Abstract
Context.—
Cell block preparation methods vary substantially across institutions and are frequently suboptimal. The growing importance of biomarker testing in the era of targeted therapies makes optimization of cell block preparation critically important.
Objective.—
To develop an improved cell block preparation method.
Design.—
Ex vivo fine-needle aspirates and scrapes from surgically resected tumors were used to develop an improved HistoGel (Thermo Fisher Scientific, Waltham, Massachusetts)-based cell block preparation method. Cellularity yield with the new versus the standard method was assessed in ex vivo split samples and in consecutive clinical fine-needle aspirates processed before (n = 100) and after (n = 100) the new method was implemented in our laboratory. Sufficiency of cell block material for potential molecular studies was estimated by manual cell quantitation.
Results.—
The key modification in the new method was pretreatment of the pelleted cells with 95% ethanol before the addition of HistoGel (HistoGel + ethanol method). In addition, we optimized the melting conditions of HistoGel and added a dark, inorganic marker to the cell pellets to highlight the desired level of sectioning during microtomy. Cell blocks from ex vivo split samples showed that the HistoGel + ethanol method yielded, on average, an 8.3-fold (range, 1–20) greater cellularity compared with the standard HistoGel-only method. After the switch from the standard HistoGel method to the modified method in our clinical practice, sufficiency of positive fine-needle aspirates for some molecular studies increased from 72% to 97% (P = .002).
Conclusions.—
We describe a simple and readily adoptable modification of the HistoGel method, which results in substantial improvement in cell capture in cell blocks, leading to a significant increase in sufficiency for potential molecular and other ancillary studies.
Biomarker testing is currently required to guide the selection of a growing number of targeted therapies in patients with a wide range of malignancies. Combined with increasing use of immunostains for tumor diagnosis, this has substantially increased the demand for the amount of tissue in small specimens. For example, in patients with advanced-stage non-small cell lung carcinoma, ancillary studies routinely include a panel of immunostains to subtype poorly differentiated tumors or to confirm the site origin. In addition, routinely required tests now include molecular and cytogenetic studies to test for EGFR mutations, ALK rearrangements, and other emerging biomarkers.1,2 Recently, next-generation sequencing platforms have entered clinical practice. These platforms afford the ability to consolidate testing of multiple genes and types of alterations into a single platform; however, some next-generation sequencing platforms require substantially larger DNA input than standard molecular methods.3,4 For cytology specimens to remain a viable diagnostic modality in the era of personalized medicine, it is crucial for those specimens to consistently provide sufficient material for diagnostic and predictive ancillary studies.
Unlike surgical biopsies, cytology specimens consist of a variety of preparations, which, depending on institutional processes, may include air-dried and alcohol fixed smears; CytoSpins (Thermo Fisher Scientific, Waltham, Massachusetts), ThinPrep (Hologic, Marlborough, Massachusetts), or SurePath (Becton Dickinson, Franklin Lakes, New Jersey) liquid-based preparations; and cell blocks. In most laboratories, ancillary studies on cytology samples are performed on cell block material, in line with the current guidelines for EGFR and ALK testing in lung carcinoma specimens,5 although the utility of non–cell block cytologic material for molecular testing has been increasingly recognized.6,7 The primary advantages of using cell blocks for ancillary studies include the operational simplicity because the processes in pathology and molecular laboratories parallel those in place for surgical specimens. Additionally, material in cell blocks may also provide architectural detail in small tissue fragments, enhancing the diagnosis. Furthermore, some commercial laboratories and clinical trials only accept paraffin-embedded material for molecular studies. Thus, the ability to efficiently capture as many cells as possible in a cell block is currently a critical issue in cytology.
Unlike the fairly standardized processing and embedding procedures for surgical biopsies across institutions, cell block preparation methods are highly variable, ranging from “home-brew” techniques to commercial methods.8 In a recent survey of 90 cytopathologists and cytotechnologists, it was reported that more than 10 cell block preparation methods are currently in use.9 Notably, 44% of respondents reported dissatisfaction with their cell block preparation method and cited low cell yield as the primary reason.9
Before this study, the standard cell block preparation method in our laboratory was the conventional HistoGel (Thermo Fisher Scientific) method, which we performed according to the manufacturer’s recommendations. Together with plasma thrombin, HistoGel represents one of the most-common cell block preparation methods, which is used in approximately 30% of US laboratories.9 This method involves centrifugation of cell suspensions and the addition of HistoGel—a modified agar—to amalgamate the cell pellet. In our clinical experience, we found that the cellularity in cell blocks prepared with this method is highly inconsistent, particularly in scant samples lacking visible tissue fragments. Given that several other widely used cell block preparation methods were also reported to have high rates of user dissatisfaction,9 we embarked on experiments to improve cell capture in cell blocks in our laboratory. Here, we describe our institutionally developed cell block protocol resulting in enhanced cellularity in cell blocks processed with this new method, as well as the subsequent validation process of this protocol in our clinical practice.
