Abstract
Giant cell tumor of bone (GCTB) is an intermediate (locally aggressive) bone tumor with a recurrence rate of >30% following surgery. GCTB recurrence is ultimately due to the proliferation of neoplastic stromal (NS) cells. However, the precise mechanism underlying the regulation of NS cell proliferation remains unknown. p62 protein is a multifunctional adaptor protein that exerts a positive role in bone tumors and metabolic bone diseases. In the present study, the mRNA and protein expression levels of p62 were detected by reverse transcription-quantitative PCR and western blotting, respectively, in 8 paired fresh GCTB tumor tissues and adjacent normal cancellous bone tissues. The association between p62 expression level and patient prognosis was subsequently analyzed in 54 paraffin-embedded tumor specimens by immunohistochemistry assay. NS cells were isolated from GCTB primary cell culture, and the role of p62 was evaluated using in vitro cell proliferation, migration and invasion assays. The results revealed that p62 mRNA and protein were overexpressed in tumor tissues. High p62 expression levels were significantly associated with the recurrence of GCTB (P=0.001). The patients in the high p62 expression group had shorter 5-year recurrence-free survival rates compared with the patients in the low p62 expression group (P<0.001). Cox regression analysis identified p62 expression as an independent prognostic indicator of the recurrence-free survival of patients with GCTB (P<0.001). The in vitro experiments revealed that p62 downregulation inhibited NS cell proliferation, invasion and migration, and promoted apoptosis. In conclusion, it was found that p62 overexpression is associated with the recurrence of GCTB via the promotion of NS cell proliferation. Therefore, p62 could be a novel prognostic indicator, and a potential therapeutic target for GCTB.
Keywords: p62/sequestosome 1, bone neoplasm, giant cell tumor of bone, recurrence, neoplastic stromal cells
Introduction
Giant cell tumor of bone (GCTB) is a common intermediate bone tumor that is locally aggressive and rarely metastasizes. Recurrence ranges from 16.7–39% after surgery alone, although the recurrence rate is very high (>80%) following drug withdrawal, and lung metastasis may occur in 1–3.2% of cases (1,2). In pathology, GCTB tissue is composed primarily of neoplastic stromal (NS) cells and multinucleated giant (MNG) cells. NS cells secrete receptor activator of nuclear factor κB ligand (RANKL), which binds to its receptor, RANK, leading to MNG cell fusion and excessive activation of osteoclasts (3). NS cells have tumor proliferation characteristics, which is the ultimate cause of GCTB recurrence (4). However, the mechanism by which NS cell proliferation is regulated has yet to be elucidated.
p62 (also known as sequestosome-1, SQSTM-1 or A170) is a crucial molecule in the regulation of cell growth, survival and proliferation (5). The p62 protein contains different types of protein-protein interaction domains, a characteristic that is consistent with its role as a versatile adaptor, especially for bone tumors and metabolic bone diseases (6) p62 is overexpressed in bone tumors, such as osteosarcoma (7) and myeloma (8). It is also associated with tumor invasion and metastasis (9). Moreover, p62 gene mutations are considered to be the main cause of Paget's disease of bone, which is a skeletal disorder characterized by excessive activation of osteoclasts, similar to GCTB (10). However, to date, the role of p62 in the mechanisms underlying the biology of GCTB has not been elucidated.
In the present study, mRNA and protein expression levels of p62 were detected by reverse transcription-quantitative PCR (RT-qPCR) and western blotting, respectively, in 8 paired fresh GCTB tumor tissues and adjacent normal cancellous bone tissues. The association between p62 expression levels and patient prognosis was subsequently analyzed in 54 paraffin-embedded GCTB specimens via immunohistochemistry assay. NS cells were isolated from GCTB primary cell cultures, and the role of p62 was evaluated with respect to the regulation of cell proliferation, invasion, migration and apoptosis in vitro. The results revealed that p62 mRNA and protein were overexpressed in tumor tissues, and that high p62 expression levels were significantly associated with the recurrence of GCTB. Furthermore, p62 downregulation led to the inhibition of NS cell proliferation, invasion and migration, and an increase in the rate of apoptosis. Therefore, it has been demonstrated that p62 overexpression is associated with GCTB recurrence via the promotion of NS cell proliferation, and p62 may be both a novel prognostic indicator and a potential therapeutic target for GCTB.
