spike (GenBank accession number MN908947.3), SARS‐CoV BJ01 spike (GenBank accession number AY278488.2) and pangolin‐CoV GD1 spike (GISAID accession number: EPI_ISL_410721) were all synthesized by Sangon and subcloned into the pcDNA3.1 vectors with a C‐terminal HA tag. The cDNAs encoding different ACE2 proteins (Table S1) were synthesized by Sangon and subcloned into pcDNA3.1 vectors with a C‐terminal 6XHis tag. All the plasmids were verified by Sanger sequencing.
2.2. Western blot analysis
Lysates of cells or filtered supernatants containing pseudoviruses were separated by SDS‐PAGE, followed by transfer to a nitrocellulose membrane (Millipore). For detection of S protein, the membrane was incubated with anti‐HA tag mouse monoclonal antibody (Bimake, 1:2,000), and the bound antibodies were detected by horseradish peroxidase (HRP)‐conjugated goat anti‐mouse IgG (Abbkine, 1:5,000). For detection of HIV‐1 p24 in supernatants, monoclonal antibody against HIV p24 (p24 MAb) was used as the primary antibody at a dilution of 1:8,000, followed by incubation with HRP‐conjugated goat anti‐mouse IgG at the same dilution. To detect the expression of 20 ACE2s in HeLa cells, mouse anti‐6XHis tag monoclonal antibody (BioWorld, 1:5,000) was used as the primary antibody, followed by incubation with HRP‐conjugated goat anti‐mouse IgG at the same dilution.
All the Western blot analyses were repeated for at least three times. The results were subjected to densitometric measurement to quantify the intensity of the bands by ImageJ program.
2.3. Pseudovirus preparation and cell‐entry assay
The pseudovirus cell‐entry assay was performed as described previously (Yang et al., 2014). In brief, HEK293T cells were co‐transfected with a luciferase‐expressing HIV‐1 plasmid (pNL4‐3.Luc.R‐E‐) and a plasmid encoding HA‐tagged SARS‐CoV‐BJ01 spike, SARS‐CoV‐2 spike or pangolin‐CoV spike. The supernatant containing pseudoviruses was collected 48 hr after transfection, and the remaining cell pellet was lysed for Western blot detection of HA‐tagged spike proteins. In cell‐entry assay, pseudoviruses were incubated with recipient cells at 37°C for 6 hr, the medium was changed and cells were incubated for an additional 42 hr. Cells were then washed with PBS buffer and lysed. Lysates were tested for luciferase activity (Promega). Each infection experiment was carried out on for three times.
2.4. Phylogenetic analysis
Multiple sequence alignment was performed for the whole aa sequences of ACEs using MAFFT with a local alignment strategy FFT‐NS‐2. The phylogenetic tree was constructed by MEGA7 using the neighbour‐joining (NJ) method with 1,000 bootstrap replicates and visualized using FigTree.
3. RESULTS AND DISCUSSION
3.1. Validation and normalization of pseudovirus stocks and ACE2 expression
In this study, we investigated the ACE2 utilization of three SARSr‐CoVs, including SARS‐CoV‐BJ01, SARS‐CoV‐2 and pangolin‐CoV. Pangolin‐CoV was tested since pangolins were suspected to be a potential intermediate host of SARS‐CoV‐2 (Zhang et al., 2020). Notably, the RBM of pangolin‐CoV spike is almost identical with SARS‐CoV‐2 spike except for the 498th (484th for SARS‐CoV) aa residue, but they are different from SARS‐CoV spike on multiple aa sites (Figure 1a).
FIGURE 1.