MATERIALS AND METHODS
In the first phase of this study, we tested multiple modifications of the cell block preparation protocol to identify a method with the greatest cell capture using split fine-needle aspirations (FNAs) and scrapes from fresh, surgically resected tumors (ex vivo samples). Ex vivo FNAs were prepared using a 25-gauge needle, and scrapes were prepared by gently scraping the cut surface of a tumor with a surgical blade. The samples were collected in CytoLyt (Hologic) solution, analogous to clinical cytology samples in our practice. CytoLyt fluid was then evenly split, and for all cases in which there were visible tissue fragments or visible sediment after centrifugation, cell blocks were prepared using different methods. Key tested modifications included addition of 95% ethanol (EtOH) step before HistoGel, titrating HistoGel liquefaction conditions, titrating the volume of added HistoGel, testing variations in centrifugation steps, and testing various methods of enhancing the visualization of the cell pellet in paraffin blocks. The details of the developed method are described in the first part of the Results section.
The first step in validating the selected new protocol consisted of quantifying the cellularity in cell blocks from split-samples prepared from 10 ex vivo FNAs and scrapes. These were prepared by collecting FNAs or scrapes in a CytoLyt, which were then evenly split and processed by the standard versus the modified cell block protocols. Cellularity was compared by manually estimating the fold difference in cell number in the matched cell blocks. To assure comparable cellularity in the split CytoLyt containers, a ThinPrep monolayer slide was prepared and reviewed for each of the split containers. ThinPrep slides were prepared for each specimen similar to our clinical practice. After the initial centrifugation of the CytoLyt fluid, cell pellets were vortexed, an aliquot from that material was transferred into PreservCyt container (Hologic), and ThinPrep slides were prepared following the manufacturer’s recommendations. The aliquot volume was 100 μL in pelleted material that had a cloudy appearance or 50 μL if the material was dense, mucoid, or bloody (Figure 1, A through F).
Figure 1.
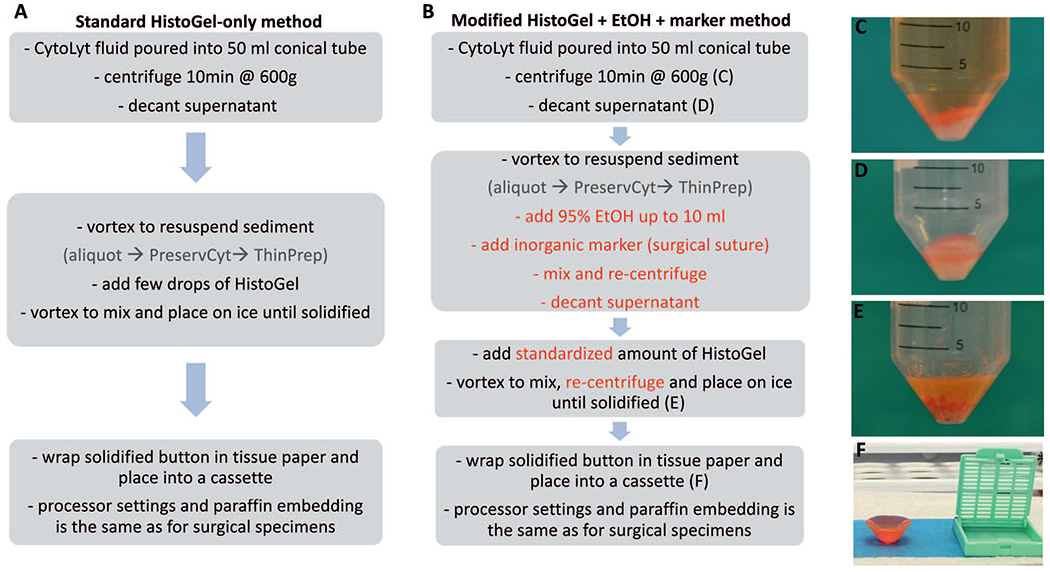
Protocol flow charts for the standard HistoGel method (A) versus the modified HistoGel + ethanol (EtOH) method (B). Key steps in each protocol are outlined. The modified steps are in a red font. The photographs illustrate initial sediment before (C) and after (D) decanting of the supernatant, sediment mixed with HistoGel after solidification on ice (E), and the typical appearance of the resultant solidified button before placement into a cassette (F).
