Abstract
Background
Metastatic breast cancer remains incurable. Next-generation sequencing (NGS) offers the ability to identify actionable genomic alterations in tumours which may then be matched with targeted therapies, but the implementation and utility of this approach is not well defined for patients with metastatic breast cancer.
Methods
We recruited patients with advanced breast cancer of any subtype for prospective targeted NGS of their most recent tumour samples, using a panel of 108 breast cancer-specific genes. Genes were classified as actionable or non-actionable using the European Society of Medical Oncology Scale for Clinical Actionability of Molecular Targets (ESCAT) guidelines.
Results
Between February 2014 and May 2019, 322 patients were enrolled onto the study, with 72% (n = 234) of patients successfully sequenced (n = 357 samples). The majority (74%, n = 171) of sequenced patients were found to carry a potentially actionable alteration, the most common being a PIK3CA mutation. Forty-three percent (n = 74) of patients with actionable alterations were referred for a clinical trial or referred for confirmatory germline testing or had a change in therapy outside of clinical trials. We found alterations in AKT1, BRCA2, CHEK2, ESR1, FGFR1, KMT2C, NCOR1, PIK3CA and TSC2 to be significantly enriched in our metastatic population compared with primary breast cancers. Concordance between primary and metastatic samples for key driver genes (TP53, ERBB2 amplification) was > 75%. Additionally, we found that patients with a higher number of mutations had a significantly worse overall survival.
Conclusion
Genomic profiling of patients with metastatic breast cancer can have clinical implications and should be considered in all suitable patients.
Keywords: Breast cancer, Metastasis, Genomic profiling, Personalised medicine
Background
Breast cancer remains the most common malignancy and cause of cancer-associated death amongst women worldwide [1, 2]. For patients with metastatic breast cancer (MBC) the disease is almost always fatal, with 5-year survival rates of approximately 26% [3, 4]. In early-stage breast cancer, treatment decisions are guided by the clinical subtypes of oestrogen receptor positive (ER+HER2−), human epidermal growth factor receptor 2 amplified (HER2+) and triple-negative breast cancer (TNBC), which are defined by the presence or absence of the oestrogen receptor and progesterone receptor and HER2 overexpression or gene amplification. This broad classification does not account for significant tumour evolution during disease progression, driven by selective pressures [1, 5]. Additionally, germline mutation testing for breast cancer patients, which could identify targetable alterations across all patients, is only common in the cases of young women, male breast cancer, or individuals with a strong family history [6].
There are an increasing number of agents that target specific genomic alterations, especially mutated protein kinases [7, 8]. Identification of suitable patients and gaining access to the corresponding targeted therapy has been challenging due to the cost of sequencing technology, the requirement in the past for non-formalin-fixed material, the inherent intra-tumoural heterogeneity of breast cancer and the lack of frequently occurring genomic alterations that can be targeted. In MBC, few genes are mutated at a frequency above 10%, leaving a wide array of infrequently altered potential targets [8]. Due to the rarity of these targets, clinical trials of targeted therapies are challenging to conduct.
In the era of precision oncology, few large-scale genomic studies have focused on metastatic breast cancer, and hence, our understanding of the breast cancer genome has relied on large-scale studies in primary tumours [7, 9, 10]. However, it is now well established that early breast cancer and MBC may have differing genomic profiles even in the same patient. More data surrounding the genomic landscape of MBC will be useful to identify oncogenic drivers specific for advanced disease with potential therapeutic implications, such as ESR1 and resistance to endocrine therapies [10, 11].
Here we describe such an initiative, from an academic tertiary referral cancer centre, where individuals with MBC had their tumour samples prospectively profiled using a targeted next generation sequencing (NGS) assay. The aim of this study was to explore the feasibility and clinical relevance of this approach.
Methods
Patient recruitment and samples
From January 2014 to May 2019, 322 patients were recruited from across Australia. All patients signed informed consent at Peter MacCallum Cancer Centre outpatient clinics (ethics committee approved study 13/123). Key inclusion criteria included age 18 years or over, histological confirmation of breast cancer and a life expectancy greater than 3 months. Patients of any gender, and with any breast cancer subtype or sites of disease, were allowed on study. Following successful enrolment, baseline information including relevant medical history, Eastern Cooperative Oncology Group performance status, past treatment and intended treatment were collected. Updates to the clinical history including survival follow-up were evaluated every 3 months.
Tumour tissue was requested for all recruited patients from various anatomical pathology service providers. Wherever possible, both primary and metastatic samples were obtained. Tissues were received as formalin-fixed, paraffin-embedded (FFPE) unstained sections at 5-μM thickness with up to 20 sections requested per sample. Prior to DNA extraction, a single slide was stained with haematoxylin and eosin to confirm the presence of cancer and to evaluate for tumour cellularity and tumour-infiltrating lymphocytes (TIL) in the sample [12].
Next-generation sequencing genomic profiling
Tumour DNA was extracted after micro- or macro-dissection (sample dependent) on sections stained with methyl green and quantified using the Fluorometric Qubit High Sensitivity system (ThermoFisher Scientific, USA). A minimum of 100 ng of DNA was required to proceed to sequencing.
A custom gene panel targeting all exons of genes recurrently altered in breast cancer was designed for the Agilent SureSelectXT Target Enrichment System (Agilent Technologies, Santa Clara, CA, USA) using the SureDesign tool (Table 1). This platform utilises biotinylated RNA baits for hybrid capture of desired DNA. Prior to library preparation with the KAPA Hyper Prep Kit (KAPA Biosystems, USA), DNA for each sample was randomly fragmented using ultrasonication (Covaris Inc., USA). Libraries passing quality control underwent hybrid capture with the custom RNA baits using the Bravo automated liquid handling robot (Agilent Technologies, Australia). Captured libraries were indexed and pooled prior to paired end sequencing on the Illumina MiSeq or NextSeq. A process matched non-tumour control sample was included in each run for use in downstream analysis. A total of 20 runs have been performed over the study, with a range of 10–40 samples per run.
Table 1.