Materials and methods
Tissue specimens and patients
For RT-qPCR and western blotting assays, 8 paired GCTB tumor tissues and the adjacent normal cancellous bone tissues (at a distance >2 cm from the tumor) were collected during surgery between May 2017 and July 2018. In addition, 54 paraffin-embedded specimens of GCTB tumor tissues were collected between January 2008 and December 2014 for immunohistochemical testing. All cases were histologically and clinically diagnosed at The Third Affiliated Hospital of Kunming Medical University, Tumor Hospital of Yunnan Province (China). The median follow-up time of the patients was 77 months (ranging from 7–128 months). The protocol for the present study was approved by the Medical Institutional and Clinical Research Ethics Committee of Tumor Hospital of Yunnan Province. All patients included in the present study provided informed verbal consent.
RT-qPCR detection of p62 expression and point mutation
Total RNA was extracted using TRIzol (Thermo Fisher Scientific, Inc.) from fresh GCTB tissues. RT-qPCR was performed using the All-in-One™ First-Strand cDNA Synthesis kit (GeneCopoeia, Inc.). The temperature protocol was as follows: 42°C for 60 min, 70°C for 5 min. The quantitative primer sequences for p62 and GAPDH primer sequences were as follows: p62 forward, 5′-TCAGGCTGTGGATGAAGTGGAA-3′ and reverse, 5′-CAGAAGATGTTTGTGGCGAGGA-3′; and GAPDH forward, 5′-TGACTTCAACAGCGACACCCA-3′ and reverse, 5′-CACCCTGTTGCTGTAGCCAAA-3′. The 7th and 8th exon sequencing were as follows: Exon 7 primer forward, 5′-AGACCCCTGCAGCCTTAACT-3′ and reverse, 5′-CCAACTCCTAACCTCCCACA-3′; and exon 8 primer forward, 5′-AGTTGAGCAGTGTGAAAAAGA-3′ and reverse, 5′-GCAGGGAGGGGTCAGGAGCGC-3′. RT-qPCR sequencing was performed using the SYBR-Green Master with Rox kit (GeneCopoeia, Inc.). The reaction conditions for qPCR were 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec, 60°C for 30 sec and 72°C for 30 sec. The mRNA expression levels in each group were quantified using the 2−ΔΔCq method (11). Sequencing of the 7 and 8th exon amplification products was performed.
Western blot assay
Fresh GCTB tissues were harvested using RIPA lysis buffer (Beyotime Institute of Biotechnology). Protein concentrations were measured using the bicinchoninic acid protein assay kit (Beyotime Institute of Biotechnology). The total protein of each specimen (30 mg/lane) were separated by SDS-PAGE (10% gels), and then transferred onto a polyvinylidene difluoride membrane (EMD Millipore). Β-actin was used as a loading control. The membrane was blocked with 5% bovine serum albumin (BSA, Beijing Solarbio Science & Technology Co., Ltd.) at room temperature for 40 min, and subsequently incubated with mouse anti-p62 (1:1,500; cat. no. ab56416; Abcam), mouse anti-RANKL (1:1,000; cat. no. ab45039; Abcam), mouse anti-RANK (1:1,000; cat. no. ab13918; Abcam) and mouse anti-β-actin (1:5,000; cat. no. ab6276; Abcam) primary antibodies overnight at 4°C. The membranes were then incubated with peroxidase-conjugated goat anti-mouse IgG (1:20,000; cat. no. A4416; Sigma-Aldrich; Merck KGaA) at room temperature for 1 h. Protein bands were visualized using SuperSignal™ West Femto Maximum Sensitivity Substrate reagents (Thermo Fisher Scientific, Inc.). The relative gray value of the immune reactive bands was compared using ImageJ software (version 1.46, National Institutes of Health).
Immunohistochemistry assay
In brief, GCTB tissues were fixed in 10% formaldehyde for 12 h at room temperature. Then paraffin-embedded specimens of GCTB tumor tissues were cut into 4-µm sections and baked at 65°C for 30 min. The sections were washed with xylene and rehydrated with 70, 80, 90 and 100% graded ethanol solutions. Tissue sections were submerged for 2 min into an EDTA buffer a 95°C and 90 kPa for antigen retrieval. Subsequently, the sections were treated with 3% hydrogen peroxide in methanol, followed by incubation with 1% rabbit serum albumin (Cell Signaling Technology, Inc.) at room temperature for 10 min. The specimens were incubated overnight at 4°C with an anti-p62 antibody (1:800; cat. no. 16177S; Cell Signaling Technology, Inc.). The specimens were then incubated with SignalStain® Boost IHC Detection Reagent (1:1,000; cat. no. 8114P; Cell Signaling Technology, Inc.) at 37°C for 30 min. The degree of immunostaining of sections was reviewed by light microscope and scored by two independent pathologists. p62 cytoplasmic staining was scored from 0–3 as follows: i) 0, no or faint staining; ii) 1, weak staining; iii) 2, moderate staining; and iv) 3, strong staining. p62 nuclear staining was scored as follows: i) 0, nuclear staining visible in <10% of nuclei; ii) 1, nuclear staining visible in >10% of nuclei. The total score was calculated, and the samples were divided into low expression (0–2) or high expression (3–4) groups (12).