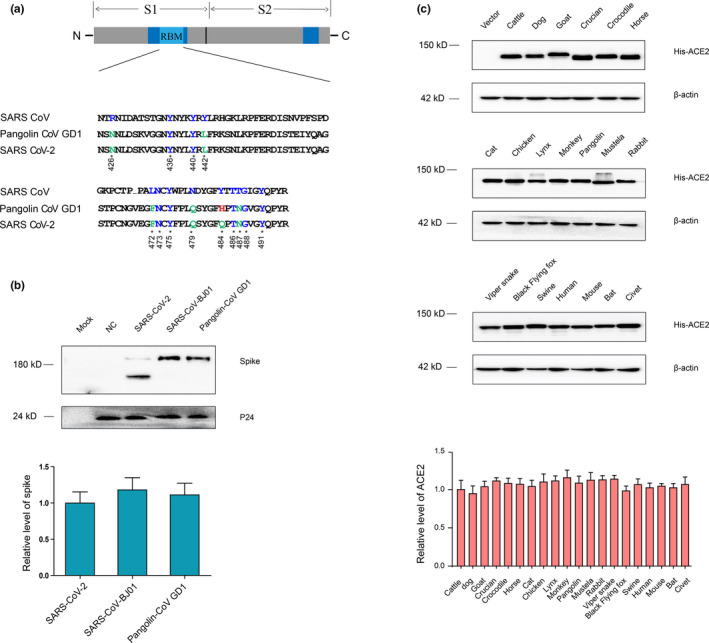
Validation of pseudovirus preparation and ACE2 expression. (a) Schematic structure of the spike protein of SARSr‐CoVs (upper panel) and alignment of the amino acid sequences of the receptor‐binding motifs (RBMs) of SARS‐CoV, SARS‐CoV‐2 and pangolin‐CoV spike proteins (lower panel). (b) Western blot detection of spike proteins of SARS‐CoV‐BJ01, SARS‐CoV‐2 and pangolin‐CoV in the pseudovirus stock solutions using an antibody against the HA tag conjugated to the viral spike proteins. HIV‐1 p24, a protein of the carrier pseudovirus, was detected as the loading control. "Mock" indicates the cells without any treatment. "NC" indicates the cells packaging the negative‐control pseudovirus that does not carry any spike. (c) Detection of different ACE2 orthologs in HeLa cells after transfecting the corresponding plasmids using an antibody against the 6XHis tag conjugated to the ACE2 proteins. β‐actin was detected as the loading control. All the Western blots were repeated for three times and the results were subjected to densitometric measurement to quantify the intensity of the bands using ImageJ program. [Colour figure can be viewed at wileyonlinelibrary.com]
In order to validate the successful preparation of the three pseudoviruses and normalize their amount, same volume of pseudovirus stocks were loaded to SDS‐PAGE and subjected to Western blot detection of HA‐tagged spike proteins. As shown in the upper panel of Figure 1b, SARS‐CoV and pangolin‐CoV showed typical bands (about 180 kDa) of SARSr‐CoV spike proteins, and SARS‐CoV‐2 showed a smaller band since SARS‐CoV‐2 spike contains a unique cleavage site of furin protease and was cleaved during package of the pseudovirus (Hoffmann et al., 2020; Shang, Wan, et al., 2020; Wang et al., 2020). These results indicated the success of pseudovirus preparation. Meanwhile, the levels of all the three spikes were measured and normalized for the following cell‐entry assays (Figure 1b, lower panel).
Then, we continued to validate the ectopic expression of 20 ACE2s from different animals in HeLa cells, an ACE2‐negative human cell line. We transfected the HeLa cells with plasmids harbouring the coding gene of ACE2s from different animals. At 48 hr post‐transfection, the cells were lysed and subjected to Western blot detection of 6XHis‐tagged ACE2 protein. As shown in the upper panel of Figure 1c, bands of all ACE2s (about 150 kDa) could be observed, indicating the successful expression of all ACE2s in HeLa cells. Meanwhile, the levels of all ACEs were measured and normalized for the cell‐entry assays (Figure 1c, lower panel).
