Abstract
Objective:
Nobiletin treatment on MDA-MB 231 cells reduces the expression of CXC chemokine receptor type 4 (CXCR4), which is highly expressed in cancer stem cell populations in tumor patients. However, the mechanisms of nobiletin in cancer stem cells (CSCs) remain elusive. This study was aimed to explore the potential target and mechanisms of nobiletin in cancer stem cells using bioinformatics approaches.
Methods:
Gene expression profiles by public COMPARE predicting the sensitivity of tumor cells to nobiletin. Functional annotations on gene lists are carried out with The Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8, and WEB-based GEne SeT Analysis Toolkit (WebGestalt). The protein-protein interaction (PPI) network was analyzed by STRING-DB and visualized by Cytoscape.
Results:
Microarray analyses reveal many genes involved in protein binding, transcriptional and translational activity. Pathway enrichment analysis revealed breast cancer regulation of estrogen signaling and Wnt/ß-catenin by nobiletin. Moreover, three hub genes, i.e. ESR1, NCOA3, and RPS6KB1 and one significant module were filtered out and selected from the PPI network.
Conclusion:
Nobiletin might serve as a lead compound for the development of CSCs-targeted drugs by targeting estrogen and Wnt/ß-catenin signaling. Further studies are needed to explore the full therapeutic potential of nobiletin in cancer stem cells.
Key Words: Nobiletin, anticancer, bioinformatics, cancer stem cells, signaling pathway
Introduction
Recent studies have shown that the ability of tumors to develop and propagate depends on a small population of cells called cancer stem cells (CSCs) (Pan et al., 2018; Zhu and Fan, 2018). CSCs are responsible for resistance to chemotherapy and radiotherapy (Toledo-Guzman et al., 2018). Conventional chemotherapy has proved to be able to reduce tumor size; however most of the tumor relapsed because the population of CSCs that were able to survive and grow into tumor bulk (Zhu and Fan, 2018). The CSC-targeted therapy will target the CSCs population whose slowly growth (Moltzahn et al., 2008), and thus the effectiveness of cancer therapy will be achieved. Collectively, CSC-targeted therapy is needed to prevent relapse after chemotherapy.
Flavonoid compounds have been shown to overcome chemoresistance (Meiyanto et al., 2012) and to inhibit CSCs (Hermawan and Putri, 2018). One potential flavonoid compound to be developed as CSC-targeted drugs is nobiletin (Figure 1A). Previous studies showed that polymetoxiflavone citrus flavonoids namely nobiletin exhibits cytotoxic effects on several cancer cells, e.g. TMK-1, MKN-45, MKN-74 and KATO-III stomach cancer cells (Yoshimizu et al., 2004), MH1C1 and HepG2 human hepatocellular carcinoma (Ohnishi et al., 2004), MDA-MB-435 breast cancer cells, MCF-7 and in HT-29 colon cancer cells (Morley et al., 2007). Studies on the combination of nobiletin and conventional chemotherapy agents have also been carried out. Nobiletin is reported to increase the uptake of chemotherapy vinblastine through inhibition of P-gp in Caco-2 cells (Takanaga et al., 2000). Nobiletin also increased doxorubicin cytotoxicity in MCF-7 breast cancer cells but not T47D cells (Meiyanto et al., 2011). In addition, nobiletin showed the effect of inhibiting metastasis by downregulating CXC chemokine receptor type 4 (CXCR4) and matrix metallopeptidase-9 on MDA-MB 231 breast cancer cells (Baek et al., 2012). Therefore, it has been proven that nobiletin is able to overcome chemoresistance and also inhibit CXCR4 which is one of the regulators of CSCs, but its molecular mechanism on CSCs need to be clarified further.
Figure 1.
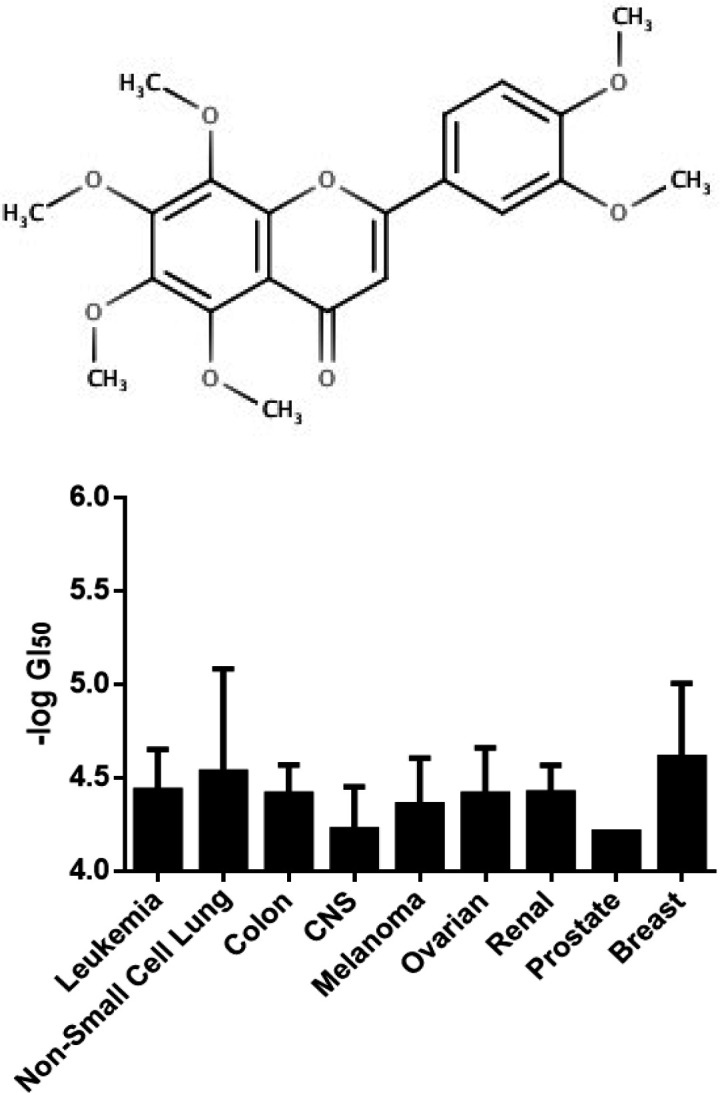
(A) Chemical Structure of Nobiletin. (B) Cytotoxicity of Nobiletin on the NCI-60 Tumor Cell Line Panel
In this study, we used comprehensive bioinformatics analysis to explore nobiletin cytotoxicity and mechanism in CSCs. Analysis of the public library from the COMPARE database was done to produce a list of drugs that have similarities with nobiletin, as well as a gene list that was influenced by nobiletin on the NCI 60 cell line panel. From the microarray data, functional annotations are then carried out to predict molecular mechanisms, functions and roles of these genes. Furthermore, an analysis of protein-protein interaction was performed from the gene list. Hence we provide information about the possible molecular mechanisms of the nobiletin and its molecular targets against cancer stem cells.
Materials and Methods
Data collection and processing
Cytotoxicity and mRNA arrays data were obtained from the NCI 60 DTP website (http.dtp.nci.nih.gov) (Monks et al., 1997). COMPARE analysis with the public library produces a list of drugs that have similarities with nobiletin, as well as a list of gene expressions on the NCI 60 cell line panel (Mahmoud et al., 2018). The similarity pattern is expressed as the Pearson correlation coefficient. In this study, the list of compounds and genes was limited to the Pearson correlation coefficient <-0.5 and> 0.5.