GCTB NS cells isolation and culture
GCTB fresh tissue was placed in 40 ml HANK's balanced salt solution and stored at 0–4°C. The tissue was washed three times with saline containing 3% penicillin-streptomycin double antibody (cat. no. 516106, diluted in 10,000 IU penicillin/ml and 10,000 µg streptomycin/ml; EMD Millipore). Twice the tissue weight of pancreatin (1 g/ml; Thermo Fisher Scientific, Inc.) was added to digest tissue at 37°C for 3 min, aspirate the solution, neutralize with an equal volume of 15% fetal bovine serum (FBS, Gibco; Thermo Fisher Scientific, Inc.), mix well, and incubated at 4°C for 2 h. 0.25%. In total, 20 ml Trypsin (EMD Millipore) was continuously added to the tissue fragments and the aforementioned steps were repeated six times. The remaining tissue was then resuspended with collagenase digestion solution (DMEM, 10% FBS, 3% double antibody, 5 mg/ml collagenase II, 0.22 µm filtration, all Biosharp Life Sciences). The tissues were then shaken at 200 rpm on Rocking Device for 2 h at 37°C (CellNest Shaker; http://www.shbiotech.com/). The digested cells were filtered with a 70-µm filter and washed twice with PBS. NS cells were cultured in DMEM/F12 supplemented with 15% FBS (Gibco; Thermo Fisher Scientific) and 2% streptomycin/penicillin (EMD Millipore).
Cell senescence assay
First, the NS cells were seeded in 6-well plates (EMD Millipore). Then culture fluid was aspirated, washed once with PBS and 1 ml β-galactosidase staining fixative was added (Beijing Solarbio Science & Technology Co., Ltd.), and cells were fixed at room temperature for 15 min. Then the cell fixative was aspirated and the cells were washed three times with PBS, each for 3 min. Then PBS was aspirated and 1 ml staining working solution (Beijing Solarbio Science & Technology Co., Ltd.) was added to each well. Then 300 µl dyeing working fluid was added in each well and incubated at 37°C overnight. The 6-well plate was sealed with parafilm or plastic wrap to prevent evaporation. Lastly, cells were observed under a light microscope at 200× magnification.
Lentivirus infection of NS cells
p62 (NM_003900) overexpression plasmid (pcDNA3.1-p62) was purchased from GeneCopoeia Co., Ltd. (cat. no. EX-M0245-Lv201). Lv201 empty lentiviral vector was purchased from GeneCopoeia (cat. no. EX-NEG-Lv201) for negative control. For p62 knockdown, short hairpin RNA (p62-shRNA, cat. no. HSH021660-LVRU6GP) and scrambled shRNA (cat. no. SHCTR001-LVRU6GP) were both purchased from GeneCopoeia. The shRNA sequences were as follows: p62-shRNA, 5′-CCATCCAGTATTCAAAGCATC-3′ and scrambled shRNA, 5′-GCTTCGCGCCGTAGTCTTA-3′. Transfections of the NS cells with plasmids (1 µg/well) and shRNA (50 nm) were performed using a Lipofectamine® 3000 kit (Gibco; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocols. Cell density was 106 cells/25 cm dish. Puromycin (1 µg/ml) was used to kill any cells that were not successfully transfected. NS cells either stably expressing, or silenced for p62 were constructed. Subsequent experimentation was conducted after transfection for 48 h. RT-qPCR was used to verify the expression of p62 mRNA, and western blotting was performed to evaluate the expression of p62 protein. NS cells were split into five groups, including: GCTB control, normal NS cells; shp62, silenced p62 gene lentivirus-transfected NS cells; NCshp62, shp62 scrambled control; mp62, exogenous p62 gene expression lentivirus-transfected NS cells and NCmp62, mp62 negative control (Lv201 empty vector).
Cell proliferation assay
NS cells stably expressing p62 were plated in 96-well plates in DMEM with 15% FBS (both Gibco; Thermo Fisher Scientific, Inc.) at a density of 5,000 cells/well. To quantify cell viability, cultures were stained after 1–4 and 5 days; 10 µl Cell Counting Kit-8 (CCK-8, Dojindo Molecular Technologies, Inc.) working solution was then added into the wells for 2 h at 37°C according to the manufacturer's instructions, after which the absorbance was measured at 450 nm using an Epoch Multi-Volume Spectrophotometer system (BioTek Instruments, Inc.).