3.2. Difference in the ACE2 utilization by SARS‐CoV and SARS‐CoV‐2
We transfected the HeLa cells with the 20 plasmids expressing different ACE2s individually or the empty vector as a control. At 48 hr post‐transfection, the cells were infected with SARS‐CoV‐BJ01, SARS‐CoV‐2 or pangolin‐CoV pseudovirus. After 48 hr of infection, the cells were lysed and subjected to luciferase assay to evaluate the cell‐entry efficiency of the pseudoviruses mediated by different ACE2s. As shown in Figure 2, little luminescence signals could be observed in samples from HeLa cells transfected with the empty vector and infected by any of the three pseudoviruses, indicating that native HeLa without ACE2 could not mediate the pseudovirus entry. Luminescence signals from cells expressing crucian, crocodile or viper snake ACE2 were also low, indicating that fish and reptilian ACE2s could barely mediate the pseudovirus entry. Cells expressing chicken or mouse ACE2 showed a strong luminescence signal when infected by the SARS‐CoV‐BJ01 pseudovirus but not by the SARS‐CoV‐2 pseudovirus, indicating that SARS‐CoV‐BJ01 could use chicken or mouse ACE2 for cell entry but SARS‐CoV‐2 could not. Pangolin‐CoV was capable of utilizing both chicken and mouse ACE2s, but its utilizing efficiency of chicken ACE2 was much lower than SARS‐CoV‐BJ01. On the contrary, SARS‐CoV‐2 pseudovirus ignited a stronger luminescence than SARS‐CoV‐BJ01 or pangolin‐CoV pseudovirus in cells expressing bat ACE2, indicating the highest utilizing capability of bat ACE2 by SARS‐CoV‐2. For the other ACE2s, infection of all the three pseudoviruses led to strong luminescence signals, implying that all the three SARSr‐CoV were capable of utilizing a broad range of ACE2s.
FIGURE 2.
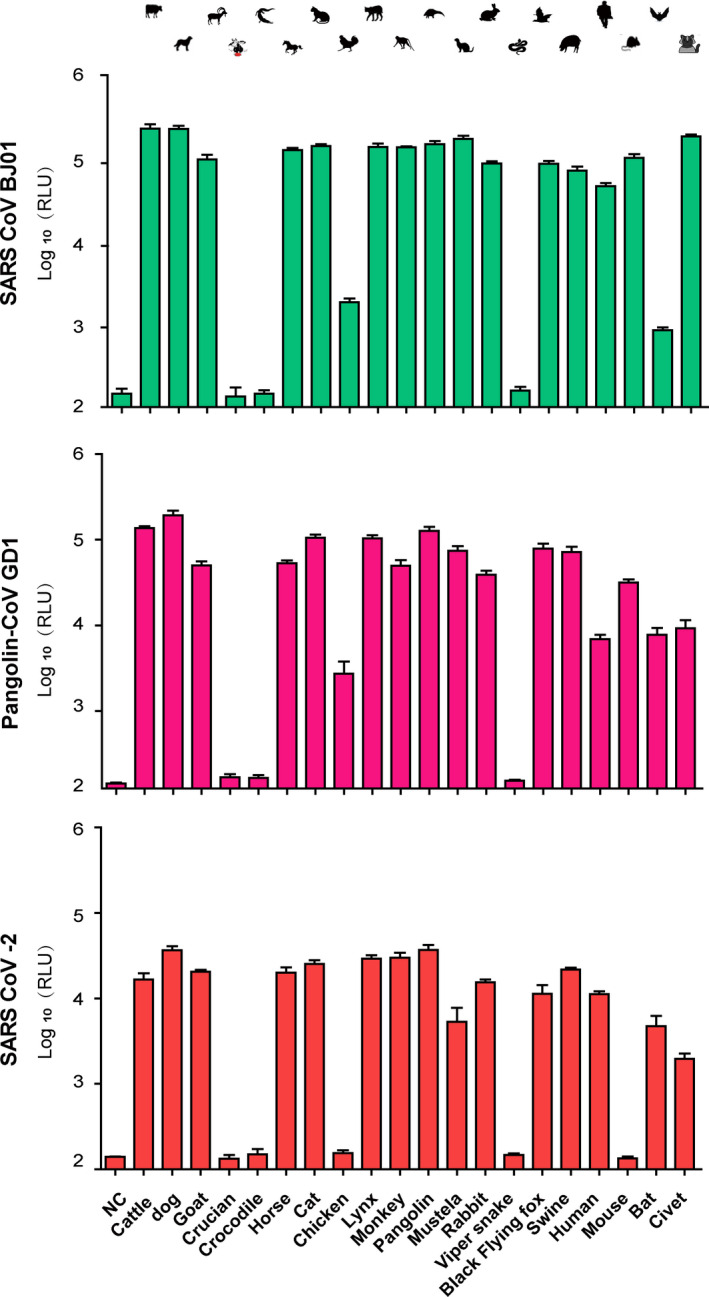
Entry efficiency of SARS‐CoV‐BJ01, SARS‐CoV‐2 and pangolin‐CoV pseudoviruses into ACE2‐expressing cells. HeLa cells expressing different ACE2 orthologs were infected by SARS‐CoV, SARS‐CoV‐2 or pangolin‐CoV pseudoviruses. At 48 hr post‐infection, pseudovirus entry efficiency was determined by measuring luciferase activity in cell lysates. The results were presented as the logarithm (base 10) of the mean relative luminescence units (RLUs) and the error bars indicated the logarithm (base 10) of the standard deviations of the RLUs (n = 9) [Colour figure can be viewed at wileyonlinelibrary.com]
