Abstract
Recent studies have displayed that circular RNA plays a key regulatory role in tumorigenesis and development. However, evidence supporting the critical role of circular RNA in the prognosis of colorectal cancer (CRC) is still limited. This study was designed to screen and identify novel circular RNA biomarkers in CRC. The microarray analysis of circular RNA expression profile was performed in three matched CRC and normal tissues. Compared with normal mucosa, a total of 208 differentially expressed circular RNAs were found in CRC, of which 95 were upregulated and 113 were downregulated. Then, the top 10 circular RNAs with the most significant differential expression were selected for verification by quantitative polymerase chain reaction (qPCR) in the above samples. The results of qPCR basically coincided with the findings of microarray analysis. The hsa_circ_022382 expression was the most significant upregulation in CRC among the 10 circular RNAs validated by qPCR. Because hsa_circ_022382 is derived from the human FADS2 gene, we named it circFADS2. Next, the increased expression of circFADS2 was further confirmed by qPCR, and its correlation with clinicopathologic parameters was analyzed in 200 CRC tissues. Herein, the expression of circFADS2 was shown to increase in 187 cases and decrease in 13 cases of CRC, and the elevated circFADS2 expression was closely related to the size, differentiation, infiltration depth, lymphatic and distant metastasis, and tumor/node/metastasis (TNM) stage of CRC. The survival analyses by Kaplan–Meier method demonstrated that patients with higher circFADS2 expression levels had shorter overall survival time and vice versa. Cox regression and area under ROC curve analyses revealed that the circFADS2 expression may be a promised biomarker for prognosis of CRC patients and had better prediction value when combined with TNM stage. In conclusion, our study identifies circFADS2 as an oncogenic biomarker and a potential prognostic factor in CRC.
Impact statement
Colorectal cancer (CRC) is the third most common malignancy worldwide with the second highest mortality rate. Although multidisciplinary cooperative therapies are helpful for improving the survival of CRC patients, the prognosis remains poor. Therefore, it is imperative to seek new biomarkers for the development of individualized treatment for each CRC patient. Circular RNA, an endogenous transcript with specific covalent closed loop, exhibits higher stability, conservation and expression abundance than the corresponding linear component and thus may be utilized as a promised biomarker. Although the majority of studies have focused on circular RNA expression profiling in various types of cancers, evidence supporting their critical role in the diagnosis and prognosis of CRC is limited. This study aimed to screen and identify novel circular RNA biomarkers of CRC by chip analysis and qPCR verification, and to highlight their potential as targets for CRC prognosis, and therapy.
Keywords: circFADS2, biomarkers, colorectal cancer, cancer progression, prognosis
Introduction
Colorectal cancer (CRC) is the third most common diagnosis and second deadliest malignancy worldwide.1 Although multidisciplinary cooperative therapies, including radiotherapy, neo-adjuvant chemotherapy, and surgery, were helpful to improve the patients’ survival,2 the prognosis of CRC patients remains poor.3 Therefore, it is urgent to seek novel biomarkers to develop individualized treatment for each CRC patient.
Circular RNA is a specific endogenous transcript with covalent closed loop.4 Due to their circular structures, circular RNAs can resist exonuclease digestion and are more conservative and stable than the corresponding linear components.5 These characters indicate their obvious advantages as biomarkers.6 Currently, circular RNAs have been shown to play crucial regulatory roles in multiple physiological or pathological processes, such as development, differentiation, and tumorigenesis.7–9 Aberrant expression profiling of circular RNAs has been utilized to characterize colorectal cancer at the molecular level, and several circular biomarkers have been identified in various studies.10,11 These biomarkers significantly contributed to elucidating the molecular mechanism of CRC carcinogenesis. However, their role in evaluating the prognosis of colorectal cancer is unclear. And so far, circular RNA biomarkers for CRC prognosis have rarely been identified. Therefore, a continuous search for novel specific circular RNA biomarkers for CRC progression and prognosis would be vital to improve treatment effect and prognostic assessment of CRC.
In this study, the circular RNAs expression profiling of three pairs of CRC and normal tissues was assessed by microarray technology. Real-time fluorescence quantitative polymerase chain reaction (qPCR) determination was then used to test and verify the findings of microarray analysis. And then, the expression of circFADS2, which was the one with the most upregulation, was further validated in 200 pairs of CRC and normal tissues, and the correlations between circFADS2 expression and individual clinicopathologic parameter of CRC patients were also investigated.
Materials and methods
Patients and tissue samples
A total of 200 patients with primary CRC who underwent radical resection without preoperative radiotherapy and chemotherapy from January 2013 to September 2013 at Jiangxi Provincial People’s Hospital Affiliated to Nanchang University (Nanchang, China) were included in the study. The participants were followed up for five years. The paired primary CRC and matched mucosal tissues were obtained and immediately frozen in liquid nitrogen. Histological diagnosis was independently made by two experienced pathologists in a double-blind way according to World Health Organization criteria. The relevant clinical data, including gender, age, tumor size, differential grade, the status of invasion, lymphatic and distant metastasis, and tumor/node/metastasis (TNM) stage were presented in Table 1. Written informed consent was obtained from all participants, and the research protocols were approved by the Ethics Committee of Jiangxi Provincial People’s Hospital Affiliated to Nanchang University.