Cell apoptosis assay
Cell early and late apoptosis assays were performed using an Annexin V-Fluorescein Isothiocyanate Apoptosis Detection kit (BD Biosciences) and NS cells stably expressing p62. All cells were collected, resuspended in 1X binding buffer, and stained with Annexin V-FITC and propidium iodide at room temperature for 5 min, followed by flow cytometry detection (Attune NxT; Thermo Fisher Scientific, Inc.). The rate of early and late apoptosis was analyzed using CellQuest™ software (version 3.3; BD Biosciences).
Cell invasion and migration assays
Cells (1×105), suspended in DMEM containing 0.1% BSA were added to the top of the Boyden chamber (EMD Millipore) at 37°C for 2 h, which was coated with 50 µl Matrigel (1 mg/ml final concentration). The lower chamber contained 10% serum-containing medium. After incubation for 24 h at 37°C, a Transwell chamber (EMD Millipore) was used to determine cell invasion and migration ability. A total of 1×105 lentivirus-infected NS cells were suspended in DMEM containing 10% FBS at a density of 5,000 cells/well. Cells were subsequently placed onto the top of each chamber. Medium containing 10% FBS was added to the bottom of the chamber. The cells were incubated for 24 h, and then cells on the upside of the membrane were wiped off to remove the non-migrated cells. Cells that had migrated to the underside of the membrane were stained with crystal violet at room temperature for 20 min and visualized under Nikon Eclipse TE2000-U microscope. A total of 4 random fields (magnification, ×100) were scanned and analyzed using the aforementioned ImageJ software.
Statistics analysis
Statistical analyses of immunohistochemistry assay were performed using the SPSS 17.0 software package (SPSS, Inc.). The significance of the differences between groups was estimated using the χ2 test. Recurrence-free survival curves were plotted according to the Kaplan-Meier method, and compared using the log-rank test. The significance of survival variables was evaluated using a multivariate Cox proportional hazards regression analysis. Statistical graphs were drawn using GraphPad Prism v.6.0 (GraphPad Software, Inc.). The differences of p62 mRNA and protein expression levels between two groups of GCTB tumor tissues and the adjacent normal cancellous bone tissues were tested with a Student's t-test. The differences of p62 protein expression levels, NS cell proliferation rates and invasion cell numbers between multiple groups were tested by one-way ANOVA with post-hoc Tukey's test. All data are presented as the mean ± standard deviation of three independent experiments. P<0.05 was considered to indicate a statistically significant difference.
Results
p62 mRNA and protein are overexpressed in fresh GCTB tumor tissues
In 8 paired fresh GCTB tumor tissues and adjacent normal cancellous bone tissues, p62 mRNA and protein were overexpressed in tumors compared with normal tissues, as determined by RT qPCR and western blot assays, respectively (Fig. 1A). Gene sequencing revealed that the G1340A mutation occurred at the 1,340 bp position in two tumor specimens between exons 7 and 8 of the p62 gene (Fig. 1B). However, the amino acid R (arginine) at position 415 was not changed.
Figure 1.
p62 expression in GCTB tissues. (A) Both p62 mRNA and protein expression levels were higher in GCTB tissues compared with adjacent normal cancellous bone tissues. (B) Gene sequencing revealed that the G1340A mutation occurred at the 1,340 bp position (red arrow) in two GCTB specimens. *P<0.05, **P<0.01. N, adjacent normal tissue; T, tumor tissue; GCTB, giant cell tumor of bone.
p62 overexpression is associated with poor prognosis of patients with GCTB
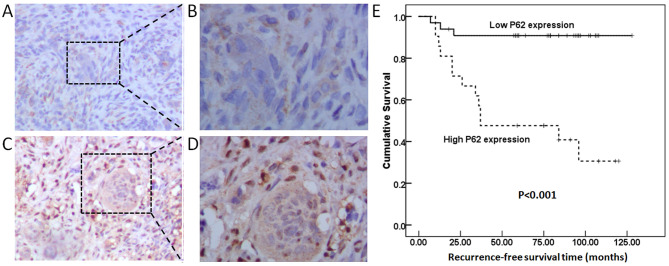
As shown in Fig. 2A-D, and as demonstrated by strong cytoplasmic and nuclear staining, the p62 protein was widely expressed in NS and MNG cells. To assess the association between p62 expression levels and clinicopathological features, the GCTB specimens were classified into a high p62 expression level group (n=20) and a low p62 expression level group (n=34). High p62 expression levels were significantly associated with GCTB recurrence (P=0.001; Table I). The patients in the high p62 expression level group had shorter 5-year recurrence-free survival rates compared with patients in the low p62 expression level group (P<0.001; Fig. 2E). Cox regression analysis identified p62 expression as an independent prognostic indicator for recurrence-free survival of patients with GCTB (P<0.001; Table II).