A broad host range is supposed to lead to effective inter‐species transmission of virus and more potential to cause a pandemic. The COVID‐19 pandemic caused by SARS‐CoV‐2 is the most severe worldwide pandemic in the recent years, surpassing the SARS pandemic in 2003, so it is likely to speculate that SARS‐CoV‐2 has a broader host range. Surprisingly, our cell‐entry result showed that SARS‐CoV‐2 had a smaller range of ACE2 utilization than SARS‐CoV. SARS‐CoV‐2 could not utilize mouse or chicken ACE2 which could be used by SARS‐CoV, indicating a narrower host range of SARS‐CoV‐2, especially in murine and birds. The reason is probably that the host range of SARS‐CoV‐2 is broad enough to support its transmission from bats to humans, and lack of infection to some kinds of animals does not affect such transmission due to redundant routes. Thus, it is not suggested to over‐interpret the determination of the host range on the possibility of a virus to cause pandemics, especially for the viruses with broad host ranges.
Notably, SARS‐CoV‐2 has a better utilization of bat ACE2 than SARS‐CoV. Though SARS‐CoV originates from bat‐SARSr‐CoV, the utilization of bat ACE2 by SARS‐CoV is quite limited which is supported by the previous reports (Ge et al., 2013; Ren et al., 2008). According our current results, SARS‐CoV‐2 utilizes Chinese horseshoe bat ACE2 much better than SARS‐CoV, indicating a higher homology between SARS‐CoV‐2 and its ancestor. This speculation is supported by the phylogenetic analysis of viral genomes in the previous study (Zhou et al., 2020).
Our cell‐entry result showed that SARS‐CoV and SARS‐CoV‐2 could use a wide variety of mammalian ACE2s, which is further supported by the reports about the susceptibility of various mammals to SARS‐CoV‐2 infection (Shi et al., 2020). However, the utilization of fish and reptilian ACE2s was quite poor for both SARS‐CoV and SARS‐CoV‐2. This can be explained by the remote phylogenetic relationship between fish/reptilian ACE2s and mammalian ACE2s. By comparison, bird ACE2s have closer phylogenetic relationship with mammalian ACE2s, and thus, bird ACE2s could be used by some but not all SARSr‐CoVs, such as SARS‐CoV. This indicates that SARSr‐CoVs are more likely to be transmitted by mammals and birds but not fish and reptiles, and more attention should be paid to domestic mammals and birds to prevent CoV pandemic.
