Abstract
Tumor necrosis factor-related apoptosis inducing ligand (TRAIL)-mediated apoptosis is a safe method for the treatment of various types of cancer. However, TRAIL therapy is less effective in certain types of cancer, including cervical cancer. To address this problem, a combinatorial approach was employed to sensitize cervical cancer at low dosages. YM155, a survivin inhibitor, was used at low dosages along with TRAIL to induce apoptosis in HeLa cells. The effects of the individual treatment with TRAIL and YM155 on apoptosis were assessed by propidium iodide assay. In addition, to validate the DNA damage exhibited by the combination treatment, the phosphorylation status of γH2A histone family member X was investigated by immunofluorescence and western blot analysis. TRAIL or YM155 alone had no significant effect on DNA damage and apoptosis. However, the TRAIL/YM155 combination triggered a synergistic pro-apoptotic stimulus in HeLa cells. The mRNA and protein levels of CASP8- and FADD-like apoptosis regulator (cFLIP), death receptor 5 (DR5) and survivin were monitored using RT-PCR and western blot analysis, respectively. This combinatorial approach downregulated both mRNA and protein expression levels of cFLIP and survivin. Further experimental results suggested that the combination treatment significantly reduced cell viability, invasion and migration of HeLa cells. Overall, the present findings indicated that the low dosage of YM155 sensitized HeLa cells to TRAIL-induced apoptosis via a mechanism involving downregulation of cFLIP and survivin. The results indicated the importance of combination drug treatment and reveal an effective therapeutic alternative for TRAIL therapy in human cervical cancer.
Keywords: tumor necrosis factor-related apoptosis inducing ligand therapy, YM155, HeLa cells, apoptosis, CASP8- and FADD-like apoptosis regulator, death receptor 5, survivin
Introduction
Cervical cancer is the fourth most prevalent type of cancer in females globally and a significant cause of mortality in developing countries (1). Cervical cancer may develop due to persistent high-risk human papillomavirus (HPV) infection (2). Progress has been made in understanding the HPV genome replication during the viral life cycle; however, accurate surveillance strategies and targeted therapies are still required to eradicate this disease (3).
Tumor necrosis factor-related apoptosis inducing ligand (TRAIL) is a powerful cancer cell apoptosis-inducing factor that can induce apoptosis in a p53-independent manner (4). However, TRAIL resistance in cancer cells is a significant barrier to improve TRAIL-based clinical therapies (5). Therefore, agents are required to either improve the impact of TRAIL or overcome resistance to it (6,7). Several studies have demonstrated that the cervical cancer HeLa cell line is resistant to TRAIL-induced apoptosis, and the mechanism of resistance is not fully understood (8,9). Various mechanisms have been proposed for TRAIL resistance (10,11). Upregulation of CASP8- and FADD-like apoptosis regulator (cFLIP) and downregulation of death receptors (DR4 and DR5) and the Bcl-2 family (Bcl-2, Bcl-xL and MCL1 apoptosis regulator, BCL2 family member) are the most commonly proposed TRAIL resistance mechanisms (12).
Survivin is an anti-apoptotic molecule that is generally overexpressed in malignant cells (13). Survivin is a member of the inhibitor of apoptosis gene family, which regulates apoptosis and the cell cycle (13,14). Survivin has been reported to interact with and inhibit caspases to decrease apoptosis (15). Survivin is expressed in various types of cancer, including breast cancer (16), cervical cancer (17), non-small cell lung cancer (18) and osteosarcoma (19). Its upregulation is associated with survival and resistance to TRAIL-based therapy (20,21).
Small molecule inhibitor YM155, a survivin suppressor, inhibits cell proliferation and mediates apoptosis in breast (22), gastric (23), liver (24), and pancreatic (25) cancer models. Other than suppressing the levels of survivin in various types of cancer, YM155 has also been reported to have strong apoptogenic properties (26,27). Additionally, it has been reported that YM155 has an anticancer effect on breast (22), esophageal (28), colon (29) and bone marrow (30) cancer and sensitizes different types of cancer cells to TRAIL-mediated apoptosis (31). However, a growing body of evidence has reported that survivin has a strong inhibitory effect on TRAIL therapy (32).
The present study investigated the impact of a low dosage of YM155 on TRAIL-mediated apoptosis and molecular mechanisms underlying TRAIL sensitization in human cervical cancer cells.
Materials and methods
Cell culture
HeLa cells were procured from the Korean Cell Line Bank; Korean Cell Line Research Foundation. Cells were maintained in DMEM (PAN-Biotech GmbH) with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.). A total of 100 µg/ml penicillin-streptomycin (HyClone; Cytiva) was added to make complete media. Cells were maintained in a CO2 cell culture incubator (Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2.
Reagents and antibodies
YM155 and TRAIL were purchased from Selleck Chemicals and PeproTech, Inc., respectively. Western blot and immunofluorescence analyses were performed using antibodies against cleaved-poly (ADP-ribose) polymerase (PARP; dilution, 1:1,000; cat. no. 5625S) from Cell Signaling Technology, Inc., cFLIP (dilution, 1:1,000; cat. no. 10394-1-AP) and DR5 (dilution, 1:1,000; cat. no. 15497-1-AP) from ProteinTech Group, Inc., γH2A histone family member X (γH2AX; dilution, 1:1,000 for western blot and 1:100 for immunofluorescence; cat. no. 05-636) from EMD Millipore, cleaved caspase-3 (dilution, 1:100 for immunofluorescence; cat. no. 9661S) from Cell Signaling Technology, Inc., GAPDH (dilution, 1:2,000; cat. no. sc-32233) and β-tubulin (1:1,000; cat. no. sc-5274) from Santa Cruz Biotechnology, Inc., and HRP-conjugated secondary anti-mouse (1:10,000; cat. no. 31430) and anti-rabbit (1:10,000; cat. no. 31460) antibodies from Thermo Fisher Scientific, Inc., and fluorescence-conjugated secondary antibodies goat anti-mouse IgG (H+L) Alexa Fluor 488 (1:250; cat. no. A32723) and goat anti-rabbit IgG (H+L) Alexa Fluor 488 (1:250; cat. no. A27034) from Invitrogen, Thermo Fisher Scientific, Inc.
Western blotting
HeLa cells were treated with 25 ng/ml TRAIL, 25 nM YM155 or a combination of both for 24 h and subjected to western blot analysis. Cells were collected after 24-h incubation at 37°C and 5% CO2 and lysed with a protein extraction buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% NP-40, 1% sodium deoxycholate, 2.5 mM sodium pyrophosphate, 1 mM b-glycerophosphate, 1 mM Na3VO4, 1 µg/ml leupeptin) with a PMSF protease inhibitor (Cell Signaling Technology, Inc.). The protein concentration was determined by Bradford assay (Bio-Rad Laboratories, Inc.). Then, 50 µg protein was loaded on 8 or 12% SDS-PAGE and transferred to PVDF membrane (EMD Millipore) activated by methanol. Following transfer, non-specific binding was blocked with 5% skim milk and incubated at room temperature for 1 h. The membranes were probed with primary antibodies and incubated at 4°C for overnight. Subsequently, the membranes were incubated with a HRP-conjugated secondary antibody for 1 h at room temperature and washed thrice for 5 min with TBS-Tween 20 (0.05%) to remove unbound probes. The membranes were subjected to chemiluminescence-based detection (Pierce ECL Plus; Thermo Fisher Scientific Inc.). Imaging was performed using a ChemiDoc system with Image Lab software version 5.2 (Bio-Rad Laboratories, Inc.).
