Abstract
Human coronaviruses (HCoVs) such as HCoV‐229E or OC43 are responsible for mild upper airway infections, whereas highly pathogenic HCoVs, including SARS‐CoV, MERS‐CoV and SARS‐CoV‐2, often evoke acute, heavy pneumonias. They tend to induce immune responses based on interferon and host inflammatory cytokine production and promotion of T1 immune profile. Less is known about their effect on T2‐type immunity. Unlike human rhinoviruses (HRV) and Respiratory Syncytial Virus (RSV), HCoVs are not considered as a dominant risk factor of severe exacerbations of asthma, mostly T2‐type chronic inflammatory disease. The relationship between coronaviruses and T2‐type immunity, especially in asthma and allergy, is not well understood. This review aims to summarize currently available knowledge about the relationship of HCoVs, including novel SARS‐CoV‐2, with asthma and allergic inflammation.
Keywords: asthma, coronavirus, COVID‐19, SARS‐CoV‐2, Th2 response
1. INTRODUCTION
Coronaviruses are a group of related viruses that infect birds and mammals. Seven strains are known that cause disease in humans. Low pathogenic HCoVs: 229E, OC43, NL63 and HKU1 commonly circulate in the environment and usually evoke mild, self‐limiting upper airway infections, such as common cold. 1 Two of 3 novel highly pathogenic HCoVs – SARS‐CoV (Severe Acute Respiratory Syndrome‐CoV), described in Guangdong, China, in 2002, and MERS‐CoV (Middle East respiratory syndrome‐CoV) found in Saudi Arabia, in 2012, more often tend to invade lower respiratory tracts. Both viruses were blamed for epidemics of pneumonia and severe acute respiratory distress, with case fatality rate reaching 11% and 36%, for SARS‐CoV and MERS‐CoV, respectively. 1 , 2 , 3 The last one SARS‐CoV‐2 was identified recently in 2019 in Wuhan, Hubei province in China, and is being responsible for currently ongoing pandemic of coronavirus‐induced disease‐19 (COVID‐19). 4 Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU) reported 7 394 801 COVID‐19 cases caused by SARS‐CoV‐2 world‐wide since December 2019 until 10 June 2020. 5 Although the mortality for SARS‐CoV‐2 infection is lower, as it reaches 5.2%, the new coronavirus seems to be more contagious and is characterized by higher morbidity. 5
The aim of this review is to summarize currently available knowledge about the relationship of HCoVs, including novel SARS‐CoV‐2, with T2‐type immunity, asthma and allergic inflammation.
2. HUMAN CORONAVIRUSES INDUCE MODERATE INTERFERON AND ROBUST INFLAMMATORY CYTOKINE PRODUCTION
HCoVs are positive‐sense, single‐stranded RNA viruses belonging to a subfamily of the Coronaviridae. 1 Like many other respiratory RNA viruses, including human rhinovirus (HRV), Respiratory Syncytial Virus (RSV) or influenza virus, they elicit universal innate immune response in human airways, targeting primarily epithelium followed by the attachment to cellular receptors by surface spike proteins. Upon internalization to the cytoplasm, viral RNA is sensed by innate immune pattern recognition receptors (PRRs), such as membranous Toll‐like receptor 3 (TLR3), cytosolic retinoic‐acid inducible gene I (RIGI) and melanoma differentiation‐associated protein 5 (MDA‐5), leading to interferon regulatory factor 3 (IRF3) and IRF7 activation. Subsequent induction of type I (IFN‐α, IFN‐β) and type II (IFN‐γ) interferon synthesis results in an activation of immune antiviral effectors such as T cytotoxic lymphocytes (Tc), natural killer cells (NK) and macrophages, allowing viral clearance. 2 Parallel activation of NF‐κB results in a range of inflammatory mediator production.
