Abstract
Patients with inflammatory bowel disease are at increased risk of colorectal cancer, which has worse prognosis than sporadic colorectal cancer. Until recently, understanding of pathogenesis in inflammatory bowel disease-associated colorectal cancer was restricted to the demonstration of chromosomic/microsatellite instabilities and aneuploidy. The advance of high-throughput sequencing technologies has highlighted the complexity of the pathobiology and revealed recurrently mutated genes involved in the RTK/RAS, PI3K, WNT, and TGFβ pathways, leading to potentially new targetable mutations. Moreover, alterations of mitochondrial DNA and the dysregulation of non-coding sequences have also been described, as well as several epigenetic modifications. Although recent studies have brought new insights into pathobiology and raised the prospect of innovative therapeutic approaches, the understanding of colorectal carcinogenesis in inflammatory bowel disease and how it differs from sporadic colorectal cancer remains not fully elucidated. Further studies are required to better understand the pathogenesis and molecular alterations leading to human inflammatory bowel disease-associated colorectal cancer.
Keywords: Inflammatory bowel disease, colorectal cancer, inflammatory bowel disease-associated cancer, genomics, epigenetics, molecular pathways
Introduction
Crohn’s disease (CD) and ulcerative colitis (UC) are chronic and idiopathic disorders defined as inflammatory bowel diseases (IBDs).1 In both diseases, patients have an increased risk of developing colorectal cancer (CRC).2 IBD-associated CRC (IBD-CRC) is diagnosed at a younger age and has worse prognosis than sporadic CRC (S-CRC). For this reason, developing knowledge about the pathobiological mechanisms contributing to this disease is essential. Such an understanding may enable the initiation of targeted preventive strategies.
The molecular events in the initiation of oncogenesis in IBD and the progression to IBD-CRC in humans have been a subject of interest for decades. Until recently, knowledge in this field was restricted to the demonstration of chromosomic instability (CIN), aneuploidy, microsatellite instability,3 and few genetic alterations. Among these altered genes, the most commonly described are p53 tumor suppressor gene mutations and p53 loss of heterozygosity (LOH),4 which are found in >80% of IBD-CRC patients.5 p53 loss of function appears to be an early event in the initiation of IBD-CRC pathogenesis,6 whereas it is a late event in S-CRC. Other genetic alterations, such as in the APC tumor suppressor gene or KRAS oncogene,7 have been identified in IBD-CRC, but at a lower frequency than in S-CRC.
More recently, the genomic landscape of IBD-CRC has been explored by means of new high-throughput sequencing technologies.5,8,9 Extensive genomic analyses have revealed new recurrent genomic alterations.
Moreover, the emergence of new fields of research (such as epigenomics and the tumoral microenvironment) highlights the complex network of factors involved in IBD-CRC. The gut microbiota is also a subject of interest and intense research.
In this review, we describe current knowledge on the molecular alterations involved in human IBD-CRC oncogenesis.
Genetic alterations in DNA coding sequences
Somatic genetic alterations
The accumulation of molecular events and mutations in somatic cells, followed by their clonal expansion, have been studied for a role in IBD-CRC pathogenesis (Figure 1). This process gives rise to a widespread preneoplastic “field change” in colonic mucosa before any histological evidence of dysplasia.10
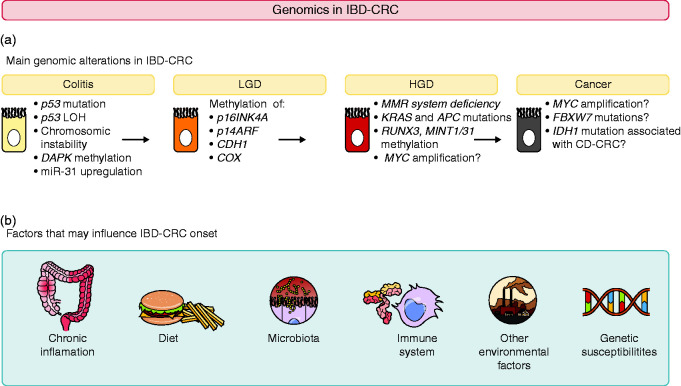
Figure 1.
Update of the main genomic alterations described in IBD-CRC and their chronology of appearance in the colitis/cancer sequence.
The main genetic alterations associated with IBD-CRC were first described in the early 1990s.6 Similar chromosomal alterations are found in S-CRC and IBD-CRC, yet there are differences in their frequency and timing of occurrence.
The first genetic investigations in IBD-CRC revealed that CIN was a major feature in the initiation of CRC in IBD, being found in 80% of cases.11 CIN is associated with the progressive accumulation of mutations in oncosuppressor genes and oncogenes.3 Aneuploidy is a consequence of CIN and occurs before dysplasia in mucosal colonic biopsy samples from patients with IBD.12 Moreover, there is a statistically significant relationship between increased aneuploidy and histological progression to dysplasia/carcinoma in UC.6 This observation highlights the progressive genomic instability during neoplastic progression of IBD-CRC.
