Abstract
Objectives
To design and evaluate 3D‐printed nasal swabs for collection of samples for SARS‐CoV‐2 testing.
Design
An iterative design process was employed. Laboratory evaluation included in vitro assessment of mock nasopharyngeal samples spiked with two different concentrations of gamma‐irradiated SARS‐CoV‐2. A prospective clinical study compared SARS‐CoV‐2 and human cellular material recovery by 3D‐printed swabs and standard nasopharyngeal swabs.
Setting, participants
Royal Melbourne Hospital, May 2020. Participants in the clinical evaluation were 50 hospital staff members attending a COVID‐19 screening clinic and two inpatients with laboratory‐confirmed COVID‐19.
Intervention
In the clinical evaluation, a flocked nasopharyngeal swab sample was collected with the Copan ESwab and a mid‐nasal sample from the other nostril was collected with the 3D‐printed swab.
Results
In the laboratory evaluation, qualitative agreement with regard to SARS‐CoV‐2 detection in mock samples collected with 3D‐printed swabs and two standard swabs was complete. In the clinical evaluation, qualitative agreement with regard to RNase P detection (a surrogate measure of adequate collection of human cellular material) in samples collected from 50 hospital staff members with standard and 3D‐printed swabs was complete. Qualitative agreement with regard to SARS‐CoV‐2 detection in three pairs of 3D‐printed mid‐nasal and standard swab samples from two inpatients with laboratory‐confirmed SARS‐CoV‐2 was also complete.
Conclusions
Using 3D‐printed swabs to collect nasal samples for SARS‐CoV‐2 testing is feasible, acceptable to patients and health carers, and convenient.
Keywords: COVID‐19, Microbiology, Infectious diseases, Respiratory tract infections
The known: The surge in laboratory SARS‐CoV‐2 testing has caused an acute global shortage of nasal swabs. Innovative approaches to sustaining diagnostic testing capacity are required. 3D‐printed medical devices may provide a scalable, local solution to this problem.
The new: We describe the design and evaluation of 3D‐printed swabs manufactured in Australia. SARS‐CoV‐2 recovery in vitro was similar for 3D‐printed swabs and two commercially available swabs. In a clinical validation study, 3D‐printed swabs captured the same quantity of human cellular material as normal swabs.
The implications: 3D‐printed swabs provide one solution to the shortage of nasal swabs needed for SARS‐CoV‐2 testing.
The pandemic of coronavirus disease 2019 (COVID‐19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), is a global public health emergency. Diagnostic testing is critical for detecting cases of COVID‐19 and, ultimately, for reducing viral transmission.1, 2 However, the unprecedented surge in laboratory testing has placed extraordinary demands on health systems, causing global imbalances in supply and demand for laboratory consumables.3 Diagnostic testing for SARS‐CoV‐2 chiefly employs the conventional paradigm of sample collection, nucleic acid extraction, and detection of viral RNA by reverse transcriptase polymerase chain reaction (RT‐PCR).4 The Australian Public Health Laboratory Network (PHLN) guidelines recommend combined deep nasal and oropharyngeal swabbing as the preferred method for collecting diagnostic specimens for SARS‐CoV‐2 RT‐PCR.5 However, the shortage of appropriate swabs for diagnostic sampling is a major bottleneck that restricts testing capacity.6 In addition, international travel and transport restrictions in response to COVID‐19 further reduce access by geographically remote countries such as Australia to swabs manufactured overseas.
Three‐dimensional (3D) printing is the construction of physical objects from three‐dimensional renderings with a printer. The use of 3D‐printed medical devices in a range of applications has increased over the past decade, including uses as custom implants and moulds for prosthetic devices.7 In this article we describe the design and laboratory and clinical evaluation of a locally manufactured 3D‐printed swab. Our work highlights one possible solution to the extreme shortage of swabs, and illustrates the value of rapid partnerships between industry, academia, and clinicians in a public health crisis.
Methods
Swab design and manufacture
We employed an iterative design process that incorporated feedback from clinical and engineering researchers. Based on initial specifications published by investigators in the United States,8 four initial prototype designs were printed. Printing was undertaken using selective laser sintering (SLS) technology (feature resolution, 80 μm) and PA2200 medical grade biocompatible nylon 12 as source material. In subsequent design iterations, we:
optimised tip geometry for maximum cell collection;
improved shaft geometry for flexibility, clinical safety (breakpoint position), and ease of use; and
ensured overall design parameters were compatible with patient comfort.