Targeted sequencing panel and actionable genes as per ESMO guidelines
| Gene | Alteration type | |
|---|---|---|
| LEVEL 1: “known actionable alteration” | ||
| AKT1 | E17K, other pathogenic mutations | |
| BRCA1/ BRCA2 | Germline mutations | |
| ESR1 | Somatic mutations, previously reported | |
| ERBB2 | Amplification and mutation | |
| PIK3CA | Somatic mutations | |
| PTEN | Homozygous deletions, loss of function mutations | |
| LEVEL 2: “potentially actionable alteration” | ||
| ARID1A | Somatic mutations | |
| ATM | Somatic mutations | |
| BRCA1/BRCA2 | Somatic mutations | |
| CDH1 | Somatic mutations | |
| ERBB3 | Somatic mutations | |
| HRAS | Somatic mutations | |
| IGF1R | Somatic mutations | |
| INPP4B | Somatic mutations | |
| MAP2K4 | Somatic mutations | |
| MAP3K1 | Somatic mutations | |
| MDM2 | Amplifications | |
| PIK3R1 | Somatic mutations | |
| RB1 | Somatic mutations | |
| Non-actionable genes | ||
| AKT2 | FANCC | NRAS |
| AKT3 | FANCG | PALB2 |
| ALK | FBXW7 | PDGFRA |
| APC | FGFR1 | PIK3R3 |
| AXIN2 | FGFR2 | PMS2 |
| BAP1 | FGFR3 | PRKAR1A |
| BLM | FGFR4 | PTCH1 |
| BMPR1A | FH | PTPN11 |
| BRAF | FLCN | RAD51C |
| BRIP1 | FOXA1 | RAD51D |
| BUB1B | GATA3 | RECQL4 |
| CASP8 | KIT | RET |
| CBFB | KRAS | RUNX1 |
| CCND1 | MAP2K1 | SDHA |
| CCND2 | MAX | SDHB |
| CDC73 | MCL1 | SDHC |
| CDK4 | MEN1 | SDHD |
| CDKN2A | MET | SF3B1 |
| CHEK2 | MLH1 | SMO |
| CTNNB1 | KMT2D | STK11 |
| DDB2 | KMT2C | TBX3 |
| EGFR | MSH2 | TMEM127 |
| ERBB4 | MSH6 | TP53 |
| ERCC2 | MUTYH | TSC1 |
| ERCC3 | NBN | TSC2 |
| ERCC4 | NCOR1 | VHL |
| ERCC5 | NF2 | WT1 |
| EXT1 | NOTCH1 | XPA |
| FANCA | NOTCH2 | XPC |
Level 1 actionable alterations are equivalent to ESMO ESCAT LOE I and II, whilst level 2 actionable alterations are equivalent to LOE III and IV as described in [13]
Assessment of gene actionability
Table 1 is adapted from the European Society for Medical Oncology (ESMO) and their European Society of Medical Oncology Scale for Clinical Actionability of Molecular Targets (ESCAT) guidelines [13]. These classify the actionability of genes according to various levels of evidence (LOE). As indicated in Table 1, level 1 actionable corresponds to ESCAT LOE I/II, whilst level 2 actionable genes is equivalent to LOE III/IV. Although amplification of ERBB2 is considered to be a level 1 actionable alteration, because it is the defining factor of the HER2+ breast cancer subtype, it is not included in the analysis. All patients classified as HER2+ would therefore have a level 1 actionable alteration; it is excluded from the analysis because it defines the HER2+ breast cancer subtype and is part of standard diagnostic testing.
Tumour-infiltrating lymphocyte (TIL) quantity evaluation
TIL analysis was performed wherever possible on available H&E slides as per the International TILs Working Group guidelines by a single pathologist (RS) [14].
Bioinformatics analysis
Raw sequencing data had adapters trimmed with cutadapt (v1.6–1.9.1) [15]. The Burrows-Wheeler Aligner algorithm was used to align sequencing reads to the human genome (GRCh37), with no altered parameters [16]. The Depth of Coverage tool in the Genome Analysis Tool Kit (GATK 3) was used to calculate the mean coverage accounting for overlapping read pair ends [17]. Duplicate reads were removed with Picard Tools (v1.77–v1.141) [18]. VarDict (v1.3.0–1.5.7) was used to call variants that were then annotated with ANNOVAR [19, 20]. CopywriteR (v2.0.1–2.0.6) was used to call copy number (CN) alterations from off-target reads with a bin width of 50 kb [21]. A mean log ratio > 0.7 was considered amplified, and < 0.7 indicative of a heterozygous or homozygous deletion.
Manual curation was then used to identify the variants most likely pathogenic and relevant to the patient’s disease. This included removing samples with a read depth of less than 100, a variant allele frequency less than 5% and variants with strand bias. Additionally, only variants that led to a protein change (non-synonymous), those located in the exonic region of the gene and those not found in either the 1000 genomes population or the ESP650 cohorts were included in the analysis [22, 23]. Integrative Genomics Viewer was then utilised to confirm the presence of the mutation and exclude sequencing error or variants in highly repetitive regions (either homopolymers > 4 bases in length or triplet nucleotide repeats) [24]. Any mutation in TP53, BRCA1, or BRCA2 that had a variant allele frequency greater than 20% was referred for consideration of germline testing.
A report was generated on the sequencing results of each patient, including an estimation of the likelihood of the mutation’s oncogenic relevance and potential therapeutic options including clinical trials. This estimation of oncogenic relevance was calculated using various resources including the ClinVar, the Catalogue of Somatic Mutations in Cancer, OncoKB and medical literature [25–27].
Statistical analysis
Survival analysis was performed using Cox regression models. In all cases, overall survival was calculated from date of study consent to either date of death or date of last follow-up. Time of diagnosis for both primary and metastatic disease was calculated from the first biopsy confirmation of cancer, or if a biopsy was unsuitable, first imaging to confirm the presence of cancer. Somatic interaction analysis and calculation was performed using the pair-wise Fisher’s exact test to detect co-occurring gene pairs and the CoMet ExactTest algorithm [28–30]. Concordance scores were calculated by dividing the number of concordant observations by the total number of observations per genetic alteration. The same principle was applied to calculate individual patient concordance scores. Calculations for mutation numbers were performed using one sample from every patient, ideally a metastatic sample; however, when no metastatic sample was available, a primary sample was included instead. Adjustment for multiple testing was performed using the method of Benjamini and Hochberg. All statistical analysis was performed using R software (R version 3.6.2 (2019-12-12)).
Results
Between February 2014 and May 2019, 322 patients with MBC were recruited. The majority of these patients were ER+HER2− (68%, n = 219), with HER2+ and TNBC representing 12% (n = 38) and 20% (n = 65) respectively. The median age of these patients was 46 years (23–74) at primary diagnosis and 52 years (24–78) at metastatic diagnosis (Table 2).
Table 2.