Another primary genetic alteration involved in IBD-CRC development is mutation of the p53 tumor suppressor gene, suspected to typically occur before development of any dysplasia.13 p53 has a pivotal influence on cell proliferation and prevents clonal expansion of mutant cells.14 There is also a strong correlation between histological progression from low grade dysplasia (LGD) to carcinoma and p53 LOH.6 In tissue from UC-CRC, p53 LOH was found in 6% of samples without dysplasia, in 33% with LGD, and in 85% with UC-CRC.6 p53 LOH was only found in aneuploid cells, suggesting these events are early in IBD-CRC pathogenesis. By contrast, p53 alteration is a late event in S-CRC oncogenesis.15
Loss of function of adenomatous polyposis coli (APC), another tumor suppressor gene, is also an early event in IBD-CRC,7 but occurs much less frequently than in S-CRC (6% versus 74%).16 Such observations support the hypothesis that IBD-CRC does not emerge from the classical APC inactivated adenoma pathway, as seen in S-CRC.17 The role of APC in IBD-CRC carcinogenesis is not clearly understood.6
Oncogene alterations in IBD-CRC are less described than tumor suppressor gene alterations. Mutations in the KRAS oncogene, which are believed to occur early in IBD-CRC oncogenesis,6,7 are less frequent in IBD-CRC than in S-CRC (9% versus 50%).18
Oxidative stress contributes to oncogenesis by impairing the DNA mismatch repair (MMR) system,19 leading to an accumulation of replicative mistakes and a microsatellite instability (MSI) phenotype in colonic mucosa in 15% of patients with IBD-CRC,20 a rate similar to S-CRC.
The 2000s have been characterized by significant evolution in genetic knowledge, exemplified by the completion of the Human Genome Project and the ENCODE project. These advances have been accompanied by the emergence of advanced DNA sequencing technologies, allowing low-cost genomic discoveries. Three studies have explored the genomic features of IBD-CRC through high-throughput sequencing.5,8,9
In a first study,5 next-generation sequencing was used to investigate coding sequence alterations in 315 cancer-related genes in 47 IBD-CRC samples and compare them with S-CRC alterations reported in The Cancer Genome Atlas (TCGA). Approximately six genetic alterations per tumor were identified for IBD-CRC; among these, three genes were recurrently altered (≥ 10% of cases). p53 was the most commonly altered gene, being altered in 89% of IBD-CRC tumors compared with 52% of S-CRC tumors in TCGA. APC was altered significantly less frequently in IBD-CRC than in S-CRC (21% versus 76%), and notably more frequently in CD-associated CRC (CD-CRC) than UC-CRC (39% versus 10%). MYC proto-oncogene amplification occurred in 26% of IBD-CRC samples versus 4% of S-CRC samples. Moreover, isocitrate dehydrogenase 1 (IDH1) mutations were significantly more frequent in IBD-CRC (11% of cases), whereas they are rare in S-CRC (affecting just 1%). IDH1 alterations at the R132 hotspot were significantly more common in CD-CRC than UC-CRC. Potentially targetable mutations were found in 36% of IBD-CRC samples, and included mutations in the PIK3/mTOR and RTK/RAS/RAF pathways.
Whole-exome sequencing from paired CRC and non-neoplastic tissue samples from 31 well-characterized IBD patients has also been performed.8 Confirming earlier observations, p53 was the most commonly mutated gene (63% of cases) whereas both APC and KRAS were significantly less frequently mutated in IBD-CRC than what was observed in S-CRC from TCGA (APC: 13% versus 81%, KRAS: 20% versus 43%). Several recurrently mutated genes were identified in IBD-CRC involving RTK/RAS (BRAF), PI3K, WNT (APC, CTNNB1, SOX9), and TGFβ (SMAD 2/4, TGFβR2) pathways. Other recurrent alterations involved the Rho GTPase network, cell communication/adhesion, cytokine signaling (IL16) and chromatin remodeling (TRRAP, EP300).8
More recently, 55 tumors from 48 IBD-CRC patients (35 tumors from 30 UC patients, 18 from 16 CD patients, two from patients with indeterminate colitis) were studied through a 50 gene hot-spot solid tumor panel.9 IBD-CRC tumor genomic profiles were compared with S-CRC TCGA data. Similar results were found to the previous genomic studies, including the presence of IDH1 mutations in a subset of IBD-CRC (7% in IBD-CRC versus 1% in S-CRC from TCGA, with difference reaching statistical significance). Other genes were also altered in >5% of IBD-CRC but the difference with TGCA data did not reach statistical significance (PIK3CA, SMAD4, and FBXW7, which was only mutated in UC-CRC).
In summary, IBD-CRC exhibits differences in genetic alterations in comparison with S-CRC, suggesting that different molecular pathways contribute to the oncogenic processes (Table 1).
Table 1.
Main genetic alterations in IBD-CRC compared with S-CRC.
| Gene | Frequency of mutations in IBD-CRC | Frequency of mutations in S-CRCa | Statisticalsignificance |
|---|---|---|---|
| p53 4 | 60–85% | 58.25–82.48% | S* |
| KRAS 8 | 4–20% | 41.65–43.5% | S* |
| TGFBR2 20 | 17% | 0.73–5% | NS |
| SMAD4 8 | 13% | 13.5–19.71% | NS |
| TRRAP 8 | 13% | 8.03–14.75% | NS |
| SOX9 8 | 10% | 7.3–16% | NS |
| PIK3CA 8 | 10% | 16–31% | NS |
| SMAD2 8 | 10% | 5.11–6.5% | NS |
| EP300 8 | 10% | 5.11–8.75% | NS |
| IDH1 5 | 11% | 1.46–2% | S* |
| FBXW7 8 | 7% | 19–20.44% | NS |
| APC 16 | 6% | 77.25–88.32% | S* |
| BRAF 9 | 4% | 4.38–15% | NS |
| CTNNB1 8 | 3% | 5.11–8.75% | NS |
aData from The Cancer Genome Atlas Project (https://portal.gdc.cancer.gov).