Functional and pathway enrichment analysis
Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were carried out by The Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8 (Huang da et al., 2009), with p<0.05 was selected as the cutoff value. Moreover, pathway enrichment was also conducted busing Overrepresentation Enrichment Analysis (ORA) from WEB-based GEne SeT AnaLysis Toolkit (WebGestalt) with FDR<0.05 was selected as the cutoff value (Wang et al., 2017a).
Construction of PPI network and module analysis
Protein-protein interaction (PPI) network was constructed with STRING-DB v11.0 (Szklarczyk et al., 2015). Confidence scores >0.7 were considered significant. PPI network was visualized by Cytoscape software. Genes with a degree score more than 5, analyzed by CytoHubba plugin, were selected as hub genes.
Results
COMPARE analysis reveals mRNA target list and standard agent
This study explored the molecular mechanism of nobiletin in CSCs. Analysis of cytotoxicity with a public database of COMPARE showed that nobiletin exhibits cytotoxicity at the same level in the NCI-60 cells panel showing by similar IC50 value (Figure 1B). COMPARE analysis identified 11 standard agents which have a correlation with nobiletin (Table 1). Tamoxifen, triciribine phosphate and 4-ipomeanol are standard drugs with the highest score of a Pearson correlation coefficient.
Table 1.
Correlation of Nobiletin to Standard Agent by COMPARE Analyses with Log IC50 of Nobiletin
| No | Correlation coefficient | NSC Code | Drugs |
|---|---|---|---|
| 1 | 0.525 | S180973 | Tamoxifen |
| 2 | 0.501 | S280594 | Triciribine Phosphate |
| 3 | 0.453 | S349438 | 4-ipomeanol |
| 4 | 0.386 | S95580 | Hexamethylenebisace Tamide |
| 5 | 0.339 | S180973 | Tamoxifen |
| 6 | 0.331 | S118994 | Diglycoaldehyde |
| 7 | 0.327 | S73754 | Fluorodopan |
| 8 | 0.31 | S51143 | Impy |
| 9 | 0.309 | S349156 | Pancratiastatin |
| 10 | 0.307 | S141540 | VP-16 (Etoposide) |
| 11 | 0.301 | S357704 | Cyanomorpholino- ADR |
Level of mRNA expression analysed by COMPARE showed 108 genes regulated by nobiletin (Table 2), which 104 and 4 genes with positive and negative Pearson correlation coefficient, respectively. FRAT2, AANAT, DYM and SNHG8 are genes with direct correlation, whereas BLOC1S6, NR2F6, VANGL1 and SEPT2 are genes with inverse correlation. Direct correlation indicates that the higher mRNA expression, the higher the chemoresistance, while inverse correlation indicates that the higher mRNA expression, the higher the chemosensitivity. FRAT2 has the highest Pearson correlation coefficient (0.612) while FRAT1 shows the Pearson correlation coefficient of 0.543. Both FRAT1 and FRAT2 are regulatory genes of Wnt/ß-catenin signaling. ESR1 shows the Pearson correlation coefficient of 0.525.
Table 2.
mRNA Expression Analysed by COMPARE with log IC50 of Nobiletin on the NCI-60 Cell Line Panel
| No | Pearson Correlation Coefficient | Gene Symbol | Gene Name |
|---|---|---|---|
| 1 | 0.612 | FRAT2 | Frequently Rearranged In Advanced T-Cell Lymphomas 2 |
| 2 | 0.586 | AANAT | Aralkylamine N-Acetyltransferase |
| 3 | 0.582 | DYM | Dymeclin |
| 4 | 0.576 | SNHG8 | Small Nucleolar RNA Host Gene 8 |
| 5 | 0.574 | LETMD1 | LETM1 Domain Containing 1 |
| 6 | 0.571 | ATXN7L3B | Ataxin 7 Like 3B |
| 7 | 0.559 | EPB41L5 | Erythrocyte Membrane Protein Band 4.1 Like 5 |
| 8 | 0.557 | PISD | Phosphatidylserine Decarboxylase |
| 9 | 0.557 | ALDH3B2 | Aldehyde Dehydrogenase 3 Family Member B2 |
| 10 | 0.555 | VPS37C | VPS37C, ESCRT-I Subunit |
| 11 | 0.555 | HEATR6 | HEAT Repeat Containing 6 |
| 12 | 0.554 | LARP4B | La Ribonucleoprotein Domain Family Member 4B |
| 13 | 0.554 | FBP1 | Fructose-Bisphosphatase 1 |
| 14 | 0.554 | AIF1L | Allograft Inflammatory Factor 1 Like |
| 15 | 0.55 | SMARCD2 | SWI/SNF Related, Matrix Associated, Actin Dependent Regulator of Chromatin, Subfamily D, Member 2 |
| 16 | 0.548 | GRTP1 | Growth Hormone Regulated TBC Protein 1 |
| 17 | 0.546 | C21orf33 | Chromosome 21 Open Reading Frame 33 |
| 18 | 0.545 | WDR25 | WD Repeat Domain 25 |
| 19 | 0.544 | TEAD2 | TEA Domain Transcription Factor 2 |
| 20 | 0.544 | EIF4B | Eukaryotic Translation Initiation Factor 4B |
| 21 | 0.543 | FRAT1 | Frequently Rearranged In Advanced T-Cell Lymphomas 1 |
| 22 | 0.542 | TREH | Trehalase |
| 23 | 0.542 | NCOA3 | Nuclear Receptor Coactivator 3 |
| 24 | 0.541 | RMND1 | Required for Meiotic Nuclear Division 1 Homolog |
| 25 | 0.541 | ALOX15 | Arachidonate 15-Lipoxygenase |
| 26 | 0.54 | TRIM37 | Tripartite Motif Containing 37 |
| 27 | 0.538 | TMEM241 | Transmembrane Protein 241 |
| 28 | 0.538 | APRT | Adenine Phosphoribosyltransferase |
| 29 | 0.537 | SP1 | Sp1 Transcription Factor |
| 30 | 0.536 | USP32 | Ubiquitin Specific Peptidase 32 |
| 31 | 0.535 | RNF44 | Ring Finger Protein 44 |
| 32 | 0.535 | BRIP1 | BRCA1 Interacting Protein C-Terminal Helicase 1 |
| 33 | 0.534 | RLN2 | Relaxin 2 |
| 34 | 0.534 | NPY1R | Neuropeptide Y Receptor Y1 |
| 35 | 0.534 | ITGA2B | Integrin Subunit Alpha 2b |
| 36 | 0.533 | ZNF282 | Zinc Finger Protein 282 |
| 37 | 0.533 | RHPN1 | Rhophilin Rho Gtpase Binding Protein 1 |
| 38 | 0.533 | MEPCE | Methylphosphate Capping Enzyme |