The final design (design G) is depicted in the Box, and the specifications are included in the online Supporting Information; the standard triangle language (STL) file is available on request (https://www.3dmeditech.com/contact-us), as are the full design specifications. Swabs were autoclaved and individually packaged in the Central Sterile Supply Department of the Royal Melbourne Hospital before use.
Box 1. Design G 3D‐printed swab. A. Side profile. B. Flexibility of upper part of the swab shaft. C. Breakpoint of swab. D. Microscope image of swab head.
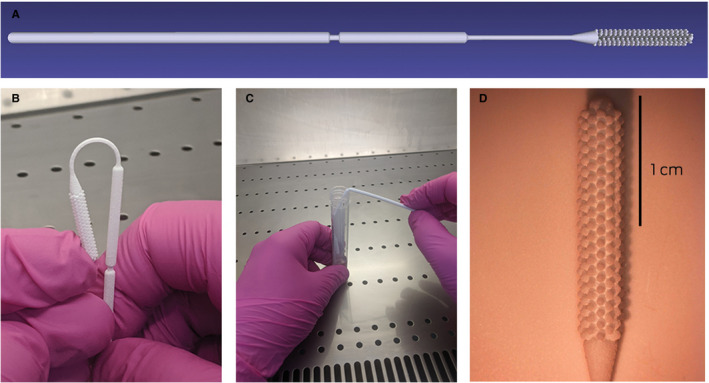
In vitro validation study
We assessed the recovery of SARS‐CoV‐2 from different transport media using the design G swabs, and compared their ability to recover SARS‐CoV‐2 with that of two swabs currently used in Australia to collect specimens for SARS‐CoV‐2 testing. We assessed the following swab/transport medium combinations:
flocked Copan ESwabs and liquid Amies medium (Copan; catalogue number 480CE);
flocked Kang Jian swabs and viral transport medium (Kang Jian Medical; catalogue number KJ502‐19);
design G 3D‐printed swabs and viral transport medium (University of Melbourne Media Preparation Unit; product number 2512);
design G swabs and liquid Amies medium (University of Melbourne Media Preparation Unit; product number 2162); and
design G swabs and normal saline.
Virus stock (SARS‐CoV‐2 strain VIC001;9 circa 107 copies/mL) was prepared in minimum essential medium (Sigma) containing 2% fetal bovine serum (Gibco), and gamma‐irradiated to allow subsequent handling in PC2 (physical containment level 2) conditions.
Our mock sample matrix of nasopharyngeal swabs consisted of 20 mL pooled nasopharyngeal swab samples collected in liquid Amies medium from patients negative for SARS‐CoV‐2 by RT‐PCR; 10 mL aliquots of the pooled sample were spiked with gamma‐irradiated SARS‐CoV‐2 to produce two different viral concentrations (16 or 160 plaque‐forming unit [PFU] equivalents/mL). Two swabs for each swab/medium combination listed above were swizzled in 500 μL aliquots of spiked mock sample at each concentration for five seconds, then immediately placed into 2 mL of the corresponding transport medium (liquid Amies, viral transport medium, or normal saline). Samples were tested for SARS‐CoV‐2 by RT‐PCR at time zero, 24 hours, and 48 hours; between assays, samples were stored at 4°C. Viral RNA was extracted with the QIAamp 96 virus QIAcube HT kit (QIAGEN); RT‐PCR for the envelope protein (E) gene was performed with previously published primers and probes.10
Clinical evaluation
We compared the performance and tolerability of 3D‐printed swabs with those of standard swabs used in our institution (Copan ESwabs). The participants were hospital staff members attending a COVID‐19 screening clinic at the Royal Melbourne Hospital during 1–5 May and inpatients with laboratory‐confirmed COVID‐19 during 1–18 May 2020. Participants received a study information sheet and provided verbal consent. A flocked nasopharyngeal swab sample was collected with the Copan ESwab and a mid‐nasal sample from the other nostril with the 3D‐printed swab; each swab was placed into 1 mL liquid Amies transport medium. The order of collection was randomised 1:1. Participants were asked to complete a brief survey about their level of discomfort with each swab.
