Abstract
Lung cancer is the leading cause of cancer-associated death worldwide. In recent years, the advancement of epidermal growth factor receptor-tyrosine kinase inhibitor (EGFR-TKI) targeted therapies has provided clinical benefits for lung cancer patients with EGFR mutations. The response to EGFR-TKI varies in patients with lung cancer, and resistance typically develops during the course of the treatment. Therefore, understanding biomarkers which can predict resistance to EGFR-TKI is important. Overexpression of GLI causes activation of the Hedgehog (Hh) signaling pathway and plays a critical role in oncogenesis in numerous types of cancer. In the present study, the role of GLI1 in erlotinib resistance was investigated. GLI1 mRNA and protein expression levels were determined using reverse transcription-quantitative PCR and immunohistochemistry (IHC) in lung cancer cell lines and tumor specimens, respectively. GLI1 mRNA expression levels were found to be positively correlated with the IC50 of erlotinib in 15 non-small cell lung cancer (NSCLC) cell lines. The downregulation of GLI1 using siRNA sensitized lung cancer cells to the erlotinib treatment, whereas the overexpression of GLI1 increased the survival of lung cancer cells in the presence of erlotinib, indicating that Hh/GLI activation may play a critical role in the development of TKI resistance in lung cancer. Combined treatment with erlotinib and a GLI1 inhibitor reduced the cell viability synergistically. A retrospective study of patients with NSCLC treated with erlotinib revealed that those with a high IHC score for GLI1 protein expression had a poorer prognosis. These results indicated that GLI1 is a key regulator for TKI sensitivity, and patients with lung cancer may benefit from the combined treatment of TKI and GLI1 inhibitor.
Keywords: GLI1, erlotinib, TKI, drug resistance, lung cancer
Introduction
Lung cancer is the most common cause of cancer-associated mortality worldwide in the 2018 global survey (1–4), and 85% of all lung cancers are non-small cell lung cancer (NSCLC) (3). As >70% of patients with lung cancer in the 2018 global survey have metastases to the regional lymph nodes or to distant sites, systemic therapies such as chemotherapy and radiotherapy played a major part in the treatment of these patients (3). The benefit of the these therapies, however, is modest in terms of controlling the proliferation of the tumor cells, and the five-year survival rate is only ~19% in the US (3,5,6). Cancer cells develop drug resistance during the course of the treatment and therefore, novel therapies are required based from the understanding of molecular mechanisms and pathways that contribute to tumor resistance, and thereby enhancing the clinical efficacy of the current therapeutic agents.
The epidermal growth factor receptor (EGFR) signaling pathway plays a critical role in cell proliferation and survival in NSCLC (7). Drugs, such as the EGFR tyrosine kinase inhibitors (TKIs), gefitinib and erlotinib, have demonstrated clinical benefit of reduced tumor size in patients with NSCLC that harbored sensitizing mutations, such as L858R and exon 19 deletion in the tyrosine kinase domain of the EGFR gene (8–10). The overall clinical efficacy from these drugs in patients with relapsed NSCLC is limited; however, due to drug resistance.
Analysis of EGFR-TKI clinical data in patients with NSCLC revealed that the clinical benefits from these types of drugs are variable. The acquired drug resistance mutation EGFR T790M for gefitinib or erlotinib was detected in ~50% of patients with clinical resistance (11–14). The third generation TKI, osimertinib, was approved for treating patients with sensitizing mutations in EGFR who are also T790M-positive, which partially alleviated the problem. The development of TKI resistance by other mechanisms, such as activating KRAS mutations (for instance, G12D or G12V), HER2 (15) or MET gene copy number amplification (16) is still challenging to overcome in clinical practice (14,17–20). Therefore, a further understanding of the tumor molecular markers, which can predict resistance to EGFR-TKIs, is important for the identification of drugs to overcome resistance (20).
The Hedgehog (Hh) signaling pathway controls multiple cellular functions, such as embryonic development, tissue patterning and wound healing (21). Aberrantly increased Hh pathway activation has been implicated in various types of inherited and sporadic malignancies (21), including lung cancer (22–25). In the quiescent state of the Hh signaling pathway, the transmembrane receptor Patched homolog-1 (PTCH1; which spans the membrane 12 times) restricts the activity of the transmembrane receptor smoothened, which is a frizzled class receptor (SMO; which spans the membrane 7 times) (21,26). The binding of the Hh ligands to PTCH1 reverses its inhibitory effect on SMO, and subsequently the activated SMO produces a complex series of cytoplasmic signal transductions, which results in the activation of zinc-finger protein GLI (GLI) family of transcription factors (21,26). The GLI transcription factors regulate the transcription of specific genes, which is dependent on the overall stimuli received by the cells as well as the type of cells (27–30).
There are three members of the GLI protein family in humans (GLI1, GLI2 and GLI3). GLI1 contains a transactivation domain and acts as a transcriptional activator (28–30). GLI2 and GLI3 both contain the transactivation and repressor domains, and function as either activators or repressors (31). GLI2 and GLI3 are the primary mediators of signal transduction upon the activation of the Hedgehog pathway, and regulate the expression of GLI1 (28). GLI1 functions in a positive feedback manner to reinforce its activity, with its levels reflecting the activation status of the GLI family proteins (29).
The importance of the Hh signaling pathway in the development of cancer has provided novel targets for oncological interventions (25). A majority of the studies have focused on inhibiting Hh signal transduction at the cell membrane level, i.e., inhibitors targeting Smo and Hh (32). For example, the SMO inhibitor, vismodegib, was approved by the Food and Drug Administration for the treatment of advanced and metastatic basal cell carcinoma (BCC) (33,34). Vismodegib and numerous other SMO inhibitors, such as sonidegib (35–38) and saridegib (39,40), are currently undergoing clinical trials in the treatment of BCC and many types of solid tumors (25).
Previous studies have revealed additional mechanisms of GLI activation, which are independent of Hh/SMO regulation and are stimulated by the cross-talk between components downstream of the Hh/SMO signaling pathway and several other oncogenic signaling pathways, such as the EGFR pathway (41,42). For example, EGFR signaling and the stimulation of its downstream components, such as RAS/MAPK and PI3K/Akt, lead to the activation of the GLI transcription factors. The importance of this non-canonical GLI activation is accentuated, as RAS/MAPK and PI3K/Akt also mediate stimuli from other growth factor pathways such as ALK (43) and HER2 (44), and both PI3K and Ras are hyper-activated in tumors (41). These findings suggest the potential importance of GLI as a key target for cancer therapeutics (27,30). Recently, pre-clinical studies have found that GLI inhibitors are more effective in inhibiting tumor cell growth in vivo and in reducing tumor growth in animal models when compared with upstream SMO inhibition (39,45,46).
In the present study, we hypothesize that GLI1, which functions as an integration point for the canonical activation from the Hh/SMO signalling pathway (28,29) and for the non-canonical activation from the EGFR signaling pathway (25,41), may be a key regulator of TKI resistance in NSCLC. The association between GLI1 and erlotinib resistance was determined in NSCLC cell lines and tumor specimens. The results indcated that GLI1 was critical for TKI sensitivity, and patients with lung cancer may benefit from the combined treatment of TK and GLI1 inhibitors.