Figure 2.
p62 protein expression as determined by immunohistochemistry assay. Low p62 expression at magnification (A) ×100 and (B) ×200. High p62 expression at magnification (C) ×100 and (D) ×200. (E) High p62 expression was associated with shorter recurrence-free survival rates in patients with GCTB (P<0.001). GCTB, giant cell tumor of bone.
Table I.
Association of p62 expression with clinicopathological features of patients with giant cell tumor of bone.
| p62 expression | ||||
|---|---|---|---|---|
| Clinicopathological features | Number of cases | Low, n (%) | High, n (%) | P-value |
| Age, years | ||||
| ≤40 | 43 | 29 (67.4) | 14 (32.6) | 0.178 |
| >40 | 11 | 5 (45.5) | 6 (54.5) | |
| Sex | ||||
| Male | 28 | 20 (71.4) | 8 (28.6) | 0.181 |
| Female | 26 | 14 (53.8) | 12 (46.2) | |
| Recurrence | ||||
| Yes | 16 | 3 (18.8) | 13 (81.2) | 0.001 |
| No | 38 | 31 (81.6) | 7 (18.4) | |
| Pathological fracture | ||||
| Yes | 40 | 26 (65) | 14 (35) | 0.600 |
| No | 14 | 8 (57.1) | 6 (42.9) | |
| Campanacci grade | ||||
| I | 2 | 1 (50) | 1 (50) | 0.906 |
| II | 9 | 6 (66.7) | 3 (33.3) | |
| III | 43 | 27 (62.8) | 16 (37.25) | |
Table II.
Cox regression analysis of recurrence-free survival in 54 patients with giant cell tumor of bone.
| 95% CI for Exp (B) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Clinicopathological features | B | SE | Wald | df | Sig | Exp(B) | Lower | Upper |
| Age | −0.073 | 0.625 | 0.014 | 1 | 0.907 | 0.930 | 0.273 | 3.165 |
| Sex | −0.048 | 0.534 | 0.008 | 1 | 0.929 | 0.954 | 0.335 | 2.716 |
| Pathological fracture | 0.136 | 0.817 | 0.028 | 1 | 0.868 | 1.146 | 0.231 | 5.678 |
| Campanacci grade | 0.342 | 0.769 | 0.198 | 1 | 0.657 | 1.407 | 0.312 | 6.349 |
| p62 expression | −2.227 | 0.650 | 11.753 | 1 | 0.001 | 0.108 | 0.030 | 0.385 |
B, regression coefficient; SE, standard error; Wald, χ2 value; df, degree of freedom; Sig, significance; Exp(B), odds ratio; CI, confidence interval.
p62 downregulation inhibits the proliferation, invasion and migration of NS cells
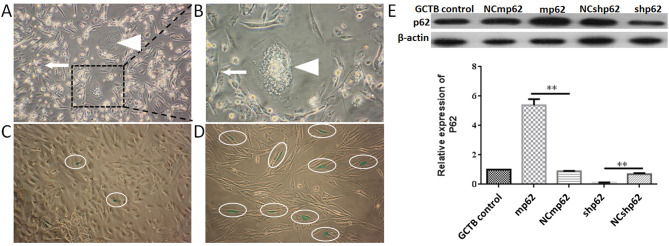
GCTB primary cells were isolated, and it was observed that the primary cells were mainly composed of spindle-shaped NS cells and osteoclast-like MNG cells. After 2–3 passage generations, it was found that the population of MNG cells gradually decreased. In the 4th passage generation, only NS cells remained (Fig. 3A-B). To further ensure that only NS cells remained in the 4th passage generation, western blotting was used to measure the expression of two markers (RANKL and RANK; Fig. S1). A cell senescence assay indicated more and more aging NS cells emerged from 4th generation (Fig. 3C and D). Therefore, 4th generation cells were used for subsequent experiments.
Figure 3.