| 39 | 0.532 | SPTSSB | Serine Palmitoyltransferase Small Subunit B |
| 40 | 0.53 | SPDEF | SAM Pointed Domain Containing ETS Transcription Factor |
| 41 | 0.53 | PIK3R2 | Phosphoinositide-3-Kinase Regulatory Subunit 2 |
| 42 | 0.53 | EIF3E | Eukaryotic Translation Initiation Factor 3 Subunit E |
| 43 | 0.529 | NUP210L | Nucleoporin 210 Like |
| 44 | 0.528 | ELP2 | Elongator Acetyltransferase Complex Subunit 2 |
| 45 | 0.527 | GATA3 | GATA Binding Protein 3 |
| 46 | 0.526 | PPM1D | Protein Phosphatase, Mg2+/Mn2+ Dependent 1D |
| 47 | 0.526 | IRX5 | Iroquois Homeobox 5 |
| 48 | 0.525 | TFF1 | Trefoil Factor 1 |
| No | Pearson Correlation Coefficient | Gene Symbol | Gene Name |
| 49 | 0.525 | RAD51C | RAD51 Paralog C |
| 50 | 0.525 | ESR1 | Estrogen Receptor 1 |
| 51 | 0.524 | RFX1 | Regulatory Factor X1 |
| 52 | 0.524 | C15orf59 | Chromosome 15 Open Reading Frame 59 |
| 53 | 0.523 | ZNF277 | Zinc Finger Protein 277 |
| 54 | 0.523 | PABPC1 | Poly(A) Binding Protein Cytoplasmic 1 |
| 55 | 0.523 | CYB561 | Cytochrome B561 |
| 56 | 0.522 | SCAMP1 | Secretory Carrier Membrane Protein 1 |
| 57 | 0.522 | PLEKHF2 | Pleckstrin Homology And FYVE Domain Containing 2 |
| 58 | 0.522 | KIAA1324 | Kiaa1324 |
| 59 | 0.522 | DSCAM | DS Cell Adhesion Molecule |
| 60 | 0.521 | XBP1 | X-Box Binding Protein 1 |
| 61 | 0.521 | TUBD1 | Tubulin Delta 1 |
| 62 | 0.52 | EMCN | Endomucin |
| 63 | 0.52 | APPBP2 | Amyloid Beta Precursor Protein Binding Protein 2 |
| 64 | 0.519 | TMEM18 | Transmembrane Protein 18 |
| 65 | 0.519 | ST6GALNAC4 | ST6 N-Acetylgalactosaminide Alpha-2,6-Sialyltransferase 4 |
| 66 | 0.519 | SPPL2B | Signal Peptide Peptidase Like 2B |
| 67 | 0.518 | TMEM183A | Transmembrane Protein 183A |
| 68 | 0.517 | PSMD6 | Proteasome 26S Subunit, Non-Atpase 6 |
| 69 | 0.517 | ECSIT | ECSIT Signalling Integrator |
| 70 | 0.516 | SIAH2 | Siah E3 Ubiquitin Protein Ligase 2 |
| 71 | 0.516 | POU6F2-AS2 | POU6F2 Antisense RNA 2 |
| 72 | 0.516 | MAX | MYC Associated Factor X |
| 73 | 0.516 | GNAO1 | G Protein Subunit Alpha O1 |
| 74 | 0.515 | SPATA17 | Spermatogenesis Associated 17 |
| 75 | 0.514 | STARD10 | Star Related Lipid Transfer Domain Containing 10 |
| 76 | 0.513 | PATZ1 | POZ/BTB And AT Hook Containing Zinc Finger 1 |
| 77 | 0.512 | PDZD3 | PDZ Domain Containing 3 |
| 78 | 0.512 | CYP2J2 | Cytochrome P450 Family 2 Subfamily J Member 2 |
| 79 | 0.512 | COX6C | Cytochrome C Oxidase Subunit 6C |
| 80 | 0.511 | PLXNA4 | Plexin A4 |
| 81 | 0.511 | PCDHB4 | Protocadherin Beta 4 |
| 82 | 0.51 | TBC1D30 | TBC1 Domain Family Member 30 |
| 83 | 0.51 | PREX1 | Phosphatidylinositol-3,4,5-Trisphosphate Dependent Rac Exchange Factor 1 |
| 84 | 0.51 | MKS1 | Meckel Syndrome, Type 1 |
| 85 | 0.509 | ZNF768 | Zinc Finger Protein 768 |
| 86 | 0.509 | PARD6B | Par-6 Family Cell Polarity Regulator Beta |
| 87 | 0.509 | PABPC3 | Poly(A) Binding Protein Cytoplasmic 3 |
| 88 | 0.509 | GATC | Glutamyl-Trna Amidotransferase Subunit C |
| 89 | 0.508 | TRPC5OS | TRPC5 Opposite Strand |
| 90 | 0.508 | KREMEN2 | Kringle Containing Transmembrane Protein 2 |
| 91 | 0.508 | HOOK2 | Hook Microtubule Tethering Protein 2 |
| 92 | 0.507 | RPS6KB1 | Ribosomal Protein S6 Kinase B1 |
| 93 | 0.507 | CEACAM21 | Carcinoembryonic Antigen Related Cell Adhesion Molecule 21 |
| 94 | 0.507 | ABCA12 | ATP Binding Cassette Subfamily A Member 12 |
| 95 | 0.506 | MVK | Mevalonate Kinase |
| 96 | 0.505 | DHTKD1 | Dehydrogenase E1 And Transketolase Domain Containing 1 |
| No | Pearson Correlation Coefficient | Gene Symbol | Gene Name |
| 97 | 0.504 | TDRD5 | Tudor Domain Containing 5 |
| 98 | 0.504 | ENTHD2 | Tepsin |
| 99 | 0.503 | SLC29A2 | Solute Carrier Family 29 Member 2 |
| 100 | 0.501 | MRPL4 | Mitochondrial Ribosomal Protein L4 |
| 101 | 0.501 | MATK | Megakaryocyte-Associated Tyrosine Kinase |
| 102 | 0.501 | FBXW9 | F-Box And WD Repeat Domain Containing 9 |
| 103 | 0.501 | EIF3C | Eukaryotic Translation Initiation Factor 3 Subunit C |
| 104 | 0.5 | RXRA | Retinoid X Receptor Alpha |
| 105 | -0.505 | BLOC1S6 | Biogenesis Of Lysosomal Organelles Complex 1 Subunit 6 |
| 106 | -0.51 | NR2F6 | Nuclear Receptor Subfamily 2 Group F Member 6 |
| 107 | -0.514 | VANGL1 | VANGL Planar Cell Polarity Protein 1 |
| 108 | -0.564 | SEPT2 | Septin 2 |
Positive correlation coefficients indicate direct correlation to logIC50 value whereas negative correlation coeficients showed inverse correlation.
Gene ontology analysis of potential nobiletin target genes
Gene ontology analysis was classified into biological process, cellular component and molecular function (Table 3). There are no significant GO analysis results of the genes with a negative Pearson correlation coefficient. We found that the upregulated genes mostly involved in the biological process related to negative regulation of transcription, translational initiation, phosphatidylinositol 3-kinase signaling and cellular response to insulin stimulus. Moreover, the upregulated genes located in the cellular component of nuclear chromatin, cytosol and cytoplasm (e.g. ESR1 and NCOA3), and play a role in the molecular function of protein binding, transcriptional and translational activity, as well as steroid hormone activity, e.g. ESR1.
Table 3.