Materials and methods
Tissue specimens
All the human NSCLC tissue samples used in the present study were obtained from the University of California, San Francisco Thoracic Oncology tissue bank, with approval from the Committee on Human Research (approval number: H8714-11647-10). The patient samples were collected between February 2011 and October 2016. There were 24 men and 13 women in the study cohort, with a mean age of 57.7±13.2 years (age range, 27–82 years). The patients only received erlotinib treatment during the study. When progression occurred, the patients were switched to other treatments such as chemotherapy. All specimens were snap-frozen in liquid nitrogen immediately following resection, and then stored at −170°C until further use. Formalin-fixed paraffin-embedded (FFPE) samples from the same patient were fixed in PBS buffer with 10% formalin for 24–48 h at room temperature and embedded in paraffin. Formalin-fixed paraffin-embedded (FFPE) samples (3 µm thick) were used for immunohistochemistry staining.
RNA extraction and reverse transcription quantitative PCR (RT-PCR)
Total RNA was isolated by using the RNeasy kit (Qiagen GmbH) according to the manufacturer's protocol. Genomic DNA contamination was eliminated using DNase I, and RT was performed using 500 ng RNA and the iScript cDNA synthesis kit (Bio-Rad Laboratories, Inc.) using random hexamer according to the manufacturer's protocol. The cDNA was analyzed using RT-qPCR n a 7500 Real-Time PCR machine (Thermo Fisher Scientific, Inc.) using SYBR-Green qPCR Master Mixes (Thermo Fisher Scientific, Inc.). The following thermocycling conditions were used: Initial denaturation 96°C for 5 min, followed by 40 cycles at 96°C for 15 sec and 60°C for 1 min. Gene expression was normalized to GAPDH using the 2−ΔΔCq method (47). The following primer sequences were used: GLI1 forward, 5′-CTCCCGAAGGACAGGTATGTAAC-3′ and GLI1 reverse, 5′-CCCTACTCTTTAGGCACTAGAGTTG-3′, GAPDH forward, 5′-ACAACAGCCTCAAGATCATCAG-3′ and GAPDH reverse, 5′-TCTTCTGGGTGGCAGTGATG-3′ (48).
Immunohistochemistry (IHC) staining
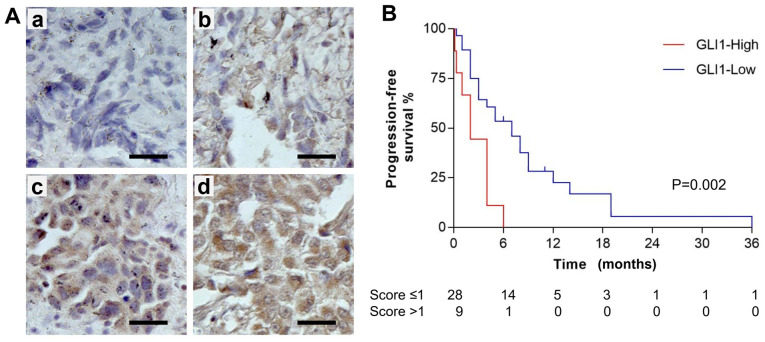
The UltraVision LP Detection System HRP DAB kit (cat. no. TL-060-HD; Thermo Fisher Scientific, Inc.) was used according to the manufacturer's protocol. All steps were carried out at room temperature except for the primary antibody incubation. Briefly, FFPE slides were deparaffinized in xylene and rehydrated in a descending ethanol series from 100–70%. Heat-mediated antigen retrieval was performed by boiling the slides for 20 min in a citrate buffer (pH 6.0). Slides were quenched in UltraVision Hydrogen Peroxide Block for 10 min at room temperature, washed three times in PBS, and incubated with UltraVision Protein Block for 5 min at room temperature. After washing once with PBS, the slides were incubated with rabbit anti-human GLI1 (1:100 diluted in PBS; Santa Cruz Biotechnology; cat. no. sc-20687) overnight at 4°C. The slides were washed three times with PBS and incubated with Primary Antibody Enhancer for 10 min at room temperature, followed by an incubation with HRP Polymer for 15 min at room temperature. The slides were re-washed three times with PBS and the color was developed following incubation with 1:30 dilution of DAB in DAB Quanto Substrate for 3 min at room temperature. The slides were subsequently washed four times with distilled water and counterstained in Mayer's hematoxylin (Thermo Fisher Scientific, Inc.) for 1 min at room temperature, washed in running tap water for 10 min and mounted. Images from representative fields were obtained using an Olympus BX43 light microscope (magnification, ×200; Olympus Corporation) and examined for positive nuclear staining. IHC staining was blindly scored by two independent pathologists. IHC score was determined using staining intensity for the majority (≥50%) of the tumor cells as shown in Fig. 5A: No staining, 0; light yellow staining, 1; yellowish/brown staining, 2; strong brown staining, 3.
Figure 5.
Association between GLI1 protein expression and progression-free survival in patients with lung cancer receiving erlotinib treatment. (A) IHC staining of GLI1 protein in lung cancer tissue samples. The images show different IHC scores: a, 0; b, 1; c, 2; d, 3. Scale bars represent 25 mm. (B) The Kaplan-Meier curves showed the association between GLI1 expression and progression-free survival in patients with lung cancer receiving erlotinib treatment. The statistical difference was evaluated using the log rank test. IHC, immunohistochemistry.
Western blotting
Cell lysates were prepared by adding 300 ml of M-PER Mammalian Protein Extraction Reagent (Thermo Fisher Scientific, Inc.) to 6-well plates. After shaking gently for 5 min at room temperature, the cell lysates were centrifuged at 14,000 × g for 5 min at 4°C. Total protein concentration was determined using a Bradford protein assay (Bio-Rad Laboratories, Inc.), 40 µg protein/lane was separated via 10% SDS-PAGE and the separated proteins were subsequently transferred onto polyvinylidene membranes (Sigma-Aldrich; Merck KGaA). The membranes were blocked with 5% non-fat dry milk for 30 min at room temperature, washed twice with TBST, and sliced right above the 50 kDa pre-stained molecular weight markers (Bio-Rad Laboratories, Inc.). The membranes were incubated with primary antibodies at room temperature for 2 h (top half: Anti-GLI1 antibody, 1:150 dilution in 5% BSA, Santa Cruz Biotechnology, sc-20687; bottom half: Anti-β-actin antibody, 1:500 dilution in 5% BSA, Sigma-Aldrich; Merck KGaA, A5441). Membranes were washed three times with TBST and subsequently incubated with secondary antibodies at room temperature for 1 h (top half: Goat anti-mouse IgG, 1:2,000 in 5% BSA, Santa Cruz Biotechnology, sc-2005; bottom half: HRP-conjugated rabbit anti-mouse IgG, 1:100,000 in 5% BSA, Sigma-Aldrich, A9044). Membranes were re-washed three times with TBST and incubated with Immobilon ECL Ultra Western HRP Substrate (Sigma-Aldrich; Merck KGaA) for 5 min at room temperature. The substrate solution was drained, and the membranes were exposed to X-ray film.
Cell cultures
The following human lung cancer cell lines were purchased from American Type Culture Collection: A549 (cat. no. CCL-185), A427 (cat. no. HTB-53), H1299 (cat. no. CRL-5803), H2170 (cat. no. CRL-5928), H1703 (cat. no. CRL-5889), H522 (cat. no. CRL-5810), H838 (cat. no. CRL-5844), H322 (cat. no. CRL-5806), H1650 (cat. no. CRL-5883), H1975 (cat. no. CRL-5908), H820 (cat. no. HTB-181), H441 (cat. no. HTB-174), H460 (cat. no. HTB-177), H1666 (cat. no. CRL-5885), HCC2935 (cat. no. CRL-2869). All the cell lines were cultured in RPMI-1640 supplemented with 10% fetal bovine serum, penicillin (100 IU/ml) and streptomycin (100 µg/ml) (all purchased from Thermo Fisher Scientific, Inc.) at 37°C in a humidified incubator with 5% CO2.