IBD-CRC: inflammatory bowel disease-associated colorectal cancer; S-CRC: sporadic colorectal cancer; S*: significant; NS: not significant.
Mitochondrial DNA (mtDNA)
The role of mitochondria in cancer is poorly understood. However, it is well-established that oxidative stress may increase mutations in mtDNA, which might correlate with precancerous status.21 In one study, mtDNA mutation rates were compared between seven UC-CRC patients, 19 UC patients without CRC and nine healthy controls.22 Patients with UC-CRC had a significantly higher number of mtDNA alterations than both healthy controls and UC patients without CRC,22 suggesting that the rate of mtDNA mutation is enhanced in mucosal cells by oxidative stress caused by chronic inflammation, leading to an accelerated oncogenic transformation.22
In another study,23 cytochrome C oxidase (COX) and peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC1α) immunostainings were used to study mitochondria protein expression in nine UC patients with colonic high grade dysplasia (HGD) or CRC compared with nine UC controls. Significant COX expression loss was found in UC colonic dysplasia compared with UC controls. Surprisingly, COX and PGC1α immunostaining intensity were significantly increased in UC-CRC, a finding supported by the increased mtDNA copy number in UC-CRC. Authors hypothesized that mitochondrial loss occurs before colonic dysplasia development in UC patients and cancer cells restore mitochondria numbers to support further proliferation.
Analyses of mtDNA could provide a new therapeutic target, and may be useful for the prediction of risk of carcinogenesis in IBD patients.
Genetic alterations in DNA non-coding sequences
Epigenetic alterations
Epigenetics refers to the heritable changes in gene expression and chromatin organization that are not due to alterations in the DNA sequence, and include DNA methylation, histone modification, RNA interference and the positioning of nucleosome. A number of studies have investigated the contribution of epigenetic mechanisms to IBD-CRC.
Promoter methylation and consequent silencing of several genes, particularly p16INK4a, p14ARF, and cadherin 1 (CDH1), is a common event. Promotor hypermethylations in p16INK4a and p14ARF are frequent and early events in IBD-CRC carcinogenesis, found in 100% and 50% of UC–CRC,24,25 respectively. Both genes play important parts in cell cycle inhibition.24,25 Methylation of the CDH1 promoter was detected in 93% of IBD patients with colorectal dysplasia, versus 6% in IBD patients without dysplastic lesions.26 This gene has an essential role in biological processes that are dysregulated in cancer, such as ordering of cell sorting, migration, and differentiation.
In IBD neoplasia, hMLH1 promoter hypermethylation occurs as frequently as in S-CRC in the setting of CRC with microsatellite instability.27 hMLH1 hypermethylation and MSI are strongly associated with diminished expression of hMLH1 protein, a component of the DNA MMR machinery, suggesting that hMLH1 hypermethylation causes defective DNA MMR in a subset of IBD neoplasms.28
A progressive increase in methylation of WNT signaling pathway genes is observed during IBD-CRC pathogenesis.29 This is in contrast with genetic mutations in this pathway, which occur late during colitis-associated tumorigenesis (i.e. APC).30
Epigenetics modification also included suppressor of cytokine signaling 3 (SOCS3) methylation. Indeed, the number of (SOCS3)-positive epithelial cells were significantly reduced during UC-CRC progression from LGD to CRC in UC patients (9.1% in UC-CRC versus 33.3% in UC-LGD),31 and this observation was correlated with prominent expression of the DNA methyltransferase gene (DMNT1).32 SOCS3 is a target gene of the IL6/STAT3 pathway that is known to regulate negative feedback of the JAK/STAT signaling cascade.33 Methylation-specific polymerase chain reaction analysis showed that SOCS3 silencing was due to its methylation. SOCS3 methylation was detected in all investigated UC-CRC samples, whereas no SOCS3 methylation was found in biopsy samples from healthy controls.31
Furthermore, the methylation status of 10 selected genes involved in tumor suppression, cell cycle regulation and aging was investigated in a large number of UC-CRC samples and compared with non-neoplastic tissue from both UC-CRC cases and UC controls.34 The degree of methylation for p16 INK4a, runt-related transcription factor 3 (RUNX3), methylated in tumor gene 1 (MINT1), MINT31, and hyperplastic polyposis protein 1 (HPP1) was significantly higher in UC-CRC tumor compared with UC controls. Conversely, unmethylated cyclooxygenase 2 (COX2) was an indicator of CRC.
Death-associated protein kinase (DAPK), a tumor suppressor gene, also seems to undergo epigenetic modification during IBD-CRC oncogenesis.35 DAPK silencing by promoter hypermethylation might be crucial for accumulation of DNA damage in UC mucosa, and therefore could contribute to initiation of the neoplastic process in UC-CRC.35
These data (Table 2) indicate that epigenetic modifications in IBD may contribute to the oncogenic transformation into CRC, and highlight the need for further research in this area.
Table 2.