For the second step of validation, we compared cellularity in 100 consecutive FNAs, performed as part of routine clinical care, and processed in our cytology laboratory using the standard HistoGel method and 100 consecutive FNAs processed after implementation of the new method. Cell blocks from positive FNAs were reviewed to determine whether they contained at least 20 tumor cells in a single hematoxylin-eosin section—the minimal cellularity threshold found to yield sufficient material for some molecular studies.3,10 The differences in cellularity of different cell block methods were analyzed by Fisher exact test.
RESULTS
Description of the Standard and Improved HistoGel-Based Methods
The key steps in the standard HistoGel-only method, recommended by the manufacturer, are summarized in Figure 1, A through F. In our practice, those steps included centrifuging the cell suspension at 600g, decanting the media by carefully poring off the supernatant, adding several drops of HistoGel, mixing, and allowing pellets to congeal on ice for 5 to 10 minutes, and checking for solidification. The resultant cone-shaped, gel-like pellet is dislodged from the conical tube with a disposable spatula, wrapped in tissue paper, and placed with the point of the cone facedown in a cassette (Figure 1, F), so that the tip of the cone is the first part of the cell pellet to become visible when being cut by the microtome. The cassette is then fixed in an automated tissue processor for 6 hours and embedded in paraffin by the standard processes used for our surgical specimens.
In optimization experiments, we found that the following modifications to the standard HistoGel-based method resulted in improved cell capture in a cell block (Figure 1, B):
Pretreatment of Cell Pellet With EtOH Before Addition of HistoGel. Pretreating the cell pellet was performed by adding 10 mL of 95% EtOH to the pelleted cells after the supernatant was decanted by carefully pouring off the additional fluid. After the addition of EtOH, the pellet was vortexed and subsequently recentrifuged.
Standardization of Liquefaction Time and the Volume of the Added HistoGel. We titrated the liquefaction time and volume of the added HistoGel. We found that the optimal liquefaction process was to place an open Eppendorf tube upright in a glass container and microwave at medium heat for 10 seconds when Eppendorf tubes were fully filled with HistoGel and for 5 seconds, with half-full tubes. We also standardized the volume of the added HistoGel to 6 drops (250 μL) for the usual sediment (1–2 mL) and 500 μL for larger (>3 mL) sediment.
Addition of a Marker for Paraffin Block Sectioning. One of the key problems with the HistoGel-only method is that the pellet formed by this method may be poorly visible in the paraffin block, thus precluding sectioning at the correct level during microtomy. After trials of several dyes and marker materials (including banana peel, as previously described11), we found that a simple and practical marker was a short (3–5 mm) segment of dark, synthetic surgical suture (2–0 Ethilon nylon suture, Johnson and Johnson, New Brunswick, New Jersey) [Figure 2, A through D].
Addition of an Extra Centrifugation Step. The final modification to the HistoGel processing procedure was the addition of an extra centrifugation step after HistoGel addition to optimally collect all material before congealing.
Figure 2.