The Top Five Gene Ontology and KEGG Pathway Enrichment of DEGs, Analysed by DAVID
| ID | Term | Count | P value | Genes |
|---|---|---|---|---|
| Biological Process | ||||
| GO:0001731 | Formation of translation preinitiation complex | 3 | 0.006970337 | EIF3C, EIF4B, EIF3E |
| GO:0032869 | Cellular response to insulin stimulus | 4 | 0.008691289 | SP1, XBP1, APRT, PIK3R2 |
| GO:0014065 | Phosphatidylinositol 3-kinase signaling | 3 | 0.009534075 | XBP1, GATA3, PIK3R2 |
| GO:0006446 | Regulation of translational initiation | 3 | 0.016576226 | EIF3C, EIF4B, EIF3E |
| GO:0009267 | Cellular response to starvation | 3 | 0.027361724 | MAX, PPM1D, KIAA1324 |
| Cellular component | ||||
| GO:0000790 | Nuclear chromatin | 6 | 0.003712494 | SP1, NCOA3, SMARCD2, RXRA, GATA3, ESR1 |
| GO:0005829 | Cytosol | 29 | 0.006647911 | RHPN1, PREX1, STARD10, RPS6KB1, BLOC1S6, EIF3C, HOOK2, XBP1, EIF3E, FRAT1, AANAT, FRAT2, PABPC1, DHTKD1, PSMD6, PIK3R2, ABCA12, MATK, PARD6B, FBP1, LARP4B, APRT, EIF4B, TRIM37, MKS1, ALOX15, MVK, SIAH2, PDZD3 |
| GO:0005654 | Nucleoplasm | 25 | 0.009918189 | RAD51C, RPS6KB1, BLOC1S6, MAX, SMARCD2, XBP1, EIF3E, GATA3, NR2F6, PATZ1, PSMD6, SCAMP1, RXRA, ESR1, SPPL2B, BRIP1, TEAD2, ECSIT, APRT, RNF44, NCOA3, SP1, TUBD1, RFX1, SIAH2 |
| Molecular function | ||||
| GO:0005515 | Protein binding | 62 | 0.007869723 | RAD51C, SEPT2, PREX1, RPS6KB1, VPS37C, HOOK2, MAX, SMARCD2, GATA3, NR2F6, FRAT1, PSMD6, RMND1, DSCAM, MATK, SCAMP1, MRPL4, VANGL1, RXRA, ESR1, FBP1, ECSIT, TRIM37, MKS1, PPM1D, ALOX15, NCOA3, MVK, SIAH2, ITGA2B, GATC, RHPN1, STARD10, EIF3C, BLOC1S6, FBXW9, XBP1, EIF3E, AANAT, LETMD1, TFF1, PABPC1, APPBP2, TMEM183A, USP32, ABCA12, PIK3R2, PARD6B, SPTSSB, SPPL2B, BRIP1, TEAD2, NPY1R, LARP4B, CYB561, EIF4B, PLEKHF2, SP1, WDR25, RFX1, DYM, PDZD3 |
| GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding | 3 | 0.017092174 | RXRA, NR2F6, ESR1 |
| GO:0001046 | Core promoter sequence-specific DNA binding | 3 | 0.02389184 | SP1, GATA3, ESR1 |
| GO:0043565 | Sequence-specific DNA binding | 8 | 0.025500996 | MAX, IRX5, SP1, XBP1, RXRA, NR2F6, SPDEF, ESR1 |
| GO:0003707 | Steroid hormone receptor activity | 3 | 0.038898918 | RXRA, NR2F6, ESR1 |
| KEGG pathway enrichment analysis | ||||
| hsa03013 | RNA transport | 6 | 0.005016166 | EIF3C, EIF4B, EIF3E, PABPC3, PABPC1, NUP210L |
| hsa05222 | Small cell lung cancer | 4 | 0.017791047 | MAX, RXRA, ITGA2B, PIK3R2 |
| hsa04915 | Estrogen signaling pathway | 4 | 0.026525517 | GNAO1, SP1, ESR1, PIK3R2 |
| hsa04919 | Thyroid hormone signaling pathway | 4 | 0.038861334 | NCOA3, RXRA, ESR1, PIK3R2 |
| hsa04150 | mTOR signaling pathway | 3 | 0.054897616* | EIF4B, RPS6KB1, PIK3R2 |
| hsa03013 | RNA transport | 6 | 0.005016166 | EIF3C, EIF4B, EIF3E, PABPC3, PABPC1, NUP210L |
*, not significant
KEGG pathway enrichment, protein-protein interaction (PPI) network construction and module selection
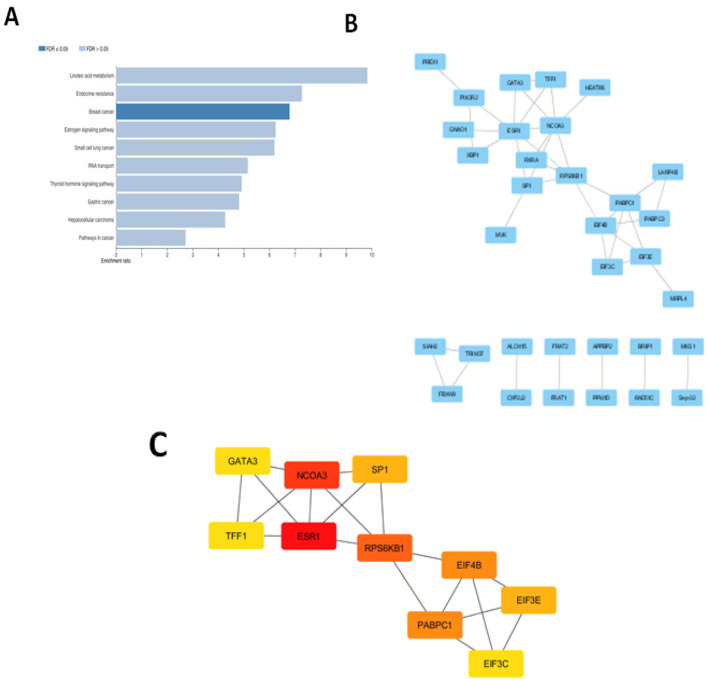
KEGG pathway enrichment indicated several pathways regulated by nobiletin (Table 3) such as RNA transport, small cell lung cancer, estrogen signaling pathway and thyroid hormone signaling pathway. Pathway enrichment analysed by WebGestalt showed breast cancer signaling regulated by nobiletin (Figure 2A). In addition, several genes involved in breast cancer regulation by targeting estrogen receptor and Wnt/ß-catenin signaling (Table 4). A total of 108 genes were constructed to PPI network complex containing 105 nodes and 40 edges, with average node degree 0.762 (Figure 2B). Three nodes with a degree score more than five were identified as hub genes, e.g. ESR1, NCOA3 and RPS6KB1 (Figure 2C and Table 5).
Figure 2.
(A), Pathway enrichment analysis of DEGs with Webgestalt; (B), Protein-protein interaction networks of DEGs, analyzed with STRING-DB and Cytoscape; (C), Top 10 hub genes with the highest degree score, analyzed by Cytoscape
Table 4.
DEGs Involved in Breast Cancer Regulation, Pathway Enrichment Analysis by WebGestalt
| User ID | Gene Symbol | Gene Name |
|---|---|---|
| RPS6KB1 | RPS6KB1 | ribosomal protein S6 kinase B1 |
| ESR1 | ESR1 | estrogen receptor 1 |
| PIK3R2 | PIK3R2 | phosphoinositide-3-kinase regulatory subunit 2 |
| SP1 | SP1 | Sp1 transcription factor |
| NCOA3 | NCOA3 | nuclear receptor coactivator 3 |
| FRAT1 | FRAT1 | FRAT1, WNT signaling pathway regulator |
| FRAT2 | FRAT2 | FRAT2, WNT signaling pathway regulator |
Table 5.
The Hub Genes Identified by PPI Networks, Possessing Degree more than 5
| Gene Symbol | Gene name | Degree score |
|---|---|---|
| ESR1 | estrogen receptor 1 | 8 |
| NCOA3 | nuclear receptor coactivator 3 | 7 |
| RPS6KB1 | ribosomal protein S6 kinase B1 | 6 |
Discussion
This study analyzed the molecular mechanism of nobiletin in CSCs using bioinformatics approaches. A pharmacological network analysis using bioinformatics approach can help to explain the potential target and mechanism of compounds in several diseases (Lee et al., 2018). Analysis of cytotoxicity with a public database of COMPARE showed that nobiletin exhibits cytotoxicity at the same level in NCI-60 cells panel showing by similar IC50 value. Nobiletin cytotoxicity does not depend on particular tissue. The low IC50 value indicates the potential of nobiletin for CSCs-targeted agents in combinatorial chemotherapy. The ideal compounds for combinatorial therapy should be potent, have low toxicity and selective (Wang et al., 2014).