RNA interference and cDNA transfection
For each well of the 6-well plates, 3×105 cells were plated in fresh media without antibiotics for 24 h prior to transfection. Cells were transfected with either 2 mg pcDNA3.1 vector containing GLI1 cDNA or pcDNA3.1 vector control (Thermo Fisher Scientific, Inc.) using Lipofectamine® 2000 transfection reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. For RNA interference, cells in 6-well plates were transfected with 100 pmole of synthesized GLI1 small interfering (si)RNA (siRNA-1, Assay ID 107670; siRNA-2, Assay ID 115641, Thermo Fisher Scientific, Inc.) or scrambled siRNA using Lipofectamine® 2000 transfection reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. After the transfection, the cells were cultured for 48 h before further analysis with drug treatment.
Cell survival assays and IC50 determination
Cells were seeded in 96-well plates at a density of 500–1,000 cells/well, and the medium was changed daily. Logarithmically growing cells were treated with increasing doses (0.01 µM to 1 mM) of erlotinib and/or with GLI inhibitor (0.01-10.00 µM), or DMSO control for 3 days. Cells were subsequently assessed for viability using CellTiter-Glo Luminescent Cell Viability Assay reagent (Promega Corporation) according to manufacturer's instructions. Luminescence was measured using a GloMax-96 Microplate Luminometer (Promega Corporation), and the percent of cell survival was calculated with the DMSO treated cells set as 100%. GraphPad Prism v6.0 (GraphPad Software, Inc.) was used to generate dose-response curves and IC50 values.
Combination index (CI)
A549 or H2170 cells were treated with 0.01–10 µM of erlotinib alone, GLI inhibitor alone, or the 1:1 combination of the two drugs for 72 h, and assayed for cell viability. The CalcuSyn software version 2.0 (Biosoft) was used to calculate the CI to identify synergistic, additive, or antagonistic drug interactions for the combination treatment as indicated in Fig. 4, where CI <1 indicates synergism, CI, 1 indicates additive effects, and CI >1 indicates antagonism. Synergism was further divided into moderate synergism (CI, 0.7–0.9), synergism (CI, 0.3–0.7) and strong synergism (CI, 0.1–0.3).
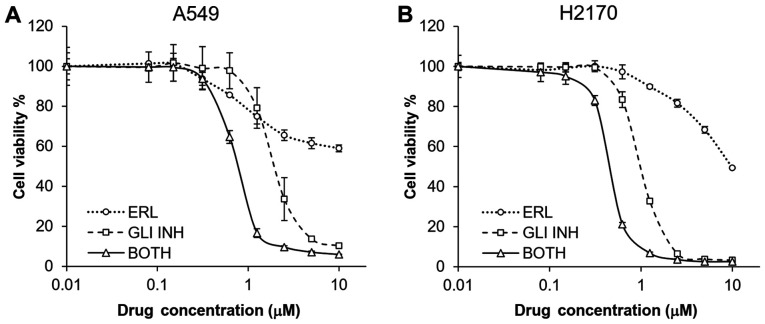
Figure 4.
Treatment with a small molecule GLI inhibitor sensitized lung cancer cell lines to erlotinib. (A) A549 cells. (B) H2170 cells. The cells were treated with either GLI inhibitor or erlotinib, or in combination at 1:1 ratio. The error bars represent SDs from three experiments. ERL, erlotinib; GLI INH, GLI1 inhibitor; BOTH, both drugs at 1:1 ratio.
Statistical analysis
The comparisons of IC50 values or cell viabilities under different experimental conditions were analyzed using the unpaired Student's t test for two groups, or by one-way ANOVA followed by Tukey's post hoc test for multiple groups. The data from three experiments were averaged and plotted as the mean ± standard deviation. The associations between GLI1 mRNA levels and IC50 for erlotinib were assessed using Pearson's correlation coefficient. The Cox proportional hazards model was used to evaluate the risks in progression-free survival (PFS) for patients with different GLI1 IHC scores. The Kaplan-Meier plot was used to illustrate the differences in PFS between the low-risk group (low GLI1 IHC staining) and high-risk group (high GLI1 IHC staining). The log-rank test was used to evaluate the differences in PFS between dichotomous groups defined by patient characteristics. The Fisher's exact test was used to analyze the association between GLI expression levels and patient characteristics. P≤0.05 was considered to indicate a statistically significant difference.
Results
Erlotinib IC50 values are not associated with driver mutations in NSCLC cell lines
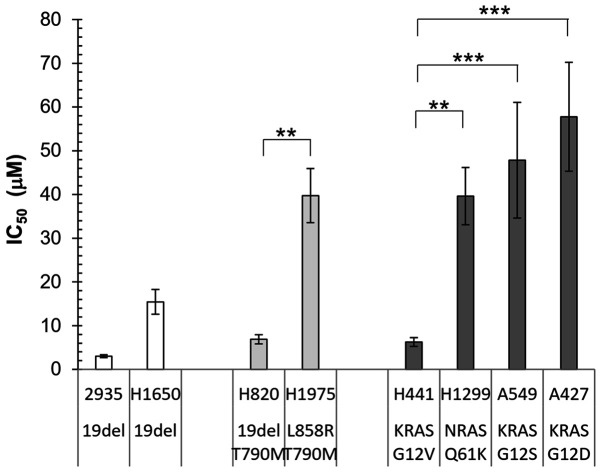
To investigate the association between gene mutations and erlotinib sensitivity, NSCLC cell lines with different EGFR and KRAS mutations as reported by COSMIC database (49,50) were examined (Fig. 1). The HCC2935 and H1650 cell lines contain the sensitizing exon 19 deletion in EGFR, while the H820 and H1975 cell lines contained both sensitizing EGFR mutation (E19del and L858R, respectively) and the T790M resistant mutation. A total of 4 cell lines with mutations in the downstream signaling protein, RAS were also investigated: A549 with KRAS G12S, H1299 with NRAS Q61K, A427 with KRAS G12D, and H441 with KRAS G12V. It was hypothesized that the cell lines containing sensitizing EGFR mutations would have a lower IC50 compared with that with EGFR T790M or RAS activating mutations.
Figure 1.
IC50 of erlotinib in non-small cell lung cancer cell lines with different mutations in either EGFR or RAS. The error bars represent SDs from three experiments. **P<0.01, ***P<0.001.
Unexpectedly, although the cell lines with a double mutation in EGFR or those with RAS mutations had on average higher IC50 values, there was a wide range of variability in the individual cell lines (Fig. 1). Both the H820 and H1975 cell lines contained the EGFR T790M mutation, as well as either the sensitizing E19del or L858R mutation; however, there was a significant difference in the IC50 values (P<0.01). In addition, for the three cell lines containing KRAS mutations at the same codon, the IC50 of H441 (KRAS G12V) was significantly lower compared with that in A549 (KRAS G12S; P<0.001) and A427 (KRAS G12D; P<0.001). Furthermore, the H1650 cell line contained the sensitizing EGFR E19del mutation; however, its IC50 was not lower compared with that in the H820 (E19del and T790M) and H441 (KRAS G12V) cell lines. These results indicated that the analysis of EGFR and RAS hotspot mutations is not sufficient to predict TKI sensitivity in cell lines, and it is likely that additional genetic or epigenetic modifications modulate their response to TKI.