GCTB primary cell culture and passage. (A) GCTB primary cells 48 h after passage (magnification, ×100). Cells are mainly composed of spindle shaped NS cells (white arrow) and osteoclast like multinucleated giant cells (white triangle). (B) GCTB primary cells 48 h after passage (magnification, ×400). NS cells (white arrow) and multinucleated giant cells (white triangle) were marked. (C) β galactosidase staining assay (green staining was indicted by white oval) of NS cells at the 4th passage (magnification, ×200). Cells grew well, and the senescence rate was 2.02±0.21%. (D) NS cells at the 7th passage became tenuous (magnification, ×200), and the senescence rate was 8.40±1.14%. (E) Lentivirus infection of NS cells, **P<0.01. GCTB control, normal NS cells; shp62, silenced p62 gene lentivirus transfected NS cells; NCshp62, shp62 scrambled control; mp62, exogenous p62 gene expression lentivirus transfected NS cells; NCmp62, mp62 negative control; GCTB, giant cell tumor of bone; NS, neoplastic stromal.
Subsequently, the expression of p62 was either downregulated or upregulated using lentiviral plasmid transfection (Fig. 3E). The differences between the silenced p62 gene lentivirus-transfected NS cells (the shp62 downregulation group) and the scrambled control (NCshp62) group were analyzed. The percentage of early and late apoptotic NS cells in the shp62 group was 47.33%, whereas the percentage of early and late apoptotic cells in the NCshp62 group was 16.58% (Fig. 4A and B). The difference in proliferation measured by relative OD values between the two groups was found to be statistically significant on day 4 (P=0.0343) and on day 5 (P=0.0305) (Fig. 4C). Downregulation of p62 also led to the inhibition of NS cell invasion (P=0.042; Fig. 5B) and migration (P=0.002; Fig. 5C).
Figure 4.
Effect of p62 downregulation on proliferation of NS cells. (A) Flow cytometry was performed to detect NS cell early and late apoptosis after p62 downregulation. (B) p62 downregulation promotes early and late apoptosis of NS cells. (C) p62 downregulation inhibits the proliferation of NS cells. *P<0.05, **P<0.01. GCTB control, normal NS cells; shp62, silenced p62 gene lentivirus-transfected NS cells; NCshp62, shp62 negative control; mp62, exogenous p62 gene expression lentivirus-transfected NS cells; NCmp62, mp62 negative control; GCTB, giant cell tumor of bone; NS, neoplastic stromal.
Figure 5.
p62 downregulation inhibits the invasion and migration of NS cells. (A) Invasion and migration of NS cells with different p62 expression (scale bar, 200 µm). (B) p62 downregulation inhibits the invasion of NS cells. (C) p62 downregulation inhibits the migration of NS cells. *P<0.05, **P<0.01. GCTB control, normal NS cells; shp62, silenced p62 gene lentivirus-transfected NS cells; NCshp62, shp62 scrambled control; mp62, exogenous p62 gene expression lentivirus-transfected NS cells; NCmp62, mp62 negative control; GCTB, giant cell tumor of bone; NS, neoplastic stromal.
Discussion
The overexpression of p62 has been detected among a melanoma and hepatocellular carcinoma (13–15), playing positive roles in both tumorigenesis and metastasis (16). Recently, p62 was demonstrated to be an effective oncotarget for solid tumors. In a multicenter phase I/IIa clinical trial, a p62 vaccine (Elenagen) alone or in combination with chemotherapy resulted in stable disease for 8–32 weeks in 44% of patients with advanced solid tumors (17). It also brought new hope for identifying potential therapeutic targets of GCTB. In the present study, overexpression of p62 mRNA and protein was observed, indicating that p62 may contribute to GCTB initiation. It was also possible to exclude the possibility of a p62 gene mutation between exon 7 and exon 8 (P392L) in GCTB, which is the most common mutation area of p62 in Paget's disease of bone (18). These results revealed that, in the GCTB tumor microenvironment, p62 overexpression may be associated with tumorigenesis that is unrelated to bone metabolism. High p62 expression levels were associated with GCTB recurrence, a finding which agreed with previous findings in other bone tumors (6,7). Finally, p62 overexpression was identified as an independent indicator of poor prognosis of GCTB. To the best of our knowledge, this is the first study that has analyzed the role of p62 in GCTB prognosis.
GCTB is not a malignant tumor, and NS cells do not have the ability to proliferate infinitely. This also explains the lack of standard GCTB cell lines that have been available to us for research purposes to date (19). To clarify the cellular and molecular mechanism of GCTB recurrence, NS cells were isolated from GCTB primary cell cultures. Fourth generation cells were used for experiments in which the cells maintained a rapid proliferation rate and stable growth characteristics during this period. Functional assays revealed that downregulation of p62 could inhibit the proliferation, invasion and migration of NS cells in GCTB. These results were consistent with most previous findings in other bone tumors (6). The present results also demonstrated that downregulation of p62 could promote apoptosis of NS cells in GCTB, a finding which provides a novel clue for the proliferative regulatory mechanism of p62.