Table 1.
The relationship between circFADS2 and clinicopathological parameters in patients with CRC.
| Variables | Total number (n = 200) | circFADS2 expressiona | P value |
|---|---|---|---|
| Ages (years) | |||
| <60 | 86 | 10.680 ± 0.505 | 0.498 |
| ≥60 | 114 | 10.143 ± 0.616 | |
| Gender | |||
| Male | 141 | 10.504 ± 0.475 | 0.829 |
| Female | 59 | 10.318 ± 0.686 | |
| Tumor size (cm) | |||
| <5 | 84 | 10.114 ± 0.967 | 0.014 |
| ≥5 | 116 | 11.261 ± 0.505 | |
| Differentiation | |||
| Well or moderately | 128 | 9.780 ± 0.512 | 0.022 |
| Poorly | 72 | 11.639 ± 0.568 | |
| Depth of tumor invasion | |||
| T1, T2 | 38 | 7.652 ± 0.956 | 0.000* |
| T3, T4 | 162 | 11.105 ± 0.412 | |
| Lymphatic metastasis | |||
| No | 55 | 7.866 ± 0.717 | 0.000* |
| Yes | 145 | 11.429 ± 0.440 | |
| Distant metastasis | |||
| No | 178 | 9.970 ± 0.394 | 0.000* |
| Yes | 22 | 14.730 ± 1.248 | |
| TNM Stage | |||
| I, II | 67 | 7.851 ± 0.662 | 0.000* |
| III, IV | 133 | 11.758 ± 0.443 | |
aIt is showed by mean ± standard deviation.
*Statistically significant difference (P < 0.05).
Circular RNA microarray
Three paired CRC and paracancerous mucosal tissues were selected for circular RNA microarray analysis and their clinic data were shown in Table 2. Comprehensive analysis of circular RNA expression profile was performed using the Arraystar Human Circular RNA Array V2.0 (8x15K, Arraystar Inc., Rockville, MD) with 13,400 probes. NanoDrop ND-1000 was used to quantify total RNA from individual sample. Sample preparation and microarray hybridization were carried out according to standard procedures of the Arraystar. To be brief, RNase R was used to remove linear RNA from total RNA (Epicentre Inc., Madison, WI). The enriched circular RNA was then amplified and transcribed into fluorescent cRNA by random primer method (Arraystar Super RNA Labeling Kit; Arraystar Inc.). The fluorescent cRNA was then hybridized with the Arraystar Human circular RNA Array V2.0. The arrays were scanned with an Agilent Scanner G2505C after washing the slides. The array images were then acquired and analyzed using Agilent Feature Extraction software (version 11.0.1.1). The R software limma package was used for Quantile normalization and subsequent data processing. The circular RNAs differentially expressed between the two groups were determined by fold-change filtration. Circular RNAs with fold-changes ≥2 and P values ≤0.05 were designed as differentially expressed ones. Hierarchical clustering was carried out to present the difference of circular RNA expression among samples. Microarray hybridization and bioinformatics analysis were completed by KangChen Bio-Tech, Inc. (Shanghai, China).
Table 2.
The clinicopathological parameters of the three pairs CRC tissues used for the analysis of circular RNAs expression profiles in microarray.
| Patient No. | Age | Gender | Tumor size | Tumor differentiation | Invasion depth | LN metastasis | Distant metastasis | TNM stage | Survival months |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 68 | Male | 2.7 | Poor | T2 | No | No | T2N0M0 | 60 |
| 2 | 63 | Male | 5.1 | Poor | T4 | Yes | No | T4N2Mo | 49 |
| 3 | 54 | Male | 7.6 | Moderate | T4 | Yes | Yes | T4N2M1 | 25 |
Validation of microarray results by real-time fluorescence qPCR
Preliminary expression verification of the top 10 dysregulated circular RNAs, including the expression of top seven upregulated and top three downregulated ones, was conducted by qPCR using the above three pairs of tissues for chip detection. To reduce experimental deviation caused by too fewer samples, we further validated the expression of the circular RNA with the most significant difference by qPCR in 200 matched CRC and normal tissues. And the correlations between the expression of the one with the most significant difference and the clinic pathological parameters were analyzed in 200 pairs of CRC and normal tissues.
Total RNAs from 200 paired CRC and paracancerous mucosa tissues were divided into two equal parts after extracting with TRIzol reagent (Life Technology., Carlsbad, CA), and one part served as control, the other was treated with RNase R to eliminate linear RNA and collect circular RNA. Then cDNA was synthesized using oligo (dT) and Random hexamer in a Prime Script® RT Reagent Kit (TaKaRa Biotech, Dalian, Liaoning, China) for the former and the latter, respectively. qPCR determination was conducted using an SYBR® Premix EX Taq real-time PCR kit (TaKaRa Biotech, Dalian, Liaoning, China) with the following reaction procedure: thermal denaturation at 95°C for 30 s, followed by 40 cycles of amplification (thermal denaturation at 95°C for 5 s, annealing at 60°C for 20 s, extension at 72°C for 40 s), and extension at 72°C for 5 min. Melt curve analyses were used to validate the amplification of the single product. Each PCR reaction system was prepared according to the instructions of the kit. The sequences of circular RNAs were from the Arraystar Human circular RNA database (Arraystar Inc.).