Study cohort clinical features
| Overall (n = 322) | ER+HER2− (n = 219, 68%) | HER2+ (n = 38, 12%) | TNBC (n = 65, 20%) | P value | |
|---|---|---|---|---|---|
| Median age at primary diagnosis (range) | 46 (23–74) | 46 (24–73) | 43.5 (34–74) | 47 (23–74) | |
| Median age at metastatic diagnosis (range) | 51 (24–78) | 52 (30–78) | 45.5 (35–67) | 50 (24–76) | |
| Median age at consent (range) | 52 (24–81) | 54 (30–78) | 48 (36–81) | 52 (24–77) | |
| Median DFI1 (range) | 40 (1–312) | 54.5 (1–312) | 25 (4–136) | 23 (1–252) | |
| De novo patients | 71 (22%) | 50 (22%) | 14 (36%) | 7 (11%) | |
| Treatment history | |||||
| Previous chemotherapy | |||||
| Yes | 309 (96%) | 208 (95%) | 37 (97%) | 64 (98%) | |
| No | 8 (2%) | 7 (3%) | 1 (3%) | 0 | |
| Unknown | 5 (2%) | 4 (2%) | 0 | 1 (2%) | |
| Previous Endocrine therapy | Chi squared: < 0.0001 | ||||
| Yes | 222 (69%) | 205 (94%) | 11 (29%) | 6 (9%) | |
| No | 95 (29%) | 10 (5%) | 27 (71%) | 58 (89%) | |
| Unknown | 5 (2%) | 4 (1%) | 0 | 1 (2%) | |
| Lines of treatment in metastatic setting | Chi squared: 0.003 | ||||
| 1 | 28 (9%) | 12 (5%) | 1 (3%) | 16 (25%) | |
| 2 | 70 (22%) | 47 (21%) | 4 (11%) | 19 (29%) | |
| 3 | 72 (22%) | 45 (21%) | 13 (34%) | 14 (22%) | |
| > 3 | 133 (41%) | 103 (47%) | 20 (53%) | 10 (15%) | |
| Unknown | 20 (6%) | 12 (5%) | 0 | 6 (9%) | |
| Sequenced patients | |||||
| Sequenced | 234 (72%) | 162 (75%) | 25 (66%) | 47 (72%) | |
| Not sequenced due to sample failure | 18 (6%) | 12 (6%) | 1 (3%) | 5 (8%) | |
| Not sequenced due to other2 | 70 (22%) | 45 (19%) | 12 (31%) | 13 (20%) | |
| Patients with actionable mutations3 | 171 (74%) | 131 (80%) | 11 (44%) | 29 (61%) | Chi squared: 0.0001 |
| Level 1: | 114 (49%) | 93 (57%) | 6 (24%) | 15 (32%) | |
| Level 2 | 57 (25%) | 38 (24%) | 5 (20%) | 14 (30%) | |
| No actionable alterations | 63 (26%) | 31 (19%) | 14 (56%) | 18 (38%) | |
DFI disease-free interval (time in months)
1Patients with de novo metastatic disease not included. 2Other reasons for not being sequenced aside from sequencing failure include: sample not received or insufficient DNA quantity for sequencing. 3Percentage calculated from number of sequenced patients. Numbers shown are reflective of the overall total and then the total within each subtype. The HER2+ subtype does not include patients with ERBB2 amplification (100%, n = 25)
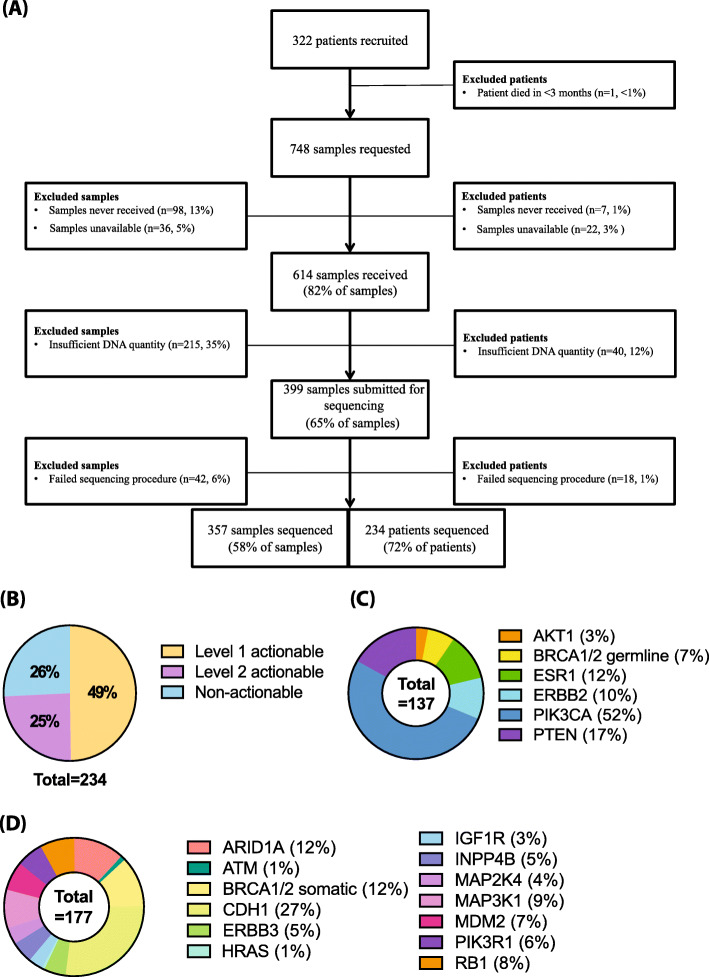
Across the cohort, 748 tumour samples were requested, with an average of 2 samples per patient (Fig. 1a). In total, 82% (n = 614) of the requested samples were received; however, the time between request and receipt varied considerably with an average of 29 days for samples stored by the parent institution and 49 days for samples retrieved from external pathology providers.
Fig. 1.
Study cohort. a Recruitment and specimen retrieval flow chart. Sample information and numbers are shown on the left of the central chart and patient information and numbers are shown on the right. b Actionable alterations. Pie chart shows the percentage breakdown of patients and the level of actionable alterations their samples contain. Patients with at least one level 1 actionable alteration amounted to 49% (n = 114). Patients with a level 2 actionable alteration accounted for 25% (n = 57). Patients without an actionable alteration amounted to 26% of the cohort (n = 60). c Level 1 actionable alteration breakdown by number of samples. AKT1 mutations (3%, n = 4), BRCA1/2 germline variants (7%, n = 9), ESR1 mutations (12%, n = 16), ERBB2 mutations (10%, n = 14), PIK3CA mutations (52%, n = 71) and PTEN mutations (17%, n = 23). ERBB2 amplifications are not included. d Level 2 actionable alteration breakdown by number of samples. ARID1A mutations (12%, n = 21), ATM mutations (1%, n = 2), BRCA1/2 non-germline mutations (12%, n = 21), CDH1 mutations (27%, n = 48), ERBB3 mutations (5%, n = 9), HRAS mutations (1%, n = 1), IGF1R mutations (3%, n = 6), INPP4B mutations (5%, n = 9), MAP2K4 mutations (4%, n = 7), MAP3K1 mutations (9%, n = 16), MDM2 amplifications (7%, n = 12), PIK3R1 mutations (6%, n = 11) and RB1 mutations (8%, n = 14)
Amongst the received samples, 35% (n = 215) had insufficient DNA quantity for sequencing, and the remaining 65% (n = 399) underwent sequencing with a success rate of 90% (n = 357). The failure rate of 10% of samples (n = 42) was largely attributed to poor quality DNA due to FFPE fixation, as neither the age of the sample nor its location were significant predictors of failure. The majority of the failed sequencing samples came from primary lesions (70%, n = 29). Metastatic samples, from sites such as lymph node (12%, n = 5), liver (5%, n = 2) and lung (5%, n = 2), accounted for less than a quarter of sequencing failures. A single specimen from the bone, ovary, brain and skin also failed sequencing (2% each). Ultimately, this led to 72% (n = 234) of recruited patients being successfully sequenced. The majority of samples sequenced successfully were primary tumours (53%, n = 189). Samples from metastatic lymph nodes (10%, n = 37) and the liver (9%, n = 31) accounted for the majority of metastatic samples (n = 168, 47%). Other metastatic sites included bone (7%, n = 26), brain (5%, n = 19), lung (3%, n = 12) and ovarian (2%, n = 6). The remaining 6 metastatic samples each came from a unique site and accounted for overall 2% (n = 6). Paired primary and metastatic tissue was available for 35% of patients (n = 81).