GLI1 expression is associated with resistance to erlotinib treatment in NSCLC cell lines
During the investigation into GLI1 function, it was found in a preliminary study that the A549 and A427 cell lines had high mRNA expression levels GLI1, whereas it was hardly detectable in H441 cells (data not shown). To investigate whether the mRNA expression level of GLI1 was correlated with erlotinib sensitivity, 15 NSCLC cell lines with a variety of mutation backgrounds (Table I) were examined. Among them, 4 cell lines contained EGFR mutations, 7 contained an activating mutation in the RAS family, and 4 did not contain either of those mutations (51–57).
Table I.
Hotspot mutations in non-small cell lung cancer cell lines.
| Gene | ||||||||
|---|---|---|---|---|---|---|---|---|
| Cell line | EGFR (Refs.) | KRAS NRAS HRAS (Refs.) | BRAF (Refs.) | PIK3CA | ALK/ROS1 MET/RET (Refs.) | TP53 | Hh/GLI signal pathway | Other mutation (Refs.) |
| H522 | – | – | – | – | RET A281Va | P191fs*b | GLI2 E538Ga | |
| H1703 | – | – | – | ALK A518Va | c.919+1G>Tb | PDGFRA ampb (29) | ||
| A549 | – | KRAS G12Sb | – | – | – | – | SMO V270I SUFU T411Ma | E2A-PBX1b (30) |
| H838 | – | KRAS G12Cb | – | – | MET I638La | p.E62*b | – | KEAP1 |
| H1299 | – | NRAS Q61Kb | – | – | – | TP53 nullb | – | p.E444*c |
| H322 | – | – | – | – | ALK S289Y Y262Ha | R248Lb | – | ERBB2 S310Fc |
| H1650 | E19delb | – | – | – | – | c.673-2A>Gb | – | |
| H1975 | L858R T790Mb | – | – | PIK3CA G118Dc | – | R273Hc | – | |
| H460 | – | KRAS Q61Hb | – | PIK3CA E545Kb | – | – | – | NEK2 G134Da |
| H820 | E19del T790Mb (31) | – | – | – | MET ampb (31) | T284Pc | – | |
| HCC 2935 | E19delb (32) | – | – | – | – | – | – | |
| H441 | – | KRAS G12Vb | – | – | – | R158Lb | – | ERBB4 K1247M, S1246Ra |
| H2170 | – | RHOA G17Vb (33) | – | PIK3CG H693Qa | – | R158Gb | – | |
| H1666 | – | – | BRAF G466Vb (34) | – | – | – | – | E2A-PBX1b (30) |
| A427 | – | KRAS G12Db | – | – | – | – | SMO R113Ga | E2A-PBX1b (30) |
The mutations shown were based on the literatures indicated in the parentheses and COSMIC database (49,50).
Mutations found in the COSMIC database but not found in ClinVar database (57), which means that the function of the mutations is currently unknown.
Hotspot driver mutations that had been published. fs*, frameshift mutation leading to premature termination; amp, gene amplification.
Mutations that are ‘pathogenic’ or ‘likely pathogenic’ according to ClinVar database. −, mutations are not present in the cell line according to COMSIC database.
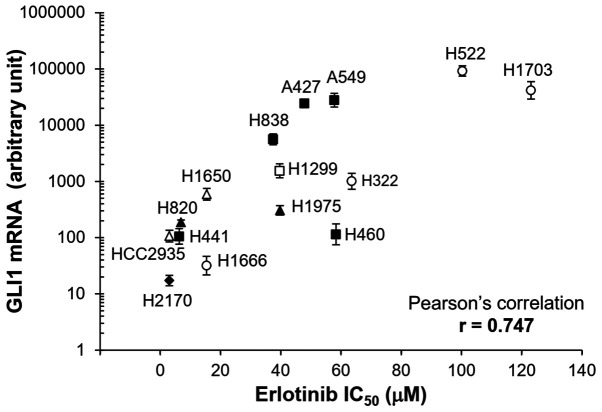
As shown in Fig. 2, a wide range of IC50 values were observed among the different cell lines. The highest IC50 values (H522 and H1703) were ~25 fold higher compared with that for the lowest one (HCC2935). The Pearson's correlation analysis showed that the mRNA expression levels of GLI1 was significantly correlated with IC50 for erlotinib (Pearson's correlation co-efficient r=0.747; P<0.01).
Figure 2.
Correlation between GLI1 mRNA expression levels and erlotinib sensitivity. GLI1 expression in the different cell lines were measured using reverse transcription-quantitative PCR and normalized to GAPDH using 2−ΔΔCq method (47). The error bars represent SDs from three experiments. Different symbols represent cell lines with different types of mutations. Open triangle, cells with EGFR sensitizing mutation (HCC2935 and H1650, both with E19del); filled triangle, cells with EGFR sensitizing mutation and resistant mutation (H820, 19del and T790M; H1975, L858R and T790M); filled square, cells with KRAS mutation (H441, G12V; H838, G12C; A427, G12D; A549, G12S; H460, Q61H); open square, cells with NRAS mutation (H1299, Q61K); filled diamond, cells with RAS family mutation (H2170, RHOA G17V); open circle, cells without common driver mutations in EGFR and RAS family (H1666, H322, H522, H1730).
GLI1 expression influences erlotinib sensitivity in lung cancer cells
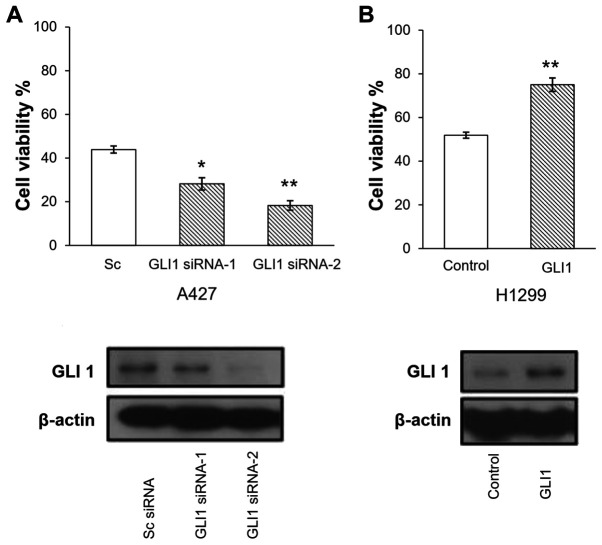
To investigate the function of GLI1 expression in erlotinib sensitivity, A427 cells, which is one of the cell lines with a high mRNA expression level of GLI1, were transfected with GLI1 siRNA-1, GLI1 siRNA-2, or scrambled control siRNA, respectively for 2 days, followed by a treatment with 25 µM erlotinib for 3 days. A total of two GLI1 siRNAs were used to ensure that the observed phenomena were due the silencing of GLI1 instead of the off-target effect a certain siRNA. As shown in Fig. 3A, the downregulation of GLI1 by siRNA sensitized A427 cells to erlotinib treatment (P<0.03 and P<0.008 for siRNA-1 and siRNA-2, respectively).
Figure 3.
GLI1 expression levels influenced resistance to erlotinib in lung cancer cell lines. (A) Reduction of GLI1 expression sensitizes lung cancer cell lines to erlotinib. A427 cells transfected with GLI1 siRNA were more sensitive to erlotinib treatment compared with those with control siRNA. (B) H1299 cells with GLI1 overexpression were more resistant to erlotinib treatment. The error bars represent SDs from three experiments. *P<0.05; **P<0.01. si, small interfering; Sc, scramble. The bottom panels depict GLI1 protein levels, with β-actin as the loading control of the corresponding samples in the upper panels.