For advanced GCTB, recurrence is often observed after denosumab withdrawal, in addition to surgery (20). The first fully humanized monoclonal antibody against RANKL, denosumab has achieved a good response in GCTB (21,22). However, the reason for the rapid recurrence after denosumab withdrawal has yet to be identified. Previous studies demonstrated that the expression of RANKL/RANK was downregulated following denosumab treatment, although the proliferation of NS cells was not inhibited; indeed, the rate even increased (23,24). In the present study, it has been demonstrated that p62 is able to promote the proliferation of NS cells. This finding provides a further explanation towards the understanding of GCTB recurrence after denosumab withdrawal.
The present study does, however, have some limitations. First, it was a single-center study, and therefore the number of clinical specimens to analyze was small. Secondly, although the positive role of p62 in the proliferation of NS cells was identified, the underlying signaling pathway associated with p62 adaptor function is not clear. Consequently, our future studies will focus on the interaction between p62 and other key regulators of GCTB, such as RANKL and RANK.
In conclusion, to the best of our knowledge, the present study is the first to have shown that p62 is overexpressed in GCTB tumor tissues. High p62 expression levels were associated with GCTB recurrence. Additionally, p62 overexpression was shown to promote NS cell proliferation by regulating apoptosis. We propose that p62 could be a novel prognostic indicator and a potential therapeutic target for GCTB.
Supplementary Material
Acknowledgements
Not applicable.
Funding
This work was supported by the National Nature Science Foundation of China (grant no. 81760486), and Science and Technology Plan of Yunnan province (grant no. 2018FB133).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JZ designed this research and revised the manuscript. SL and FY performed the western blot analysis, RT-qPCR assays and cell experiments, and wrote the manuscript. DL and CH performed the immunohistochemistry assays. HH analyzed the data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The protocol for the present study was approved by the Medical Institutional and Clinical Research Ethics Committee of Yunnan Tumor Hospital (approval no. KY202030; Kunming, China). All patients included in the present study previously provided informed verbal consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Mavrogenis AF, Igoumenou VG, Megaloikonomos PD, Panagopoulos GN, Papagelopoulos PJ, Soucacos PN. Giant cell tumor of bone revisited. SICOT J. 2017;3:54. doi: 10.1051/sicotj/2017041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Niu X, Zhang Q, Hao L, Ding Y, Li Y, Xu H, Liu W. Giant cell tumor of the extremity: Retrospective analysis of 621 Chinese patients from one institution. J Bone Joint Surg Am. 2012;94:461–467. doi: 10.2106/JBJS.J.01922. [DOI] [PubMed] [Google Scholar]
- 3.Lüke J, Hasenfratz M, Möller P, Barth TFE. New aspects on giant cell tumor of bone. Pathologe. 2018;39:125–131. doi: 10.1007/s00292-017-0391-9. (In German) [DOI] [PubMed] [Google Scholar]
- 4.Zhou Z, Li Y, Xu L, Wang X, Chen S, Yang C, Xiao J. Biological characteristics of a novel giant cell tumor cell line derived from spine. Tumour Biol. 2016;37:9681–9689. doi: 10.1007/s13277-016-4867-7. [DOI] [PubMed] [Google Scholar]
- 5.Fan L, Yin S, Zhang E, Hu H. Role of p62 in the regulation of cell death induction. Apoptosis. 2018;23:187–193. doi: 10.1007/s10495-018-1445-z. [DOI] [PubMed] [Google Scholar]
- 6.Sánchez-Martín P, Saito T, Komatsu M. p62/SQSTM1: ‘Jack of all trades’ in health and cancer. FEBS J. 2019;286:8–23. doi: 10.1111/febs.14712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu Y, Wang Q, Zhou Y, Sun L, Hu B, Xue H, Li M, Zhang K, Ren C, Duan N, et al. Overexpression of p62 is associated with poor prognosis and aggressive phenotypes in osteosarcoma. Oncol Lett. 2018;15:9889–9895. doi: 10.3892/ol.2018.8579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sha Z, Schnell HM, Ruoff K, Goldberg A. Rapid induction of p62 and GABARAPL1 upon proteasome inhibition promotes survival before autophagy activation. J Cell Biol. 2018;217:1757–1776. doi: 10.1083/jcb.201708168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J, Yang Z, Dong J. P62: An emerging oncotarget for osteolytic metastasis. J Bone Oncol. 