Flow cytometry
Apoptosis was assessed using propidium iodide (PI) staining kit (BD Biosciences) according to the manufacturer's protocol. Briefly, HeLa cells were seeded at a density of 2.5×105 per well and treated with 25 ng/ml TRAIL and 25 nM YM155 for 24 h. Subsequently, 5×104 cells were collected and washed with ice-cold PBS twice in the presence of 10% FBS, and 70% ethanol was added into the cell suspensions. A total of 2 mg/ml RNaseA was added and incubated for 15 min at 4°C. Finally, cells were stained with 10 µl PI (50 mg/ml) at room temperature for 10 min. DNA content was measured within 1 h using flow cytometry (FACS CANTO II; BD Biosciences). Data were analyzed using FACSDiva software (version 8; BD Biosciences).
Immunofluorescence
Immunofluorescence was used to assess nuclear morphology and DNA damage by γH2AX foci formation and cleaved caspase-3 activation assays. A total of 2×104 HeLa cells were seeded into a 4-well dish containing cover slips and incubated overnight. Subsequently, HeLa cells were treated with 25 ng/ml TRAIL and/or 25 nM YM155 and incubated for 24 h. The reaction was immediately stopped and cells were fixed with 4% paraformaldehyde at room temperature (Wako Chemicals USA, Inc.) for 20 min. Fixed cells were washed using 0.03% Triton-X 100 (Sigma-Aldrich; Merck KGaA) three times. To eliminate non-specific binding, 5% BSA (Bovogen Biologicals Pty Ltd.) in 1X PBS was used as a blocking reagent. After 1 h of blocking at room temperature, primary antibody was added, followed by incubation at 4°C overnight. The next day, cells were washed using PBS and incubated with relevant Alexa Fluor 48-8 and 594-conjugated secondary antibodies at room temperature for 1 h. DAPI (1 µg/ml; Invitrogen; Thermo Fisher Scientific, Inc.) was added to the cells and incubated at room temperature for 15 min to stain the nuclei, and cells were mounted on glass slides. Cells were imaged at ×40 magnification using a confocal microscope (Leica TCS SP5; Leica Microsystems GmbH).
Reverse transcription-quantitative (RT-q)PCR
HeLa cells were treated with 25 ng/ml TRAIL and/or 25 nM YM155 for 24 h and harvested for RNA extraction. TRIzol® reagent (Favorgen Biotech Corporation) was used to extract the RNA. A total of 40 ml RNase-free water was added to the extracted vial to reconstitute the RNA. Total RNA concentration was quantified using a Nanodrop (Thermo Fisher Scientific, Inc.). The SuperScript III First-Strand Synthesis System (Thermo Fisher Scientific, Inc.) was used to synthesize the cDNA according to the manufacturer's protocol. Briefly, 1 µg of total RNA was reverse transcribed using the SuperScript III kit with 25 pmol oligo-dT primer and 50 pmol random hexamer. Then, the cDNA was diluted at 1:10 with dilution buffer provided by the manufacturer. QPCR was performed using a real-time PCR system (Thermo Fisher Scientific, Inc.) and Fast SYBR® Green Master Mix (Thermo Fisher Scientific, Inc.) with the following primers: DR5 forward, 5′-CCCAACAAGACCTAGCTCCC-3′ and reverse, 5′-GACCTCCTTTTCTGCTTGCG-3′; cFLIP forward, 5′-GCCGAGGCAAGATAAGCAAG-3′ and reverse, 5′-AGTCTGTTCAAGGAGCAGGG-3′; survivin forward, 5′-AGAACTGGCCCTTCTTGGAGG-3′ and reverse, 5′-CTTTTTATGTTCCTCTATGGGGTC-3′; and GAPDH forward, 5′-CTGACTTCAACAGCGACACC-3′ and reverse, 5′-TAGCCAAATTCGTTGTCATACC-3′. The relative quantification of gene expression was determined using the 2−ΔΔCq method (33). The thermocycling conditions were as follows: Pre-incubation at 95°C for 3 min, followed by 45 cycles of 95°C for 15 sec and 60°C for 60 sec. The final amplification cycle was followed by a melt curve analysis for the specificity of the RT-qPCR.
Cell viability assay
The cytotoxicity of TRAIL and YM155 was assessed using a Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.). Following drug treatment, 10 µl CCK-8 reagent was added, and the mixture was incubated at 37°C for 2 h according to the manufacturer's instructions. Subsequently, absorbance was measured at 540 nm using a microtiter plate reader (Bio-Rad Laboratories, Inc.).
Matrigel invasion assay
HeLa cells were seeded, and cell invasion was evaluated using 0.8-µm Transwell chambers coated with Matrigel at 37°C for 1 h (Corning Inc.) according to the manufacturer's protocol. In brief, HeLa cells were suspended at a density of 2.5×104 cells in 500 µl serum-free DMEM and placed in the upper chamber. Subsequently, 700 µl complete DMEM was added to the lower chamber, followed by incubation at 37°C for 24 h. Cells on the surface of the insert were removed, and the lower surface was fixed with ice-cold methanol at room temperature for 15 min. After fixing, cells were stained with crystal violet (0.1%, diluted in methanol) for 5 min at room temperature. Cells were counted using bright field microscopy and compared among the DMSO, YM155, TRAIL and combination groups. The numbers of invaded cells were represented graphically.
In vitro scratch assay
An in vitro scratch assay was performed to assess the migratory activity of HeLa cells treated with 25 ng/ml TRAIL and/or 25 nM YM155. After 24-h treatment, the cells were harvested and re-seeded at 90% confluency and incubated at 37°C overnight. Using a sterile pipette tip, scratches were made in the monolayers of HeLa cells. The scratched monolayer was washed with PBS and incubated at 37°C with serum-free 1X DMEM. The wounded region was captured under a light microscope (IX71; Olympus Corporation) at 0, 24, 48 and 72 h, and ImageJ 1.51j8 software (National Institutes of Health) was used to calculate the percentage of wound closure as previously described (34). Data were obtained from three independent experiments.
Soft agar assay
DMEM (1X) complete medium and 1% agarose were mixed at an equal ratio of 1:1 (v/v) and plated onto 35-mm dishes. The next day, cells were seeded at a density of 1×104 per well and suspended in 0.7% agarose/1X DMEM mixture. Subsequently, 1% DMSO, 5 nM YM155, 5 ng/ml TRAIL and YM155/TRAIL combination were added to DMEM. Medium containing the aforementioned drug concentrations was added every other day for 14 days. Crystal violet (0.01% diluted in 20% methanol) was added at room temperature for 5 min to stain anchorage-independent colonies. Finally, cells were observed under a light microscope at ×4 magnification.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 8 (GraphPad Software, Inc.) and data are presented as the mean ± standard deviation of three independent experiments. Data were analyzed by one-way ANOVA. Multiple comparisons among the groups were performed by Tukey's post hoc test. For in vitro scratch assay, two-way ANOVA followed by Tukey's post hoc test was performed. P<0.05 was considered to indicate a statistically significant difference.