Main epigenetic alterations (methylation) in IBD-CRC compared with S-CRC.
| Gene | Gene methylation rate in IBD-CRC | Significance between gene’s promoter methylation and IBD-CRC | Gene methylation rate in S-CRC |
|---|---|---|---|
| p16INK4a 24 | 100% | S* | 32–40% |
| CDH1 26 | 93% | S* | 87% |
| HPP1 34 | 40–78.7% | S* | 84% |
| MINT31 34 | 59.4% | S* | 11% |
| MINT1 34 | 50.5% | S* | 15% |
| p14ARF 25 | 50% | S* | 33% |
| hMLH1 27 | 9% | NS | 5% |
| RUNX3 34 | 44.6% | S* | 23.5% |
| DAPK 35 | 27.6% | S* | 57.4% |
IBD-CRC: inflammatory bowel disease-associated colorectal cancer; S-CRC: sporadic colorectal cancer; IBD-CRC: IBD associated colorectal cancer; S-CRC: sporadic colorectal cancer; S*: significant; NS: not significant.
microRNAs
microRNAs (miRs) have recently emerged as important factors in cancer biology. These small non-coding RNAs are involved in post-transcriptional regulation and have a role in carcinogenic processes through their control of key cellular functions (apoptosis, differentiation, cell cycle progression and immune functions).36
The involvement of miRs and their dysregulation in IBD-CRC has recently been explored in two important works.37,38
In the first published study,37 175 fresh-frozen colon tissue samples from 14 healthy controls, 35 IBD patients (inflamed and non-inflamed mucosa), 11 IBD-dysplasia, 37 IBD-CRC, and 15 S-CRC were collected and their miR expression determined by miR microarrays. The expression level of miR-31 was found to increase in the progression from inflamed IBD colonic tissue to IBD-dysplasia,37 suggesting that miR-31 upregulation is an early event in colorectal neoplastic transformation in IBD patients. miR-31 expression levels were also threefold higher in IBD-CRC than in S-CRC.37
In the second study,38 the same experience was repeated, using miR microarrays to identify altered expression of miRs between IBD-CRC samples and IBD controls. Thirty differentially expressed miRs were identified between those two populations (18 upregulated and 12 downregulated in IBD-CRC). miR-224, which was overexpressed, exhibited the highest discriminatory power for distinguishing between IBD-CRC from IBD controls. miR-224 is assumed to dysregulate cell cycle control by targeting p21, a key cell cycle regulator.38
Those recent results uncover new miRs signaling pathways, suggesting an etiologic role for miRs in the development of IBD-CRC.
Proinflammatory and protumoral molecular pathways
TLR4 (Toll-like receptor 4)/NFκB (nuclear factor-kappa B) signaling pathways
TLR4/NFκB overactivation and overexpression have been observed in several inflammatory diseases, including IBD, and are suspected to promote colitis and its associated CRC.39 In humans, only two studies exploring the role of TLR4/NFκB signaling pathway in IBD-CRC have been published.40,41 Using a Western blot assay, it was demonstrated in 2007 that non-dysplastic colon samples from four patients with UC-associated CRC (UC-CRC) had low expression of TLR4 protein, whereas matched tumor protein samples from the same UC-CRC patients showed higher expression of TLR4.40 Moreover, immunostaining analysis revealed a predominantly epithelial expression of TLR4.40
More recently, TLR4/NFκB signaling was investigated in 35 Algerian patients with UC and nine with UC-CRC.41 Transcriptomic analysis demonstrated that mRNA levels of TLR4 were significantly increased in colonic mucosa of UC-CRC patients compared with UC patients.41 Circulating levels of tumor necrosis factor (TNF) and nitric oxide were increased in UC-CRC versus UC patients, with statistical significance for plasma TNF levels (374.7 pg/ml versus 291.5 pg/ml).41 This result was supported by significantly higher TNF mRNA levels in colonic mucosa of patients with UC-CRC compared with active UC patients.41 Additionally, immunofluorescent staining analyses indicated that NFκB and IκB kinase protein expression was highly increased in colonic mucosa of UC-CRC in comparison with UC controls.41
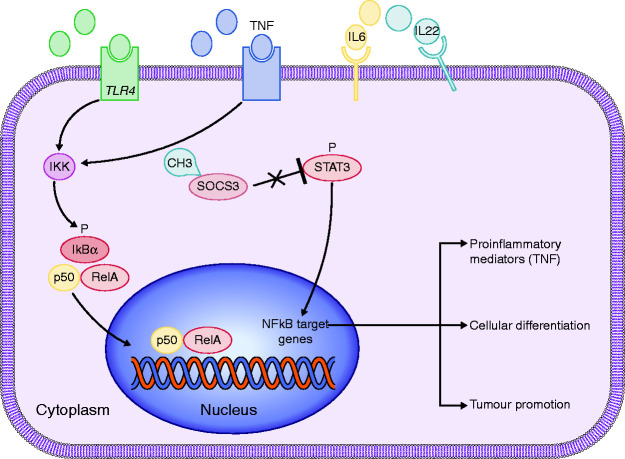
These results suggest that increased epithelial TLR4 expression occurs during the initiation of UC-CRC and might have a role in the transition from inflammation to neoplasia via the TLR4/NFκB pathway. Thus, proteins of the NFκB pathway could be useful therapeutic targets in IBD-CRC patients (Figure 2).
Figure 2.
TLR4/NFκB and STAT3 pathways in IBD-CRC.