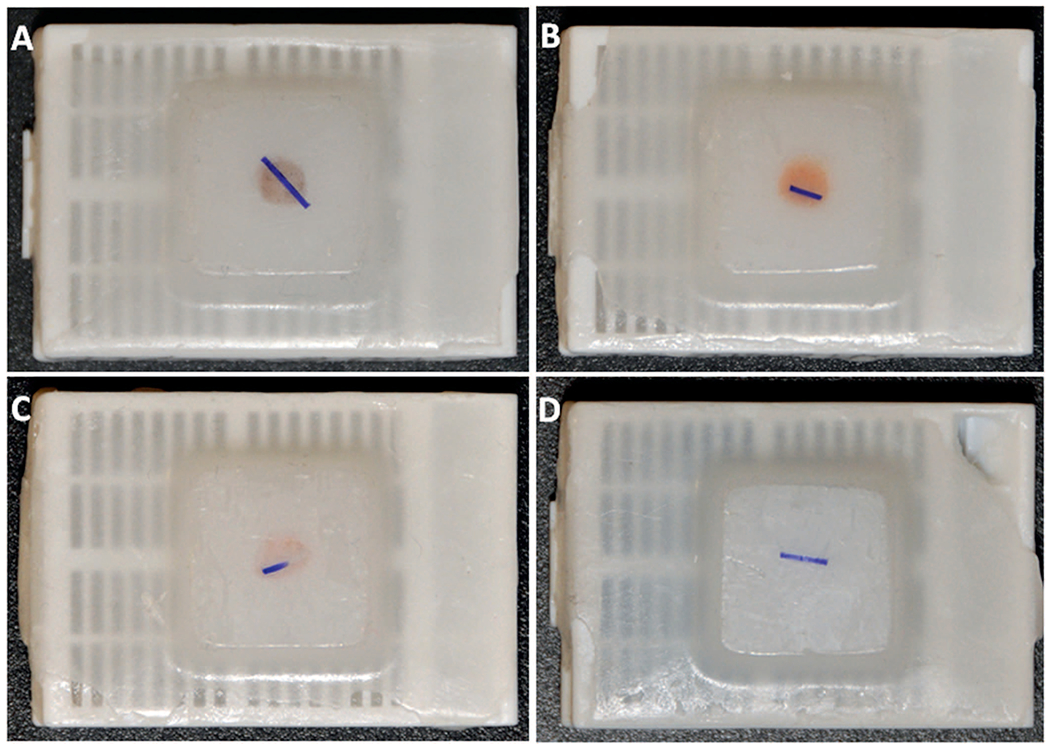
Illustration of dark suture as a marker of the cell button location in the paraffin block. Blocks A, B, and C are sufficiently visible, even in the absence of a marker, but the small button in block D is not visible, and the appropriate level of sectioning would not be identifiable without the marker.
Validation of the New Method in Split Ex Vivo Samples
As summarized in Table 1, ex vivo scant FNAs and generous scrapes (n = 10) were prepared using surgically resected tumors. Each sample was collected in CytoLyt fluid, which was mixed and evenly split into 3 conical tubes, which were processed as follows: (1) standard HistoGel-only, (2) HistoGel + EtOH, or (3) EtOH only. In 9 of 10 samples (90%), the HistoGel + EtOH method yielded substantially greater cellularity than HistoGel-only and EtOH-only methods. The mean increase in cellularity with the HistoGel + EtOH versus HistoGel-only method was an 8.3-fold (range, 1–20) increase, and there was a 136-fold (range, 1–1000) increase with the HistoGel + EtOH versus the EtOH-only method. One FNA (FNA1) sample yielded almost acellular cell blocks by all methods—that case had minimal cellularity in the CytoLyt needle rinse, as confirmed in the corresponding ThinPrep slides. The increased cellularity and the more-condensed appearance of the cell pellets with the HistoGel + EtOH method compared with other methods are illustrated in Figure 3, A through F.
Table 1.
Comparison of Cellularity in Cell Blocks Prepared From Ex Vivo Samples By 3 Methods
| Characteristic | HistoGel | EtOH | HistoGel + EtOH |
|---|---|---|---|
| Individual comparisonsa | |||
| FNA1 (liver; metastatic CRC ) | 1 (almost acellular) | 1 (almost acellular) | 1 (almost acellular) |
| FNA2 (liver; metastatic CRC2) | 50 | 1 (almost acellular) | 1000 |
| FNA3 (lung; lung adenocarcinoma) | 10 | 1 | 20 |
| FNA4 (lymph node; reactive lymph node) | 3 | 1 | 15 |
| Scraping 1 (liver; metastatic CRC ) | 10 | 1 | 50 |
| Scraping 2 (liver; metastatic CRC2) | 1 | 1 | 1.5 |
| Scraping 3 (lung; lung adenocarcinoma) | 1 | 2 | 2 |
| Scraping 4 (lymph node; reactive lymph node) | 1 | 5 | 10 |
| Fold differences | |||
| HistoGel + EtOH versus HistoGel alone, mean (range); median | 8.3 (1–20); 3.5 | ||
| HistoGel + EtOH versus EtOH alone, mean (range); median | 136 (1–1000); 8.5 | ||
Abbreviations: CRC, colorectal carcinoma; EtOH, ethanol; FNA, fine-needle aspiration.
Shown are the fold differences in total cell numbers in a single hematoxylin-eosin-stained section, with cellularity in the least cellular cell block counted as 1.
Figure 3.