Results
YM155 sensitizes HeLa cells to TRAIL-mediated apoptosis
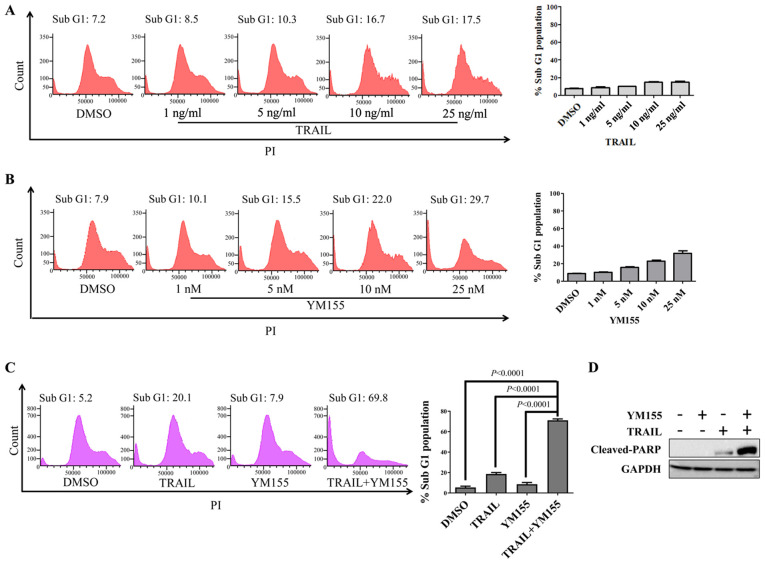
First, HeLa cells were treated with 1% DMSO, 25 ng/ml TRAIL or 25 nM YM155 for 24 h. PI staining was used to analyze apoptotic cells by flow cytometry (Fig. 1A and B). TRAIL or YM155 alone did not induce cell death in HeLa cells (Fig. 1A and B), whereas the combination of TRAIL and YM155 significantly increased the sub-G1 population compared with the DMSO control group in HeLa cells (Fig. 1C). To further validate the combinatorial effect of TRAIL-YM155, western blotting was performed to assess cleaved-PARP levels. Consistent with the sub-G1 population experimental results, combination treatment induced greater cleaved-PARP expression than individual treatment (Fig. 1D). Overall, YM155 enriched TRAIL-mediated apoptosis via an extrinsic signaling pathway in human cervical cancer cells.
Figure 1.
YM155 sensitizes HeLa cells to TRAIL-induced apoptosis. (A) HeLa cells were treated with indicated concentrations of TRAIL for 24 h and DNA content was measured by flow cytometry using PI staining. (B) HeLa cells were treated with indicated concentrations of YM155 for 24 h and PI staining was used for flow cytometry. (C) HeLa cells were treated with 25 ng/ml TRAIL and 25 nM YM155 for 24 h and subjected to PI staining and flow cytometry to assess the apoptotic population. Data are presented as the mean and standard deviation of three independent experiments. One-way ANOVA followed by Tukey's post hoc test was used and P-values are indicated. (D) HeLa cells were treated with 25 ng/ml TRAIL and 25 nM YM155 and subjected to western blotting with cleaved-PARP antibody. GAPDH was used as the loading control. PARP, poly (ADP-ribose) polymerase; PI, propidium iodide; TRAIL, tumor necrosis factor-related apoptosis inducing ligand.
Combination of TRAIL and YM155 induces DNA damage toxicity in HeLa cells
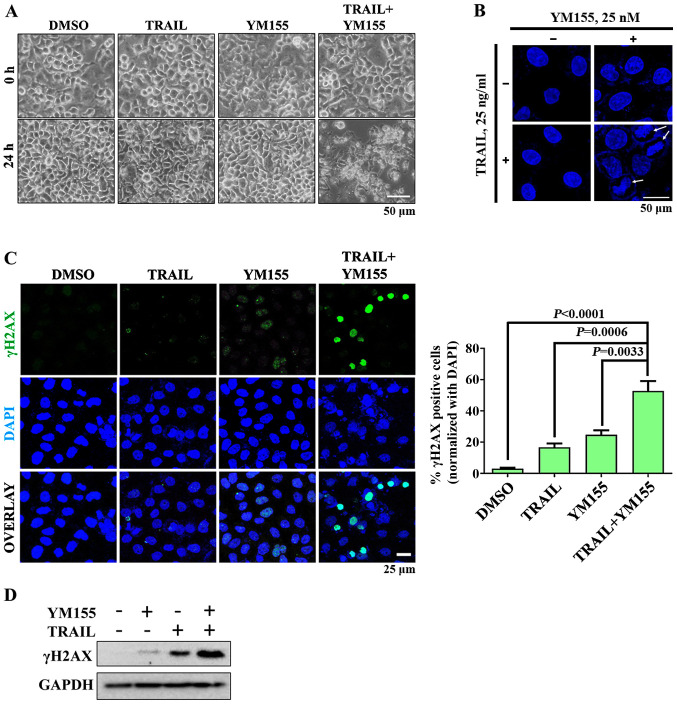
In order to investigate whether combination treatment of TRAIL and YM155 lead to DNA damage, cells were treated with 25 ng/ml TRAIL or 25 nM YM155 alone and in combination. Bright-field microscopy images revealed that cells in the combination treatment group exhibited apoptotic morphology, whereas cells in the single agent treatment groups did not (Fig. 2A). Subsequently, the shape of the nucleus of HeLa cells was assessed by staining with DAPI. As presented in Fig. 2B, chromatin condensation was observed following combination treatment but not when cells were treated with TRAIL or YM155 alone. To further confirm that combination treatment induces DNA damage in HeLa cells, γH2AX foci formation was assessed using immunofluorescence. As presented in Fig. 2C, robust H2AX phosphorylation was observed in HeLa cells treated with the combination of TRAIL and YM155 compared with in the single agent treatment and DMSO control groups. Consistent with the aforementioned results, combination treatment of TRAIL and YM155 resulted in higher protein expression levels of γH2AX than individual treatment (Fig. 2D). Overall, the combination of TRAIL and YM155 induced DNA damage in HeLa cells but individual treatment did not.
Figure 2.