3.3. Phylogenetic analysis of ACE2s and key amino acids for SARS‐CoV‐2 utilization
To evaluate the phylogenetic relationship of the 20 ACE2s assessed above, we built a phylogenetic tree based on the aa sequences of all the ACE2s (Figure 3a). In the tree, we observed no branches corresponding to the ACE2 utilization by SARS‐CoV‐BJ01 or SARS‐CoV‐2 pseudoviruses, indicating that the whole sequence analysis could hardly reveal the key factors underlying the receptor utilization by the viruses.
FIGURE 3.
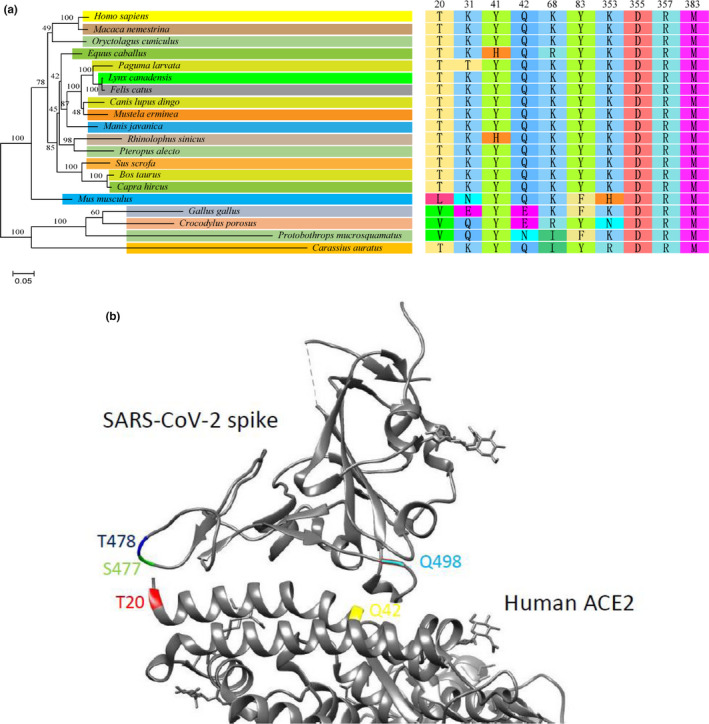
Phylogenetic analysis of the 20 ACE2 orthologs and the key amino acid residues for SARS‐CoV‐2 utilization. (a) The phylogenetic tree was constructed on the whole aa sequences of ACE2s using NJ method by MEGA7 with 1,000 bootstrap replicates (left panel) and the amino acids on the 9 critical sites predicted previously were listed (right panel). The colours in the figure do not contain any biological or technical meaning but just for easy reading. (b) The structure of the complex of SARS‐CoV‐2 spike and human ACE2 was adapted from Protein Data Bank (PDB ID: 6VW1). S477, T478 and Q498 of SARS‐CoV‐2 spike were labelled in green, blue and cyan, respectively. T20 and Q42 were labelled in red and yellow, respectively [Colour figure can be viewed at wileyonlinelibrary.com]
In our previous study, we predicted 9 key aa sites on human ACE2 potentially critical for the receptor utilization, including T20, K31, Y41, K68, Y83, K353, D355, R357 and M383, based on the SARS‐CoV‐2 utilization of human, bat, civet, swine and mouse ACE2s (Qiu et al., 2020). Here, we tested 15 more species to validate the role of the 9 sites. As shown in Figure 3a, Y/H41, D355, R357 and R383 were conserved in all ACE2s, and K68 was conserved in both mouse and chicken ACE2s which could not be used by SARS‐CoV‐2, indicating that these sites were not determining the receptor utilization by SARS‐CoV‐2. Q42 was conserved in mouse ACE2 but was substituted by E42 in chicken ACE2, indicating a complicated role of this site. On the contrary, T20, K31 and Y83 in usable ACE2s were distinct from the corresponding aa in unusable ACE2s, indicating their critical role in determining SARS‐CoV‐2 utilization.