STAT3 (signal transducer and activator of transcription 3) pathway
STAT3 pathway activation has been observed in many human carcinomas, including CRC.42 This signaling pathway promotes survival, cell proliferation and may also promote NFκB activity. Several inflammatory cytokines that activate STAT3 have been associated with IBD, including interleukin (IL) 643 and IL22 (Figures 2 and 3).44
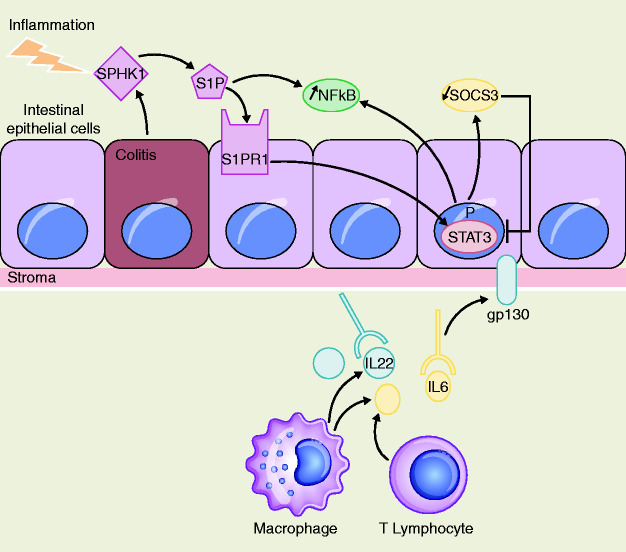
Figure 3.
STAT3 pathway and IL6/STAT3/S1PR1 amplification loop in IBD-CRC.
Three studies have explored STAT3 signaling in human IBD-related colorectal carcinogenesis;31,45,46 two investigated the IL6/STAT3 axis,31,45 whereas another studied IL22/STAT3 signaling.46
In a first study, authors used immunohistochemical scoring to determine IL6/STAT3 expression in colon biopsy specimens from patients with UC-CRC at different phases of the oncogenic process:31 nine UC patients with LGD, seven UC patients with HGD and 11 patients with UC-CRC.31 Patients with UC-CRC had significantly increased expression of IL6 (72.7% in UC dysplasia and/or UC-CRC versus 0% in healthy controls) and p-STAT3 (54.5% versus 11.1%) in colonic epithelial cells.31
Recently, the role of sphingosine kinase 1 (SPHK1) and sphingosine-1-phosphate (S1P) on IL6/STAT3 signaling in UC-CRC patients has been investigated.45 S1P is a bioactive lipid mediator generated by SPHK1, with both molecules acting as upstream mediators of IL6 and STAT3 signaling.47 Using immunohistochemistry, IL6, p-STAT3, and p-SPHK1 expression levels in S-CRC were measured in comparison with UC-CRC.45 Although there was no difference in IL6 and p-STAT3 expression between the two populations, p-SPHK1 levels were significantly higher in UC-CRC (9/10 tumors) than in S-CRC (3/10 tumors).45 Thus, upregulated SPHK1 in UC-CRC may lead to S1P increased expression and activation of IL6/STAT3 signaling via the S1P receptor (S1PR1).45 Consequently, a persistent IL6/STAT3/S1PR1 amplification loop is created that is critical to the development of UC-CRC (Figure 3).
The IL22/STAT3 axis was investigated in colon biopsies from 74 UC patients and 10 patients with UC colonic dysplasia.46 Immunohistochemical analysis showed that significantly more IL22-positive cells and p-STAT3 cells were observed in UC-dysplasia than in UC controls.
Chronic inflammation associated with IBD substantially due to reactive oxygen and nitrogen species oversynthesis contributed to the onset of IBD-CRC. Indeed, it induces a series of responses that prolong inflammation, such as matrix metalloproteinase and TLR4/NFκB, resulting in DNA damages and the activation of procarcinogenic genes and silencing of tumor suppressor pathways.48 Therefore, regulation of those proinflammatory and protumoral pathways may be important to control the progression of IBD into IBD-CRC.
STAT3 signaling is a complex network implicated in IBD-CRC pathogenesis. Overall, targeting the STAT3 pathway may offer different therapeutic options (STAT3 targeting drugs and inhibitors of IL6/IL22 and S1P/SPHK1) to prevent the progression of colitis into cancer.
Microbiota in IBD-CRC
It is established that gut microbiota plays a crucial role on the resistance against invading pathogens.49 Dysbiosis of gut microbiota, defined as a shift in the balance between commensal and pathogenic microorganisms leading to a decrease in gut microbial diversity,50 is often found in IBD.51 Resident microbes can promote carcinogenesis by inducing inflammation, increasing cell proliferation, and producing metabolites (such as butyrate),52 which might affect DNA integrity and immune regulation.53
The Enterobacteriaceae family,54 especially adherent–invasive E. coli and Sphingomonas,55 have been associated with the ileal mucosa of patients with IBD-CRC. Conversely, a decrease of Fusobacterium and Ruminococcus genus were observed in IBD-CRC versus S-CRC.55 However, to our knowledge, there is still no evidence of significant microbiota changes between IBD-CRC and S-CRC.51
Microbiota changes might also generate epimutations,56 which are currently a subject of intense research. Further investigations are needed to confirm the implication of microbiota in IBD-CRC and to determine its role in this oncogenic process.