Effect of combination treatment on DNA damage in HeLa cells. HeLa cells were treated with 25 ng/ml TRAIL and 25 nM YM155 for 24 h and subjected to subsequent analysis. (A) Morphological alterations in HeLa cells were observed using bright-field microscopy. Assays were performed in triplicate. Scale bar, 50 µM. (B) Immunofluorescence experiments were performed to assess nucleus morphology by DAPI staining (blue). Arrows indicate nucleus condensation. Assays were performed in triplicate. Scale bar, 50 µm. (C) DNA double strand breaks in HeLa cells were estimated using a γH2AX foci formation assay. Assays were performed in triplicate. Green, γH2AX; blue, nucleus stained with DAPI. Magnification, ×20. Scale bar, 25 µm. The right panel shows the calculated data, presented as the γH2AX-positive cells. Data are presented as the mean and standard deviation of three independent experiments. One-way ANOVA followed by Tukey's post hoc test was used and P-values are indicated. (D) Western blot analysis was performed to assess DNA damage using an endogenous γH2AX antibody. GAPDH was used as the loading control. γH2AX, γH2A histone family member X; TRAIL, tumor necrosis factor-related apoptosis inducing ligand.
YM155 increases TRAIL-induced apoptosis via downregulation of cFLIP and survivin levels
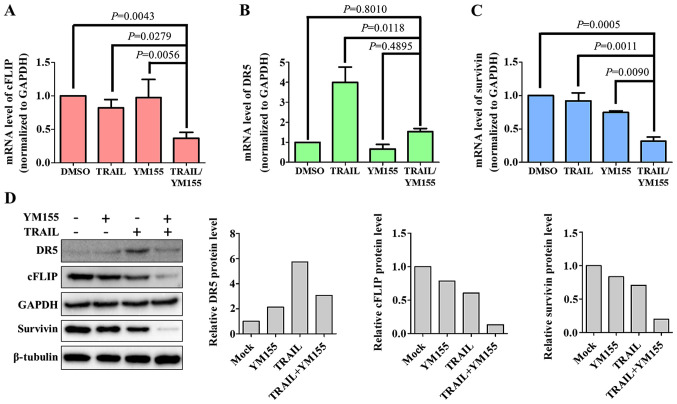
In order to determine the mechanisms underlying YM155-mediated TRAIL sensitivity in HeLa cells, apoptosis-associated proteins, including DR5 (pro-apoptotic), cFLIP (anti-apoptotic) and survivin (anti-apoptotic) were examined in the present study. HeLa cells were treated with 1% DMSO, 25 ng/ml TRAIL or 25 nM YM155 alone and in combination for 24 h and then subjected to RT-qPCR analysis. As presented in Fig. 3, HeLa cells treated with both TRAIL and YM155 exhibited downregulation of cFLIP and survivin at the mRNA and protein levels (Fig. 3A, C and D). DR5, a pro-apoptotic protein involved in TRAIL sensitivity (35), exhibited no changes following combination treatment (Fig. 3B and D). Notably, treatment with TRAIL alone induced a significant increase in the mRNA levels of DR5 (Fig. 3B). Similarly, the protein level of DR5 was also upregulated by TRAIL treatment (Fig. 3D). This suggests that DR5 upregulation was specific to TRAIL treatment alone and not in combination. The aforementioned data indicate that downregulation of cFLIP and survivin serves a major role in combination treatment of YM155 and TRAIL-mediated apoptosis.
Figure 3.
Combination treatment downregulates mRNA and protein levels of cFLIP and survivin. Reverse transcription-quantitative PCR was used to assess the mRNA expression levels of (A) cFLIP, (B) DR5 and (C) survivin following treatment with 25 ng/ml TRAIL and 25 nM YM155 for 24 h. Data are presented as the mean and standard deviation of three independent experiments. One-way ANOVA followed by Tukey's post hoc test was used, and P-values are indicated. (D) Western blotting was performed to assess protein expression levels of cFLIP, DR5 and survivin after treatment with 25 ng/ml TRAIL and 25 nM YM155 for 24 h. GAPDH and β-tubulin were used as the loading controls. cFLIP, CASP8 and FADD like apoptosis regulator; DR5, death receptor 5; TRAIL, tumor necrosis factor-related apoptosis inducing ligand.
Combination treatment decreases cancer progression in vitro
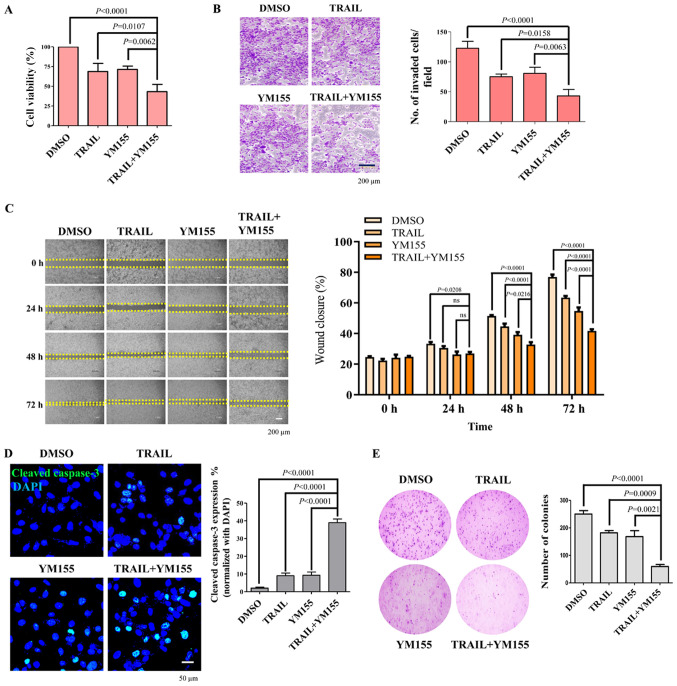
The effect of TRAIL and YM155 combination treatment in HeLa cells was next assessed using a cell viability assay. As presented in Fig. 4A, combination treatment of TRAIL and YM155 significantly decreased the viability of HeLa cells (2.08-fold) compared with the DMSO control. To test the hypothesis that the combination of TRAIL and YM155 decreases cancer progression, the effect of the combination treatment on HeLa cell migration and invasion was assessed using an in vitro scratch assay and Matrigel-coated chamber invasion assay, respectively. HeLa cells treated with both TRAIL and YM155 exhibited a significant decrease in invasive capability compared with the DMSO control and single agent treatment groups (Fig. 4B). Consistently, combination treatment also resulted in a delayed wound closure rate compared with the DMSO control and single agent treatment groups (Fig. 4C). It has been reported that caspase-3 is a cysteine-aspartic acid protease that will be cleaved by TRAIL and executes apoptosis in cancer cells (36,37). Therefore, the present study monitored the levels of cleaved caspase-3 in HeLa cells. To achieve this, HeLa cells were treated with the 25 ng/ml TRAIL or 25 nM YM155 alone or in combination for 24 h. Subsequently, cleaved caspase-3 levels in samples were evaluated by immunofluorescence. It was observed that the combination treatment of TRAIL and YM155 significantly increased the cleaved caspase-3 levels compared with DMSO and TRAIL or YM155 alone treated samples (Fig. 4D). Subsequently, cell proliferation was assessed using an anchorage-independent colony formation assay. HeLa cells were selected with the combination of 5 ng/ml of TRAIL and 5 nM of YM155 or individual treatment, and colony formation was analyzed. The number of colonies that appeared in cells treated with both TRAIL and YM155 was significantly lower compared with the DMSO control and single agent treatment groups (Fig. 4E). Overall, the results of the present study suggest that combination treatment of TRAIL and YM155 decreased the viability, invasion and migration of HeLa cells. The synergistic behavior of TRAIL/YM155 combination further induced DNA damage and apoptosis and may be an effective strategy to treat human cervical cancer.