COMPARE analysis identified 11 standard agents which have a correlation with nobiletin (Table 1). Tamoxifen, triciribine phosphate and 4-ipomeanol are standard drugs with the highest score of a Pearson correlation coefficient. Tamoxifen is a classical selective estrogen receptor modulator (SERM) for adjuvant chemotherapy of estrogen receptor-positive (Daurio et al., 2016). Tamoxifen activates tumor suppressor gene maspin in breast cancer (Liu et al., 2004). 4-ipomeanol, a lung-toxic furanoterpenoid produced by sweet potatoes (Ipomoea batatas) infected with the fungus Fusarium solani (Boyd and Wilson, 1972; Lakhanpal et al., 2001), is the first agent to undergo preclinical study at the National Cancer Institute (NCI) based on a specific biochemical-biological rationale for clinical investigation as an antineoplastic agent targeted lung cancer (Christian et al., 1989). Phase I and phase II clinical trial of 4-ipomeanol in patients with non-small cell lung cancer and advanced hepatocellular carcinoma, respectively showed that 4-ipomeanol is not recommended for those diseases (Kasturi et al., 1998; Lakhanpal et al., 2001). Triciribine, an inhibitor of Akt phosphorylation and activation, reduces CSC population in T-cell acute lymphoblastic leukemia cells (Evangelisti et al., 2011) and human breast cancer cells SKBR3 cells (Jain et al., 2015). Accordingly, nobiletin probably acts as a kinase inhibitor in inhibiting CSCs.
COMPARE analysis showed that FRAT1 and FRAT2 are genes with positive, while VANGL1 is genes with negative Pearson correlation coefficient, respectively. Those genes also involve in the Wnt/ß-catenin signaling pathway. The frequently rearranged in advanced T-cell lymphomas 1 (Frat 1) and 2 (Frat 2) are positively regulator of the Wnt signaling pathway by stabilizing ß-catenin through the association with GSK-3 (Saitoh et al., 2001). Upon binding to GSK3, Frat prevents the phosphorylation and accompanying degradation of ß-catenin and allows the activation of downstream target genes (van Amerongen and Berns, 2005; Luan et al., 2008). Wnt/ß-catenin signalling may be aberrantly activated through Frat1 overexpression in ovarian serous adenocarcinomas (Wang et al., 2006). The expression of Frat is also positively correlated with the degree of tumor differentiation and the abnormal cell expression of ß-catenin in lung cancer (Luan et al., 2008). Overexpression of Frat1 and abnormal expression of β-catenin were found to represent a poor prognosis for the non-small cell lung cancer patients (Zhang et al., 2012). Frat1 demonstrates oncogenic properties in prostate cancer by inhibiting GSK 3β against β-catenin and thus promoting cell growth (Zhang et al., 2016), while Frat2 mediates the oncogenic activation of Rac by mixed lineage leukemia fusions (Walf-Vorderwulbecke et al., 2012). VANGL1 encodes a transmembrane protein that interacts with Frizzeld a receptor of Wnt (Jenny et al., 2003) and negatively regulates canonical Wnt/β-catenin signaling in mammalian cells. FRAT1 and FRAT2 are tumor promoting genes whereas VANGL1 is a tumor suppressor gene which involved in the Wnt/ß-catenin signaling pathway and thus posses as a molecular target of nobiletin.
KEGG pathway enrichment analysis revealed that estrogen and Wnt/ß-catenin signaling are regulated by nobiletin. There is only a few studies on the role of estrogen in BCSCs. A study demonstrated that estrogen treatment reduces mammosphere formation from estrogen receptor-positive breast cancer cells (Simoes et al., 2011). Other studies showed that estrogen signaling blocking by tamoxifen induces chemoresistance due to EGFR and estrogen receptor cross talk (Shou et al., 2004). Expression of Wnt/ß-catenin signaling pathway-regulated genes correlates with estrogen receptor expression (Lamb et al., 2013). Activation of Wnt/ß-catenin signaling and CSCs properties are associated with advanced progression of ER-positive breast cancer (Sun et al., 2018). The Wnt/ß-catenin pathway is considered to be one of the most important pathways in the regulation of CSCs (Wang et al., 2016). A study showed that Wnt/ß-catenin and estrogen signaling pathways cross-talk in vivo through functional interaction between ERα and β-catenin (Kouzmenko et al., 2004). Therefore it is interesting to further explore the effect of nobiletin in estrogen and Wnt/ß-catenin signaling as well as its cross-talk in CSCs.
Pathway enrichment analysis with KEGG also showed the mTOR pathway regulated by nobiletin even the p-value is slightly greater than the cut off (p= 0.0548). The PI3K/Akt/mTOR signaling pathway is important for CSCs maintenance and could be a promising target for development of CSC-target drugs (Matsubara et al., 2013; Xia and Xu, 2015; Dandawate et al., 2016; Francipane and Lagasse, 2016). Rapamycin and triciribine target CSC population, and inhibits migration and invasion on glioblastoma and neuroblastoma cells (Bahmad et al., 2018). COMPARE analysis showed that triciribin is one of the compounds with the highest similarity to nobiletin, and therefore the effect of nobiletin in mTOR signaling is also potential to be further explored.
There are three hub genes identified from PPI networks, i.e. ESR1, NCOA3, RPS6KB1 ESR1 encodes estrogen receptor alpha which regulates estrogen signaling upon estrogen binding. Abnormal estrogen signaling leads to the development of a variety of diseases, such as cancer, metabolic and cardiovascular disease, neurodegeneration, inflammation, and osteoporosis (Jia et al., 2015). NCOA3 encodes nuclear receptor coactivator 3, a member of the nuclear receptor co-activator family known to be overexpressed in breast cancer and essentially involved in estrogen-mediated cancer cell proliferation (Wagner et al., 2013). Overexpression of NCOA3 promotes breast cancer chemoresistance to tamoxifen (Burwinkel et al., 2005) and paclitaxel (Ao et al., 2016). NCOA3 also drives the formation of cancer stem-like cells and supports tumor outgrowth in breast cancer models (Rohira et al., 2017). Moreover, NCOA3 is a selective co-activator of ERα-mediated transactivation of PLAC1, novel cancer-associated placental in MCF-7 breast cancer cells (Wagner et al., 2013). RPS6KB1 encodes ribosomal protein S6 kinase B1 which plays a key role in regulating protein translation and progression of hepatocellular carcinoma (Li et al., 2012), prostate cancer (Cai et al., 2015) and small cell lung cancer (Chen et al., 2017). S6K1 also activates ERα and promotes the proliferation of estrogen receptor-positive breast cancer cells (Holz, 2012). Taken together, those three genes regulate estrogen signaling in breast cancer and could be evaluated for further studies of marker and target genes of nobiletin in breast cancer stem cells.