Across the sequenced samples, there was a median of 3 non-synonymous somatic mutations per sample (range 0–10) and 4 focal CN alterations (range 1–28). Level 1 alterations were found in 49% (n = 114) of all patients sequenced and 25% (n = 57) of all patients were found to carry a level 2 change (Fig. 1b). This amounts to 74% (n = 171) of patients harbouring a potentially actionable alteration. The majority of these samples were of the ER+HER2− subtype (77%, n = 131), with HER2+ and TNBC subtypes accounting for 6% (n = 11) and 17% (n = 29) of the total cohort respectively. Fifty-seven percent of ER+HER2− cases (n = 93/162), 24% of HER2+ (n = 6/25) and 32% of TNBC cases (n = 15/47) had level 1 alterations.
As shown in Fig. 1c, PIK3CA was the most common level 1 actionable alteration, being found in 30% of sequenced patients and accounting for 52% of patients with level 1 actionable mutations (n = 71). Of the cohort with actionable alterations (n = 171), 43% (n = 74) were referred onto a molecularly targeted clinical trial. The majority of these patients had PIK3CA mutations (89%, n = 68), followed by 2 patients with PTEN mutations, and 4 patients with ERBB2 mutations. A further 2 patients with FGFR1 amplifications were referred for FGFR inhibitor trials, although this gene is not considered actionable according to ESMO Clinical Actionability guidelines [13]. Of ER+HER2− patients, 10% (n = 24) were found to carry an ESR1 mutation in their metastatic tumour sample. Nearly all of these patients had been treated with prior endocrine therapy (Table 2).
A total of 21 patients were found to harbour BRCA1 or BRCA2 mutations. Of these, 36% (n = 8) were previously known pathogenic germline BRCA1 or BRCA2 carriers. The remaining 64% (n = 13) were referred on to germline testing. Of these patients, 2 were confirmed to carry germline variants of unknown significance, 5 confirmed wild type and 6 patients did not attend germline testing. A previously known pathogenic germline mutation in CHEK2 was also identified in a single patient.
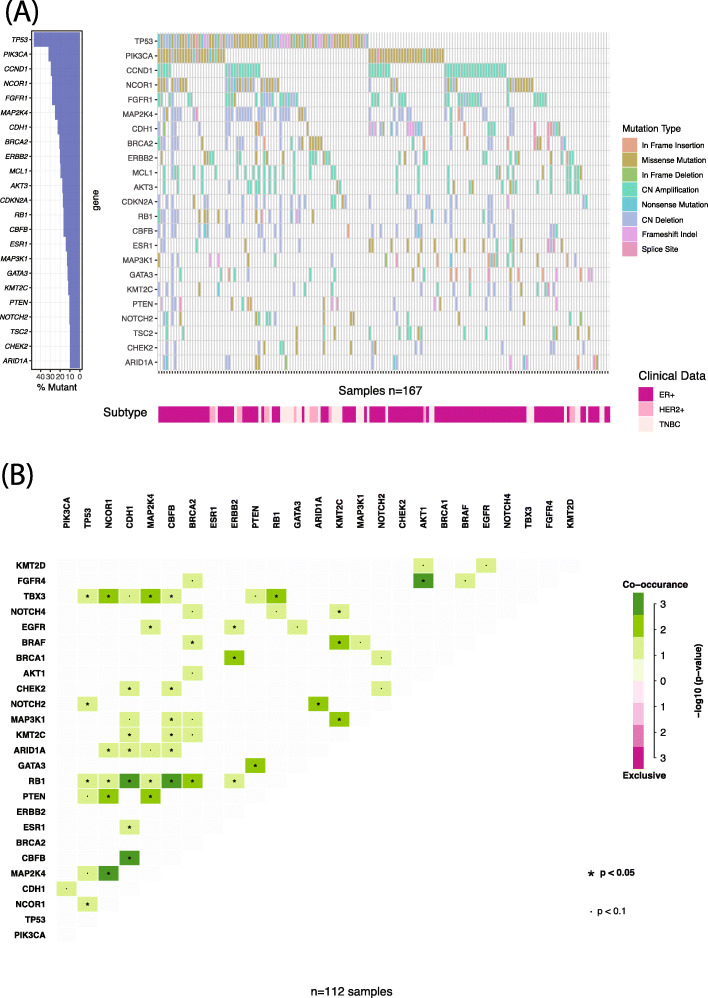
Figure 2a shows the genomic landscape of the sequenced metastatic samples (n = 167 samples). Amongst these samples, TP53 was the most commonly altered at 40%, followed by PIK3CA, CCND1, NCOR1 and FGFR1. In order to identify any gene that may be enriched in the metastatic cohort, the mutational frequencies were compared to those in the TCGA breast cancer dataset comprised of primary tumours [31]. The genes AKT1, BRCA2, CHEK2, ESR1, FGFR4, KMT2C, NCOR1, PIK3CA and TSC2 were significantly enriched in our metastatic study population compared to the TCGA cohort (Supplementary Table 1).
Fig. 2.
Metastatic mutational landscape. a Oncoplot of somatic mutations identified at a frequency of 10% or more in sequenced metastatic samples (n = 168). Percentage frequency of the genes is shown in the barchart to the left of the central plot. Clinical subtype and mutation type are indicated on their respective legend. For genes significantly enriched in this cohort compared to TCGA, see Supplementary Table 1. b Mutational co-occurrence plot for metastatic ER+HER2− samples (n = 112). Degree of significance indicated in the legend, with only results of the top 25 genes shown. No mutational exclusivity pairs were identified. For individual p values, see Supplementary Table 2
We performed an analysis to identify alterations that may be significantly co-occurring or mutually exclusive. Each subtype was analysed individually using the 25 most frequently altered genes within each cohort. In ER+HER2− metastatic samples (n = 112) (Fig. 2b and Supplementary Table 2), 43 gene-pairs were found to significantly co-occur. When this analysis was performed in the TNBC subtype (n = 40 samples), only 2 gene pairs were found to be co-occurring: RB1 and FGFR1 (p = 0.01), as well as CHEK2 and CDH1 (p = 0.025). In the ER+HER2− group, RB1 mutations were found to co-occur with 8 other genes, suggesting a significant role within this subtype (Supplementary Table 2). Overall, in our cohort TNBC metastatic samples have less significantly co-occurring genes than ER+HER2− breast cancers.