2016;5:30–37. doi: 10.1016/j.jbo.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaw B, Burrel CL, Green D, Navarro-Martinez A, Scott D, Daroszewska A, van't Hof R, Smith L, Hargrave F, Mistry S, et al. Molecular insights into an ancient form of Paget's disease of bone. Proc Natl Acad Sci USA. 2019;116:10463–10472. doi: 10.1073/pnas.1820556116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 12.Langer R, Neppl C, Keller MD, Schmid R, Tschan MP, Berezowska S. Expression analysis of autophagy related indicators LC3B, p62 and HMGB1 indicate an autophagy-independent negative prognostic impact of high p62 expression in pulmonary squamous cell carcinomas. Cancers (Basel) 2018;10:281. doi: 10.3390/cancers10090281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karras P, Riveiro-Falkenbach E, Cañón E, Tejedo C, Calvo TG, Martínez-Herranz R, Alonso-Curbelo D, Cifdaloz M, Perez-Guijarro E, Gómez-López G, et al. p62/SQSTM1 fuels melanoma progression by opposing mRNA decay of a selective set of pro-metastatic factors. Cancer Cell. 2019;35:46–63 e10. doi: 10.1016/j.ccell.2018.11.008. [DOI] [PubMed] [Google Scholar]
- 14.Xing M, Li P, Wang X, Li J, Shi J, Qin J, Zhang X, Ma Y, Francia G, Zhang JY. Overexpression of p62/IMP2 can promote cell migration in hepatocellular carcinoma via activation of the Wnt/β-catenin pathway. Cancers (Basel) 2019;12:7. doi: 10.3390/cancers12010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Umemura A, He F, Taniguchi K, Nakagawa H, Yamachika S, Font-Burgada J, Zhong Z, Subramaniam S, Raghunandan S, Duran A, et al. p62, upregulated during preneoplasia, induces hepatocellular carcinogenesis by maintaining survival of stressed HCC-Initiating cells. Cancer Cell. 2016;29:935–948. doi: 10.1016/j.ccell.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moscat J, Karin M, Diaz-Meco MT. p62 in Cancer: Signaling adaptor beyond autophagy. Cell. 2016;167:606–609. doi: 10.1016/j.cell.2016.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ponomarenko DM, Klimova ID, Chapygina YA, Dvornichenko VV, Zhukova NV, Orlova RV, Manikhas GM, Zyryanov AV, Burhanova LA, Badrtdinova II, et al. Safety and efficacy of P62 DNA vaccine ELENAGEN in a first-in-human trial in patients with advanced solid tumors. Oncotarget. 2017;8:53730–53739. doi: 10.18632/oncotarget.16574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singer FR. Paget's disease of bone-genetic and environmental factors. Nat Rev Endocrinol. 2015;11:662–671. doi: 10.1038/nrendo.2015.138. [DOI] [PubMed] [Google Scholar]
- 19.Cowan RW, Singh G. Giant cell tumor of bone: A basic science perspective. Bone. 2013;52:238–246. doi: 10.1016/j.bone.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 20.Lipplaa A, Dijkstra S, Gelderblom H. Challenges of denosumab in giant cell tumor of bone, and other giant cell-rich tumors of bone. Curr Opin Oncol. 2019;31:329–335. doi: 10.1097/CCO.0000000000000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Branstetter DG, Nelson SD, Manive JC, Blay JY, Chawla S, Thomas DM, Jun S, Jacobs I. Denosumab induces tumor reduction and bone formation in patients with giant-cell tumor of bone. Clin Cancer Res. 2012;18:4415–4424. doi: 10.1158/1078-0432.CCR-12-0578. [DOI] [PubMed] [Google Scholar]
- 22.Chawla S, Henshaw R, Seeger L, Choy E, Blay JY, Ferrari S, Kroep J, Grimer R, Reichardt P, Rutkowski P, et al. Safety and efficacy of denosumab for adults and skeletally mature adolescents with giant cell tumor of bone: Interim analysis of an open-label, parallel-group, phase 2 study. Lancet Oncol. 2013;14:901–908. doi: 10.1016/S1470-2045(13)70277-8. [DOI] [PubMed] [Google Scholar]
- 23.Mukaihara K, Suehara K, Kohsaka S, Akaike K, Tanabe T, Kubota D, Ishii M, Fujimura T, Kazuno S, Okubo T, et al. Protein expression profiling of giant cell tumors of bone treated with denosumab. PLoS One. 2016;11:e0148401. doi: 10.1371/journal.pone.0148401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lau CP, Huang L, Wong KC, Kumta SM. Comparison of the anti-tumor effects of denosumab and zoledronic acid on the neoplastic stromal cells of giant cell tumor of bone. Connect Tissue Res. 2013;54:439–449. doi: 10.3109/03008207.2013.848202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.