Previous studies showed the role of nobiletin in estrogen, Wnt/ß-catenin and mTOR signaling. In estrogen signaling, nobiletin prevents bone loss due induced by estrogen deficiency in rats (Harada et al., 2011; Matsumoto et al., 2018) and inhibits lower cytotoxicity on MCF-7 estrogen receptor-positive breast cancer cells than on SKBR3 HER2 positive and MDA-MB 468 triple-negative breast cancer cells (Chen et al., 2014). Moreover, treatment of nobiletin in lower dose decreases activity and expression of aromatase on MCF-7 cells (Rahideh et al., 2017). In Wnt/ß-catenin signaling, nobiletin inhibits its signaling pathway in hypoxia stimulated Caki-1 and 786-O renal cell carcinoma (Liu et al., 2019), and inhibits invasion via inhibition of AKT/GSK3ß/ß-catenin pathway in glioblastoma cells (Zhang et al., 2017). Nobiletin shows inhibition of mTOR signaling on MDA-MB-468 triple-negative breast cancer cells (Chen et al., 2014). On the mTOR signaling pathway, nobiletin also protects cadmium-induced neurotoxicity induced by cadmium (Qu et al., 2018) and increases the sensitivity of colorectal cancer to oxiplatin (Li et al., 2019). Accordingly, those studies support the present study and enhance the development potential of nobiletin as CSCs-drugs by targeting estrogen, Wnt/ß-catenin and mTOR signaling.
This present study showed that nobiletin target estrogen signaling and Wnt/ß-catenin signaling. Protein interaction networks showed three hub genes regulates estrogen signaling. A previous study also showed functional interaction between estrogen and Wnt/ß-catenin signaling (Kouzmenko et al., 2004). Estradiol not only stimulates the estrogen signaling pathway but also increases the cancer stem cell (CSC) population in estrogen receptor-positive breast cancer cells (Kurebayashi et al., 2017). Treatment with hormone antagonist in estrogen receptor-positive breast cancer cells may repress their estrogen receptors and be resistant to hormone therapy (Simoes et al., 2015). However, a recent study showed that tamoxifen-resistant cells exhibit increased stemness properties via activation of Wnt/ß-catenin signaling (Leung et al., 2017). The interaction of CXC chemokine receptor type 4 (CXCR4) with its ligand CXC motif ligand 12 (CXCL12) plays important roles in maintaining CSCs properties in tamoxifen-resistant breast cancer cells (Dubrovska et al., 2012), nasopharyngeal CSCs (Tian et al., 2017), esophageal CSCs (Wang et al., 2017b), and stimulates the angiogenesis in vascular endothelial cells through upregulation of the MAPK/ERK and PI3K/AKT and Wnt/β-catenin pathways. (Song et al., 2018). A study showed that nobiletin decreases the expression of CXCR4 in breast cancer cells (Baek et al., 2012). Accordingly, nobiletin is potential to target CSCs by inhibiting estrogen and Wnt/ß-catenin signaling.
This present study has several limitations, including the mRNA data used for the PPI network. This might give different results because the expression of mRNA is not always correlated to the protein level. This study is also using bioinformatics approaches, therefore further in vitro and in vivo studies are needed to validate the results as well as to explore the full therapeutic potential of nobiletin on CSCs.
In conclusion, we found that tamoxifen, triciribine phosphate and 4-ipomeanol are standard drugs with the highest score of Pearson correlation coefficient to nobiletin. Moreover, many genes involved in protein binding, transcriptional and translational activity. Importantly, pathway enrichment analysis revealed breast cancer regulation of estrogen signaling and Wnt/ß-catenin by nobiletin. In addition, three hub genes, i.e. ESR1, NCOA3, and RPS6KB1 and one significant module were filtered out and selected from the PPI network. Taken together, using a bioinformatics approach, we showed that nobiletin might serve as a lead compound for the development of cancer stem cells-targeted drugs by targeting targets estrogen and Wnt/ß-catenin signaling.
Author contribution
AH-conception and design of the study, acquisition, analysis and interpretation of data, drafting and revising the article and final approval of the version to be published, HP-acquisition and analysis of data, drafting the article and final approval of the version to be published
Availability of material
The datasets analysed during the present study are online available in the public database.
Funding statement
This research did not receive any specific grants from funding agencies in the public, commercial, or not-for-profit sectors.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no conflict of interest.
References
- Ao X, Nie P, Wu B, et al. Decreased expression of microRNA-17 and microRNA-20b promotes breast cancer resistance to taxol therapy by upregulation of NCOA3. Cell Death Dis. 2016;7:e2463. doi: 10.1038/cddis.2016.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek SH, Kim SM, Nam D, et al. Antimetastatic effect of nobiletin through the down-regulation of CXC chemokine receptor type 4 and matrix metallopeptidase-9. Pharm Biol. 2012;50:1210–8. doi: 10.3109/13880209.2012.664151. [DOI] [PubMed] [Google Scholar]
- Bahmad H, Mouhieddine T, Chalhoub R, et al. PO-283 The Akt/mTOR pathway in cancer stem/progenitor cells is a therapeutic target for glioblastoma and neuroblastoma. ESMO Open. 2018;3:A131. doi: 10.18632/oncotarget.26088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd MR, Wilson BJ. Isolation and characterization of 4-ipomeanol, a lung-toxic furanoterpenoid produced by sweet potatoes (Ipomoea batatas) J Agric Food Chem. 1972;20:428–30. doi: 10.1021/jf60180a066. [DOI] [PubMed] [Google Scholar]
- Burwinkel B, Wirtenberger M, Klaes R, et al. Association of NCOA3 polymorphisms with breast cancer risk. Clin Cancer Res. 2005;11:2169–74. doi: 10.1158/1078-0432.CCR-04-1621. [DOI] [PubMed] [Google Scholar]
- Cai C, Chen QB, Han ZD, et al. miR-195 inhibits tumor progression by targeting RPS6KB1 in human prostate cancer. Clin Cancer Res. 2015;21:4922–34. doi: 10.1158/1078-0432.CCR-15-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Yang L, Zhang R, et al. Hyperphosphorylation of RPS6KB1, rather than overexpression, predicts worse prognosis in non-small cell lung cancer patients. PLoS One. 2017;12:e0182891. doi: 10.1371/journal.pone.0182891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ono M, Takeshima M, et al. Antiproliferative and apoptosis-inducing activity of nobiletin against three subtypes of human breast cancer cell lines. Anticancer Res. 2014;34:1785–92. [PubMed] [Google Scholar]
- Christian MC, Wittes RE, Leyland-Jones B, et al. 4-Ipomeanol: a novel investigational new drug for lung cancer. J Natl Cancer Inst. 1989;81:1133–43. doi: 10.1093/jnci/81.15.1133. [DOI] [PubMed] [Google Scholar]