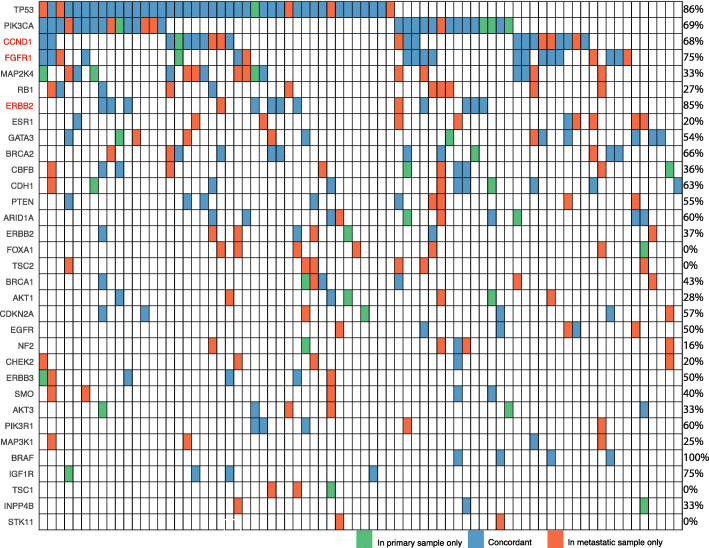
The concordance of key driver alterations was investigated between primary and metastatic samples (Fig. 3) across the 81 patients (35%) of the cohort with paired samples. Of note, PIK3CA was largely concordant with a score of 69% (n = 20/29 patients). In the 9 patients who displayed discordance, the PIK3CA mutation was found exclusively in the metastatic lesion in 5 patients and exclusively in the primary lesion in 4 patients. The only gene to show 100% concordance was BRAF (n = 4 patients); however, TP53, FGFR1 amplification, ERBB2 amplification and IFG1R showed concordance scores greater than 75%. FOXA1, STK11, TSC1 and TSC2 were found to have no concordance between tumours and were largely restricted to metastases (Fig. 3). When tumour concordance was evaluated within each patient, only 18% (n = 15 patients) were found to have 100% concordance between their primary and metastatic lesion, whilst 7% of patients (n = 6) had no concordant findings, highlighting the genomic heterogeneity of metastatic breast cancer within individual patients.
Fig. 3.
Concordance plot of primary and metastatic paired samples. Data for 76 patients and their paired sequenced samples is shown. Genetic alterations included in this figure are putative driver genes observed in 2 or more samples. A further 4 patients and their paired samples have been excluded from the figure because their pairs did not include the genes shown. Concordance scores for each gene is listed on the right of the figure. Genes listed in red indicate that it is CN amplification concordance being compared, rather than mutation concordance (shown in black text)
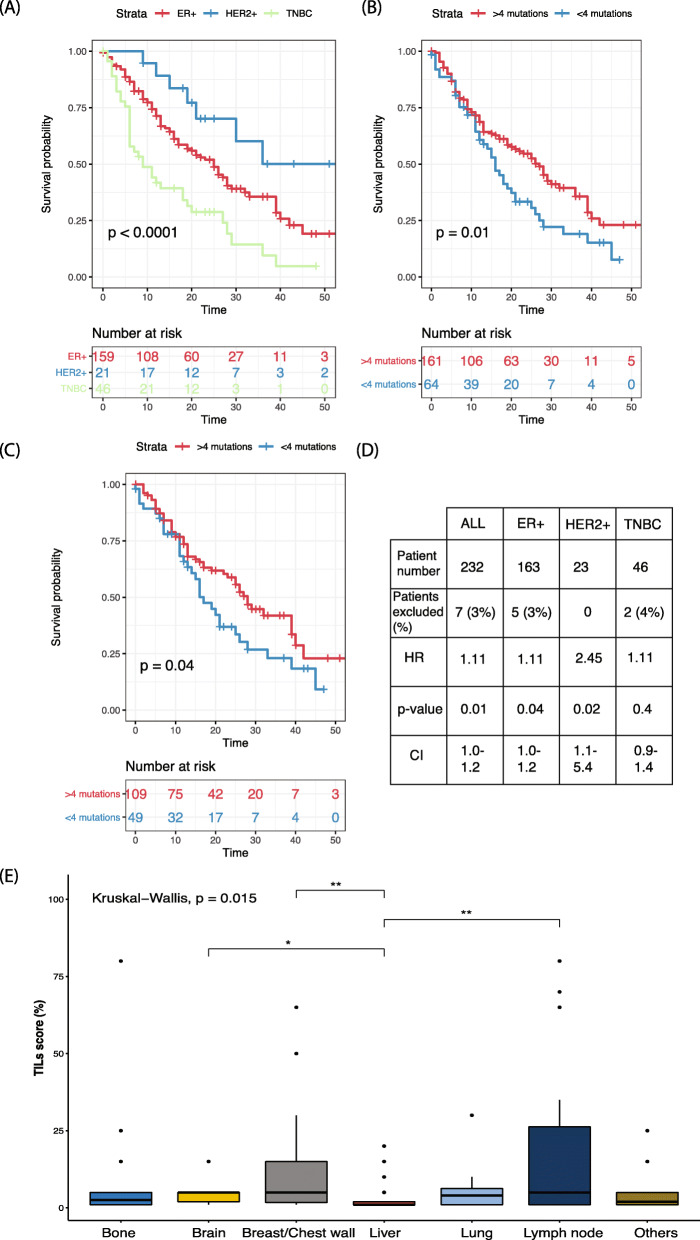
We investigated factors associated with survival, and unsurprisingly, the clinical subtype (Fig. 4a) had a significant impact on overall survival (OS). When the time between primary disease and metastatic disease diagnosis was compared amongst the patients, those with a longer disease-free interval had a significantly better OS (hazard ratio (HR) = 0.61, confidence interval (CI) = 0.39–0.94, p = 0.02). Amongst the patients with sequenced samples, we found that higher mutation number was associated with significantly worse survival (HR = 1.1 per mutation, CI = 1.0–1.2, p = 0.01, Fig. 4b). This trend was observed in both the ER+HER2− subtype (HR 1.1, CI = 1.0–1.2, p = 0.04; Fig. 4c) and the HER2+ subtype (HR = 2.5, CI = 1.1–5.3, p = 0.02), but not in the TNBC subtype (HR = 1.1, CI = 0.86–1.4, p = 0.4; Fig. 4d). The same subtype level analysis was performed using copy number; however, no significant results were observed.
Fig. 4.