To amplify the reverse splice site of each circular RNA, PCR primers were designed in divergent orientation using primer premier software version 5.0 (Premier Biosoft International, USA) and synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). To check if linear RNA species were removed completely or not, we also designed the corresponding linear FADS2 mRNA amplification primers and measured the FADS2 mRNA expression levels by qPCR before and after RNase R treatment. All the primer sequences were displayed in Table 3. The relative expression of each circular RNA in CRC compared with normal tissue was calculated according to the equation of 2−△△Ct with GAPDH as an internal control.12 Each experiment was conducted in triplicate.
Table 3.
Quantitative polymerase chain reaction primers and products sizes.
| Amplicon | Primers | Best transcript | Gene symbol | Product length |
|---|---|---|---|---|
| hsa_circ_101622 | F: 5'CAGATGGACAAAGTGGAGCA3' | NM_18689 | CEMIP | 131bp |
| R: 5'GCTGGGGAAGTCATTCTTGA3' | ||||
| hsa_circ_022382 | F: 5'GGATGGCTGCAACATGATTA3' | NM_004265 | FADS2 | 157bp |
| R: 5'TCAGCAGGGGTTTCAAGAAC3' | ||||
| hsa_circ_100833 | F: 5'GCCAAGCCTAACATCTTCCA3' | NM_004265 | FADS2 | 145bp |
| R: 5'TCAGCAGGGGTTTCAAGAAC3' | ||||
| hsa_circ_101621 | F: 5'ACCCACCCACATACATCAGG3' | NM_018689 | CEMIP | 132bp |
| R: 5'ATGTCCACGCCGTCTATCTC3' | ||||
| hsa_circ_008068 | F: 5'TGATGAAGCTTTGCGAAGAA3' | NM_032116 | KATNAL1 | 137bp |
| R: 5'CGTAATTTCCAAGAAGGGCATA3' | ||||
| hsa_circ_051749 | F: 5'TCATCTTCCTCGCCAGCTAC3' | NM_000836 | GRIN2D | 142bp |
| R: 5'AGTGAGGCACATGACGAACA3' | ||||
| hsa_circ_407279 | F: 5'CCCAGTGGAGGCTGTTCTAC3' | NM_138780 | SYTL5 | 130bp |
| R: 5'AAGATTCCCCCAAGGAATTC3' | ||||
| hsa_circ_406503 | F: 5'AGTTGTGGGGTGGTCCTCTG3' | NR_037884 | LOC100507053 | 170bp |
| R: 5'TAGCAGAAAACTGCGGGTCT3' | ||||
| hsa_circ_406549 | F: 5'CTGGTTCCTCAGCTCTCCAC3' | NM_000901 | NR3C2 | 169bp |
| R: 5'GGATCATCTGTTTGCCTGCT3' | ||||
| hsa_circ_406015 | F: 5'CGGACAGCCTCTCAGTCTCT3' | NM_015265 | SATB2 | 197bp |
| R: 5'GTTGTCGGTGTCGAGGTTTT3' | ||||
| GAPDH mRNA | F: 5'GGGAAACTGTGGCGTGAT3' | NM_001289746 | GAPDH | 299bp |
| R: 5'GAGTGGGTGTCGCTGTTGAC3' | ||||
| FADS2 mRNA | F: 5'ACGCTGGAGAAGATG3' | NM_004265 | FADS2 | 307bp |
| R: 5'GCCGTGATGAGGGTAG3' |
F: forward; R: reverse; Chr: chromosome; bp: base pairs.
Statistical analysis
SPSS software version 25.0 (SPSS, Inc., Chicago, IL) was used to statistically analyze the data of this study. Paired t-test was used to analyze the qPCR data of circular RNA expression in matched CRC and normal tissues. Independent-sample t-test was applied to evaluate the correlation between circFADS2 expression and each clinicopathologic variable. Kaplan–Meier method and Log-rank test were used for survival analyses of patients with CRC. The significance of various variables for survival was evaluated by the Cox proportional hazards model in univariate and multivariate analyses. Receiver operating characteristic (ROC) curve was employed to evaluate the prediction value of each clinical variable of patients with CRC. P value less than 0.05 was regarded as a statistically significant difference.