Figure 4.
Effect of combination treatment on cell viability, cellular invasion and migration of HeLa cells. (A) HeLa cells were treated with 25 ng/ml TRAIL and 25 nM YM155 for 24 h. Cell Counting Kit-8 reagent was added to the treated cells to assess cell viability. (B) Matrigel cell invasion assay in HeLa cells. Cells were treated with 25 ng/ml TRAIL and 25 nM YM155 for 24 h. Cells were seeded in the upper chamber of the plate. After 24 h of incubation, cells on the bottom of the chamber membrane were fixed, stained and invasive cells were counted for graphical representation. Scale bar, 200 µm. (C) HeLa cells were treated with 25 ng/ml TRAIL and 25 nM YM155 for 24 h and assessed for migratory potential using a wound healing assay. Images were captured at 0, 24, 48 and 72 h. Scale bar, 200 µm. Right panel indicates the percentage of wound closure. Data are presented as the mean and standard deviation of three independent experiments. Two-way ANOVA was used and P-values are as indicated. (D) HeLa cells were treated with either TRAIL or YM155 or the combination at 25 ng/ml or 25 nM, respectively, for 24 h. An immunofluorescence assay was performed to monitor the activation of cleaved caspase-3 (green). Nuclei were stained with DAPI (blue). Scale bar, 50 µm. Right panel is the quantified data presented as the percentage of cleaved caspase-3 expression. (E) HeLa cells were selected with 5 ng/ml of TRAIL and/or 5 nM of YM155. After 14 days of selection with 5 ng/ml TRAIL and 5 nM YM155, colonies were stained with 0.01% crystal violet, and images were captured at ×4 magnification. The assay was performed in triplicate. The right panel is the quantified data of the soft agar assay presented as colony numbers. Data are presented as the mean and standard deviation of three independent experiments. One-way ANOVA followed by Tukey's post hoc test was used, and P-values are as indicated. TRAIL, tumor necrosis factor-related apoptosis inducing ligand.
Discussion
Despite the use of aggressive and toxic treatments, patients with advanced stage cervical cancer have an unacceptably high mortality rate; an estimated 13,800 new cervical cancer will be diagnosed and 4,290 deaths will occur in the United States in 2020 (38). Epidemiological, clinical and cellular studies have reported that persistent HPV infections are involved in the development of cervical cancer (38), although cervical cancer can also develop without HPV infection. The poor survival percentage demands research into more effective novel treatment approaches. In the present study, the sensitivity of TRAIL improved with the addition of YM155 in the treatment of human cervical cancer cells. The therapeutic potential of TRAIL and YM155 combination treatment was analyzed in HeLa cells. Previous research has demonstrated that HeLa cells are less sensitive to TRAIL-induced apoptosis (39,40). To sensitize the HeLa cells to TRAIL-mediated apoptosis, YM155 was used as a combinatorial drug partner. YM155 has been reported as a survivin inhibitor developed for cancer treatment, and its effects have been demonstrated in multiple preclinical studies and phase I/II clinical trials (41–44).
Several reports have suggested that YM155 sensitizes cells to TRAIL-induced apoptosis in human renal carcinoma Caki cells (31), triple negative breast cancer (45) and a number of other types of cancer. However, to the best of our knowledge, there are currently no data on the combination of TRAIL and YM155 in human cervical cancer. Therefore, the effect of YM155 on the induction of TRAIL-mediated apoptosis in HeLa cells was assessed in the present study. It was revealed that HeLa cells were sensitized by the addition of a low concentration of YM155, but not in individually treated groups. Previous studies have suggested that YM155 induces DNA damage in several types of cancer (46,47). Therefore, phosphorylation of H2AX, which causes double strand breaks, was assessed by immunofluorescence and western blotting assays. YM155-treated cells expressed γH2AX. Notably, treatment with TRAIL alone did not induce DNA damage, whereas the combination treatment of TRAIL and YM155 was associated with marked expression of γH2AX in HeLa cells and eventually led to apoptosis. A previous report has suggested that the combination of YM155 and TRAIL in SiHa cells induced a 48% higher apoptotic rate compared with that of the DMSO group (48), whereas the present combinatorial approach demonstrated an ~71% increase in apoptosis relative to that of the DMSO group by activating cleaved caspase-3 in HeLa cells. The main difference is that the cells were first treated with YM155 alone for 24 h and then TRAIL treatment was applied for another 15 h in the previous study whereas the present study induced apoptosis via synergistic action of YM155 and TRAIL together in cervical cancer cells. This explains the importance of the synergistic behavior of YM155/TRAIL in a cellular environment to hinder the progress of cervical cancer growth. It is important to note that the use of antibiotics in drug response studies can alter gene expression (49). Both genetic and phenotypical characterizations of YM155 and TRAIL have been studied extensively (50–53). Throughout the experiments in the present study, both the control and experimental groups received equal amounts of antibiotics, and thus, their effects on the results were negligible.
Previous data revealed that DR5 is the major determinant of TRAIL-induced apoptosis (54,55), and cFLIP and survivin confer resistance to TRAIL therapy for different cancer types (56,57). Several reports have demonstrated that DR5 serves a greater role in apoptosis compared with that of DR4 (58,59). DR5 activates the extrinsic apoptotic signaling pathway by binding of the death ligand (TRAIL) (60,61). However, the action of TRAIL-mediated apoptosis is mainly associated with Fas-associated via death domain (FADD) recruitment to the DR5/TRAIL complex. cFLIP inhibits the action of FADD and prevents it from binding to the DR5/TRAIL complex (62,63). High cFLIP expression in human cervical cancer confers resistance to TRAIL therapy (64). Additionally, it has been noted that DR4 serves an antagonistic role to DR5 in TRAIL-mediated apoptosis (59). Furthermore, DR4 is responsible for the sensitization of cancer cells during the combination treatment involving irradiation (39). Therefore, regulating cFLIP is more crucial than the other genes involved in DR5/TRAIL-mediated apoptotic signaling pathway. However, elucidating the role of DR4 in TRAIL-mediated apoptosis is essential for future investigations. In our previous study, cFLIP was successfully downregulated in HeLa cells using the CRISPR-Cas9 system and its effectiveness in TRAIL-mediated apoptosis was examined (8). On the other hand, consistent with our previous study (8), the mRNA and protein expression levels of cFLIP were decreased when HeLa cells were treated with TRAIL and YM155 together. However, this combinatorial approach did not alter endogenous DR5 expression.