Prognostic associations in this cohort of sequenced metastatic breast cancer patients. a Overall survival by subtype for all recruited patients (n = 323). b Overall survival of patients based on the mutational burden of 4 mutations (75th percentile) or more (n = 234). c Overall survival of ER+HER2− patients based on the median mutation number of 4 or more (n = 163 patients). d Table of HR for all patients and all subtypes by mutation number per sample. Patients were excluded if there was incomplete survival information. e Spread of TILs across distant metastatic site (n = 123)
Although breast cancer is not commonly considered to be immunogenic, the presence of TILs has been known to impact survival in primary breast cancer [32]. In our dataset, TILs in the metastatic lesion were not significantly associated with OS for any subtype, as calculated from time of metastatic diagnosis (Supplementary Table 3). As anticipated, amongst the primary samples, TNBC patients with a higher number of TILs had a significantly longer DFI (Supplementary Table 3). This was also evident using the TNBC metastatic samples, where those with higher TILs had a significantly longer disease-free interval than those with lower TILs (HR 4.07, CI 1.2–13.6, p value: 0.02). Consistent with previous data some metastatic sites had significantly greater TIL infiltrate (Fig. 4e) [33].
Discussion
In this study, 49% (n = 114) of patients were found to harbour a level 1 actionable alteration, which corresponds to the ESCAT LOE I/II genes. Level 1 actionable alterations were of high clinical relevance with 65% (n = 74/111) of patients with alterations in this category referred for clinical trial. Level 2 actionable alterations were identified in 25% (n = 57) of patients and did not impact clinical decision making due to the unavailability of a matched therapy. This amounts to almost three-quarters (74%, n = 171) of the cohort being found to harbour at least one actionable alteration, albeit with variable clinical importance. Of ER+HER2− patients, 10% (n = 24) were found to carry ESR1 alterations, and 64% (n = 13) of patients with a BRCA1 or BRCA2 mutation with a high variant allele fraction were recommended for germline testing. Additionally, we found several genes that were enriched in the metastatic setting across the whole cohort, as well as discordant findings in patients with both primary and metastatic samples, highlighting the need to profile metastatic tissue where possible. Finally, we found that higher mutation number was also associated with worse survival. These findings were obtained using a targeted panel sequencing far fewer genes (around one-third) than some which are commercially available. The outcomes of this study suggest that a panel featuring only ESCAT LOE I/II genes, equivalent to level 1 in this study, would be sufficient for the majority of patients who could benefit from genomic interrogation. These smaller and less expensive assays may make genomic profiling more broadly accessible to breast cancer patients and clinicians.
The distribution and frequency of metastatic genomic alterations in this study are similar to the findings of other studies, including enrichment of ESR1 mutations believed to be the result of therapeutic selective pressure [7, 10, 11, 34]. Mutations in ESR1 are almost exclusively found in metastatic lesions of ER+HER2− patients, although some studies have shown they can exist prior to the onset of metastases in patients treated with endocrine therapies [35, 36]. This was observed in 2 patients, shown in Fig. 3, where the primary samples were collected post-endocrine therapy and had already developed an ESR1 alteration. ESR1 mutations arise due to prolonged treatment with tamoxifen and aromatase inhibitors; however, retrospective analysis of the PALOMA-2 and PALOMA-3 studies found that patients treated with a selective oestrogen degrader either alone or in combination with CDK4/6 inhibitors also showed development of ESR1 mutations [37–39].
As expected, TP53 alterations were the most prevalent across all samples. Of all the level 1 actionable genes, PIK3CA was found to be the most commonly altered, consistent with other studies [34]. Recently, the α-subunit specific PIK3CA inhibitor alpelisib was approved for use in ER+HER2− MBC patients with hotspot PIK3CA mutations, providing a matched therapy for this most common alteration [34, 40, 41]. Conversely, the finding that 25% of patients carried a level 2 alteration which lack proven therapeutic options highlights the difficulties in matching the majority of patients with targeted therapies. It is uncertain how many of these level 2 alterations may be clinically relevant in the near future. Nevertheless, the high prevalence of PIK3CA alterations alone means that a nearly a quarter of MBC patients could be treated with a matched targeted therapy, even accounting for those that could not have a metastatic sample sequenced as there was a high concordance rate (69% concordance) with the primary tumour.
Like other studies, we found a proportion of patients (26%) did not carry a potentially actionable alteration. Patients where no alteration was found may benefit from more comprehensive genomic analysis such as whole-genome sequencing, where mutational signatures such as homologous recombination repair deficiency or more rarely, microsatellite instability could be identified and utilised for therapeutic decisions [42, 43].
Our concordance analysis in 81 patients with metachronous primary and metastatic samples allows observation of how key driver genes change in metastatic disease. Mutations in both TSC2 and STK11 were found exclusively in metastatic samples and are possibly associated with the development of metastatic disease or resistance to therapy. The finding that TSC2 was also enriched in our metastatic cohort compared to the TCGA primary data set also supports this hypothesis [11]. The high frequency of mutations in the genes RB1, TSC1 and FOXA1 in the metastatic samples with few or no concordant cases also suggests these genes are similarly relevant to the biology of metastatic disease. Co-mutation analysis in both the ER+HER2− and TNBC subgroups suggested that RB1 may also play a key role in the emergence of metastatic and treatment-resistant subclonal elements. This finding is supported by other studies that have linked alterations in RB1 with resistance to endocrine therapies and CDK4 inhibitors [10, 44, 45].
Our study has several limitations. Firstly, the somatic landscape of the sequenced tumour may not be indicative of the whole metastatic disease burden. Novel methods such as circulating tumour DNA sequencing could be utilised to avoid sampling bias and increase the number of patients for whom genomic information is available. Nevertheless there are still cases of discordance between circulating tumour DNA and tumour DNA from sequenced samples, and plasma sampling has a higher rate of false negatives [1, 46]. Secondly, a panel of the size employed in this study cannot accurately estimate the true total mutation burden, and the mutation number per sample reported here cannot be considered a surrogate measure. Thirdly, the cohort of patients in our study may not be representative of the general metastatic breast cancer population as there was likely to be some bias towards enrolling younger and fitter patients. Fourthly, the rate at which patients receive therapies matched to their genomic profile is subject to the availability of these therapies and clinical trials, which will vary greatly across clinical contexts, and the timeframe in which the genomic profile can be delivered. The latter was heavily influenced by the time required to receive tissue samples. Reducing this further will require closer integration of pathology services with the treatment team and greater awareness of the urgency of tissue requests. Fifthly, consistent with other studies, we found that a relatively high rate of patients could not be delivered a genomic profile due to the difficulty in obtaining a suitable sample as 35% of samples had insufficient DNA. Due to the inherent difficulties in obtaining metastatic biopsies from MBC patients, these failure rates for tumour sequencing are unlikely to improve.
Conclusion
This study has found that prospective genomic sequencing for the management of MBC is both feasible and of utility as an adjunct to standard management with 49% of sequenced patients carrying a level 1 actionable alteration. PIK3CA was the most common actionable mutation, found in 52% (n = 71) of patients with a level 1 actionable alteration. Of patients referred for a clinical trial with a matched therapy, 89% (n = 68) had a PIK3CA mutation. We found that PIK3CA mutation status, and genomic profiles in general, demonstrates within-patient heterogeneity which is clinically relevant, although PIK3CA was usually concordant across primary and metastatic samples. Nevertheless, the presence of genomic heterogeneity indicates sequencing of metastatic lesions is preferred if possible. Due to the inherent difficulties in obtaining biopsies of metastatic disease, the combination of tumour and circulating tumour DNA sequencing is likely to be the optimal strategy to improve the rate of successful genomic profiling. It is expected the clinical utility of genomic profiling will increase over time with greater availability of targeted therapies, as has already transpired with the recent approval of the PIK3CA inhibitor alpelisib. We conclude that where possible, patients with MBC should undergo molecular profiling.