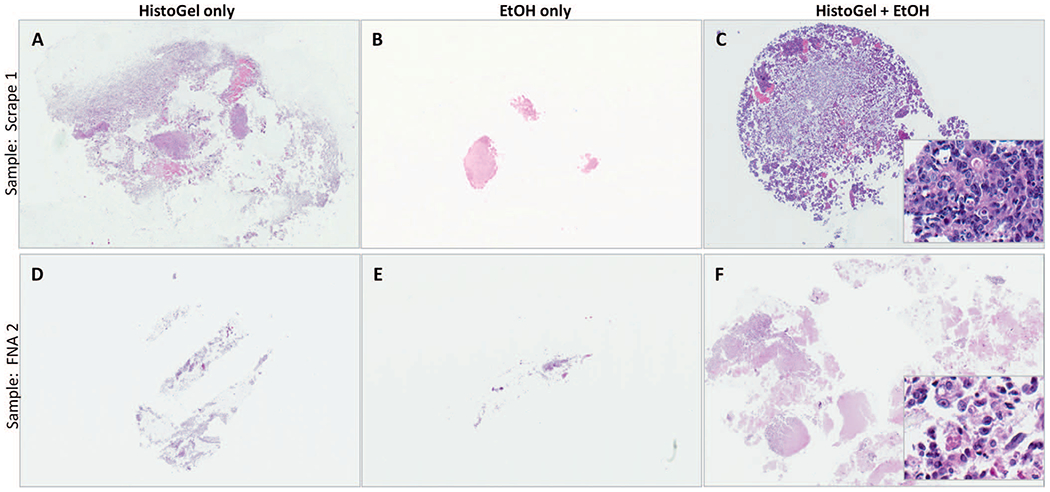
Illustration of the microscopic appearance of cell blocks prepared by the HistoGel + ethanol (EtOH) method versus HistoGel-only and the EtOH-only methods. Note the substantially greater cell capture and more-compact appearance of cell blocks prepared with the HistoGel + EtOH method. For quantitation of increased cell capture, see Table 1. The insets in C and F illustrate excellent cytomorphologic preservation with this method (hematoxylin-eosin, original magnifications ×20 [A through F] and ×400 [insets]).
Validation of the New Method in Consecutive Clinical Samples
After implementation of the HistoGel + EtOH method in our clinical practice, we reviewed 100 consecutive FNAs prepared by the new method and 100 consecutive FNAs prepared by the prior, standard HistoGel method. As shown in Table 2, we found that, for positive FNA results, the frequency of cellular cell blocks adequate for molecular studies increased from 72% (31 of 43) with the standard HistoGel method to 97% (38/39) with the modified method (P = .002).
Table 2.
Comparison of Cell Block Recovery in Consecutive Clinical Fine-Needle Aspirations (FNAs) Using the Standard HistoGel-Only Method Versus the Modified Method (HistoGel + Ethanol [EtOH])
| Characteristic | Standard HistoGel Method, n = 100 | Modified (HistoGel + EtOH) Method, n = 100 | P Value |
|---|---|---|---|
| Distribution of diagnostic categories | |||
| Positive | 43 | 39 | .67 |
| Other (NDX, NEG, ATP, SUS) | 57 | 61 | |
| Frequency of cellular cell blocks in positive FNA results | n = 43, No. (%) | n = 39, No. (%) | |
| Cell block containing >20 tumor cells on representative H&E section | 31 (72) | 38 (97) | .002 |
| No cell block or cell block containing <20 tumor cells | 12 (28) | 1/39 (3) | |
Abbreviations: ATP, atypical; H&E, hematoxylin and eosin; NDX, nondiagnostic; NEG, negative; SUS, suspicious.
DISCUSSION
Here, we describe an improved method for preparing HistoGel-based cell blocks, which results in substantial improvement of cell capture compared with the standard HistoGel method. We found that for almost all matched ex vivo samples, the new method yielded greater cellularity than the standard HistoGel method, with a mean 8.3-fold greater cellularity. After implementation in clinical practice, we estimate that the frequency of cell blocks with cellularity potentially sufficient for at least some molecular studies increased from 72% to 97%. This has important implications for the ability to perform predictive molecular and other ancillary studies on cytology specimens.