Results
Circular RNA microarray analysis
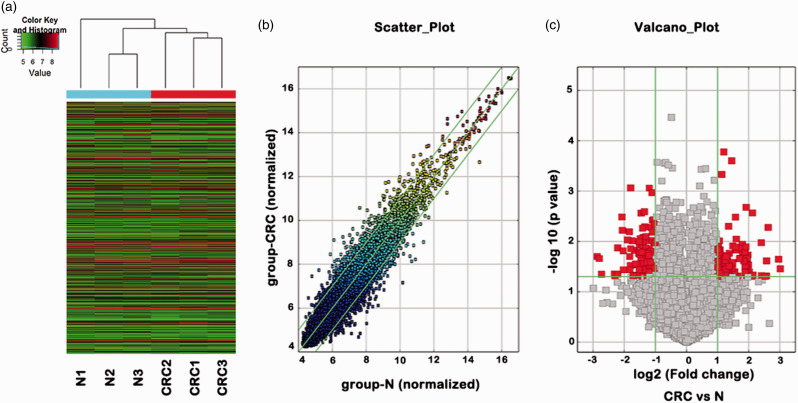
Totally 6719 circular RNAs were detected in the three matched CRC and adjacent mucosal tissues using the circular RNA microarray. Hierarchical clustering of expressions of the circular RNAs indicated the cancer tissues are separated from the normal tissues (Figure 1(a)). The normalized expression values of the circular RNAs from the normal tissues (x-axis in Figure 1(b)) are compared with those from the cancer tissues (y-axis in Figure 1(b)), indicating some circular RNAs had differential expressions between the cancer and normal tissues. Using the criteria of fold change more than 2 and P value less than 0.05, 208 circular RNAs (red points in Figure 1(c)) were determined as differentially expressed circular RNAs.
Figure 1.
Expression profile analyses of circular RNAs in colorectal cancer and normal mucosal tissues based on microarray determination. (a) Unsupervised hierarchical cluster analysis of the circular RNA expression from three matched colorectal cancer (CRC1–CRC3) and normal tissues (N1–N3). Circular RNAs in red corresponded to elevated expression and those in green corresponded to decreased expression. (b) Scatter plot of the circular RNA expression levels based on microarray analysis. The transverse and longitudinal coordinates represent the normal group and the colorectal cancer group, respectively. The diagonals on both sides represent 2.0-fold increased or decreased expression. (c) Volcano plot of the circular RNA expression levels based on microarray analysis. The squares on the left and right of the two vertical green lines represent the circular RNAs of 2.0-fold decreased or increased expression, respectively, and the squares above the horizontal green line correspond to the circular RNAs with P < 0.05. The red points represent differentially expressed circular RNAs with statistical significance. N: normal colorectal mucosal tissue, CRC: colorectal cancer tissue. (A color version of this figure is available in the online journal.)
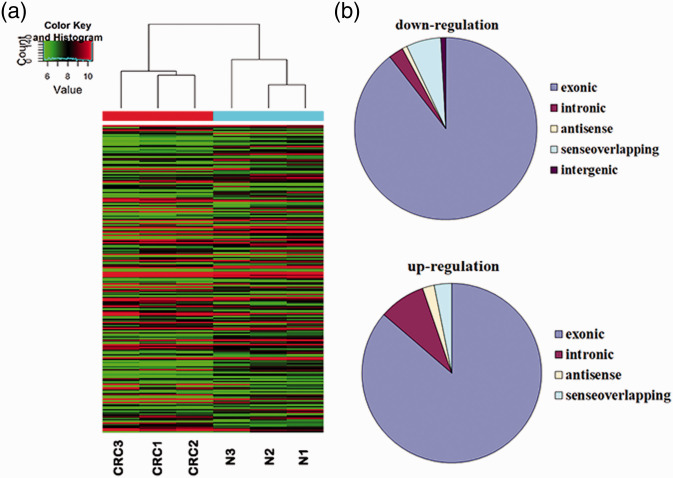
Hierarchical clustering of expressions of 208 differentially expressed circular RNAs separated colorectal cancer tissues more clearly from the normal tissues (Figure 2(a)). Of all 208 differentially expressed circular RNAs, 95 were upregulated and 113 were downregulated in CRC, including exonic, intronic, antisense, sense overlapping, and intergenic circular RNA types (Figure 2(b)). The top 30 upregulated and downregulated circular RNAs in CRC compared with adjacent tissues are displayed in Table 4. These data indicated that there was discrimination of circular RNAs between CRC and normal mucosal tissues.
Figure 2.
Differentially expressed circular RNA between CRC and normal mucosa in microarray analysis. (a) Hierarchical clustering of the differentially expressed circular RNA from three matched colorectal cancer (CRC1–CRC3) and normal mucosa (N1–N3). Circular RNA in red showed elevated expression and those in green represented decreased expression. (b) Pie chart of statistical analysis of differential expressed circular RNA types. (A color version of this figure is available in the online journal.)
Table 4.
The top 15 up- and downregulated circular RNA in CRC based on chip analysis.