Activation of inflammatory and antiviral responses strongly depends on HCoVs pathogenicity. Highly pathogenic HCoVs are more likely to cause immune dysregulation, encompassing a robust inflammatory cytokine and chemokine release, called the cytokine storm, which may be accompanied by the low interferon‐related response that consequently enables virus to evade the immune system 6 (Figure 1). Indeed, in severe patients infected with SARS‐CoV and MERS‐CoV, highly elevated concentrations of TNF‐α, IL‐6, IL‐1, IP‐10, MCP‐1 and IL‐8 have been observed. 6 , 7 , 8 , 9 HCoVs‐infected monocytes, macrophages, dendritic cells (DCs) and T cells may also produce inflammatory cytokines and chemokines. 10 , 11 , 12 , 13 Enhanced serum cytokine levels correlate with increased neutrophil and monocyte numbers in peripheral blood compared to individuals with mild to moderate disease. Extensive lung infiltration with neutrophils and macrophages, necrosis of bronchiolar epithelium, diffuse alveolar damage and oedema lead to acute respiratory distress syndrome (ARDS). 6 , 14
Figure 1.
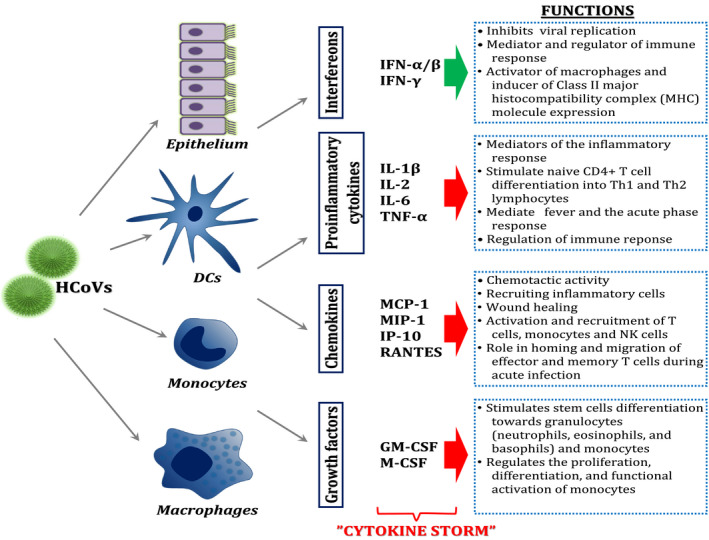
Host inflammatory cytokine immune response to HCoVs infections. Symbol explanation: green arrow, desirable antiviral effect; red arrow, immediate and excessive secretion of cytokines
Similarly to SARS‐CoV and MERS‐CoV, novel highly pathogenic SARS‐CoV‐2 has been recently shown to evoke a cytokine storm, encompassing high IP‐10, MCP‐1, MIP‐1α, M‐CSF, G‐CSF, TNF‐α, IL‐1, IL‐2, IL‐7 levels in plasma and bronchoalveolar lavage fluid of patients requiring admission to the intensive care unit and in those with severe or lethal COVID‐19. 4 , 15 , 16 , 17 , 18 , 19
3. CORONAVIRUS INFECTION, T‐TYPE IMMUNITY AND ASTHMA EXACERBATIONS
Next to a wide range of host inflammatory cytokines, markedly increased T1‐type cytokines: IFN‐γ, IL‐12 and IL‐17 concentrations were observed in severe SARS‐CoV and MERS‐CoV‐infected patients. 7 , 9 , 20 , 21 , 22 Virus‐specific Th cells of these individuals could produce large amounts of IFN‐γ. This Th1 polarization induced by the virus might persist, as specific memory T lymphocytes were shown to produce IFN‐γ several years after infection. 23 Besides, SARS‐CoV and MERS‐CoV were shown to induce IFN‐γ and IL‐12 production in DCs and monocytes/macrophages. 10 , 11 , 12 , 24 These T1‐type cytokines orchestrate cell‐mediated antiviral immune responses. Besides, they promote polarization of naive T cells towards Th1‐profile and the development T cytotoxic cells (Tc). 25 , 26 On the other hand, serum levels of T2‐type cytokines: IL‐4, IL‐5 and IL‐13 were unchanged in the above mentioned SARS and MERS patients or were slightly decreased (IL‐4), also in disease convalescent individuals 7 , 9 , 21 , 22 (Figure 2). In patients with the novel COVID19, apart from host inflammatory cytokine robust release, the increased IFN‐γ and IL‐12 serum concentrations and T17‐related responses were also observed. 17 , 19 , 27 Additionally, Huang et al and Liu et al have recently noted the surprising concomitant elevation of serum IL‐4, which might suggest the different features of SARS‐CoV‐2 with regard to T2‐profile induction from other HCoVs. 17 , 27 However, Wen at al. did not note any increase of IL‐4 during the course of infection. 28 Severe COVID‐19 individuals had lymphopenia and low Th cell numbers. 29 , 30 One should underline that instances of immunological abnormalities caused by coronaviruses mentioned above may depend on the strength of viral pathogenicity, severity of the infection and failure of the immunity.