Subsequently, the effect of GLI1 overexpression on erlotinib sensitivity was assessed. H1299 cells, which has a moderate level of GLI1 mRNA expression as shown in Fig. 2, were transfected with GLI1 expression vector or a control vector for 2 days, followed by treatment with 30 mM erlotinib for 3 days. As shown in Fig. 3B, upon GLI1 overexpression, the cells exhibited increased viability in the presence of 30 µM erlotinib.
Taken together, the results suggested that GLI1 modulates erlotinib sensitivity in lung cancer cells, and that high GLI1 mRNA levels may be a critical and independent mechanism to confer resistance to erlotinib in lung cancer.
Combination treatment of a GLI1 inhibitor and erlotinib synergistically suppressed proliferation of lung cell lines
Subsequently, two NSCLC cell lines, H2170 and A549, were treated with both a small molecule GLI inhibitor (58) and different concentrations of erlotinib for 72 h. The combination treatment synergistically suppressed proliferation of both the H2170 and A549 cell lines (Fig. 4). CalcuSyn analysis showed that the CI for the two compounds was 0.173 and 0.231 for H2170 and A549, respectively, suggesting a strong synergism of the two drugs in both cell lines.
GLI1 expression is associated with progression-free survival in NSCLC patients receiving erlotinib treatment
To investigate the clinical relevance of GLI1 as a predictive biomarker for EGFR-TKI therapy, a retrospective study was conducted in a cohort of 37 lung cancer patients that received erlotinib treatment. More than half of the patients progressed within 6 months of erlotinib treatment. Only one patient was progression-free for 36 months. The median follow-up time was 4 months, with a range of 0.13 to 36 months. FFPE tissue specimens were collected in the surgeries before erlotinib treatment and were analyzed by IHC staining. The slides were scored according to the standard protocol (Fig. 5A, with panel a-d showing representative staining of score 0, 1, 2 and 3, respectively). The mean IHC score (± standard deviation) for the cohort was 0.95 (±0.76). By using a Cox proportional hazards model, it was calculated that for every 1-point increase in the IHC score, patients were 2.1 (95% confidence interval, 1.30–3.32) times more likely to progress or die.
Subsequently, each case was asigned to either GLI1-low (IHC score ≤1) or GLI-high (IHC score >1) groups. Kaplan-Meier curves for progression-free survival (PFS) of GLI1-low (28 cases) and GLI1-high (9 cases) groups of patients with lung cancer receiving erlotinib treatment are presented in Fig. 5B, which revealed a significant difference in PFS by the log-rank test (P=0.0021). The data indicated that PFS in patients receiving erlotinib treatment was significantly improved for those with a low GLI1 expression compared with those with a high GLI1 expression. No statistically significant differences were observed between the dichotomous groups defined by other clinicopathological characteristics such as sex, smoker, age or tumor stage (Table II). Notably, more GLI1-high cases were observed in the squamous cell carcinoma subgroup in comparison with the adenocarcinoma subgroup (P=0.034), although the two pathological subgroups did not show a significant difference in PFS. When the PFS of the adenocarcinomas (25 samples) was analyzed, the association between high GLI1 expression and poor PFS was also observed (P=0.0020). The squamous cell carcinomas (9 samples) did not show a significant difference in PFS between the GLI1-high and GLI1-low groups, probably due to the small number of cases. In summary, the result suggests that GLI1 may be a predictive biomarker for patients treated with EGFR-TKI.
Table II.
Analysis of GLII levels and other clinicopathological variables for patient survival.
| Expression analysis | Survival analysis | ||||
|---|---|---|---|---|---|
| Clinicopathological variable | GLI1-high, n (%) | GLI1-low, n (%) | P-valuea | MeanPFSd, months | P-valueb |
| Sex | |||||
| Male | 6 (25.0) | 18 (75.0) | >0.999 | 5.2 | 0.575 |
| Female | 3 (23.1) | 10 (76.9) | 7.1 | ||
| Smoker | |||||
| Yes | 6 (35.3) | 11 (64.7) | 0.251 | 7.3 | 0.873 |
| No | 3 (15.0) | 17 (85.0) | 4.8 | ||
| Pathological subtypec | |||||
| Adeno | 4 (16.0) | 21 (84.0) | 0.034 | 7.0 | 0.236 |
| Squamous | 5 (55.6) | 4 (44.4) | 4.5 | ||
| Age, years | |||||
| <60 | 4 (21.1) | 15 (78.9) | 0.714 | 5.7 | 0.872 |
| >60 | 5 (27.8) | 13 (72.2) | 7.2 | ||
| Stage | |||||
| III | 3 (42.9) | 4 (57.1) | 0.327 | 3.8 | 0.241 |
| IV | 6 (20.0) | 24 (80.0) | 7.0 | ||
Fisher exact test.
Log-rank test.
Three cases were not included in the two pathological subtypes listed in the table. Two cases were adenosquamous carcinomas and one was a large cell neuroendocrine carcinoma.
PFS, progression-free survival.
Discussion
The present study found that GLI1 expression was associated with the resistance to EGFR-TKI treatment in lung cancer cell lines. To the best of our knowledge, the current study has also revealed for the first time that erlotinib-treated lung cancer patients with low GLI1 expression had significantly improved progression-free survival compared with those with high GLI1 expression.
The association between GLI1 expression and lung cancer prognosis has been investigated previously. Gialmanidis et al (59) examined 80 NSCLC cases using IHC staining of the Hh and Patched pathway proteins, and found a significant association between lymph node metastases and nuclear GLI1 immunolocalization in adenocarcinomas. The survival of patients, however, was not reported. Ishikawa et al (48) examined the mRNA expression levels of GLI1 using RT-qPCR in 102 patients with stage II–IV lung adenocarcinoma following surgical resection, and found that the top 15% ranked cases according to mRNA expression had a hazard ratio of 3.1 (95% CI, 1.5–6.2) for tumor progression. Notably, the prognosis was unrelated to EGFR mutation status, which had a hazard ratio of 1.1 (95% CI, 0.58–2.0). Bora-Singhal et al (60) analyzed GLI1 mRNA levels from a public database with 360 NSCLC cases, which had been determined using an Affymatrix microarray, and found that high GLI1 mRNA levels was associated with poorer overall survival (P=0.04). Recently, mesenchyme homeobox 2-dependent GLI1 protein expression was found to be associated with clinical progression and poorer overall survival in a cohort of 90 patients with NSCLC undergoing platinum-based oncological therapy with both EGFR-non-mutated and EGFR-mutated statuses (61). Taken together, these studies showed that in general, a higher GLI1 expression has been associated with a poorer prognosis in NSCLC (62).
To the best of our knowledge, the current study, has for the first time, investigated the association between GLI1 expression levels and TKI treatment outcome in patients with NSCLC. It was found that the high level of the IHC staining of the GLI1 protein was significantly associated with poor progression-free survival in patients treated with erlotinib. The overall survival, however, did not reach statistical significance (data not shown). This could be due to several reasons. Firstly, the cohort might have been not large enough to reach statistical significance. In future studies, a different cohort with more samples should be collected from an independent source to further validate the observed association. Secondly, tumor cells may accumulate additional mutations during the course of treatment, which might have affected GLI1 expression levels. It was well-known that resistance to TKI treatment, such as gefitinib and erlotinib, develops within a year (20,63) due to different mechanisms, including EGFR T790M mutation (13), HER2 (15) or MET amplification (16), or mutations in additional oncogenes (20), such as KRAS (14) or PIK3CA (64). The levels of GLI1 may change accordingly and differ from those in the original tumors. Thus, continuously monitoring the expression levels of GLI1 would provide additional information. However, the tissues are typically unavailable following the initial surgery or biopsy, which impedes the IHC analysis. With the development and approval of novel technology for evaluating mRNA or protein expression levels using circulating nucleic acid or exosomes, such measurements may become feasible in the near future.