| Circular RNA | Alias | Fold change | P value | circRNA type | Chr | Gene symbol |
|---|---|---|---|---|---|---|
| Upregulation | ||||||
| hsa_circRNA_101622 | hsa_circRNA_0003893 | 8.0674284 | 0.035 | Exonic | Chr15 | CEMIP |
| hsa_circRNA_022382 | hsa_circRNA_0022382 | 7.7660942 | 0.022 | Exonic | Chr11 | FADS2 |
| hsa_circRNA_100833 | hsa_circRNA_0022383 | 6.160305 | 0.005 | Exonic | Chr11 | FADS2 |
| hsa_circRNA_101621 | hsa_circRNA_0002970 | 5.9554927 | 0.024 | Exonic | Chr15 | CEMIP |
| hsa_circRNA_008068 | hsa_circRNA_0008068 | 5.929514 | 0.048 | Exonic | Chr13 | KATNAL1 |
| hsa_circRNA_051749 | hsa_circRNA_0051749 | 5.7423023 | 0.049 | Exonic | Chr19 | GRIN2D |
| hsa_circRNA_407279 | 5.2961367 | 0.047 | Exonic | ChrX | SYTL5 | |
| hsa_circRNA_100834 | hsa_circRNA_0022392 | 5.2855338 | 0.010 | Exonic | Chr11 | FADS2 |
| hsa_circRNA_104740 | hsa_circRNA_0086444 | 4.389346 | 0.046 | Exonic | Chr9 | HAUS6 |
| hsa_circRNA_406192 | 4.3474706 | 0.049 | Intronic | Chr22 | MYH9 | |
| hsa_circRNA_105039 | hsa_circRNA_0091934 | 4.3345499 | 0.003 | Exonic | ChrX | FLNA |
| hsa_circRNA_023525 | hsa_circRNA_0023525 | 4.0475354 | 0.039 | Exonic | Chr11 | UCP2 |
| hsa_circRNA_000968 | hsa_circRNA_0000968 | 3.970826 | 0.029 | Intronic | Chr19 | ZNF544 |
| hsa_circRNA_087497 | hsa_circRNA_0087497 | 3.9146361 | 0.018 | Exonic | Chr9 | IARS |
| hsa_circRNA_084900 | hsa_circRNA_0084900 | 3.8772586 | 0.021 | Exonic | Chr8 | KIAA1429 |
| Downregulation | ||||||
| hsa_circRNA_406503 | 7.3157415 | 0.020 | Sense Overlapping | Chr4 | LOC100507053 | |
| hsa_circRNA_406549 | 6.9883733 | 0.022 | Exonic | Chr4 | NR3C2 | |
| hsa_circRNA_406015 | 6.633301 | 0.045 | Exonic | Chr2 | SATB2 | |
| hsa_circRNA_071127 | hsa_circRNA_0071127 | 4.9722271 | 0.044 | Exonic | Chr4 | NR3C2 |
| hsa_circRNA_005297 | hsa_circRNA_0005297 | 4.8990873 | 0.048 | Exonic | Chr3 | MRPL47 |
| hsa_circRNA_007379 | hsa_circRNA_0007379 | 4.6240731 | 0.013 | Intergenic | Chr14 | |
| hsa_circRNA_019225 | hsa_circRNA_0019225 | 4.6210812 | 0.044 | Exonic | Chr10 | PLCE1 |
| hsa_circRNA_402150 | 4.2079122 | 0.003 | Exonic | Chr2 | SNTG2 | |
| hsa_circRNA_101631 | hsa_circRNA_0003856 | 4.1660114 | 0.006 | Exonic | Chr15 | PDE8A |
| hsa_circRNA_008882 | hsa_circRNA_0008882 | 4.0867121 | 0.015 | Sense Overlapping | chrM | MTND5 |
| hsa_circRNA_101036 | hsa_circRNA_0025765 | 4.0745242 | 0.036 | Exonic | Chr12 | TMTC1 |
| hsa_circRNA_101632 | hsa_circRNA_0005889 | 3.7135218 | 0.009 | Exonic | Chr15 | PDE8A |
| hsa_circRNA_103310 | hsa_circRNA_0064648 | 3.5704159 | 0.006 | Exonic | Chr3 | RBMS3 |
| hsa_circRNA_070521 | hsa_circRNA_0070521 | 3.5386452 | 0.020 | Exonic | Chr4 | SLC39A8 |
| hsa_circRNA_404385 | 3.5302 | 0.036 | Exonic | chrX | CHMP1B2P | |
Validation of microarray results by qPCR
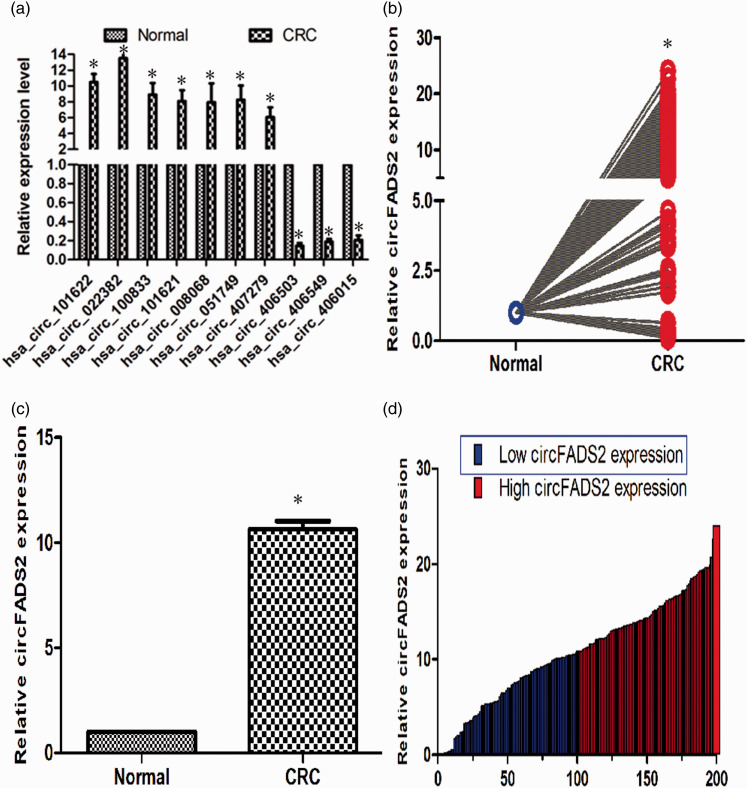
To validate the results of circular RNA microarray analysis, the top 10 dysregulated circular RNAs (using the standards of fold change more than 5 and P value less than 0.05), including seven upregulated and three downregulated ones (Table 4) were further assessed by qPCR. The very low FADS2 mRNA expression level suggested that the linear RNAs were removed completely after RNase R treatment (Figure S1). The results of circular RNAs expression by qPCR validation in the three pairs CRC and normal tissues coincided with those of the chip analysis, except that circFADS2 was the top 1 upregulated one in the results of qPCR validation (Figure 3(a)), whereas it was the top 2 in the results of chip analysis. The results of further validation in 200 pairs of tissues by qPCR showed that the elevated circFADS2 expression was detected in 93.5% (187/200) CRC