Both the mRNA and protein expression levels of survivin were not significantly decreased when the low dose of YM155 (25 nM) alone was applied. Similarly, a low concentration of YM155 (20 nM) did not exhibit any inhibitory effect on survivin expression in human renal cell carcinoma cell lines treated for 48 h; however, high concentrations of YM155 (>40 nM) decrease survivin protein levels in renal cell carcinoma cells (27). It was hypothesized that a low concentration of YM155 does not block the transcription of baculoviral IAP repeat containing 5 (BIRC5), a gene that encodes the survivin protein, and thus it has no inhibitory effect on survivin in these cell lines (27). However, further research is required at the transcriptional level to elucidate the interaction of the low dosage of YM155 and the BIRC5 gene. In the present study, the low dosage of YM155 sensitized HeLa cells to TRAIL-mediated apoptosis and decreased the mRNA and protein levels of survivin. Therefore, the combination therapy holds promise as a method to overcome the problem of side effects due to high doses and could be safe for use in patients. However, further investigations are required to validate the combination treatment in different human cervical cancer cell lines and its potential outcome in mouse xenograft models.
As available treatments have limited efficacy in human cervical cancer (64), the identification of novel therapeutic approaches to enhance patient outcomes is urgently required. The present study suggests combination treatment using YM155 and TRAIL as a therapeutic approach. Specifically, YM155 enhanced TRAIL-mediated apoptosis in HeLa cells. The results demonstrated that YM155 sensitizes human cervical cancer cells to TRAIL-induced cell death by activating the intrinsic pathway of apoptosis, and YM155/TRAIL combination downregulated anti-apoptotic proteins, such as cFLIP and survivin, in HeLa cells.
Acknowledgements
The authors would like to thank Dr Bharathi Suresh (Hanyang University, Seoul, Republic of Korea) for her assistance with statistical analysis.
Funding
The present study was supported by the Korea Health Technology R&D Project through the Korea Health Industry Development Institute, funded by the Ministry of Health & Welfare, Republic of Korea (grant no. HI18C2383) and the National Research Foundation of Korea, which is funded by the Ministry of Education (grant no. 2018M3A9H3022412).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
SJO and SR were responsible for the study design. APC and NP performed the main experiments related to apoptosis and oncogenic based assays, and drafted the manuscript. NRK performed cell viability assays and performed statistical analysis. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Burd EM. Human papillomavirus and cervical cancer. Clin Microbiol Rev. 2003;16:1–17. doi: 10.1128/CMR.16.1.1-17.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soto D, Song C, McLaughlin-Drubin ME. Epigenetic alterations in human papillomavirus-associated cancers. Viruses. 2017;9:248–265. doi: 10.3390/v9090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willms A, Schittek H, Rahn S, Sosna J, Mert U, Adam D, Trauzold A. Impact of p53 status on TRAIL-mediated apoptotic and non-apoptotic signaling in cancer cells. PLoS One. 2019;14:e0214847. doi: 10.1371/journal.pone.0214847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang S, El-Deiry WS. TRAIL and apoptosis induction by TNF-family death receptors. Oncogene. 2003;22:8628–8633. doi: 10.1038/sj.onc.1207232. [DOI] [PubMed] [Google Scholar]
- 6.Zhang L, Fang B. Mechanisms of resistance to TRAIL-induced apoptosis in cancer. Cancer Gene Ther. 2005;12:228–2237. doi: 10.1038/sj.cgt.7700792. [DOI] [PubMed] [Google Scholar]
- 7.Pitti RM, Marsters SA, Ruppert S, Donahue CJ, Moore A, Ashkenazi A. Induction of apoptosis by Apo-2 ligand, a new member of the tumor necrosis factor cytokine family. J Biol Chem. 1996;271:12687–12690. doi: 10.1074/jbc.271.22.12687. [DOI] [PubMed] [Google Scholar]
- 8.Poondla N, Chandrasekaran AP, Heese K, Kim KS, Ramakrishna S. CRISPR-mediated upregulation of DR5 and downregulation of cFLIP synergistically sensitize HeLa cells to TRAIL-mediated apoptosis. Biochem Biophys Res Commun. 2019;512:60–65. doi: 10.1016/j.bbrc.2019.03.018. [DOI] [PubMed] [Google Scholar]
- 9.Seo OW, Kim JH, Lee KS, Lee KS, Kim JH, Won MH, Ha KS, Kwon YG, Kim YM. Kurarinone promotes TRAIL-induced apoptosis by inhibiting NF-κB-dependent cFLIP expression in HeLa cells. Exp Mol Med. 2012;44:653–664. doi: 10.3858/emm.2012.44.11.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang A, Wilson NS, Ashkenazi A. Proapoptotic DR4 and DR5 signaling in cancer cells: Toward clinical translation. Curr Opin Cell Biol. 2010;22:837–844. doi: 10.1016/j.ceb.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 11.Russo M, Mupo A, Spagnuolo C, Russo GL. Exploring death receptor pathways as selective targets in cancer therapy. Biochem Pharmacol. 2010;80:674–682. doi: 10.1016/j.bcp.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 12.Malhi H, Gores GJ. TRAIL resistance results in cancer progression: A TRAIL to perdition? Oncogene. 2006;25:7333–7335. doi: 10.1038/sj.onc.1209765. [DOI] [PubMed] [Google Scholar]
- 13.Mobahat M, Narendran A, Riabowol K. Survivin as a preferential target for cancer therapy. Int J Mol Sci. 2014;15:2494–2516. doi: 10.3390/ijms15022494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukuda S, Pelus LM. Survivin, a cancer target with an emerging role in normal adult tissues. Mol Cancer Ther. 2006;5:1087–1098. doi: 10.1158/1535-7163.MCT-05-0375. [DOI] [PubMed] [Google Scholar]
- 15.Shin S, Sung BJ, Cho YS, Kim HJ, Ha NC, Hwang JI, Chung CW, Jung JK, Oh BH. An Anti-apoptotic protein human survivin is a direct inhibitor of caspase-3 and −7. Biochemistry. 2001;40:1117–1123. doi: 10.1021/bi001603q. [DOI] [PubMed] [Google Scholar]
- 16.Lv YG, Yu F, Yao Q, Chen JH, Wang L. The role of survivin in diagnosis, prognosis and treatment of breast cancer. J Thorac Dis. 2010;2:100–110. [PMC free article] [PubMed] [Google Scholar]
- 17.Cao XQ, Lu HS, Zhang L, Chen LL, Gan MF. MEKK3 and survivin expression in cervical cancer: Association with clinicopathological factors and prognosis. Asian Pacific J Cancer Prev. 2014;15:5271–5276. doi: 10.7314/APJCP.2014.15.13.5271. [DOI] [PubMed] [Google Scholar]