Discussion
Although IBD-CRC represents <1% of all CRC cases, patients with IBD are among the highest risk groups for developing CRC. Moreover, IBD-CRC typically has an earlier occurrence and a worse prognosis than S-CRC.2 Therefore, it is important to identify molecular mechanisms involved in IBD-CRC carcinogenesis to prevent IBD-CRC and to develop targeted therapies for patients. Such an understanding might also help to better understand the role played by inflammation in colonic cancer more generally.
IBD-CRC is a complex disease associated with multifactorial causes. It is now well established that chronic inflammation associated with IBD contributes to the onset of IBD-CRC via increased expression of genes involved in cell cycle regulation and cancer-relevant signaling pathways.57 Therefore, studying somatic genomic events in IBD-CRC is of major interest in understanding the molecular mechanisms responsible for the initiation and the progression of IBD-CRC. However, despite the major advances in genetics made these last ten years, very few genomic discoveries have been made about IBD-CRC. In this review, we reported all the studies about genetic and molecular alterations described in human IBD-CRC.
Three studies have investigated the genetic alterations in IBD-CRC using high-throughput sequencing technologies,5,8,9 the results of which support the theory that most IBD-CRCs arise through different pathways than S-CRC. They have also expanded existing knowledge of the molecular alterations in IBD-CRC, especially the identification of potentially targetable mutations and some relevant recurrent alterations (MYC amplification, IDH1, FBXW7), even though most of these findings did not reach statistical significance.
However, there are important limitations for these studies, mainly due to restricted genomic analysis (such as the hot-spot panel), retrospective designs and small sample sizes. Most of the published articles were conducted in UC patients, yet it could be hypothesized that the molecular alterations are similar in CD patients because they are at the same risk of CRC as UC patients of similar disease duration and extension. Moreover, limited data regarding epigenomics and microarrays analysis are available. Overall, there is no evidence of distinct genomic signatures in IBD-CRC and S-CRC.
The gut microbiota is of growing interest in IBD-CRC. Nowadays, IBD has become a global disease with accelerating incidence in newly industrialized countries, whose societies have become more westernized, and this might lead to an increased incidence of IBD-CRC in the near future. Because gut microbiota modifications are directly linked to environmental factors (especially diet), its role in IBD-CRC carcinogenesis should be considered an important component to better understand IBD-CRC development and to explore new therapeutic strategies such as fecal microbiota transplantation to prevent CRC initiation.
The tumoral microenvironment is a complex network and an emerging research field. The role of the tumor microenvironment in IBD-CRC has never been studied in humans but warrants further attention as it may provide new insights into malignant transformation, tumor progression, and combination therapy strategies for IBD-CRC.
Even though further investigations are required to understand the role of these factors in IBD-CRC oncogenesis, this comprehensive review highlights the major molecular and somatic genomic alterations found in IBD-CRC. The described alterations in IBD-CRC would deserve further attention and should be considered as potential biomarkers to prevent CRC in patients suffering from IBD or as potentially new molecular targets for personalized medicine in IBD-CRC.
In conclusion, IBD-CRC is a complex disease. Although recent studies have brought new insights into IBD-CRC pathobiology and revealed potentially targetable molecular alterations, the understanding of colorectal carcinogenesis in IBD, which differs from S-CRC, remains not fully elucidated. Further studies are required to better understand the molecular alterations leading to human IBD-CRC and to propose precision medicine for these patients, including personalized endoscopic surveillance programs.
Declaration of conflicting interests
The author(s) have no conflicts of interest to declare.
Funding
The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iDs
Marie Muller https://orcid.org/0000-0001-6412-932X
Franck Hansmannel https://orcid.org/0000-0003-1661-2362
References
- 1.Molodecky NA, Soon IS, Rabi DM, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology 2012; 142: 46–54. [DOI] [PubMed] [Google Scholar]
- 2.Peyrin-Biroulet L, Lepage C, Jooste V, et al. Colorectal cancer in inflammatory bowel diseases: a population-based study (1976–2008). Inflamm Bowel Dis 2012; 18: 2247–2251. [DOI] [PubMed] [Google Scholar]
- 3.Scarpa M. Inflammatory colonic carcinogenesis: a review on pathogenesis and immunosurveillance mechanisms in ulcerative colitis. World J Gastroenterol 2014; 20: 6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brentnall TA, Crispin DA, Rabinovitch PS, et al. Mutations in the p53 gene: an early marker of neoplastic progression in ulcerative colitis. Gastroenterology 1994; 107: 369–378. [DOI] [PubMed] [Google Scholar]