The improved cell block method described here represents a modification of a standard HistoGel method and comprises 4 key modifications: (1) pretreatment of the cell pellet with 95% EtOH before the addition of HistoGel, (2) standardization of the volume and liquefaction time for the HistoGel, (3) addition of a visual microtomy marker, and (4) addition of an extra centrifugation step after the addition of the HistoGel. Although uncommon, some laboratories use 95% EtOH as the sole reagent in cell block preparation.9 We found that EtOH alone yielded substantially lower cell capture than HistoGel alone. However, we hypothesized that combining the HistoGel with the 95% EtOH step might improve the cell capture, which, indeed, we found to be the case. It is likely that EtOH changes osmolarity, leading to increased dehydration of the cells, and more effective decantation. It is also possible that EtOH clears mucoid and other extracellular substances from the cell pellet, thus allowing for greater cell aggregation when HistoGel is added. The second aspect of the improved process in our protocol was the standardization of the volume and melting time of the HistoGel. The manufacturer’s instructions do not specify those parameters, but we found that inconsistent HistoGel liquefaction or added volume may result in suboptimal cell block yield, which is in agreement with experience from other laboratories.8 The third modification—addition of a short, dark surgical suture to mark the pellet in the paraffin block—does not lead to greater cell capture, but it solves another common problem with HistoGel cell blocks—the poor visibility of the cell pellet in the paraffin block during microtome sectioning. This issue is most pronounced for pellets prepared from scant specimens lacking visible tissue fragments, and may lead to either too-shallow or too-deep sectioning of the block, resulting in either acellular slides or inadvertent cutting through the pellet. In initial experiments, we tested a variety of dyes to highlight the pellet, but they did not yield improved results. We also attempted to use small cuts of dried banana peel—an interesting idea, suggested by Varsegi and Shidham.11 Indeed, we also found banana peel to work well as a visual marker. However, we found that a short piece of dark surgical suture was a good practical alternative because it is readily available in hospital settings. In addition, the use of an inorganic substance avoids at least the theoretical concern that nucleic acids found in plant cells might interfere with some molecular assays. The selected suture size (2–0 gauge) does not interfere with cutting by the blade of the microtome, and it is easily visible. It is worth noting that, in fact, most cell blocks prepared by the HistoGel + EtOH method had pellets that were readily visible in the paraffin block, even without the marker (Figure 3, A through C). This is likely because the pellets prepared by this method are more condensed than those prepared with HistoGel-only method. Nonetheless, in some instances, the pellets were not readily visible (Figure 3, D). In such cases, suture material provided the essential guidance for the initial level to start sectioning.
In addition to the standard HistoGel method, multiple other cell block preparation methods are currently used in clinical practice. As reported by Crapanzano et al,9 a recent survey revealed that more than 10 cell block preparation methods are currently employed in cytopathology laboratories in the United States, with the most-common methods being plasma thrombin (33%), HistoGel (27%), and Cellient (Hologic) automated cell block system (27%). Notably, in that survey, it was found that HistoGel is associated with the lowest degree of satisfaction, which is in line with our experience with the unmodified HistoGel method. However, a substantial rate of dissatisfaction was reported for all other methods as well. Of the common non-HistoGel cell block preparation methods—plasma thrombin—is one of the earliest methods used to create cell blocks, which works analogously to HistoGel by congealing the cell pellet. The plasma thrombin method has been reported to have a comparable cell yield to the HistoGel method.12 The limitation with the plasma thrombin method is its dependence on the blood bank supply. In addition, there is at least a theoretical concern that those preparations could be contaminated with cell-free nucleic acids. In recent years, it has become well established that patients with malignant tumors may have circulating cell-free tumor DNA, which is becoming increasingly used for clinical disease screening and monitoring.13,14 Thus, it is theoretically possible that, in a donor with an occult malignant tumor, plasma thrombin could be contaminated with cell-free tumor DNA, which could be detected by high-sensitivity methods, such as next-generation sequencing. Another common cell block preparation method—Cellient—is reported to achieve good results for specimens with low cellularity.15 The limitations for the Cellient method include the cost of equipment and consumables. In addition, because of longer processing time, multiple machines are required in a large-volume setting; although, anecdotally, we are aware that some laboratories reserve use of Cellient processing for hypocellular specimens only. Another technique advocated by some laboratories is the collodion bag method.12,16 In a recent study, the collodion method was found to be superior to the unmodified HistoGel and plasma thrombin methods.12 However, collodion is suspended in ether-based solvent, and it must be handled in a laminar flow hood and stored in small volumes in a flame-proof enclosure.8,12 Other selected cell block preparation methods include gelatin embedding, agar embedding, the “Shidham method,” simple sedimentation, pregelatinized starch, and more recently, the so-called “tissue coagulum clot” method, among others.8,9,11,17 Interlaboratory comparison of the methods used by various laboratories, including the method we developed, would be of great interest.