Among the three key sites, substitutions of K31 and Y83 have been reported to abolish or strongly inhibit SARS‐CoV binding and the mechanism has been well documented (Li et al., 2005; Wan, Shang, Shang, Graham, Baric, & Li, 2020). Remarkably, our study revealed T20 of ACE2 as a potential key aa residue for SARS‐CoV‐2 utilization. T20 has not been reported to affect SARS‐CoV utilization but mouse ACE2 with T20L and chicken ACE2 with T20V could not be used by SARS‐CoV‐2. T20 is located at the N terminus of most mature mammalian ACE2s. According to the structure of SARS‐CoV‐2 receptor‐binding domain complexed with human ACE2, the N‐terminal T20 of ACE2 is close to S477 and T478 of SARS‐CoV‐2 RBM (Figure 3b; Shang, Ye, et al., 2020). Both threonine and serine contain hydroxyl radicals that allow them to form hydrogen bonds with each other. Thus, T20 of human ACE2 is likely to bridge with S477 and T478 of SARS‐CoV‐2 spike via hydrogen bonds, which stabilizes the ACE2‐spike binding. However, L20 on mouse ACE2 and V20 on chicken ACE2 are both aliphatic aa that cannot form hydrogen bonds to support the ACE2‐spike binding, which may impair the utilization of these two ACE2s by SARS‐CoV‐2. In SARS‐CoV spike, the two aa residues are substituted by G463 and K464. K464 can form hydrogen bonds with threonine and G463 is a non‐polar aa that can interact with valine or leucine via hydrophobic bond together with the adjacent A461. Therefore, SARS‐CoV spike can interact with various N‐terminal aa of ACE2s, and this may be the reason why SARS‐CoV can utilize mouse and chicken ACE2s while SARS‐CoV‐2 cannot. Pangolin CoV spike shares high similarity with SARS‐CoV‐2 which keeps S477 and T478. However, pangolin‐CoV spike harbours alkaline H498 that can interact with acidic Q42 and E42 of mouse and chicken ACE2, respectively, via ionic affinity, which may complementally support the ACE2–spike binding and allow the utilization of mouse and chicken ACE2 by pangolin‐CoV. On the contrary, SARS‐CoV‐2 spike substitutes H498 with acidic Q498, leading to ionic repulsion with Q42 and E42, blocking its binding with mouse and chicken ACE2s.
Nevertheless, these sites were identified only by the sequence analysis, which was not enough to substantiate their roles in ACE2 utilization by the coronaviruses. The actual roles of them should be verified by further studies, such as single mutation on these sites and functional assays, which are our major direction in the future. Before further verification, any over‐interpretation about the roles of these sites in ACE2 utilization should be avoided.
In summary, our results showed less ACE2 utilization by SARS‐CoV‐2 compared to SARS‐CoV and pangolin‐CoV, especially for murine and bird ACE2s, indicating narrower host range of SARS‐CoV‐2. Meanwhile, we found that the N‐terminal T20 and Q42 might be critical in determining the difference of ACE2 utilization by the three SARSr‐CoVs. Our findings deepen the understanding about the receptor utilization and the host range of SARS‐CoV‐2, providing useful information for tracing virus transmission routes and preventing pandemics caused by CoVs in the future.
CONFLICT OF INTEREST
The authors declare no conflict of interests.
ETHICAL APPROVAL
Ethical statement is not applicable since no sample collection or questionnaires from animals/human have been gathered.
Supporting information
Table S1
ACKNOWLEDGEMENTS
This work was jointly funded by the National Natural Science Foundation of China (grant numbers 32041001 and 81902070), the National Key Research and Development Program of China (grant number 2017YFD0500104) and the Provincial Natural Science Foundation of Hunan Province (grant numbers 2019JJ20004, 2019JJ50035 and 2020SK3001).