- 18.Falleni M, Pellegrini C, Marchetti A, Oprandi B, Buttitta F, Barassi F, Santambrogio L, Coggi G, Bosari S. Survivin gene expression in early-stage non-small cell lung cancer. J Pathol. 2003;200:620–626. doi: 10.1002/path.1388. [DOI] [PubMed] [Google Scholar]
- 19.Trieb K, Lehner R, Stulnig T, Sulzbacher I, Shroyer KR. Survivin expression in human osteosarcoma is a marker for survival. Eur J Surg Oncol. 2003;29:379–82. doi: 10.1053/ejso.2002.1415. [DOI] [PubMed] [Google Scholar]
- 20.Park SH, Park SJ, Kim JO, Shin J-H, Kim ES, Jo YK, Kim JS, Park SJ, Jin DH, Hwang JJ, et al. Down-regulation of survivin by nemadipine-a sensitizes cancer cells to TRAIL-induced apoptosis. Biomol Ther (Seoul) 2013;21:29–34. doi: 10.4062/biomolther.2012.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hwang JS, Lee HC, Oh SC, Lee DH, Kwon KH. Shogaol overcomes TRAIL resistance in colon cancer cells via inhibiting of survivin. Tumor Biol. 2015;36:8819–8829. doi: 10.1007/s13277-015-3629-2. [DOI] [PubMed] [Google Scholar]
- 22.Véquaud E, Séveno C, Loussouarn D, Engelhart L, Campone M, Juin P, Barillé-Nion S. YM155 potently triggers cell death in breast cancer cells through an autophagy-NF-kB network. Oncotarget. 2015;6:13476–13486. doi: 10.18632/oncotarget.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng XJ, Lin JC, Ding YF, Zhu L, Ye J, Tu SP. Survivin inhibitor YM155 suppresses gastric cancer xenograft growth in mice without affecting normal tissues. Oncotarget. 2016;7:7096–7109. doi: 10.18632/oncotarget.6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang C, Cao x, Gei Y, Wang Y, Liu G, Cheng G, Liu Q. Silencing of survivin by YM155 induces apoptosis and growth arrest in hepatocellular carcinoma cells. Oncol Lett. 2015;10:1627–1631. doi: 10.3892/ol.2015.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao X, Puszyk WM, Lu Z, Ostrov DA, George TJ, Robertson KD, Liu C. Small molecule inhibitor YM155-mediated activation of death receptor 5 is crucial for chemotherapy-induced apoptosis in pancreatic carcinoma. Mol Cancer Ther. 2015;14:80–89. doi: 10.1158/1535-7163.MCT-14-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jane EP, Premkumar DR, Sutera PA, Cavaleri JM, Pollack IF. Survivin inhibitor YM155 induces mitochondrial dysfunction, autophagy, DNA damage and apoptosis in Bcl-xL silenced glioma cell lines. Mol Carcinog. 2017;56:1251–1265. doi: 10.1002/mc.22587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sim MY, Huynh H, Go ML, Yuen JSP. Action of YM155 on clear cell renal cell carcinoma does not depend on survivin expression levels. PLoS One. 2017;12:e0178168. doi: 10.1371/journal.pone.0178168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao N, Mao Y, Han G, Ju Q, Zhou L, Liu F, Xu Y, Zhao X. YM155, a survivin suppressant, triggers PARP-dependent cell death (parthanatos) and inhibits esophageal squamous-cell carcinoma xenografts in mice. Oncotarget. 2015;6:18445–18459. doi: 10.18632/oncotarget.4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li WL, Lee MR, Cho MY. The small molecule survivin inhibitor YM155 may be an effective treatment modality for colon cancer through increasing apoptosis. Biochem Biophys Res Commun. 2016;471:309–314. doi: 10.1016/j.bbrc.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 30.Feng W, Yoshida A, Ueda T. YM155 induces caspase-8 dependent apoptosis through downregulation of survivin and Mcl-1 in human leukemia cells. Biochem Biophys Res Commun. 2013;435:52–57. doi: 10.1016/j.bbrc.2013.04.036. [DOI] [PubMed] [Google Scholar]
- 31.Woo SM, Min K, Seo BR, Kwon TK. YM155 sensitizes TRAIL-induced apoptosis through cathepsin S-dependent down-regulation of Mcl-1 and NF-κB-mediated down-regulation of c-FLIP expression in human renal carcinoma Caki cells. Oncotarget. 2016;7:61520–61532. doi: 10.18632/oncotarget.11137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Azuhata T, Scott D, Griffith TS, Miller M, Sandler AD. Survivin inhibits apoptosis induced by TRAIL, and the ratio between survivin and TRAIL receptors is predictive of recurrent disease in neuroblastoma. J Pediatr Surg. 2006;41:1431–1440. doi: 10.1016/j.jpedsurg.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 33.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 34.Latifi-Pupovci H, Kuçi Z, Wehner S, Bönig H, Lieberz R, Klingebiel T, Bader P, Kuçi S. In vitro migration and proliferation (‘wound healing’) potential of mesenchymal stromal cells generated from human CD271+ bone marrow mononuclear cells. J Transl Med. 2015;13:315–323. doi: 10.1186/s12967-015-0676-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuang AA, Diehl GE, Zhang J, Winoto A. FADD is required for DR4- and DR5-mediated apoptosis. J Biol Chem. 2000;275:25065–25068. doi: 10.1074/jbc.C000284200. [DOI] [PubMed] [Google Scholar]
- 36.Ponder KG, Boise LH. The prodomain of caspase-3 regulates its own removal and caspase activation. Cell Death Discov. 2019;5:56. doi: 10.1038/s41420-019-0142-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salvesen GS. Caspases: Opening the boxes and interpreting the arrows. Cell Death Differ. 2002;9:3–5. doi: 10.1038/sj.cdd.4400963. [DOI] [PubMed] [Google Scholar]
- 38.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 39.Maduro JH, de Vries EGE, Meersma GJ, Hougardy BMT, van der Zee AGJ, de Jong S. Targeting pro-apoptotic TRAIL receptors sensitizes HeLa cervical cancer cells to irradiation-induced apoptosis. Int J Radiat Oncol Biol Phys. 2008;72:543–552. doi: 10.1016/j.ijrobp.2008.06.1902. [DOI] [PubMed] [Google Scholar]
- 40.Nakahara T, Kita A, Yamanaka K, Mori M, Amino N, Takeuchi M, Tominaga F, Hatakeyama S, Kinoyama I, Matsuhisa A, et al. YM155, a novel small-molecule survivin suppressant, induces regression of established human hormone-refractory prostate tumor xenografts. Cancer Res. 2007;67:8014–8021. doi: 10.1158/0008-5472.CAN-07-1343. [DOI] [PubMed] [Google Scholar]