Figure 2.
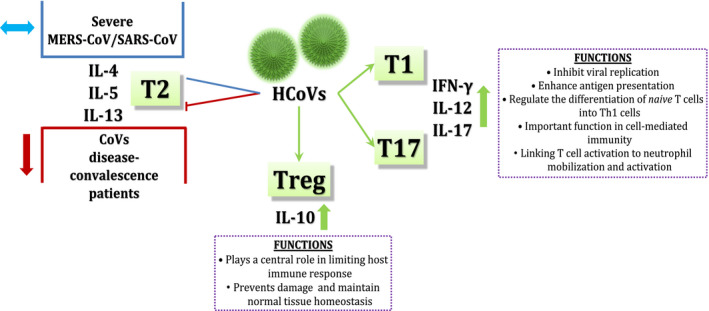
Different types of immune response in HCoVs infections. Symbol explanation: ⬆, increase; ⬇, decrease; ⬌, constants; ├, blocking; ↘, promoting
Interestingly, other RNA respiratory viruses, including human rhinovirus (HRV) and respiratory syncytial virus (RSV), tend to induce T2‐related immune responses, especially in allergic patients, as they facilitate IL‐33 and IL‐25 epithelial production and favour mucosal innate lymphoid cells 2 (ILC2) and Th2 cell predominance providing high amounts of IL‐4 and IL‐5, thus leading to eosinophil attraction into the lungs and triggering exacerbations in majority of asthma patients. 31 , 32 , 33 , 34 , 35 , 36
Lack of significant activation of T2‐type immunity by HCoVs suggests that these viruses may not have particular capacity to enhance asthmatic T2‐like inflammation as compared to other several respiratory viruses. Indeed, unlike HRV or RSV, low pathogenic HCoVs, SARS‐CoV and MERS‐CoV are not considered as significant risk factors for the development of asthma exacerbations. Meta‐analysis of 63 studies from 1971 to 2017 revealed that HCoVs, mostly 229E, OC43 and NL63, were related on average to 8.4% of asthma flares, which depended on age, type of respiratory infection and region: in Europe—9.6%, Asia—5.4%, America—14.2% and Oceania—1.8%. 37 In West Indies, only in 1.3% of children with all asthma exacerbations, HCoV‐OC43 was detected, whereas HRV and RSV were most prevalent. 38 In Leung's et al study, HCoV‐229E or OC43 were observed only in 4.8% of all viral asthma exacerbations in children living in Hong Kong, China. 39 Virus carriers, including asthmatics, may, however, be asymptomatic. Stable asthmatic individuals were shown to more likely carry respiratory viruses, including HCoV (8.3%) and HRV (25%), without any symptoms. Furthermore, these viruses were also found in up to 18% of healthy individuals. 40
Based on above‐mentioned results in cohorts of patients with airway infections with low pathogenic HCoVs or severe SARS‐CoV or MERS‐CoV, individuals with asthma and/or allergy are not noted or they comprise very small numbers. T cell profiles of immune response in HCoVs‐related asthma exacerbated patients were not intensively investigated.