Although our laboratory currently uses CytoLyt as needle-rinse collection medium for most FNAs, the cell block preparation method described here can be used with any collection media. Indeed, it was confirmed in a separate study18 at our institution that, for needle rinses collected in formalin, the HistoGel + EtOH method resulted in superior cellularity with better tumor cell aggregation than the standard HistoGel method.
The described protocol has 2 potential limitations that should be acknowledged. One is a longer cell block processing time. In our experience, for a batch of 10 cases, the HistoGel + EtOH protocol takes approximately 50 consecutive minutes, whereas the standard HistoGel protocol takes approximately 30 minutes. Given the substantially superior results and the ability to process the specimens in batches, we believe that this relatively small increase in processing time and labor is justified. The second issue, which we have encountered in a few cells blocks prepared with the HistoGel + EtOH method, is that the cell button in the paraffin block occasionally has a brittle appearance. Therefore, the center of the cell button may recess, resulting in only the peripheral circle of the cells being represented in a cut section. However, after using the HistoGel + EtOH method in clinical practice now for approximately 3 years, we find that such instances were uncommon, and when they did occur, deeper levels usually helped provide a more-complete cross-section of the cell pellet. A third potential limitation is the concern that pretreating the cell pellet with 95% EtOH could have some effect on the immunohistochemistry. It is important to emphasize that the most important effect on antigenicity is imparted by the initial fixative in the collection media, and we anticipate that the short subsequent EtOH step is unlikely to have a significant effect on antigenicity. We performed a limited side-by-side comparison of several commonly used antibodies in split, ex vivo samples, prepared with and without EtOH step, and found immunoreactivity to be identical (data not shown). Nevertheless, if a laboratory elects to adopt this method, revalidation of immunostains should be considered.
With regard to sufficiency for potential molecular studies, the actual cellularity requirements vary, depending on the molecular platform used. In this study, we defined sufficiency as the total number of cells that were shown in our laboratory19 and in other laboratories3 to provide sufficient material for some molecular studies (a few 100 to 1000 cells). In contrast, some next-generation sequencing assays, such as the MiSeq platform (Illumina, San Diego, California) currently used at our institution, have substantially higher DNA input requirements (5000–15 000 cells). Sufficiency of various cytologic preparations for those high-DNA input assays is currently under investigation at our institution.
Even though the method described resulted in substantially improved cell capture, in our clinical practice we still occasionally encounter cases in which we fail to obtain a cellular cell block despite ample cellularity in the needle rinse, as confirmed by a matched ThinPrep slide, which might be related to the specifics of a particular sample or to unavoidable human factors involved in the multistep manual process of cell block preparation. Even though instances of cell block failure are currently uncommon in our practice, they may have drastic implications for an individual patient. To mitigate such instances, we have recently developed and implemented a novel process in our cytology laboratory, in which material from the needle rinse fluid that is not captured in a cell pellet (which includes both unpelleted cells and cell-free DNA) is entirely precipitated for DNA extraction, and none of the material that escaped cell block preparation is wasted. This process is described in detail in Tian et al.20 In combination, the new cell block preparation method and collection of the unpelleted material substantially improved the sufficiency of cytology samples for diagnostic and predictive ancillary studies in our practice.20
In summary, we describe a modification of the HistoGel-based cell block preparation method that leads to substantial improvement in cell recovery from FNA needle rinses compared with the standard HistoGel method. We show the validation of this method both in ex vivo split samples and in routine clinical FNAs before and after the implementation of the new method in our laboratory. This protocol is simple and readily adoptable and leads to substantially increased sufficiency of FNA samples for molecular testing.
Acknowledgments
This study was supported in part by a Cancer Center Support Grant from the US National Institutes of Health/National Cancer Institute (grant P30CA008748).
Footnotes
The authors have no relevant financial interest in the products or companies described in this article.