cases, and reduced expression was observed in the other 6.5% (13/200) CRC cases. The highest and lowest relative expression levels of circFADS2 in colorectal cancer were 24.091 and 0.103 times that of normal tissues, respectively (P < 0.01, Figure 3(b)). And the average and median relative expression levels of circFADS2 in colorectal cancer were 10.600 ± 0.383 and 10.697 times that of normal tissues, respectively (P < 0.01, Figure 3(c) and (d)). The comparison between high circFADS2 expression (folds > 10.697) and low circFADS2 expression (folds < 10.697) was shown in Figure 3(d). The above results indicate that the expression of circFADS2 in CRC tissues is significantly higher than that in normal tissues.
Figure 3.
qPCR validation analysis of the circular RNA expression in matched CRC and adjacent tissues. (a) qPCR verification analysis of the top 10 differential expressed circular RNA from microarray data. A total of seven upregulated circular RNA and three downregulated ones, which fold change >5 and P < 0.05, were validated (n = 3, *P < 0.01). (b) The average relative hsa_circ_022382 (named circFADS2) expression of CRC compared with corresponding normal tissues by qPCR (n = 200, *P < 0.01). (c) Relative circFADS2 level in CRC tissues compared with normal tissues (n = 200). (d) Relative circFADS2 expression of individual paired CRC and normal tissue (n = 200, *P < 0.01). The blue and red bar represent low and high circFADS2 expression, respectively. All experiments were performed in triplicates. GAPDH was used to normalize as reference gene. The data statistical significance is assessed by paired t-test. That P value less than 0.05 is considered statistically significant. (A color version of this figure is available in the online journal.)
Analyses of the correlations between circFADS2 expression and clinicopathological parameters
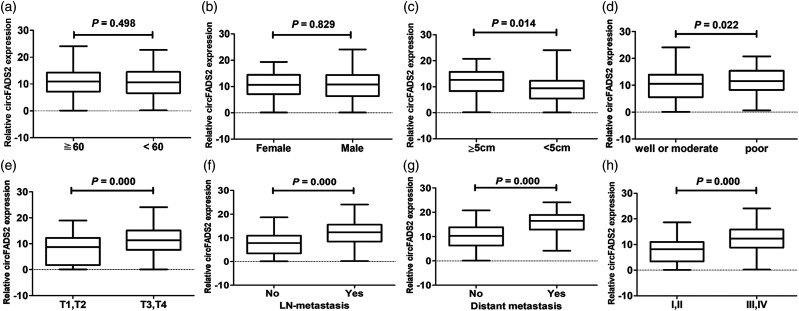
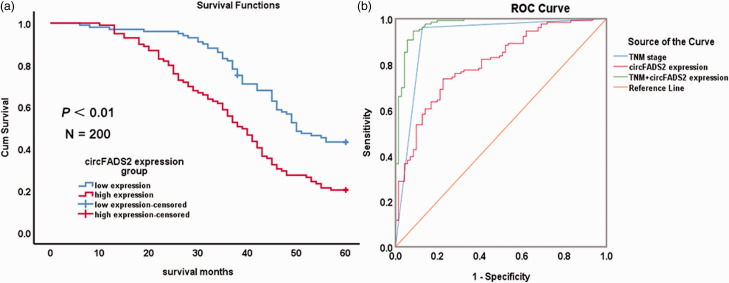
The correlations between circFADS2 expression and clinic pathological parameters of patients with CRC are summarized in Table 1. Among 200 cases of colorectal cancer, 145 were lymph node metastasis and 22 were liver metastasis. The circFADS2 expression had no significant correlation with age (P = 0.843, Figure 4(a)) and gender (P = 0.696, Figure 4(b)). However, there were significant correlation of the circFADS2 expression with tumor size (P = 0.014, Figure 4(c)), differentiation grade (P = 0.022, Figure 4(d)), depth of invasion (P = 0.000, Figure 4(e)), lymphatic metastasis (P = 0.000, Figure 4(f)), distant metastasis (P = 0.000, Figure 4(g)), and TNM stage (P = 0.000, Figure 4(h)). Kaplan–Meier survival curve showed that the patients with high circFADS2 expression had shorter overall survival time than those with low circFADS2 expression and vice versa, as compared with the log-rank statistical method (P = 0.000; Figure 5(a)). Univariate Cox regression analysis demonstrated that high circFADS2 expression is closely correlated with high risk of death due to CRC (Table 5). Moreover, multivariate Cox regression analysis revealed that in addition to distant metastasis (P = 0.000) and TNM stage (P = 0.016), circFADS2 expression (P = 0.023) was another independent predictor for CRC prognosis (Table 5). ROC curve analyses indicated that the area under the ROC curve of circFADS2 expression (0.803) was smaller than that of TNM stage (0.917) and lymphatic metastasis (0.876), but apparently higher than that of invasion depth (0.691) and distant metastasis (0.585; Table S1). However, the area under the ROC curve of the combined circFADS2 expression and TNM stage model (0.973) was significantly greater than that