- 41.Rauch A, Hennig D, Schäfer C, Wirth M, Marx C, Heinzel T, Schneider G, Krämer OH. Survivin and YM155: How faithful is the liaison? Biochim Biophys Acta. 2014;1845:202–220. doi: 10.1016/j.bbcan.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 42.Kelly RJ, Thomas A, Rajan A, Chun G, Lopez-Chavez A, Szabo E, Spencer S, Carter CA, Guha U, Khozin S, et al. A phase I/II study of sepantronium bromide (YM155, survivin suppressor) with paclitaxel and carboplatin in patients with advanced non-small-cell lung cancer. Ann Oncol. 2013;24:2601–2606. doi: 10.1093/annonc/mdt249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giaccone G, Zatloukal P, Roubec J, Floor K, Musil J, Kuta M, van Klaveren RJ, Chaudhary R, Gunther A, Shamsili S. Multicenter phase II trial of YM155, a small-molecule suppressor of survivin, in patients with advanced, refractory, non-small-cell lung cancer. J Clin Oncol. 2009;27:4481–4486. doi: 10.1200/JCO.2008.21.1862. [DOI] [PubMed] [Google Scholar]
- 44.Pennati M, Sbarra S, Cesare MD, Lopergolo A, Locatelli SL, Campi E, Daidone MG, Carlo-Stella C, Gianni AM, Zaffaroni N. YM155 sensitizes triple-negative breast cancer to membrane-bound TRAIL through p38 MAPK- and CHOP-mediated DR5 upregulation. Int J Cancer. 2015;136:299–309. doi: 10.1002/ijc.28993. [DOI] [PubMed] [Google Scholar]
- 45.Iwasa T, Okamoto I, Suzuki M, Nakahara T, Satoh T, Fukuoka M, Ono K, Nakagawa K. Radiosensitizing effect of YM155, a novel Small-molecule survivin suppressant, in Non-small cell lung cancer cell lines. Clin Cancer Res. 2008;14:6496–6504. doi: 10.1158/1078-0432.CCR-08-0468. [DOI] [PubMed] [Google Scholar]
- 46.Hong M, Ren MQ, Silva J, Paul A, Wilson WD, Schroeder C, Weinberger P, Janik J, Hao Z. YM155 inhibits topoisomerase function. Anticancer Drugs. 2017;28:142–152. doi: 10.1097/CAD.0000000000000441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakamura H, Taguchi A, Kawana K, Baba S, Kawata A, Yoshida M, Fujimoto A, Ogishima J, Sato M, Inoue T, et al. Therapeutic significance of targeting survivin in cervical cancer and possibility of combination therapy with TRAIL. Oncotarget. 2018;9:13451–13461. doi: 10.18632/oncotarget.24413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ryu AH, Eckalbar WL, Kreimer A, Yosef N, Ahituv N. Use antibiotics in cell culture with caution: Genome-wide identification of antibiotic-induced changes in gene expression and regulation. Sci Rep. 2017;7:7533. doi: 10.1038/s41598-017-07757-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winter GE, Radic B, Mayor-Ruiz C, Blomen VA, Trefzer C, Kandasamy RK, Huber KVM, Gridling M, Chen D, Klampfl T, et al. The solute carrier SLC35F2 enables YM155-mediated DNA damage toxicity. Nat Chem Biol. 2014;10:768–773. doi: 10.1038/nchembio.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Griffith T, Stokes B, Kucaba T, Earel J, Jr, van Oosten R, Brincks E, Norian L. TRAIL Gene therapy: From preclinical development to clinical application. Curr Gene Ther. 2009;9:9–19. doi: 10.2174/156652309787354612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamanaka K, Nakahara T, Yamauchi T, Kita A, Takeuchi M, Kiyonaga F, Kaneko N, Sasamata M. Antitumor activity of YM155, a selective Small-molecule survivin suppressant, alone and in combination with docetaxel in human malignant melanoma models. Clin Cancer Res. 2011;17:5423–5431. doi: 10.1158/1078-0432.CCR-10-3410. [DOI] [PubMed] [Google Scholar]
- 52.Wiley SR, Schooley K, Smolak PJ, Din WS, Huang CP, Nicholl JK, Sutherland GR, Smith TD, Rauch C, Smith CA, et al. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity. 1995;3:673–682. doi: 10.1016/1074-7613(95)90057-8. [DOI] [PubMed] [Google Scholar]
- 53.Kim K, Fisher MJ, Xu SQ, El-Deiry WS. Molecular determinants of response to TRAIL in killing of normal and cancer cells. Clin Cancer Res. 2000;6:335–346. [PubMed] [Google Scholar]
- 54.Kang Z, Sun SY, Cao L. Activating Death receptor DR5 as a therapeutic strategy for rhabdomyosarcoma. ISRN Oncol. 2012;2012:395952. doi: 10.5402/2012/395952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Day TW, Najafi F, Wu CH, Safa AR. Cellular FLICE-like inhibitory protein (c-FLIP): A novel target for Taxol-induced apoptosis. Biochem Pharmacol. 2006;71:1551–1561. doi: 10.1016/j.bcp.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 56.Zong H, Yin B, Chen J, Ma B, Cai D, He X. Over-Expression of c-FLIP confers the resistance to TRAIL-induced apoptosis on gallbladder carcinoma. Tohoku J Exp Med. 2009;217:203–208. doi: 10.1620/tjem.217.203. [DOI] [PubMed] [Google Scholar]
- 57.Surget S, Chiron D, Gomez-Bougie P, Descamps G, Ménoret E, Bataille R, Moreau P, Gouill SL, Amiot M, Pellat-Deceunynck C. Cell death via DR5, but not DR4, is regulated by p53 in myeloma cells. Cancer Res. 2012;72:4562–4573. doi: 10.1158/0008-5472.CAN-12-0487. [DOI] [PubMed] [Google Scholar]
- 58.Chen S, Fu L, Raja SM, Yue P, Khuri FR, Sun SY. Dissecting the roles of DR4, DR5 and c-FLIP in the regulation of Geranylgeranyltransferase I inhibition-mediated augmentation of TRAIL-induced apoptosis. Mol Cancer. 2010;9:23. doi: 10.1186/1476-4598-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Refaat A, Abd-Rabou A, Reda A. TRAIL combinations: The new ‘trail’ for cancer therapy (Review) Oncol Lett. 2014;7:1327–1332. doi: 10.3892/ol.2014.1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gasparian ME, Chernyak BV, Dolgikh DA, Yagolovich AV, Popova EN, Sycheva AM, Moshkovskii SA, Kirpichnikov MP. Generation of new TRAIL mutants DR5-A and DR5-B with improved selectivity to death receptor 5. Apoptosis. 2009;14:778–787. doi: 10.1007/s10495-009-0349-3. [DOI] [PubMed] [Google Scholar]
- 61.Kischkel FC, Lawrence DA, Chuntharapai A, Schow P, Kim KJ, Ashkenazi A. Apo2L/TRAIL-dependent recruitment of endogenous FADD and caspase-8 to death receptors 4 and 5. Immunity. 2000;12:611–620. doi: 10.1016/S1074-7613(00)80212-5. [DOI] [PubMed] [Google Scholar]
- 62.Thomas LR, Henson A, Reed JC, Salsbury FR, Thorburn A. Direct binding of Fas-associated death domain (FADD) to the tumor necrosis Factor-related Apoptosis-inducing ligand receptor DR5 is regulated by the death effector domain of FADD. J Biol Chem. 2004;279:32780–32785. doi: 10.1074/jbc.M409578200. [DOI] [PubMed] [Google Scholar]
- 63.Wang W, Wang S, Song X, Sima N, Xu X, Luo A, Chen G, Deng D, Xu Q, Meng L, et al. The relationship between c-FLIP expression and human papillomavirus E2 gene disruption in cervical carcinogenesis. Gynecol Oncol. 2007;105:571–577. doi: 10.1016/j.ygyno.2007.01.051. [DOI] [PubMed] [Google Scholar]
- 64.Hu Z, Ma D. The precision prevention and therapy of HPV-related cervical cancer: New concepts and clinical implications. Cancer Med. 2018;7:5217–5236. doi: 10.1002/cam4.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.