Nucleic acid was extracted and SARS‐CoV‐2 RT‐PCR performed as described above. In addition, samples from inpatients with laboratory‐confirmed COVID‐19 were assessed for SARS‐CoV‐2 by RT‐PCR using the Xpert Xpress SARS‐CoV‐2 assay (Cepheid) according to the manufacturers’ instructions.11 Semi‐quantitative real time RT‐PCR detection of a human housekeeping gene (RNase P) served as a surrogate marker for the amount of cellular material derived from each swab.12
Statistical analysis
Differences between samples collected with control and 3D‐printed swabs in RNAse P cycle threshold (Ct) values (a lower Ct value indicates fewer PCR cycles were required for detection, and therefore higher target concentrations) were assessed in a Wilcoxon matched pairs rank test undertaken in R 3.5.1; plots were prepared with GraphPad Prism 8.4.2.
Ethics approval
Ethics approval for this project was provided by the Melbourne Health Human Research Ethics Committee (reference, RMH HREC QA2020059).
Results
In vitro performance
Qualitative agreement with respect to detecting SARS‐CoV‐2 using Copan ESwabs, Kang Jian swabs, and design G 3D‐printed swabs in the three transport media was complete; SARS‐CoV‐2 was detected at both concentrations, at all three time points, and with each swab/medium combination (Supporting Information, table 1). Differences between the three swab types in Ct values for E gene detection at each concentration and each of the three time points were negligible (Supporting Information, figures 1A,B). In addition, Ct values for E gene detection using the 3D‐printed swab were similar for each of the three transport media (Supporting Information, figures 1C,D).
Clinical evaluation and acceptability
SARS‐CoV‐2 E gene was not detected by RT‐PCR in nasal swabs (one nostril each with a Copan ESwab and a design G 3D‐printed swab) collected from 50 hospital staff who attended a COVID‐19 screening clinic at Royal Melbourne Hospital during 1–5 May 2020. Qualitative agreement between the Copan ESwab and 3D‐printed swab for RNase P detection by RT‐PCR was complete, and the distribution of Ct values for the two swab types were similar (ESwabs: median, 27.2; interquartile range [IQR], 26.4–29.0; design G 3D swabs: median, 27.1; IQR, 25.7–28.3; Supporting Information, figure 2).
Three paired swabs were collected from two patients with laboratory‐confirmed COVID‐19 admitted to Royal Melbourne Hospital between 1 May and 18 May. Qualitative agreement between the Copan ESwab and design G 3D‐printed swab for SARS‐CoV‐2 E gene detection by RT‐PCR and with the Xpert Xpress SARS‐CoV‐2 assay was complete (Supporting Information, table 2).
The 52 study participants scored the discomfort experienced with the 3D‐printed swab (median, 5 points on a 10‐point scale; IQR, 4–6 points) and the Copan ESwab (median, 5 points; IQR, 3–6) similarly; 35 participants (67%) preferred the 3D‐printed swab, ten (19%) the Copan ESwab, and eight had no preference (15%). Health care providers described the swabs as easy to use, moderately easy to snap at the breakpoint, and as providing a good balance between flexibility and rigidity. Two of the four health care providers involved preferred the 3D‐printed swab, and two had no preference.
Discussion
Given the critical shortage of laboratory consumables required for SARS‐CoV‐2 testing, innovative solutions are required to sustain diagnostic capacity. We have reported the feasibility, acceptability, and utility of 3D‐printed swabs for collecting samples for SARS‐CoV‐2 testing. We also report that 3D‐printed swabs can be combined with various transport media (including widely available normal saline) without markedly affecting SARS‐CoV‐2 detection by RT‐PCR.
The widespread availability of 3D‐printing technology may enable many countries to ensure local swab supplies, and its scalability means that thousands of swabs can be produced each day. This may provide local manufacturing solutions to swab shortages in an unpredictable international market for both high and low income countries. Further, the nylon 2 used for swab production can be autoclaved, ensuring a sterile swab supply. We have included with this article the specifications for our final design so that other groups can adopt and modify our design.
Our work builds on recent work in the United States, where evaluation of four 3D‐printed swab prototypes found no differences between control and 3D‐printed swabs with respect to the detection of SARS‐CoV‐2.8 The authors of the US study also employed an iterative design process, with close collaboration between academic, clinical, and industrial partners, but health care providers and participants both preferred standard swabs to the 3D‐printed swabs. This divergence from our finding may reflect differences in sampling strategies; we used nasopharyngeal sampling as the default collection method (consistent with PHLN guidelines at the time of the study),13 whereas the 3D‐printed swab was used for mid‐nasal swabs. That is, the default method was more invasive than that used with the 3D‐printed swab.