of the circFADS2 expression alone and TNM stage alone models (Figure 5(b) and Table 6). For CRC prognosis, the specificity and sensitivity of circFADS2 expression were 77.5% and 73.6%, which was markedly lower than those of TNM stage (96.1% and 99.87%, respectively). The specificity and sensitivity of the combined circFADS2 expression and TNM stage model (96.1% and 99.87%, respectively) were the same as that of TNM stage alone model. The above findings suggest that high circFADS2 expression may be closely related to the progress of CRC and poor prognosis, and that the combination of circFADS2 expression and TNM stage can better predict the prognosis of CRC patients than TNM stage alone.
Figure 4.
Comparison of relative circFADS2 expression between the two stratifications of each clinic pathologic parameter (n = 200). (a) Comparison between the two age stratifications as ≥ 60 years and <60 years. (b) Comparison between the two gender stratifications as male and female. (c) Comparison between the two tumor size stratifications as ≥ 5 cm and < 5 cm. (d) Comparison between the two tumor differentiation stratifications as well/moderate and poor. (e) Comparison between the two tumor invasion depth stratifications as T1, T2 and T3, T4. (f) Comparison between the two lymph node metastasis stratifications as yes and no. (g) Comparison between the two distal metastasis stratifications as yes and no. (h) Comparison between the two TNM stratifications as I, II and III, IV. All experiments were performed in triplicates. GAPDH was used to normalize as reference gene. The data statistical significance is assessed by independent t-test. That P value less than 0.05 is considered statistically significant.
Figure 5.
Comparison of the sensitivity and specificity of various parameter of patients with colorectal cancer for survival prediction. (a) Kaplan–Meier survival analysis of hsa_circ_022382 (circFADS2) expression of CRC patients (N = 200, log-rank test, P = 0.000). Red curve represents the survival of patients with high circFADS2 expression, blue curve represents the survival of patients with low expression of circFADS2. (b) Comparisons of the predictive values of CRC prognosis by the TNM stage and/or circFADS2 expression model. The red line represents ROC curve of circFADS2 expression model, the blue line represents ROC curve of TNM model, the green line represents ROC curve of the combined model of circFADS2 expression and TNM stage. (A color version of this figure is available in the online journal.)
Table 5.
Univariate and multivariate Cox regression analyses of the individual parameter for the correlations with overall survival rate of patients with CRC (n = 200).
| Parameters |
Univariate |
Multivariate |
||||
|---|---|---|---|---|---|---|
| RR | 95% CI for RR | P value | RR | 95% CI for RR | P value | |
| Gender | 1.007 | 0.992–1.023 | 0.355 | 0.998 | 0.978–1.019 | 0.881 |
| Age | 1.408 | 0.958–2.068 | 0.081 | 1.152 | 0.755–1.758 | 0.512 |
| Tumor size | 1.551 | 1.367–1.758 | 0.000* | 1.008 | 0.859–1.181 | 0.925 |
| Differentiation | 1.918 | 1.363–2.298 | 0.000* | 1.025 | 0.682–1.542 | 0.904 |
| Depth of invasion | 7.570 | 3.529–16.238 | 0.000* | 1.399 | 0.406–4.821 | 0.595 |
| LN metastasis | 70.668 | 17.003–293.710 | 0.000* | 113,161.467 | 0.000–7.696E+59 | 0.857 |
| Distant metastasis | 24.300 | 13.926–42.402 | 0.000* | 9.407 | 4.816–18.373 | 0.000* |
| TNM stage | 1.118 | 1.078–1.160 | 0.000* | 1.054 | 1.010–1.099 | 0.016* |
| circFADS2 expression | 62.734 | 22.508–174.852 | 0.000* | 6.228 | 1.287–30.131 | 0.023* |
RR: relative risk; CI: confidence interval.
*Statistically significant difference (P < 0.05).
Table 6.