4. ALLERGIC INFLAMMATION AND SUSCEPTIBILITY TO CORONAVIRUS INFECTIONS
What is now worth paying particular attention is that according to preliminary epidemiological data, asthma and allergies do not seem to be a risk factor for SARS‐CoV‐2 infection. 41 Existing studies have not shown an expected prevalence of asthmatic individuals among COVID‐19 patients. 42 A hypothesis has recently appeared that asthma or allergy might surprisingly decrease the risk of SARS‐CoV‐2 infection, which tends to be associated with the decreased airway epithelial angiotensin‐converting enzyme 2 (ACE‐2) expression in allergic asthma patients as compared to healthy individuals. 43 , 44 ACE‐2 is used by SARS‐CoV‐2 as a entry receptor. Moreover, allergen provocation of the upper and lower respiratory tract inducing allergic airway inflammation was shown to lead to a significant decrease in ACE‐2 expression, suggesting possible relationship between allergic inflammation and assumed lower susceptibility to SARS‐CoV‐2 infection. 44 In addition, vast majority of patients with lethal disease outcome were noted to have low numbers of peripheral blood eosinophils, which are predictor cells of T2‐type immune activity. 43 The key T2 cytokine which is presumed to be responsible for deep ACE‐2 down‐regulation is IL‐13. 44 These data suggest that certain aspects of type 2 immune response and accumulation of eosinophils might provide potential protective effects against COVID‐19. 42 However, another study did not find any differences in ACE‐2 expression in sputum cells between asthma and healthy individuals. 45 This work did not, however, distinguish between allergic and non‐allergic asthma background.
What may have a clinical relevance, inhaled glucocorticosteroids (GCS) intake, with an exception of triamcinolone and nasal corticosteroids, has been shown to be associated with reduced expression of both ACE2 and TMPRSS2 protease. 44 , 45 One should also consider a significance of a biological treatments of asthma and other allergic diseases in susceptibility to CARS‐CoV‐2 infection. Primary data show that only 2 of 245 patients with atopic dermatitis treated with anti‐IL‐4/13 therapy developed COVID‐19. 46 However, the effect of other anti‐T2 immune response biological is, yet, unknown.
As far as other HCoVs are concerned, regulation of their entry receptor expression by T2‐type cytokines appears to be different. Research results show that IL‐4, IL‐13, but also IFN‐γ—the representative of T1‐immunity—may increase expression of both aminopeptidase N (APN/CD13, a receptor for HCoV‐229E), and dipeptidyl‐peptidase IV (DPPIV/CD26, a receptor for MERS‐CoV). 47 , 48 , 49 , 50 Moreover, contrary to the inhaled GCS, systemic GCS, including hydrocortisone and dexamethasone, tested, to note, several years ago, were shown to up‐regulate APN/CD13, but to reduce DPPIV/CD26 expression. 49 No effect of other inflammatory cytokines IL‐1, IL‐2, IL‐6, IL‐7, IL‐12, IL‐15 on APN/CD13 and DPPIV/CD26 was observed. 48 , 49 , 51 Finally, both TGF‐β and TNF‐α reduced DPPIV/CD26 expression, but did not affect APN/CD13. 48 However, not all studies confirm these observations. 50 The research in this field was mostly conducted many years ago and only one concerned epithelial cells (B2Bs), whereas the others were based mainly on fibroblasts and immortalized cell lines, which actually makes their interpretation in susceptibility virus infection difficult.
5. SUMMARY
The relationship between coronaviruses and T2‐type immunity, especially in asthma and allergy, is not well understood and still poses unresolved dilemma. Considering limited and not coherent data, the establishment of the effect of HCoVs on T2‐type immunity and the role of T‐type profiles in determining susceptibility to coronavirus infection and asthma exacerbations needs intense investigation. The threat of inevitable exposure of human population to HCoVs world‐wide, including SARS‐CoV‐2, in up‐coming years should, therefore, mobilize the scientific society to take measures to research HCoVs, in the field of asthma and allergy.