- 5.Yaeger R, Shah MA, Miller VA, et al. Genomic alterations observed in colitis-associated cancers are distinct from those found in sporadic colorectal cancers and vary by type of inflammatory bowel disease. Gastroenterology 2016; 151: 278–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burmer GC, Rabinovitch PS, Haggitt RC, et al. Neoplastic progression in ulcerative colitis: Histology, DNA content, and loss of a p53 allele. Gastroenterology 1992; 103: 1602–1610. [DOI] [PubMed] [Google Scholar]
- 7.Redston MS, Papadopoulos N, Caldas C, et al. Common occurrence of APC and K-ras gene mutations in the spectrum of colitis-associated neoplasias. Gastroenterology 1995; 108: 383–392. [DOI] [PubMed] [Google Scholar]
- 8.Robles AI, Traverso G, Zhang M, et al. Whole-exome sequencing analyses of inflammatory bowel disease−associated colorectal cancers. Gastroenterology 2016; 150: 931–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alpert L, Yassan L, Poon R, et al. Targeted mutational analysis of inflammatory bowel disease–associated colorectal cancers. Hum Pathol 2019; 89: 44–50. [DOI] [PubMed] [Google Scholar]
- 10.Choi C-HR, Bakir IA, Hart AL, et al. Clonal evolution of colorectal cancer in IBD. Nat Rev Gastroenterol Hepatol 2017; 14: 218–229. [DOI] [PubMed] [Google Scholar]
- 11.Itzkowitz S. Colon carcinogenesis in inflammatory bowel disease: applying molecular genetics to clinical practice. J Clin Gastroenterol 2003; 36: S70–S74. [DOI] [PubMed] [Google Scholar]
- 12.Rubin CE, Haggitt RC, Burmer GC, et al. DNA aneuploidy in colonic biopsies predicts future development of dysplasia in ulcerative colitis. Gastroenterology 1992; 103: 1611–1620. [DOI] [PubMed] [Google Scholar]
- 13.Bressenot A. Microscopic features of colorectal neoplasia in inflammatory bowel diseases. World J Gastroenterol 2014; 20: 3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levine AJ, Momand J, Finlay CA. The p53 tumour suppressor gene. Nature 1991; 351: 453–456. [DOI] [PubMed] [Google Scholar]
- 15.Raskov H. Colorectal carcinogenesis-update and perspectives. World J Gastroenterol 2014; 20: 18151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomlinson I, Ilyas M, Johnson V, et al. A comparison of the genetic pathways involved in the pathogenesis of three types of colorectal cancer. J Pathol 1998; 184: 148–152. [DOI] [PubMed] [Google Scholar]
- 17.Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014; 505: 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Umetani N, Sasaki S, Watanabe T, et al. Genetic alterations in ulcerative colitis-associated neoplasia focusing on APC, K-ras gene and microsatellite instability. Jpn J Cancer Res 1999; 90: 1081–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brentnall TA, Crispin DA, Bronner MP, et al. Microsatellite instability in nonneoplastic mucosa from patients with chronic ulcerative colitis. Cancer Res 1996; 56: 1237–1240. [PubMed] [Google Scholar]
- 20.Fujiwara I, Yashiro M, Kubo N, et al. Ulcerative colitis-associated colorectal cancer is frequently associated with the microsatellite instability pathway. Dis Colon Rectum 2008; 51: 1387–1394. [DOI] [PubMed] [Google Scholar]
- 21.Croteau DL, Bohr VA. Repair of oxidative damage to nuclear and mitochondrial DNA in mammalian cells. J Biol Chem 1997; 272: 25409–25412. [DOI] [PubMed] [Google Scholar]
- 22.Nishikawa M, Oshitani N, Matsumoto T, et al. Accumulation of mitochondrial DNA mutation with colorectal carcinogenesis in ulcerative colitis. Br J Cancer 2005; 93: 331–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ussakli CH, Ebaee A, Binkley J, et al. Mitochondria and tumor progression in ulcerative colitis. J Natl Cancer Inst 2013; 105: 1239–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsieh CJ, Klump B, Holzmann K, et al. Hypermethylation of the p16INK4a promoter in colectomy specimens of patients with long-standing and extensive ulcerative colitis. Cancer Res 1998; 58: 3942–3945. [PubMed] [Google Scholar]
- 25.Sato F, Harpaz N, Shibata D, et al. Hypermethylation of the p14(ARF) gene in ulcerative colitis-associated colorectal carcinogenesis. Cancer Res 2002; 62: 1148–1151. [PubMed] [Google Scholar]
- 26.Azarschab P, Porschen R, Gregor M, et al. Epigenetic control of the E-cadherin gene (CDH1) by CpG methylation in colectomy samples of patients with ulcerative colitis. Genes Chromosomes Cancer 2002; 35: 121–126. [DOI] [PubMed] [Google Scholar]
- 27.Sanchez JA, DeJulius KL, Bronner M, et al. Relative role of methylator and tumor suppressor pathways in ulcerative colitis-associated colon cancer: Inflamm Bowel Dis 2011; 17: 1966–1970. [DOI] [PubMed] [Google Scholar]
- 28.Fleisher AS, Esteller M, Harpaz N, et al. Microsatellite instability in inflammatory bowel disease-associated neoplastic lesions is associated with hypermethylation and diminished expression of the DNA mismatch repair gene, hMLH1. Cancer Res 2000; 60: 4864–4868. [PubMed] [Google Scholar]
- 29.Dhir M, Montgomery EA, Glöckner SC, et al. Epigenetic regulation of WNT signaling pathway genes in inflammatory bowel disease (IBD) associated neoplasia. J Gastrointest Surg 2008; 12: 1745–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Itzkowitz SH. Molecular biology of dysplasia and cancer in inflammatory bowel disease. Gastroenterol Clin North Am 2006; 35: 553–571. [DOI] [PubMed] [Google Scholar]
- 31.Li Y, de Haar C, Chen M, et al. Disease-related expression of the IL6/STAT3/SOCS3 signalling pathway in ulcerative colitis and ulcerative colitis-related carcinogenesis. Gut 2010; 59: 227–235. [DOI] [PubMed] [Google Scholar]