- Dandawate PR, Subramaniam D, Jensen RA, et al. Targeting cancer stem cells and signaling pathways by phytochemicals: Novel approach for breast cancer therapy. Semin Cancer Biol. 2016;40-41:192–208. doi: 10.1016/j.semcancer.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daurio NA, Tuttle SW, Worth AJ, et al. AMPK Activation and metabolic reprogramming by Tamoxifen through Estrogen receptor-independent mechanisms suggests new uses for this therapeutic modality in cancer treatment. Cancer Res. 2016;76:3295–306. doi: 10.1158/0008-5472.CAN-15-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovska A, Hartung A, Bouchez LC, et al. CXCR4 activation maintains a stem cell population in tamoxifen-resistant breast cancer cells through AhR signalling. Br J Cancer. 2012;107:43. doi: 10.1038/bjc.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelisti C, Ricci F, Tazzari P, et al. Preclinical testing of the Akt inhibitor triciribine in T-cell acute lymphoblastic leukemia. J Cell Physiol. 2011;226:822–31. doi: 10.1002/jcp.22407. [DOI] [PubMed] [Google Scholar]
- Francipane MG, Lagasse E. Therapeutic potential of mTOR inhibitors for targeting cancer stem cells. Br J Clin Pharmacol. 2016;82:1180–8. doi: 10.1111/bcp.12844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada S, Tominari T, Matsumoto C, et al. Nobiletin, a polymethoxy flavonoid, suppresses bone resorption by inhibiting NFkappaB-dependent prostaglandin E synthesis in osteoblasts and prevents bone loss due to estrogen deficiency. J Pharmacol Sci. 2011;115:89–93. doi: 10.1254/jphs.10193sc. [DOI] [PubMed] [Google Scholar]
- Hermawan A, Putri H. Current report of natural product development against breast cancer stem cells. Int J Biochem Cell Biol. 2018;104:114–32. doi: 10.1016/j.biocel.2018.09.012. [DOI] [PubMed] [Google Scholar]
- Holz MK. The role of S6K1 in ER-positive breast cancer. Cell Cycle. 2012;11:3159–65. doi: 10.4161/cc.21194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain MV, Jangamreddy JR, Grabarek J, et al. Nuclear localized Akt enhances breast cancer stem-like cells through counter-regulation of p21(Waf1/Cip1) and p27(kip1) Cell Cycle. 2015;14:2109–20. doi: 10.1080/15384101.2015.1041692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny A, Darken RS, Wilson PA, et al. Prickle and Strabismus form a functional complex to generate a correct axis during planar cell polarity signaling. EMBO J. 2003;22:4409–20. doi: 10.1093/emboj/cdg424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia M, Dahlman-Wright K, Gustafsson JA. Estrogen receptor alpha and beta in health and disease. Best Pract Res Clin Endocrinol Metab. 2015;29:557–68. doi: 10.1016/j.beem.2015.04.008. [DOI] [PubMed] [Google Scholar]
- Kasturi VK, Dearing MP, Piscitelli SC, et al. Phase I study of a five-day dose schedule of 4-Ipomeanol in patients with non-small cell lung cancer. Clin Cancer Res. 1998;4:2095. [PubMed] [Google Scholar]
- Kouzmenko AP, Takeyama K, Ito S, et al. Wnt/beta-catenin and estrogen signaling converge in vivo. J Biol Chem. 2004;279:40255–8. doi: 10.1074/jbc.C400331200. [DOI] [PubMed] [Google Scholar]
- Kurebayashi J, Koike Y, Ohta Y, et al. Anti-cancer stem cell activity of a hedgehog inhibitor GANT61 in estrogen receptor-positive breast cancer cells. Cancer Sci. 2017;108:918–30. doi: 10.1111/cas.13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakhanpal S, Donehower RC, Rowinsky EK. Phase II study of 4-ipomeanol, a naturally occurring alkylating furan, in patients with advanced hepatocellular carcinoma. Invest New Drugs. 2001;19:69–76. doi: 10.1023/a:1006408803734. [DOI] [PubMed] [Google Scholar]
- Lamb R, Ablett MP, Spence K, et al. Wnt pathway activity in breast cancer sub-types and stem-like cells. PLoS One. 2013;8:e67811–e. doi: 10.1371/journal.pone.0067811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AY, Park W, Kang T-W, et al. Network pharmacology-based prediction of active compounds and molecular targets in Yijin-Tang acting on hyperlipidaemia and atherosclerosis. J Ethnopharmacol. 2018;221:151–9. doi: 10.1016/j.jep.2018.04.027. [DOI] [PubMed] [Google Scholar]
- Leung EY, Askarian-Amiri ME, Sarkar D, et al. Endocrine therapy of Estrogen receptor-positive breast cancer cells: Early differential effects on stem cell markers. Front Oncol. 2017;7:184. doi: 10.3389/fonc.2017.00184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Zhang Z, Jiang G, et al. Nobiletin sensitizes colorectal cancer cells to oxaliplatin by PI3K/Akt/MTOR pathway. Front Biosci. 2019;24:303–12. doi: 10.2741/4719. [DOI] [PubMed] [Google Scholar]
- Li PD, Zhang WJ, Zhang MY, et al. Overexpression of RPS6KB1 predicts worse prognosis in primary HCC patients. Med Oncol. 2012;29:3070–6. doi: 10.1007/s12032-012-0268-y. [DOI] [PubMed] [Google Scholar]
- Liu F, Zhang S, Yin M, et al. Nobiletin inhibits hypoxia-induced epithelial-mesenchymal transition in renal cell carcinoma cells. J Cell Biochem. 2019;120:2039–46. doi: 10.1002/jcb.27511. [DOI] [PubMed] [Google Scholar]
- Liu Z, Shi HY, Nawaz Z, et al. Tamoxifen induces the expression of maspin through estrogen receptor-alpha. Cancer Lett. 2004;209:55–65. doi: 10.1016/j.canlet.2003.11.018. [DOI] [PubMed] [Google Scholar]
- Luan L, Zhang Y, Guo N, et al. Expressions of Frat and beta-catenin in lung cancer and their clinicopathological correlations. Zhongguo Fei Ai Za Zhi. 2008;11:62–6. doi: 10.3779/j.issn.1009-3419.2008.01.015. [DOI] [PubMed] [Google Scholar]
- Mahmoud N, Saeed MEM, Sugimoto Y, et al. Cytotoxicity of nimbolide towards multidrug-resistant tumor cells and hypersensitivity via cellular metabolic modulation. Oncotarget. 2018;9:35762–79. doi: 10.18632/oncotarget.26299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara S, Ding Q, Miyazaki Y, et al. mTOR plays critical roles in pancreatic cancer stem cells through specific and stemness-related functions. Sci Rep. 2013;3:3230. doi: 10.1038/srep03230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto S, Tominari T, Matsumoto C, et al. Effects of Polymethoxyflavonoids on bone loss induced by Estrogen deficiency and by LPS-dependent inflammation in Mice. Pharmaceuticals. 2018:11. doi: 10.3390/ph11010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiyanto E, Hermawan A, Anindyajati Natural products for cancer-targeted therapy: citrus flavonoids as potent chemopreventive agents. Asian Pac J Cancer Prev. 2012;13:427–36. doi: 10.7314/apjcp.2012.13.2.427. [DOI] [PubMed] [Google Scholar]
- Meiyanto E, Hermawan A, Junedi S, et al. Nobiletin increased cytotoxic activity of doxorubicin on MCF-7 cells but not on T47D cells. Int J Phytomedicine. 2011;3:129–37. [Google Scholar]
- Moltzahn FR, Volkmer JP, Rottke D, et al. “Cancer stem cells”-lessons from Hercules to fight the Hydra. Urol Oncol. 2008;26:581–9. doi: 10.1016/j.urolonc.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Monks A, Scudiero DA, Johnson GS, et al. The NCI anti-cancer drug screen: a smart screen to identify effectors of novel targets. Anticancer Drug Des. 1997;12:533–41. [PubMed] [Google Scholar]