References
- 1.Travis WD, Rekhtman N. Pathological diagnosis and classification of lung cancer in small biopsies and cytology: strategic management of tissue for molecular testing. Semin Respir Crit Care Med. 2011;32(1):22–31. [DOI] [PubMed] [Google Scholar]
- 2.Sholl LM. The molecular pathology of lung cancer. Surg Pathol Clin. 2016; 9(3):353–378. [DOI] [PubMed] [Google Scholar]
- 3.Roy-Chowdhuri S, Stewart J. Preanalytic variables in cytology: lessons learned from next-generation sequencing—the MD Anderson experience [published online ahead of print June 22, 2016]. Arch Pathol Lab Med. 2016; 140(11):1191–1199. [DOI] [PubMed] [Google Scholar]
- 4.Hyman DM, Solit DB, Arcila ME, et al. Precision medicine at Memorial Sloan Kettering Cancer Center: clinical next-generation sequencing enabling next-generation targeted therapy trials. Drug Discov Today. 2015;20(12):1422–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindeman NI, Cagle PT, Beasley MB, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guidelinefrom the College ofAmerican Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology [published correction appears in Arch Pathol Lab Med. 2013;137(9):1174]. Arch Pathol Lab Med. 2013;137(6):828–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roy-Chowdhuri S, Aisner DL, Allen TC, et al. Biomarker testing in lung carcinoma cytology specimens: a perspective from members of the Pulmonary Pathology Society. Arch Pathol Lab Med. 2016;140(11):1267–1272. [DOI] [PubMed] [Google Scholar]
- 7.Rekhtman N, Roy-Chowdhuri S. Cytology specimens: a goldmine for molecular testing. Arch Pathol Lab Med. 2016;140(11):1189–1190. [DOI] [PubMed] [Google Scholar]
- 8.Saqi A The state of cell blocks and ancillary testing: past, present, and future. Arch Pathol Lab Med. 2016;140(12):1218–1322. [DOI] [PubMed] [Google Scholar]
- 9.Crapanzano JP, Heymann JJ, Monaco S, Nassar A, Saqi A. The state of cell block variation and satisfaction in the era of molecular diagnostics and personalized medicine. Cytojournal. 2014;11:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rekhtman N, Brandt SM, Sigel CS, et al. Suitability of thoracic cytology for new therapeutic paradigms in non-small cell lung carcinoma: high accuracy of tumor subtyping and feasibility of EGFR and KRAS molecular testing. J Thorac Oncol. 2011;6(3):451–58. [DOI] [PubMed] [Google Scholar]
- 11.Varsegi GM, Shidham V. Cell block preparation from cytology specimen with predominance of individually scattered cells. J Vis Exp. 2009;(29):1316. doi: 10.3791/1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balassanian R, Wool GD, Ono JC, et al. A superior method for cell block preparation for fine-needle aspiration biopsies. Cancer Cytopathol. 2016;124(7): 508–518. [DOI] [PubMed] [Google Scholar]
- 13.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11(6):426–437. [DOI] [PubMed] [Google Scholar]
- 14.Oxnard GR, Paweletz CP, Sholl LM. Genomic analysis of plasma cell-free DNA in patients with cancer. JAMA Oncol. 2016;3(6):740–741. [DOI] [PubMed] [Google Scholar]
- 15.Prendeville S, Brosnan T, Browne TJ, McCarthy J. Automated Cellient™ cytoblocks: better, stronger, faster? Cytopathology. 2014;25(6):372–380. [DOI] [PubMed] [Google Scholar]
- 16.Kalhor N, Wistuba II. Perfecting the fine-needle aspirate cell block. Cancer Cytopathol. 2013;121 (3):109–110. [DOI] [PubMed] [Google Scholar]
- 17.Yung RC, Otell S, Illei P, et al. Improvement of cellularity on cell block preparations using the so-called tissue coagulum clot method during endobronchial ultrasound-guided transbronchial fine-needle aspiration. Cancer Cytopathol. 2012;120(3):185–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bullock N, Rudomina D, Rekhtman N. Optimizing cytology cell blocks for more cellular and reliable immunohistochemical results in breast cancer specimens [abstract 136]. Mod Pathol. 2015;28(suppl 2):36A. [Google Scholar]
- 19.Brandt SM, Tafe LJ, Arcila ME, et al. Performance of cytologic specimens in EGFR and KRAS molecular testing with a focus on minimal cellularity requirements [abstract 1726]. Mod Pathol. 2011;91(suppl 1):405A–06A. [Google Scholar]
- 20.Tian SK, Killian JK, Rekhtman N, et al. Optimizing workflows and processing of cytologic samples for comprehensive analysis by next-generation sequencing: Memorial Sloan Kettering Cancer Center Experience [published online ahead of print September 2, 2016]. Arch Pathol Lab Med. 2016;140(11): 1200–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]