Supplementary information
Additional file 1: Supplementary Table 1. List of genes enriched in study dataset compared to the TCGA dataset. Supplementary Table 2. Genes-pairs that co-occur in ER+ metastatic breast cancer. Supplementary Table 3. Hazard ratios and confidence intervals for TILs samples.
Acknowledgements
We would like to thank the following people: the breast care nurses and staff of the Breast unit at the Peter MacCallum Cancer Centre; Catherine Zilberg and Emily Humphrey; Eveline Niedermayr; and Timothy Semple, Sreeja Gadipally and Gisela Mir Arnau.
Abbreviations
- CI
Confidence interval
- CN
Copy number
- COSMIC
Catalogue of somatic mutations in cancer
- DFI
Disease-free interval
- ER +
Oestrogen-receptor positive
- ESCAT
European Society of Medical Oncology Scale for Clinical Actionability of Molecular Targets
- ESMO
European Society of Medical Oncology
- FFPE
Formalin-fixed paraffin-embedded
- HER2+
Human epidermal growth factor receptor 2 amplified
- HR
Hazard ratio
- LOE
Levels of evidence
- NGS
Next generation sequencing
- OS
Overall survival
- TIL
Tumour-infiltrating lymphocytes
- TNBC
Triple-negative breast cancer
Authors’ contributions
PS, ZLT and SL designed the study, assisted by AF and SF. PS and SL were principal investigators for the study. PS piloted the study and built and tested the analysis pipeline with AF and SF. PS, SJL, PF, SJD and SL recruited patients to the study. RS scored TILs on all samples. CVG, PS, CFW, AK and KK collected study materials and data. CVG performed all analyses in the paper and wrote the manuscript. All authors reviewed and approved the final manuscript.
Funding
We thank the Breast Cancer Trials Group Australia and New Zealand for a seed grant that funded the main study as well as the Peter MacCallum Foundation Women’s Health Initiative.
PS was supported by the Royal Australian College of Physicians Shields Research Entry Scholarship, the NHMRC and NBCF co-funded post-graduate scholarship and Cancer Therapeutics CRC scholarship. SL is supported by the National Breast Cancer Foundation of Australia Endowed Chair and the Breast Cancer Research Foundation, New York.
CVG is supported though an Australian Government Research Training Program (RTP) Scholarship.
Availability of data and materials
De-identified genomic variant data is available upon request to the corresponding author.
Ethics approval and consent to participate
Ethics approval for this study was granted by the Peter MacCallum Cancer Centre ethics board (#13/123).
Consent for publication
Not applicable.
Competing interests
PS acts as a consultant (not compensated) for Roche-Genentech.
SL receives research funding to her institution from Novartis, Bristol Meyers Squibb, Merck, Roche-Genentech, Puma Biotechnology, Pfizer, Eli Lilly and Seattle Genetics. She has acted as consultant (not compensated) to Seattle Genetics, Pfizer, Novartis, BMS, Merck, AstraZeneca and Roche-Genentech. She has acted as consultant (paid to her institution) to Aduro Biotech, Novartis and G1 Therapeutics.
PF has received travel and honoraria for lectures from Novartis and AstraZeneca.
SJD has received research funding to her institution from Roche-Genentech and Cancer Therapeutics CRC.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Courtney van Geelen and Peter Savas are co-first authors.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13058-020-01328-0.
References
- 1.Arnedos M, Vicier C, Loi S, Lefebvre C, Michiels S, Bonnefoi H, et al. Precision medicine for metastatic breast cancer--limitations and solutions. Nat Rev Clin Oncol. 2015;12(12):693–704. doi: 10.1038/nrclinonc.2015.123. [DOI] [PubMed] [Google Scholar]
- 2.Harbeck N, Gnant M. Breast cancer. Lancet (London) 2017;389(10074):1134–1150. doi: 10.1016/S0140-6736(16)31891-8. [DOI] [PubMed] [Google Scholar]
- 3.Institute. NC . SEER Stat Fact Sheets: Breast Cancer. 2018. [Google Scholar]
- 4.El Sayed R, El Jamal L, El Iskandarani S, Kort J, Abdel Salam M, Assi H. Endocrine and targeted therapy for hormone-receptor-positive, HER2-negative advanced breast cancer: insights to sequencing treatment and overcoming resistance based on clinical trials. Front Oncol. 2019;9:510. doi: 10.3389/fonc.2019.00510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rivenbark AG, O'Connor SM, Coleman WB. Molecular and cellular heterogeneity in breast cancer: challenges for personalized medicine. Am J Pathol. 2013;183(4):1113–1124. doi: 10.1016/j.ajpath.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellis MJ. Mutational analysis of breast cancer: guiding personalized treatments. Breast (Edinburgh) 2013;22(Suppl 2):S19–S21. doi: 10.1016/j.breast.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vasan N, Yelensky R, Wang K, Moulder S, Dzimitrowicz H, Avritscher R, et al. A targeted next-generation sequencing assay detects a high frequency of therapeutically targetable alterations in primary and metastatic breast cancers: implications for clinical practice. Oncologist. 2014;19(5):453–458. doi: 10.1634/theoncologist.2013-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yates LR, Knappskog S, Wedge D, Farmery JHR, Gonzalez S, Martincorena I, et al. Genomic evolution of breast cancer metastasis and relapse. Cancer Cell. 2017;32(2):169–84.e7. doi: 10.1016/j.ccell.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Basho RK, de Melllo D, Ueno NT, Wathoo C, Chen H, Shariati M, et al. Clinical outcome based on multigene profiling in metastatic breast cancer patients. Oncotarget. 2016;7(47):76362–76373. doi: 10.18632/oncotarget.12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bertucci F, Ng CKY, Patsouris A, Droin N, Piscuoglio S, Carbuccia N, et al. Genomic characterization of metastatic breast cancers. Nature. 2019;569(7757):560–564. doi: 10.1038/s41586-019-1056-z. [DOI] [PubMed] [Google Scholar]
- 11.Lefebvre C, Bachelot T, Filleron T, Pedrero M, Campone M, Soria JC, et al. Mutational profile of metastatic breast cancers: a retrospective analysis. PLoS Med. 2016;13(12):e1002201. doi: 10.1371/journal.pmed.1002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salgado R, Loi S. Tumour infiltrating lymphocytes in breast cancer: increasing clinical relevance. Lancet Oncol. 2018;19(1):3–5. doi: 10.1016/S1470-2045(17)30905-1. [DOI] [PubMed] [Google Scholar]
- 13.Condorelli R, Mosele F, Verret B, Bachelot T, Bedard PL, Cortes J, et al. Genomic alterations in breast cancer: level of evidence for actionability according to ESMO Scale for Clinical Actionability of molecular Targets (ESCAT) Ann Oncol. 2019;30(3):365–373. doi: 10.1093/annonc/mdz036. [DOI] [PubMed] [Google Scholar]
- 14.Salgado R, Denkert C, Demaria S, Sirtaine N, Klauschen F, Pruneri G, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol. 2015;26(2):259–271. doi: 10.1093/annonc/mdu450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17(1):3.