- Morley KL, Ferguson PJ, Koropatnick J. Tangeretin and nobiletin induce G1 cell cycle arrest but not apoptosis in human breast and colon cancer cells. Cancer Lett. 2007;251:168–78. doi: 10.1016/j.canlet.2006.11.016. [DOI] [PubMed] [Google Scholar]
- Ohnishi H, Asamoto M, Tujimura K, et al. Inhibition of cell proliferation by nobiletin, a dietary phytochemical, associated with apoptosis and characteristic gene expression, but lack of effect on early rat hepatocarcinogenesis in vivo. Cancer Sci. 2004;95:936–42. doi: 10.1111/j.1349-7006.2004.tb03180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Ma S, Cao K, et al. Therapeutic approaches targeting cancer stem cells. J Cancer Res Ther. 2018;14:1469–75. doi: 10.4103/jcrt.JCRT_976_17. [DOI] [PubMed] [Google Scholar]
- Qu Y, Liu Y, Chen L, et al. Nobiletin prevents cadmium-induced neuronal apoptosis by inhibiting reactive oxygen species and modulating JNK/ERK1/2 and Akt/mTOR networks in rats. Neurol Res. 2018;40:211–20. doi: 10.1080/01616412.2018.1424685. [DOI] [PubMed] [Google Scholar]
- Rahideh ST, Keramatipour M, Nourbakhsh M, et al. Comparison of the effects of nobiletin and letrozole on the activity and expression of aromatase in the MCF-7 breast cancer cell line. Biochem Cell Biol. 2017;95:468–73. doi: 10.1139/bcb-2016-0206. [DOI] [PubMed] [Google Scholar]
- Rohira AD, Yan F, Wang L, et al. Targeting SRC coactivators blocks the tumor-initiating capacity of cancer stem-like cells. Cancer Res. 2017;77:4293–304. doi: 10.1158/0008-5472.CAN-16-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Moriwaki J, Koike J, et al. Molecular cloning and characterization of FRAT2, encoding a positive regulator of the WNT signaling pathway. Biochem Biophys Res Commun. 2001;281:815–20. doi: 10.1006/bbrc.2001.4421. [DOI] [PubMed] [Google Scholar]
- Shou J, Massarweh S, Osborne CK, et al. Mechanisms of tamoxifen resistance: increased estrogen receptor-HER2/neu cross-talk in ER/HER2-positive breast cancer. J Natl Cancer Inst. 2004;96:926–35. doi: 10.1093/jnci/djh166. [DOI] [PubMed] [Google Scholar]
- Simoes BM, Alferez DG, Howell SJ, et al. The role of steroid hormones in breast cancer stem cells. Endocr Relat Cancer. 2015;22:177–86. doi: 10.1530/ERC-15-0350. [DOI] [PubMed] [Google Scholar]
- Simoes BM, Piva M, Iriondo O, et al. Effects of estrogen on the proportion of stem cells in the breast. Breast Cancer Res Treat. 2011;129:23–35. doi: 10.1007/s10549-010-1169-4. [DOI] [PubMed] [Google Scholar]
- Song Z-Y, Wang F, Cui S-X, et al. Knockdown of CXCR4 inhibits CXCL12-induced angiogenesis in HUVECs through downregulation of the MAPK/ERK and PI3K/AKT and the Wnt/β-Catenin pathways. Cancer Invest. 2018;36:10–8. doi: 10.1080/07357907.2017.1422512. [DOI] [PubMed] [Google Scholar]
- Sun X, Xu C, Xiao G, et al. Breast cancer stem-like cells are sensitized to tamoxifen induction of self-renewal inhibition with enforced Let-7c dependent on Wnt blocking. Int J Mol Med. 2018;41:1967–75. doi: 10.3892/ijmm.2018.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:447–52. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takanaga H, Ohnishi A, Yamada S, et al. Polymethoxylated flavones in orange juice are inhibitors of P-glycoprotein but not cytochrome P450 3A4. J Pharmacol Exp Ther. 2000;293:230–6. [PubMed] [Google Scholar]
- Tian Y, Song Y, Bai W, et al. CXCR4 knockdown inhibits the growth and invasion of nasopharyngeal cancer stem cells. Oncol Lett. 2017;13:2253–9. doi: 10.3892/ol.2017.5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Guzman ME, Bigoni-Ordonez GD, Ibanez Hernandez M, et al. Cancer stem cell impact on clinical oncology. World J Stem Cells. 2018;10:183–95. doi: 10.4252/wjsc.v10.i12.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Amerongen R, Berns A. Re-evaluating the role of Frat in Wnt-signal transduction. Cell Cycle. 2005;4:1065–72. [PubMed] [Google Scholar]
- Wagner M, Koslowski M, Paret C, et al. NCOA3 is a selective co-activator of estrogen receptor alpha-mediated transactivation of PLAC1 in MCF-7 breast cancer cells. BMC Cancer. 2013;13:570. doi: 10.1186/1471-2407-13-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walf-Vorderwulbecke V, de Boer J, Horton SJ, et al. Frat2 mediates the oncogenic activation of Rac by MLL fusions. Blood. 2012;120:4819–28. doi: 10.1182/blood-2012-05-432534. [DOI] [PubMed] [Google Scholar]
- Wang J, Vasaikar S, Shi Z, et al. WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017a;45:W130–w7. doi: 10.1093/nar/gkx356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Wang Z, Peng C, et al. Dietary compound isoliquiritigenin targets GRP78 to chemosensitize breast cancer stem cells via beta-catenin/ABCG2 signaling. Carcinogenesis. 2014;35:2544–54. doi: 10.1093/carcin/bgu187. [DOI] [PubMed] [Google Scholar]
- Wang R, Sun Q, Wang P, et al. Notch and Wnt/beta-catenin signaling pathway play important roles in activating liver cancer stem cells. Oncotarget. 2016;7:5754–68. doi: 10.18632/oncotarget.6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Cao Y, Zhang S, et al. Stem cell autocrine CXCL12/CXCR4 stimulates invasion and metastasis of esophageal cancer. Oncotarget. 2017b;8:36149–60. doi: 10.18632/oncotarget.15254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Hewitt SM, Liu S, et al. Tissue microarray analysis of human FRAT1 expression and its correlation with the subcellular localisation of beta-catenin in ovarian tumours. Br J Cancer. 2006;94:686–91. doi: 10.1038/sj.bjc.6602988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P, Xu X-Y. PI3K/Akt/mTOR signaling pathway in cancer stem cells: from basic research to clinical application. Am J Cancer Res. 2015;5:1602–9. [PMC free article] [PubMed] [Google Scholar]
- Yoshimizu N, Otani Y, Saikawa Y, et al. Anti-tumour effects of nobiletin, a citrus flavonoid, on gastric cancer include: antiproliferative effects, induction of apoptosis and cell cycle deregulation. Aliment Pharmacol Ther. 2004;20:95–101. doi: 10.1111/j.1365-2036.2004.02082.x. [DOI] [PubMed] [Google Scholar]
- Zhang W, Xiong H, Zou Y, et al. Frequently rearranged in advanced Tcell lymphomas1 demonstrates oncogenic properties in prostate cancer. Mol Med Rep. 2016;14:3551–8. doi: 10.3892/mmr.2016.5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zheng K, Li C, et al. Nobiletin inhibits invasion via inhibiting AKT/GSK3beta/beta-catenin signaling pathway in Slug-expressing glioma cells. Oncol Rep. 2017;37:2847–56. doi: 10.3892/or.2017.5522. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Han Y, Zheng R, et al. Expression of Frat1 correlates with expression of beta-catenin and is associated with a poor clinical outcome in human SCC and AC. Tumour Biol. 2012;33:1437–44. doi: 10.1007/s13277-012-0394-3. [DOI] [PubMed] [Google Scholar]
- Zhu P, Fan Z. Cancer stem cells and tumorigenesis. Biophys Rep. 2018;4:178–88. doi: 10.1007/s41048-018-0062-2. [DOI] [PMC free article] [PubMed] [Google Scholar]