It had been reported that the squamous cell carcinomas (SCC) of the lung, which accounts for ~30% of the NSCLC cases, has a worse prognosis than the adenocarcinoma subtype (65,66). Notably, in the present study the SCC subtype had a higher percentage of cases with high GLI1 expression (Table II), but no significant difference in the PFS of SCC was observed in comparison with that of the adenocarcinoma subtype. This may have been due to the small sample size used in the current study (9 SCC cases). The contribution of GLI1 in the prognosis of SCC will be investigated by using a larger cohort in prospective studies.
As further evidence to support the critical role of GLI1 in resistance to erlotinib, it was found that changes in the GLI1 levels affected the sensitivity to the erlotinib treatment in lung cancer cell lines (Fig. 3). Downregulation of GLI1 expression using siRNA transfection sensitized lung cancer cells to erlotinib treatment, while upregulation of GLI1 increased resistance to erlotinib treatment. Together, the data suggests that a high GLI1 level may be a critical mechanism for erlotinib resistance, and that GLI1 may serve as an independent predictive biomarker for the efficacy of EGFR-TKIs in lung cancer.
If GLI1 is involved in TKI sensitivity, it may be envisioned that pharmacological inhibition of GLI1 function may sensitize the cells to TKI treatment. Indeed, the combination treatment of a GLI inhibitor and erlotinib synergistically suppressed proliferation of the H2170 and A549 lung cancer cell lines in vitro compared with that in each single agent alone (Fig. 4). Similarly, Bai et al (67) reported that A549 and H1975 cells, which had high levels of GLI1 expression, were resistant to another EGFR TKI, gefitinib, and the use of the SMO inhibitor, SANT-1, showed a synergistic effect with gefitinib in A549 and H1975 cell lines. Recently, it was reported that the downregulation of GLI1 using microRNA-873 in the PC9 lung cancer cell line enhanced its sensitivity to gefitinib (68), which is consistent with the current study. In the future, with the development of safe and efficient GLI1 inhibitors, combination treatment of different EGFR-TKIs and GLI inhibitors may be investigated.
In summary, the present study addressed the critical role that GLI1 may play in EGFR-TKI resistance in lung cancer and provides a novel molecular basis to develop novel strategies for the treatment of lung cancer. As GLI activation has been suggested to be a putative key control point of both canonical Hh signaling (29) and non-canonical oncogenic pathways, such as EGFR (41,42), combinations of GLI inhibitors with EGFR-TKIs could overcome the ineffectiveness of single agent treatments and may prolong the duration of clinical benefits from these agents.
Acknowledgements
The authors would like to acknowledge the contributions of Ms Pu Xue and Ms Fang Wang regarding the cell cultures (Zhejiang Provincial Key Laboratory of Applied Enzymology and Precision medicine Center of the Yangze Delta Region Institute of Tsinghua University Zhejiang).
Funding
This study was supported by the National Science Foundation for Young Scientists of China (grant no. 81902910), Zhejiang Provincial Natural Science Foundation of China (grant no. LY15H160048), Jiaxing Science and Technology Project (grant no. 2015BZ12001) and Jiaxing Nanhu District Science and Technology Project (grant no. 2018QC03).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Authors' contributions
ZD, YW and PZ performed IHC staining of clinical samples and analyzed the IHC data. XY, YW and ZD analyzed clinical outcome data. VD, MZ, CH, and YL performed experiments involving cell culture, drug treatments, and viability assays. VD and FZ performed RT-qPCR. ZD, FD and HS conceived the study, participated in its design and coordination, and drafted the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This investigation has been conducted in accordance to the ethical standards of the Declaration of Helsinki, as well as national and international guidelines, with the approval of the institutional review board of the University of California, San Francisco (UCSF). Studies involving patient tissues were approved by the Committee on Human Research (approval number: H8714-11647-10) at UCSF, and written informed consent was obtained from each patient prior to tissue specimen collection.
Patient consent for publication
Written consent for publishing results of the study involving tissue specimens was obtained from each patient prior to the study.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Boloker G, Wang C, Zhang J. Updated statistics of lung and bronchus cancer in United States (2018) J Thorac Dis. 2018;10:1158–1161. doi: 10.21037/jtd.2018.03.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Duma N, Santana-Davila R, Molina JR. Non-small cell lung cancer: Epidemiology, screening, diagnosis, and treatment. Mayo Clin Proc. 2019;94:1623–1640. doi: 10.1016/j.mayocp.2019.01.013. [DOI] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 5.Cronin KA, Lake AJ, Scott S, Sherman RL, Noone AM, Howlader N, Henley SJ, Anderson RN, Firth AU, Ma J, et al. Annual report to the nation on the status of cancer, part I: National cancer statistics. Cancer. 2018;124:2785–2800. doi: 10.1002/cncr.31551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng RM, Zong YN, Cao SM, Xu RH. Current cancer situation in China: Good or bad news from the 2018 global cancer statistics? Cancer Commun (Lond) 2019;39:22. doi: 10.1186/s40880-019-0368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.da Cunha Santos G, Shepherd FA, Tsao MS. EGFR mutations and lung cancer. Annu Rev Pathol. 2011;6:49–69. doi: 10.1146/annurev-pathol-011110-130206. [DOI] [PubMed] [Google Scholar]
- 8.Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 9.Selvaggi G, Novello S, Torri V, Leonardo E, De Giuli P, Borasio P, Mossetti C, Ardissone F, Lausi P, Scagliotti GV. Epidermal growth factor receptor overexpression correlates with a poor prognosis in completely resected non-small-cell lung cancer. Ann Oncol. 2004;15:28–32. doi: 10.1093/annonc/mdh011. [DOI] [PubMed] [Google Scholar]
- 10.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 11.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi S, Boggon TJ, Dayaram T, Jänne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, Halmos B. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 13.Gazdar AF. Activating and resistance mutations of EGFR in non-small-cell lung cancer: Role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene. 2009;28(Suppl 1):S24–S31. doi: 10.1038/onc.2009.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pao W, Wang TY, Riely GJ, Miller VA, Pan Q, Ladanyi M, Zakowski MF, Heelan RT, Kris MG, Varmus HE. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takezawa K, Pirazzoli V, Arcila ME, Nebhan CA, Song X, de Stanchina E, Ohashi K, Janjigian YY, Spitzler PJ, Melnick MA, et al. HER2 amplification: A potential mechanism of acquired resistance to EGFR inhibition in EGFR-mutant lung cancers that lack the second-site EGFRT790M mutation. Cancer Discov. 2012;2:922–933. doi: 10.1158/2159-8290.CD-12-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 17.Massarelli E, Varella-Garcia M, Tang X, Xavier AC, Ozburn NC, Liu DD, Bekele BN, Herbst RS, Wistuba II. KRAS mutation is an important predictor of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. Clin Cancer Res. 2007;13:2890–2896. doi: 10.1158/1078-0432.CCR-06-3043. [DOI] [PubMed] [Google Scholar]