- 16.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Broad Institute . Picard Toolkit.: GitHub Repository. 2019. [Google Scholar]
- 19.Lai Z, Markovets A, Ahdesmaki M, Chapman B, Hofmann O, McEwen R, et al. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 2016;44(11):e108. doi: 10.1093/nar/gkw227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuilman T, Velds A, Kemper K, Ranzani M, Bombardelli L, Hoogstraat M, et al. CopywriteR: DNA copy number detection from off-target sequence data. Genome Biol. 2015;16:49. doi: 10.1186/s13059-015-0617-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.NHLBI GO Exome Sequencing Project (ESP). Exome variant server. Seattle: National Heart, Lung and Blood Institute. Available from: http://evs.gs.washington.edu/EVS/. Accessed 10 Apr 2019.
- 24.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46(D1):D1062–D10d7. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43(Database issue):D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017;2017. 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed]
- 28.Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28(11):1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dees ND, Zhang Q, Kandoth C, Wendl MC, Schierding W, Koboldt DC, et al. MuSiC: identifying mutational significance in cancer genomes. Genome Res. 2012;22(8):1589–1598. doi: 10.1101/gr.134635.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leiserson MD, Wu HT, Vandin F, Raphael BJ. CoMEt: a statistical approach to identify combinations of mutually exclusive alterations in cancer. Genome Biol. 2015;16:160. doi: 10.1186/s13059-015-0700-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, et al. Oncogenic signaling pathways in The Cancer Genome Atlas. Cell. 2018;173(2):321–37.e10. doi: 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loi S, Drubay D, Adams S, Pruneri G, Francis PA, Lacroix-Triki M, et al. Tumor-infiltrating lymphocytes and prognosis: a pooled individual patient analysis of early-stage triple-negative breast cancers. J Clin Oncol. 2019;37(7):559–569. doi: 10.1200/JCO.18.01010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luen SJ, Salgado R, Fox S, Savas P, Eng-Wong J, Clark E, et al. Tumour-infiltrating lymphocytes in advanced HER2-positive breast cancer treated with pertuzumab or placebo in addition to trastuzumab and docetaxel: a retrospective analysis of the CLEOPATRA study. Lancet Oncol. 2017;18(1):52–62. doi: 10.1016/S1470-2045(16)30631-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23(6):703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dustin D, Gu G, Fuqua SAW. ESR1 mutations in breast cancer. Cancer. 2019;125:3714–28. [DOI] [PMC free article] [PubMed]
- 36.Zundelevich A, Dadiani M, Kahana-Edwin S, Itay A, Sella T, Gadot M, et al. ESR1 mutations are frequent in newly diagnosed metastatic and loco-regional recurrence of endocrine-treated breast cancer and carry worse prognosis. Breast cancer Res. 2020;22(1):16. doi: 10.1186/s13058-020-1246-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loibl S, Turner NC, Ro J, Cristofanilli M, Iwata H, Im SA, et al. Palbociclib combined with fulvestrant in premenopausal women with advanced breast cancer and prior progression on endocrine therapy: PALOMA-3 results. Oncologist. 2017;22(9):1028–1038. doi: 10.1634/theoncologist.2017-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cristofanilli M, DeMichele A, Giorgetti C, Turner NC, Slamon DJ, Im SA, et al. Predictors of prolonged benefit from palbociclib plus fulvestrant in women with endocrine-resistant hormone receptor-positive/human epidermal growth factor receptor 2-negative metastatic breast cancer in PALOMA-3. Eur J Cancer. 2018;104:21–31. doi: 10.1016/j.ejca.2018.08.011. [DOI] [PubMed] [Google Scholar]
- 39.Dieras V, Rugo HS, Schnell P, Gelmon K, Cristofanilli M, Loi S, et al. Long-term pooled safety analysis of palbociclib in combination with endocrine therapy for HR+/HER2- advanced breast cancer. J Natl Cancer Inst. 2019;111(4):419–430. doi: 10.1093/jnci/djy109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mayer IA, Abramson VG, Formisano L, Balko JM, Estrada MV, Sanders ME, et al. A phase Ib study of alpelisib (BYL719), a PI3Kalpha-specific inhibitor, with letrozole in ER+/HER2- metastatic breast cancer. Clin Cancer Res. 2017;23(1):26–34. doi: 10.1158/1078-0432.CCR-16-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andre F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS, et al. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N Engl J Med. 2019;380(20):1929–1940. doi: 10.1056/NEJMoa1813904. [DOI] [PubMed] [Google Scholar]
- 42.Pellegrino B, Musolino A, Llop-Guevara A, Serra V, De Silva P, Hlavata Z, et al. Homologous recombination repair deficiency and the immune response in breast cancer: a literature review. Transl Oncol. 2020;13(2):410–422. doi: 10.1016/j.tranon.2019.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen H, Yang ES, Conry M, Fiveash J, Contreras C, Bonner JA, et al. Predictive biomarkers for immune checkpoint blockade and opportunities for combination therapies. Genes Dis. 2019;6(3):232–246. doi: 10.1016/j.gendis.2019.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Condorelli R, Spring L, O'Shaughnessy J, Lacroix L, Bailleux C, Scott V, et al. Polyclonal RB1 mutations and acquired resistance to CDK 4/6 inhibitors in patients with metastatic breast cancer. Ann Oncol. 2018;29(3):640–645. doi: 10.1093/annonc/mdx784. [DOI] [PubMed] [Google Scholar]
- 45.O'Leary B, Cutts RJ, Liu Y, Hrebien S, Huang X, Fenwick K, et al. The genetic landscape and clonal evolution of breast cancer resistance to palbociclib plus fulvestrant in the PALOMA-3 trial. Cancer Discov. 2018;8(11):1390–1403. doi: 10.1158/2159-8290.CD-18-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oliveira KCS, Ramos IB, Silva JMC, Barra WF, Riggins GJ, Palande V, et al. Current perspectives on circulating tumor DNA, precision medicine, and personalized clinical management of cancer. Mol Cancer Res. 2020;18:517-28. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Table 1. List of genes enriched in study dataset compared to the TCGA dataset. Supplementary Table 2. Genes-pairs that co-occur in ER+ metastatic breast cancer. Supplementary Table 3. Hazard ratios and confidence intervals for TILs samples.
Data Availability Statement
De-identified genomic variant data is available upon request to the corresponding author.