Supporting information
Supplementary Material
Chałubiński M, Gajewski A, Kowalski ML. The relationship between human coronaviruses, asthma and allergy—An unresolved dilemma. Clin Exp Allergy. 2020;50:1122–1126. 10.1111/cea.13718
REFERENCES
- 1. Jonsdottir HR, Dijkman R. Coronaviruses and the human airway: a universal system for virus‐host interaction studies. Virol J. 2016;13:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Greenberg SB. Update on human rhinovirus and coronavirus infections. Semin Respir Crit Care Med. 2016;37:555‐571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. WHO . Middle East respiratory syndrome coronavirus (MERS CoV). https://www.who.int/emergencies/mers‐cov/en/ 2017.
- 4. Guo YR, Cao QD, Hong ZS, et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID‐19) outbreak ‐ an update on the status. Mil Med Res. 2020;7:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. "Coronavirus COVID‐19 Global Cases by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU)" ArcGIS. Johns Hopkins CSSE. https://gisanddata.maps.arcgis.com/apps/opsdashboard/index.html#/bda7594740fd40299423467b48e9ecf6 2020.
- 6. Channappanavar R, Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol. 2017;39:529‐539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mahallawi WH, Khabour OF, Zhang Q, Makhdoum HM, Suliman BA. MERS‐CoV infection in humans is associated with a pro‐inflammatory Th1 and Th17 cytokine profile. Cytokine. 2018;104:8‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Faure E, Poissy J, Goffard A, et al. Distinct immune response in two MERS‐CoV‐infected patients: can we go from bench to bedside? PLoS One. 2014;9:e88716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chien JY, Hsueh PR, Cheng WC, Yu CJ, Yang PC. Temporal changes in cytokine/chemokine profiles and pulmonary involvement in severe acute respiratory syndrome. Respirology. 2006;11:715‐722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhou J, Chu H, Chan JF, Yuen KY. Middle East respiratory syndrome coronavirus infection: virus‐host cell interactions and implications on pathogenesis. Virol J. 2015;12:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chu H, Zhou J, Wong BH, et al. Productive replication of Middle East respiratory syndrome coronavirus in monocyte‐derived dendritic cells modulates innate immune response. Virology. 2014;454–455:197‐205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Law HK, Cheung CY, Ng HY, et al. Chemokine up‐regulation in SARS‐coronavirus‐infected, monocyte‐derived human dendritic cells. Blood. 2005;106:2366‐2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Funk CJ, Wang J, Ito Y, et al. Infection of human alveolar macrophages by human coronavirus strain 229E. J Gen Virol. 2012;93:494‐503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ng DL, Al Hosani F, Keating MK, et al. Clinicopathologic, immunohistochemical, and ultrastructural findings of a fatal case of middle east respiratory syndrome Coronavirus infection in the United Arab Emirates, April 2014. Am J Pathol. 2016;186:652‐658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Runfeng L, Yunlong H, Jicheng H, et al. Lianhuaqingwen exerts anti‐viral and anti‐inflammatory activity against novel coronavirus (SARS‐CoV‐2). Pharmacol Res. 2020;156:104761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Conti P, Ronconi G, Caraffa A, et al. Induction of pro‐inflammatory cytokines (IL‐1 and IL‐6) and lung inflammation by Coronavirus‐19 (COVI‐19 or SARS‐CoV‐2): anti‐inflammatory strategies. J Biol Regul Homeost Agents. 2020;34(2):327–331. [DOI] [PubMed] [Google Scholar]