Wang Q, Qiu Y, Li J‐Y, Liao C‐H, Zhou Z‐J, Ge X‐Y. Receptor utilization of angiotensin‐converting enzyme 2 (ACE2) indicates a narrower host range of SARS‐CoV‐2 than that of SARS‐CoV. Transbound Emerg Dis.2021;68:1046–1053. 10.1111/tbed.13792
Qiong Wang and Ye Qiu contributed equally to this work.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Bulut, C. , & Kato, Y. (2020). Epidemiology of COVID‐19. Turkish Journal of Medical Sciences, 50(SI‐1), 563–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit, E. , van Doremalen, N. , Falzarano, D. , & Munster, V. J. (2016). SARS and MERS: Recent insights into emerging coronaviruses. Nature Reviews Microbiology, 14(8), 523–534. 10.1038/nrmicro.2016.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue, M. , Hsieh, F. , Baronas, E. , Godbout, K. , Gosselin, M. , Stagliano, N. , … Acton, S. (2000). A novel angiotensin‐converting enzyme‐related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1–9. Circulation Research, 87(5), E1–E9. 10.1161/01.res.87.5.e1 [DOI] [PubMed] [Google Scholar]
- Ferrario, C. M. , Trask, A. J. , & Jessup, J. A. (2005). Advances in biochemical and functional roles of angiotensin‐converting enzyme 2 and angiotensin‐(1–7) in regulation of cardiovascular function. American Journal of Physiology. Heart and Circulatory Physiology, 289(6), H2281–2290. 10.1152/ajpheart.00618.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, X. Y. , Li, J. L. , Yang, X. L. , Chmura, A. A. , Zhu, G. , Epstein, J. H. , … Shi, Z. L. (2013). Isolation and characterization of a bat SARS‐like coronavirus that uses the ACE2 receptor. Nature, 503(7477), 535–538. 10.1038/nature12711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, Y. , Zheng, B. J. , He, Y. Q. , Liu, X. L. , Zhuang, Z. X. , Cheung, C. L. , … Poon, L. L. (2003). Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science, 302(5643), 276–278. 10.1126/science.1087139 [DOI] [PubMed] [Google Scholar]
- Hoffmann, M. , Kleine‐Weber, H. , Schroeder, S. , Kruger, N. , Herrler, T. , Erichsen, S. , … Pohlmann, S. (2020). SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell, 181(2), 271–280.e8. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon, C. C. , Lam, T. Y. , Shi, Z. L. , Drummond, A. J. , Yip, C. W. , Zeng, F. , … Leung, F. C. (2008). Evidence of the recombinant origin of a bat severe acute respiratory syndrome (SARS)‐like coronavirus and its implications on the direct ancestor of SARS coronavirus. Journal of Virology, 82(4), 1819–1826. 10.1128/JVI.01926-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, T.‐Y. , Jia, N. A. , Zhang, Y.‐W. , Shum, M.‐H. , Jiang, J.‐F. , Zhu, H.‐C. , … Cao, W.‐C. (2020). Identifying SARS‐CoV‐2 related coronaviruses in Malayan pangolins. Nature, 583(7815), 282–285. 10.1038/s41586-020-2169-0 [DOI] [PubMed] [Google Scholar]
- Li, W. , Zhang, C. , Sui, J. , Kuhn, J. H. , Moore, M. J. , Luo, S. , … Farzan, M. (2005). Receptor and viral determinants of SARS‐coronavirus adaptation to human ACE2. EMBO Journal, 24(8), 1634–1643. 10.1038/sj.emboj.7600640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, R. , Zhao, X. , Li, J. , Niu, P. , Yang, B. , Wu, H. , … Tan, W. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet, 395(10224), 565–574. 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meo, S. A. , Alhowikan, A. M. , Al‐Khlaiwi, T. , Meo, I. M. , Halepoto, D. M. , Iqbal, M. , … Ahmed, N. (2020). Novel coronavirus 2019‐nCoV: Prevalence, biological and clinical characteristics comparison with SARS‐CoV and MERS‐CoV. European Review for Medical and Pharmacological Sciences, 24(4), 2012–2019. 10.26355/eurrev_202002_20379 [DOI] [PubMed] [Google Scholar]