The urgent need for laboratory consumables for SARS‐CoV‐2 testing has catalysed the development of novel approaches to diagnostic testing. The compatibility of 3D‐printed swabs with the wide range of available commercial and in‐house SARS‐CoV‐2 RT‐PCR platforms must be investigated. Moreover, 3D‐printed swabs could also be used in the diagnosis of other common upper respiratory tract pathogens, including influenza virus, respiratory syncytial virus, and Streptococcus pyogenes.
Competing interests
No relevant disclosures.
Supporting information
Design specifications for the prototype G swab; 2. Supplementary tables and figures
Appendix
Acknowledgements
Deborah Williamson is supported by an Investigator Grant from the National Health and Medical Research Council (NHMRC) (APP1174555). Benjamin Howden is supported by an NHMRC Practitioner Fellowship (APP1105905). Katherine Bond is supported by an NHMRC Postgraduate Scholarship (GNT1191321). Our work was supported by a grant from the NHMRC Medical Research Future Fund (APP2002317). 3DMEDitech had no role in the design or conduct of the laboratory or clinical study.
We thank the staff and patients who contributed to this investigation. We also thank the staff in the Department of Microbiology at the Royal Melbourne Hospital, as well as Julie McAuley from the University of Melbourne Department of Microbiology and Immunology for technical assistance with SARS‐CoV‐2 viral quantification for the in vitro study.
References
- 1. Wang CJ, Ng CY, Brook RH. Response to COVID‐19 in Taiwan: big data analytics, new technology, and proactive testing. JAMA 2020; 323: 1341–1342. [DOI] [PubMed] [Google Scholar]
- 2. Lee VJ, Chiew CJ, Khong WX. Interrupting transmission of COVID‐19: lessons from containment efforts in Singapore. J Travel Med 2020; 27: taaa039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sharfstein JM, Becker SJ, Mello MM. Diagnostic testing for the novel coronavirus. JAMA 2020; 323: 1437–1438. [DOI] [PubMed] [Google Scholar]
- 4. Hadaya J, Schumm M, Livingston EH. Testing individuals for coronavirus disease 2019 (COVID‐19). JAMA 2020; 323: 1981. [DOI] [PubMed] [Google Scholar]
- 5. Public Health Laboratory Network . PHLN guidance on laboratory testing for SARS‐CoV‐2 (the virus that causes COVID‐19), version 1.11. Updated 16 June 2020. https://www.health.gov.au/resources/publications/phln-guidance-on-laboratory-testing-for-sars-cov-2-the-virus-that-causes-covid-19 (viewed June 2020).
- 6. Vermeiren C, Marchand‐Senécal X, Sheldrake E, et al. Comparison of Copan Eswab and FLOQswab for COVID‐19 PCR diagnosis: working around a supply shortage. J Clin Microbiol 2020; 58: e00669–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tack P, Victor J, Gemmel P, et al. 3D‐printing techniques in a medical setting: a systematic literature review. Biomed Eng Online 2016; 15: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Callahan CJ, Lee R, Zulauf KE, et al. Open development and clinical validation of multiple 3D‐printed nasopharyngeal collection swabs: rapid resolution of a critical COVID‐19 testing bottleneck. J Clin Microbiol 2020; 58: e00876‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Caly L, Druce J, Roberts J, et al. Isolation and rapid sharing of the 2019 novel coronavirus (SARS‐CoV-2) from the first patient diagnosed with COVID‐19 in Australia. Med J Aust 2020; 212: 459–462. https://www.mja.com.au/journal/2020/212/10/isolation-and-rapid-sharing-2019-novel-coronavirus-sars-cov-2-first-patient [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019‐nCoV) by real‐time RT‐PCR. Euro Surveill 2020; 25: 2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wolters F, van de Bovenkamp J, van den Bosch B, et al. Multi‐center evaluation of Cepheid Xpert® Xpress SARS‐CoV‐2 point‐of-care test during the SARS‐CoV‐2 pandemic. J Clin Virol 2020; 128: 104426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boddicker JD, Rota PA, Kreman T, et al. Real‐time reverse transcription‐PCR assay for detection of mumps virus RNA in clinical specimens. J Clin Microbiol 2007; 45: 2902–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Public Health Laboratory Network . PHLN guidance on laboratory testing for SARS‐CoV‐2 (the virus that causes COVID‐19), version 1.7. Updated 28 April 2020. https://www.health.gov.au/resources/publications/phln-guidance-on-laboratory-testing-for-sars-cov-2-the-virus-that-causes-covid-19 (viewed Apr 2020).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Design specifications for the prototype G swab; 2. Supplementary tables and figures
Appendix