- 32.Li Y, Deuring J, Peppelenbosch MP, et al. IL-6-induced DNMT1 activity mediates SOCS3 promoter hypermethylation in ulcerative colitis-related colorectal cancer. Carcinogenesis 2012; 33: 1889–1896. [DOI] [PubMed] [Google Scholar]
- 33.Linossi EM, Babon JJ, Hilton DJ, et al. Suppression of cytokine signaling: the SOCS perspective. Cytokine Growth Factor Rev 2013; 24: 241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garrity-Park MM, Loftus EV, Sandborn WJ, et al. Methylation status of genes in non-neoplastic mucosa from patients with ulcerative colitis-associated colorectal cancer. Am J Gastroenterol 2010; 105: 1610–1619. [DOI] [PubMed] [Google Scholar]
- 35.Kuester D, Guenther T, Biesold S, et al. Aberrant methylation of DAPK in long-standing ulcerative colitis and ulcerative colitis-associated carcinoma. Pathol Res Pract 2010; 206: 616–624. [DOI] [PubMed] [Google Scholar]
- 36.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 2008; 9: 102–114. [DOI] [PubMed] [Google Scholar]
- 37.Olaru AV, Selaru FM, Mori Y, et al. Dynamic changes in the expression of MicroRNA-31 during inflammatory bowel disease-associated neoplastic transformation. Inflamm Bowel Dis 2011; 17: 221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olaru AV, Yamanaka S, Vazquez C, et al. MicroRNA-224 negatively regulates p21 expression during late neoplastic progression in inflammatory bowel disease. Inflamm Bowel Dis 2013; 19: 471–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Atreya R, Neurath M. Signaling molecules: the pathogenic role of the IL-6/STAT-3 trans signaling pathway in intestinal inflammation and in colonic cancer. Curr Drug Targets 2008; 9: 369–374. [DOI] [PubMed] [Google Scholar]
- 40.Fukata M, Chen A, Vamadevan AS, et al. Toll-like receptor-4 promotes the development of colitis-associated colorectal tumors. Gastroenterology 2007; 133: 1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rafa H, Benkhelifa S, AitYounes S, et al. All-trans retinoic acid modulates TLR4/NF-κB signaling pathway targeting TNF-α and nitric oxide synthase 2 expression in colonic mucosa during ulcerative colitis and colitis associated cancer. Mediators Inflamm 2017; 2017: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Farooqi AA, de la Roche M, Djamgoz MBA, et al. Overview of the oncogenic signaling pathways in colorectal cancer: mechanistic insights. Semin Cancer Biol 2019; 58: 65–79. [DOI] [PubMed] [Google Scholar]
- 43.Carey R, Jurickova I, Ballard E, et al. Activation of an IL-6:STAT3-dependent transcriptome in pediatric-onset inflammatory bowel disease: Inflamm Bowel Dis 2008; 14: 446–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang MS, Bowcutt R, Leung JM, et al. Integrated analysis of biopsies from inflammatory bowel disease patients identifies SAA1 as a link between mucosal microbes with TH17 and TH22 cells. Inflamm Bowel Dis 2017; 23: 1544–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuza K, Nagahashi M, Shimada Y, et al. Upregulation of phosphorylated sphingosine kinase 1 expression in colitis-associated cancer. J Surg Res 2018; 231: 323–330. [DOI] [PubMed] [Google Scholar]
- 46.Yu L-Z. Expression of interleukin-22/STAT3 signaling pathway in ulcerative colitis and related carcinogenesis. World J Gastroenterol 2013; 19: 2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takabe K, Paugh SW, Milstien S, et al. ‘ Inside-out’ signaling of sphingosine-1-phosphate: therapeutic targets. Pharmacol Rev 2008; 60: 181–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beaugerie L, Itzkowitz SH. Cancers complicating inflammatory bowel disease. N Engl J Med 2015; 372: 1441–1452. [DOI] [PubMed] [Google Scholar]
- 49.Kang M, Martin A. Microbiome and colorectal cancer: unraveling host-microbiota interactions in colitis-associated colorectal cancer development. Semin Immunol 2017; 32: 3–13. [DOI] [PubMed] [Google Scholar]
- 50.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 2009; 9: 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ni J, Wu GD, Albenberg L, et al. Gut microbiota and IBD: causation or correlation? Nat Rev Gastroenterol Hepatol 2017; 14: 573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lopez A, Hansmannel F, Kokten T, et al. Microbiota in digestive cancers: our new partner? Carcinogenesis 2017; 38: 1157–1166. [DOI] [PubMed] [Google Scholar]
- 53.Abreu MT, Peek RM. Gastrointestinal malignancy and the microbiome. Gastroenterology 2014; 146: 1534–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Darfeuille-Michaud A, Neut C, Barnich N, et al. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn’s disease. Gastroenterology 1998; 115: 1405–1413. [DOI] [PubMed] [Google Scholar]
- 55.Richard ML, Liguori G, Lamas B, et al. Mucosa-associated microbiota dysbiosis in colitis associated cancer. Gut Microbes 2018; 9: 131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ventham NT, Kennedy NA, Nimmo ER, et al. Beyond gene discovery in inflammatory bowel disease: the emerging role of epigenetics. Gastroenterology 2013; 145: 293–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Waldner MJ, Neurath MF. Mechanisms of immune signaling in colitis-associated cancer. Cell Mol Gastroenterol Hepatol 2015; 1: 6–16. [DOI] [PMC free article] [PubMed] [Google Scholar]