- 17. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xiong Y, Liu Y, Cao L, et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID‐19 patients. Emerg Microbes Infect. 2020;9:761‐770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pedersen SF, Ho YC. SARS‐CoV‐2: A storm is raging. J Clin Invest. 2020;130(5):2202‐2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang CH, Liu CY, Wan YL, et al. Persistence of lung inflammation and lung cytokines with high‐resolution CT abnormalities during recovery from SARS. Respir Res. 2005;6:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wong CK, Lam CW, Wu AK, et al. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin Exp Immunol. 2004;136:95‐103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang Y, Li J, Zhan Y, et al. Analysis of serum cytokines in patients with severe acute respiratory syndrome. Infect Immun. 2004;72:4410‐4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fan YY, Huang ZT, Li L, et al. Characterization of SARS‐CoV‐specific memory T cells from recovered individuals 4 years after infection. Arch Virol. 2009;154:1093‐1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hu W, Yen YT, Singh S, Kao CL, Wu‐Hsieh BA. SARS‐CoV regulates immune function‐related gene expression in human monocytic cells. Viral Immunol. 2012;25:277‐288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Saha B, Jyothi Prasanna S, Chandrasekar B, Nandi D. Gene modulation and immunoregulatory roles of interferon gamma. Cytokine. 2010;50:1‐14. [DOI] [PubMed] [Google Scholar]
- 26. Watford WT, Moriguchi M, Morinobu A, O'Shea JJ. The biology of IL‐12: coordinating innate and adaptive immune responses. Cytokine Growth Factor Rev. 2003;14:361‐368. [DOI] [PubMed] [Google Scholar]
- 27. Liu YZC, Huang F, Yang Y, Wang F, Yuan J, et al.2019‐novel coronavirus (2019‐nCoV) infections trigger an exaggerated cytokine response aggravating lung injury. http://www.chinaxiv.org/abs/202002.00018 Accessed 18 Feb 2020. 2020.
- 28. Wen W, Su W, Tang H, et al. Immune cell profiling of COVID‐19 patients in the recovery stage by single‐cell sequencing. Cell Discov. 2020;6:31. 10.1038/s41421-020-0168-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang JJ, Dong X, Cao YY, et al. Clinical characteristics of 140 patients infected with SARS‐CoV‐2 in Wuhan, China. Allergy. 2020. [DOI] [PubMed] [Google Scholar]
- 30. Qin C, Zhou L, Hu Z, et al. Dysregulation of immune response in patients with COVID‐19 in Wuhan, China. Clin Infect Dis. 2020;71(15):762–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hansel TT, Tunstall T, Trujillo‐Torralbo MB, et al. A comprehensive evaluation of nasal and bronchial cytokines and chemokines following experimental rhinovirus infection in allergic asthma: increased interferons (IFN‐gamma and IFN‐lambda) and Type 2 inflammation (IL‐5 and IL‐13). EBioMedicine. 2017;19:128‐138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miyauchi K, Helper T. Cell Responses to Respiratory Viruses in the Lung: Development, Virus Suppression, and Pathogenesis. Viral Immunol. 2017;30:421‐430. [DOI] [PubMed] [Google Scholar]
- 33. Beale J, Jayaraman A, Jackson DJ, et al. Rhinovirus‐induced IL‐25 in asthma exacerbation drives type 2 immunity and allergic pulmonary inflammation. Sci Transl Med. 2014;6:256ra134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. You D, Marr N, Saravia J, et al. IL‐4Ralpha on CD4+ T cells plays a pathogenic role in respiratory syncytial virus reinfection in mice infected initially as neonates. J Leukoc Biol. 2013;93:933‐942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Legg JP, Hussain IR, Warner JA, Johnston SL, Warner JO. Type 1 and type 2 cytokine imbalance in acute respiratory syncytial virus bronchiolitis. Am J Respir Crit Care Med. 2003;168:633‐639. [DOI] [PubMed] [Google Scholar]