- Parry, J. (2003). WHO warns that death rate from SARS could reach 10%. BMJ, 326(7397), 999. 10.1136/bmj.326.7397.999/a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, Y. , Zhao, Y. B. , Wang, Q. , Li, J. Y. , Zhou, Z. J. , Liao, C. H. , & Ge, X. Y. (2020). Predicting the angiotensin converting enzyme 2 (ACE2) utilizing capability as the receptor of SARS‐CoV‐2. Microbes and Infection, 22(4–5), 221–225. 10.1016/j.micinf.2020.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, W. , Qu, X. , Li, W. , Han, Z. , Yu, M. , Zhou, P. , … Shi, Z. (2008). Difference in receptor usage between severe acute respiratory syndrome (SARS) coronavirus and SARS‐like coronavirus of bat origin. Journal of Virology, 82(4), 1899–1907. 10.1128/JVI.01085-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang, J. , Wan, Y. , Luo, C. , Ye, G. , Geng, Q. , Auerbach, A. , & Li, F. (2020). Cell entry mechanisms of SARS‐CoV‐2. Proceedings of the National Academy of Sciences, 117(21), 11727–11734. 10.1073/pnas.2003138117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang, J. , Ye, G. , Shi, K. , Wan, Y. , Luo, C. , Aihara, H. , … Li, F. (2020). Structural basis of receptor recognition by SARS‐CoV‐2. Nature, 581(7807), 221–224. 10.1038/s41586-020-2179-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J. , Wen, Z. , Zhong, G. , Yang, H. , Wang, C. , Huang, B. , … Bu, Z. (2020). Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS‐coronavirus 2. Science, 368(6494), 1016–1020. 10.1126/science.abb7015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler, K. , Masignani, V. , Eickmann, M. , Becker, S. , Abrignani, S. , Klenk, H. D. , & Rappuoli, R. (2003). SARS–beginning to understand a new virus. Nature Reviews Microbiology, 1(3), 209–218. 10.1038/nrmicro775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, Y. , Shang, J. , Graham, R. , Baric, R. S. , & Li, F. (2020). Receptor recognition by novel coronavirus from Wuhan: An analysis based on decade‐long structural studies of SARS. Journal of Virology, 94(7), e00127‐20. 10.1128/JVI.00127-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q. , Qiu, Y. , Li, J. Y. , Zhou, Z. J. , Liao, C. H. , & Ge, X. Y. (2020). A unique protease cleavage site predicted in the spike protein of the novel pneumonia coronavirus (2019‐nCoV) potentially related to viral transmissibility. Virologica Sinica, 35(3), 337–339. 10.1007/s12250-020-00212-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, F. , Zhao, S. , Yu, B. , Chen, Y. M. , Wang, W. , Song, Z. G. , … Zhang, Y. Z. (2020). A new coronavirus associated with human respiratory disease in China. Nature, 579(7798), 265–269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, X. , Chakraborti, S. , Dimitrov, A. S. , Gramatikoff, K. , & Dimitrov, D. S. (2003). The SARS‐CoV S glycoprotein: Expression and functional characterization. Biochemical and Biophysical Research Communications, 312(4), 1159–1164. 10.1016/j.bbrc.2003.11.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y. , Du, L. , Liu, C. , Wang, L. , Ma, C. , Tang, J. , … Li, F. (2014). Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat‐to‐human transmission of MERS coronavirus. Proceedings of the National Academy of Sciences, 111(34), 12516–12521. 10.1073/pnas.1405889111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, T. , Wu, Q. , & Zhang, Z. (2020). Probable pangolin origin of SARS‐CoV‐2 associated with the COVID‐19 outbreak. Current Biology, 30(7), 1346–1351.e2. 10.1016/j.cub.2020.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, P. , Yang, X. L. , Wang, X. G. , Hu, B. , Zhang, L. , Zhang, W. , … Shi, Z. L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270–273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, N. , Zhang, D. , Wang, W. , Li, X. , Yang, B. , Song, J. , … Tan, W. ; China Novel Coronavirus Investigating and Research Team . (2020). A Novel Coronavirus from Patients with Pneumonia in China, 2019. New England Journal of Medicine, 382(8), 727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