- 18.Dragnev KH, Ma T, Cyrus J, Galimberti F, Memoli V, Busch AM, Tsongalis GJ, Seltzer M, Johnstone D, Erkmen CP, et al. Bexarotene plus erlotinib suppress lung carcinogenesis independent of KRAS mutations in two clinical trials and transgenic models. Cancer Prev Res (Phila) 2011;4:818–828. doi: 10.1158/1940-6207.CAPR-10-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Linardou H, Dahabreh IJ, Kanaloupiti D, Siannis F, Bafaloukos D, Kosmidis P, Papadimitriou CA, Murray S. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: A systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol. 2008;9:962–972. doi: 10.1016/S1470-2045(08)70206-7. [DOI] [PubMed] [Google Scholar]
- 20.Wu SG, Shih JY. Management of acquired resistance to EGFR TKI-targeted therapy in advanced non-small cell lung cancer. Mol Cancer. 2018;17:38. doi: 10.1186/s12943-018-0777-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merchant AA, Matsui W. Targeting hedgehog-a cancer stem cell pathway. Clin Cancer Res. 2010;16:3130–3140. doi: 10.1158/1078-0432.CCR-09-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Velcheti V, Govindan R. Hedgehog signaling pathway and lung cancer. J Thorac Oncol. 2007;2:7–10. doi: 10.1097/JTO.0b013e31802c0276. [DOI] [PubMed] [Google Scholar]
- 23.Amakye D, Jagani Z, Dorsch M. Unraveling the therapeutic potential of the hedgehog pathway in cancer. Nat Med. 2013;19:1410–1422. doi: 10.1038/nm.3389. [DOI] [PubMed] [Google Scholar]
- 24.Yuan Z, Goetz JA, Singh S, Ogden SK, Petty WJ, Black CC, Memoli VA, Dmitrovsky E, Robbins DJ. Frequent requirement of hedgehog signaling in non-small cell lung carcinoma. Oncogene. 2007;26:1046–1055. doi: 10.1038/sj.onc.1209860. [DOI] [PubMed] [Google Scholar]
- 25.Giroux-Leprieur E, Costantini A, Ding VW, He B. Hedgehog signaling in lung cancer: From oncogenesis to cancer treatment resistance. Int J Mol Sci. 2018;19:2835. doi: 10.3390/ijms19092835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niyaz M, Khan MS, Mudassar S. Hedgehog signaling: An achilles' heel in cancer. Transl Oncol. 2019;12:1334–1344. doi: 10.1016/j.tranon.2019.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sabol M, Trnski D, Musani V, Ozretić P, Levanat S. Role of GLI transcription factors in pathogenesis and their potential as new therapeutic targets. Int J Mol Sci. 2018;19:2562. doi: 10.3390/ijms19092562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruiz i Altaba A. Gli proteins encode context-dependent positive and negative functions: Implications for development and disease. Development. 1999;126:3205–3216. doi: 10.1242/dev.126.14.3205. [DOI] [PubMed] [Google Scholar]
- 29.Hui CC, Angers S. Gli proteins in development and disease. Annu Rev Cell Dev Biol. 2011;27:513–537. doi: 10.1146/annurev-cellbio-092910-154048. [DOI] [PubMed] [Google Scholar]
- 30.Niewiadomski P, Niedziółka SM, Markiewicz Ł, Uśpieński T, Baran B, Chojnowska K. Gli proteins: Regulation in development and cancer. Cells. 2019;8:147. doi: 10.3390/cells8020147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasaki H, Nishizaki Y, Hui C, Nakafuku M, Kondoh H. Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: Implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development. 1999;126:3915–3924. doi: 10.1242/dev.126.17.3915. [DOI] [PubMed] [Google Scholar]
- 32.Yang L, Xie G, Fan Q, Xie J. Activation of the hedgehog-signaling pathway in human cancer and the clinical implications. Oncogene. 2010;29:469–481. doi: 10.1038/onc.2009.392. [DOI] [PubMed] [Google Scholar]
- 33.Sekulic A, Migden MR, Oro AE, Dirix L, Lewis KD, Hainsworth JD, Solomon JA, Yoo S, Arron ST, Friedlander PA, et al. Efficacy and safety of vismodegib in advanced basal-cell carcinoma. N Engl J Med. 2012;366:2171–2179. doi: 10.1056/NEJMoa1113713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Basset-Séguin N, Hauschild A, Kunstfeld R, Grob J, Dréno B, Mortier L, Ascierto PA, Licitra L, Dutriaux C, Thomas L, et al. Vismodegib in patients with advanced basal cell carcinoma: Primary analysis of STEVIE, an international, open-label trial. Eur J Cancer. 2017;86:334–348. doi: 10.1016/j.ejca.2017.08.022. [DOI] [PubMed] [Google Scholar]
- 35.Lear JT, Migden MR, Lewis KD, Chang ALS, Guminski A, Gutzmer R, Dirix L, Combemale P, Stratigos A, Plummer R, et al. Long-term efficacy and safety of sonidegib in patients with locally advanced and metastatic basal cell carcinoma: 30-month analysis of the randomized phase 2 BOLT study. J Eur Acad Dermatol Venereol. 2018;32:372–381. doi: 10.1111/jdv.14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodon J, Tawbi HA, Thomas AL, Stoller RG, Turtschi CP, Baselga J, Sarantopoulos J, Mahalingam D, Shou Y, Moles MA, et al. A phase I, multicenter, open-label, first-in-human, dose-escalation study of the oral smoothened inhibitor Sonidegib (LDE225) in patients with advanced solid tumors. Clin Cancer Res. 2014;20:1900–1909. doi: 10.1158/1078-0432.CCR-13-1710. [DOI] [PubMed] [Google Scholar]
- 37.Kieran MW, Chisholm J, Casanova M, Brandes AA, Aerts I, Bouffet E, Bailey S, Leary S, MacDonald TJ, Mechinaud F, et al. Phase I study of oral sonidegib (LDE225) in pediatric brain and solid tumors and a phase II study in children and adults with relapsed medulloblastoma. Neuro Oncol. 2017;19:1542–1552. doi: 10.1093/neuonc/nox109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Minami H, Ando Y, Ma BB, Hsiang Lee J, Momota H, Fujiwara Y, Li L, Fukino K, Ito K, Tajima T, et al. Phase I, multicenter, open-label, dose-escalation study of sonidegib in Asian patients with advanced solid tumors. Cancer Sci. 2016;107:1477–1483. doi: 10.1111/cas.13022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ko AH, LoConte N, Tempero MA, Walker EJ, Kate Kelley R, Lewis S, Chang WC, Kantoff E, Vannier MW, Catenacci DV, et al. A Phase I study of FOLFIRINOX plus IPI-926, a hedgehog pathway inhibitor, for advanced pancreatic adenocarcinoma. Pancreas. 2016;45:370–375. doi: 10.1097/MPA.0000000000000458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jimeno A, Weiss GJ, Miller WH, Jr, Gettinger S, Eigl BJ, Chang AL, Dunbar J, Devens S, Faia K, Skliris G, et al. Phase I study of the hedgehog pathway inhibitor IPI-926 in adult patients with solid tumors. Clin Cancer Res. 2013;19:2766–2774. doi: 10.1158/1078-0432.CCR-12-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lauth M, Toftgård R. Non-canonical activation of GLI transcription factors: Implications for targeted anti-cancer therapy. Cell Cycle. 2007;6:2458–2463. doi: 10.4161/cc.6.20.4808. [DOI] [PubMed] [Google Scholar]