- 36. Jackson DJ, Makrinioti H, Rana BM, et al. IL‐33‐dependent type 2 inflammation during rhinovirus‐induced asthma exacerbations in vivo. Am J Respir Crit Care Med. 2014;190:1373‐1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zheng XY, Xu YJ, Guan WJ, Lin LF. Regional, age and respiratory‐secretion‐specific prevalence of respiratory viruses associated with asthma exacerbation: a literature review. Arch Virol. 2018;163:845‐853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Matthew J, Pinto Pereira LM, Pappas TE, et al. Distribution and seasonality of rhinovirus and other respiratory viruses in a cross‐section of asthmatic children in Trinidad. West Indies. Ital J Pediatr. 2009;35:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Leung TF, To MY, Yeung AC, Wong YS, Wong GW, Chan PK. Multiplex molecular detection of respiratory pathogens in children with asthma exacerbation. Chest. 2010;137:348‐354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Turchiarelli V, Schinkel J, Molenkamp R, et al. Repeated virus identification in the airways of patients with mild and severe asthma during prospective follow‐up. Allergy. 2011;66:1099‐1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wu Z, McGoogan JM. Characteristics of and important lessons from the Coronavirus disease 2019 (COVID‐19) outbreak in China: summary of a report of 72314 cases from the Chinese center for disease control and prevention. JAMA. 2020. 10.1001/jama.2020.2648 [Online ahead of print] [DOI] [PubMed] [Google Scholar]
- 42. Liu S, Zhi Y, Ying S. COVID‐19 and asthma: reflection during the pandemic. Clin Rev Allergy Immunol. 2020;59(1):78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sajuthi SP, DeFord P, Jackson ND, et al. Type 2 and interferon inflammation strongly regulate SARS‐C oV‐2 related gene expression in the airway epithelium. bioRxiv The Preprint Server for Biology https://www.biorxiv.org/content/10.1101/2020.04.09.034454v1 2020
- 44. Jackson DJ, Busse WW, Bacharier LB, et al. Association of respiratory allergy, asthma, and expression of the SARS‐CoV‐2 receptor ACE2. J Allergy Clin Immunol. 2020;146(1):203–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Peters MC, Sajuthi S, Deford P, et al. COVID‐19 related genes in sputum cells in asthma: relationship to demographic features and corticosteroids. Am J Respir Crit Care Med. 2020;202(1):83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ferrucci S, Romagnuolo M, Angileri L, Berti E, Tavecchio S. Safety of dupilumab in severe atopic dermatitis and infection of Covid‐19: two case reports. J Eur Acad Dermatol Venereol. 2020;34(7):e303–e304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gerbaud P, Guibourdenche J, Jarray R, et al. APN/CD13 is over‐expressed by Psoriatic fibroblasts and is modulated by CGRP and IL‐4 but not by retinoic acid treatment. J Cell Physiol. 2018;233:958‐967. [DOI] [PubMed] [Google Scholar]
- 48. Kehlen A, Gohring B, Langner J, Riemann D. Regulation of the expression of aminopeptidase A, aminopeptidase N/CD13 and dipeptidylpeptidase IV/CD26 in renal carcinoma cells and renal tubular epithelial cells by cytokines and cAMP‐increasing mediators. Clin Exp Immunol. 1998;111:435‐441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sorrell JM, Brinon L, Baber MA, Caplan AI. Cytokines and glucocorticoids differentially regulate APN/CD13 and DPPIV/CD26 enzyme activities in cultured human dermal fibroblasts. Arch Dermatol Res. 2003;295:160‐168. [DOI] [PubMed] [Google Scholar]
- 50. van der Velden VH, Naber BA, van der Spoel P, Hoogsteden HC, Versnel MA. Cytokines and glucocorticoids modulate human bronchial epithelial cell peptidases. Cytokine. 1998;10:55‐65. [DOI] [PubMed] [Google Scholar]
- 51. Kunii R, Nemoto E, Kanaya S, Tsubahara T, Shimauchi H. Expression of CD13/aminopeptidase N on human gingival fibroblasts and up‐regulation upon stimulation with interleukin‐4 and interleukin‐13. J Periodontal Res. 2005;40:138‐146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