- 42.Mimeault M, Batra SK. Frequent deregulations in the hedgehog signaling network and cross-talks with the epidermal growth factor receptor pathway involved in cancer progression and targeted therapies. Pharmacol Rev. 2010;62:497–524. doi: 10.1124/pr.109.002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ou SI, Shirai K. Anaplastic lymphoma kinase (ALK) signaling in lung cancer. Adv Exp Med Biol. 2016;893:179–187. doi: 10.1007/978-3-319-24223-1_9. [DOI] [PubMed] [Google Scholar]
- 44.Garrido-Castro AC, Felip E. HER2 driven non-small cell lung cancer (NSCLC): Potential therapeutic approaches. Transl Lung Cancer Res. 2013;2:122–127. doi: 10.3978/j.issn.2218-6751.2013.02.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benvenuto M, Masuelli L, De Smaele E, Fantini M, Mattera R, Cucchi D, Bonanno E, Di Stefano E, Frajese GV, Orlandi A, et al. In vitro and in vivo inhibition of breast cancer cell growth by targeting the Hedgehog/GLI pathway with SMO (GDC-0449) or GLI (GANT-61) inhibitors. Oncotarget. 2016;7:9250–9270. doi: 10.18632/oncotarget.7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Srivastava RK, Kaylani SZ, Edrees N, Li C, Talwelkar SS, Xu J, Palle K, Pressey JG, Athar M. GLI inhibitor GANT-61 diminishes embryonal and alveolar rhabdomyosarcoma growth by inhibiting Shh/AKT-mTOR axis. Oncotarget. 2014;5:12151–12165. doi: 10.18632/oncotarget.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 48.Ishikawa M, Sonobe M, Imamura N, Sowa T, Shikuma K, Date H. Expression of the GLI family genes is associated with tumor progression in advanced lung adenocarcinoma. World J Surg Oncol. 2014;12:253. doi: 10.1186/1477-7819-12-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, Boutselakis H, Cole CG, Creatore C, Dawson E, et al. COSMIC: The catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47:D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Forbes SA, Tang G, Bindal N, Bamford S, Dawson E, Cole C, Kok CY, Jia M, Ewing R, Menzies A, et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): A resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 2010;38:D652–D657. doi: 10.1093/nar/gkp995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramos AH, Dutt A, Mermel C, Perner S, Cho J, Lafargue CJ, Johnson LA, Stiedl AC, Tanaka KE, Bass AJ, et al. Amplification of chromosomal segment 4q12 in non-small cell lung cancer. Cancer Biol Ther. 2009;8:2042–2050. doi: 10.4161/cbt.8.21.9764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mo ML, Chen Z, Zhou HM, Li H, Hirata T, Jablons DM, He B. Detection of E2A-PBX1 fusion transcripts in human non-small-cell lung cancer. J Exp Clin Cancer Res. 2013;32:29. doi: 10.1186/1756-9966-32-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bean J, Brennan C, Shih JY, Riely G, Viale A, Wang L, Chitale D, Motoi N, Szoke J, Broderick S, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci USA. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Minakata K, Takahashi F, Nara T, Hashimoto M, Tajima K, Murakami A, Nurwidya F, Yae S, Koizumi F, Moriyama H, et al. Hypoxia induces gefitinib resistance in non-small-cell lung cancer with both mutant and wild-type epidermal growth factor receptors. Cancer Sci. 2012;103:1946–1954. doi: 10.1111/j.1349-7006.2012.02408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cortes JR, Ambesi-Impiombato A, Couronné L, Quinn SA, Kim CS, da Silva Almeida AC, West Z, Belver L, Martin MS, Scourzic L, et al. RHOA G17V induces T follicular helper cell specification and promotes lymphomagenesis. Cancer Cell. 2018;33:259–273.e7. doi: 10.1016/j.ccell.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pratilas CA, Hanrahan AJ, Halilovic E, Persaud Y, Soh J, Chitale D, Shigematsu H, Yamamoto H, Sawai A, Janakiraman M, et al. Genetic predictors of MEK dependence in non-small cell lung cancer. Cancer Res. 2008;68:9375–9383. doi: 10.1158/0008-5472.CAN-08-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, Maglott DR. ClinVar: Public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bosco-Clément G, Zhang F, Chen Z, Zhou HM, Li H, Mikami I, Hirata T, Yagui-Beltran A, Lui N, Do HT, et al. Targeting Gli transcription activation by small molecule suppresses tumor growth. Oncogene. 2014;33:2087–2097. doi: 10.1038/onc.2013.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gialmanidis IP, Bravou V, Amanetopoulou SG, Varakis J, Kourea H, Papadaki H. Overexpression of hedgehog pathway molecules and FOXM1 in non-small cell lung carcinomas. Lung Cancer. 2009;66:64–74. doi: 10.1016/j.lungcan.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 60.Bora-Singhal N, Perumal D, Nguyen J, Chellappan S. Gli1-mediated regulation of Sox2 facilitates self-renewal of stem-like cells and confers resistance to EGFR inhibitors in non-small cell lung cancer. Neoplasia. 2015;17:538–551. doi: 10.1016/j.neo.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Armas-López L, Piña-Sánchez P, Arrieta O, de Alba EG, Ortiz-Quintero B, Santillán-Doherty P, Christiani DC, Zúñiga J, Ávila-Moreno F. Epigenomic study identifies a novel mesenchyme homeobox2-GLI1 transcription axis involved in cancer drug resistance, overall survival and therapy prognosis in lung cancer patients. Oncotarget. 2017;8:67056–67081. doi: 10.18632/oncotarget.17715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dimou A, Bamias A, Gogas H, Syrigos K. Inhibition of the hedgehog pathway in lung cancer. Lung Cancer. 2019;133:56–61. doi: 10.1016/j.lungcan.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 63.Nguyen KS, Kobayashi S, Costa DB. Acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancers dependent on the epidermal growth factor receptor pathway. Clin Lung Cancer. 2009;10:281–289. doi: 10.3816/CLC.2009.n.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, Bergethon K, Shaw AT, Gettinger S, Cosper AK, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cooke DT, Nguyen DV, Yang Y, Chen SL, Yu C, Calhoun RF. Survival comparison of adenosquamous, squamous cell, and adenocarcinoma of the lung after lobectomy. Ann Thorac Surg. 2010;90:943–948. doi: 10.1016/j.athoracsur.2010.05.025. [DOI] [PubMed] [Google Scholar]
- 66.Kawase A, Yoshida J, Ishii G, Nakao M, Aokage K, Hishida T, Nishimura M, Nagai K. Differences between squamous cell carcinoma and adenocarcinoma of the lung: Are adenocarcinoma and squamous cell carcinoma prognostically equal? Jpn J Clin Oncol. 2012;42:189–195. doi: 10.1093/jjco/hyr188. [DOI] [PubMed] [Google Scholar]
- 67.Bai XY, Zhang XC, Yang SQ, An SJ, Chen ZH, Su J, Xie Z, Gou LY, Wu YL. Blockade of hedgehog signaling synergistically increases sensitivity to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer cell lines. PLoS One. 2016;11:e0149370. doi: 10.1371/journal.pone.0149370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jin S, He J, Li J, Guo R, Shu Y, Liu P. MiR-873 inhibition enhances gefitinib resistance in non-small cell lung cancer cells by targeting glioma-associated oncogene homolog 1. Thorac Cancer. 2018;9:1262–1270. doi: 10.1111/1759-7714.12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.