Area under the curve of TNM or/and circFADS2.
| Test result variable(s) | Area | SEa | Asymptotic Sig.b |
Asymptotic 95% Confidence interval |
|
|---|---|---|---|---|---|
| Lower bound | Upper bound | ||||
| TNM stage | 0.917 | 0.025 | 0.000 | 0.868 | 0.967 |
| circFADS2 | 0.803 | 0.031 | 0.000 | 0.742 | 0.865 |
| TNM stage + circFADS2 | 0.973 | 0.012 | 0.000 | 0.950 | 0.996 |
The test result variable(s): TNM stage has at least one tie between the positive actual state group and the negative actual state group. Statistics may be biased.
(a) Under the nonparametric assumption.
(b) Null hypothesis: true area = 0.5.
Discussion
Mounting evidence has demonstrated that circular RNAs are linked to multiple human cancers, such as colorectal, bladder, lung, and hepatocellular cancer.11,13–15 Aberrant expression of circular RNA plays a vital role in various pathological processes of cancer by regulating multiple signaling pathways such as the Wnt/β-catenin, MAPK/ERK, AKT1/mTOR, and EGFR/STAT3 signaling pathways.16–19 They function as ceRNAs that regulate cancer cell proliferation, invasion, EMT, and metastasis because of their capability of sponging cancer-related miRNAs.15–21 In addition, some circular RNAs can “sponge” RNA binding proteins,22 whereas others may regulate parental gene transcription.5,23 To date, some synthetic circular RNAs have been shown to impart remarkable anti-cancer effects,24,25 suggesting that they may also be potentially utilized in the treatment of specific chronic diseases.26
Recent researches have demonstrated the involvement of multiple circular RNAs in the occurrence and development of CRC.27,28 Xie et al. reported the important role of hsa_circ_001569 in the proliferation and invasion of CRC.29 The downregulation of hsa_circ_0003906 is significantly correlated with lymphatic metastasis and poor differentiation of CRC.30 However, little is known about the prognostic circular RNAs biomarkers of CRC.
In present study, the landscape of circular RNAs in CRC tissues was obtained by circular chip analysis. More than 208 differentially expressed circular RNAs, including 113 downregulated and 95 upregulated ones, whose fold changes more than 2 and P values less than 0.05, were identified between paired CRC and adjacent normal tissues. Among these, 11 circular RNAs with differentially expressed fold changes >5 were found, and the top 7 upregulated as well as the top 3 downregulated ones were chosen for qPCR validation. The results of qPCR coincided with those of microarray, except that circFADS2 was the top 2 upregulated one in chip analysis, while it was the top 1 upregulated one in qPCR validation. Further analysis demonstrated a remarkable increase of circFADS2 expression in 93.5% (187/200) of the CRC cases, and the increased circFADS2 expression was notably correlated with large size, poor differentiation, deep invasive depth, lymphatic and distant metastasis, and late TNM stage of CRC. The results of Kaplan–Meier survival analyses suggested that the patients with high circFADS2 expression had shorter overall survival time than those with low circFADS2 expression and vice versa. Univariate and multivariate cox regression analyses indicated that circFADS2 expression can be utilized as an independent predictor for CRC prognosis, and ROC curve analysis revealed that circFADS2 expression has better prognostic value when combined with the TNM stage. So, circFADS2 is a novel potential biomarker of CRC prognosis. Regrettably, a limitation in this study is that the circFADS2 expression in other tumors should be tested to investigate whether it is specific in colorectal cancer.
Conclusion
Our chip analysis and qPCR verification of circular RNAs revealed that many circular RNAs are dysregulated in CRC tissues, as compared with that of the corresponding normal mucosa. Moreover, circFADS2 is the most significant upregulated circular RNA in CRC tissues. The elevated circFADS2 expression was closely related to large size, poor differentiation, deep invasive depth, lymphatic and distant metastasis, late TNM stage, and poor outcome of CRC. Therefore, the increased circFADS2 expression may play a key regulatory role in CRC malignant progress, and circFADS2 could be utilized as a novel prognostic biomarker and a potential therapeutic target for CRC. Further work is required for clarifying the underlying regulatory mechanism of the involvement of circFADS2 in the progress and prognosis of CRC.
Supplemental Material
Supplemental material, sj-pdf-1-ebm-10.1177_1535370220929965 for CircFADS2: A potential prognostic biomarker of colorectal cancer by Yi-Sheng Xiao, Hua-Zhang Tong, Xi-Hong Yuan, Chang-Hui Xiong, Xiao-Yang Xu and Yuan-Feng Zeng in Experimental Biology and Medicine
Supplemental material, sj-pdf-2-ebm-10.1177_1535370220929965 for CircFADS2: A potential prognostic biomarker of colorectal cancer by Yi-Sheng Xiao, Hua-Zhang Tong, Xi-Hong Yuan, Chang-Hui Xiong, Xiao-Yang Xu and Yuan-Feng Zeng in Experimental Biology and Medicine
Authors’ contributions
All authors were involved in the project design, interpretation of the results, and review of the manuscript; YSX, HZT, XHY, and XYX conducted the experiments. CHX performed the statistical analyses of experimental data. YFZ wrote and revised the manuscript. All authors gave their final approval of the submitted and published versions. YSX and HZT contributed equally to this work. YFZ is the corresponding author.
DECLARATION OF CONFLICTING INTERESTS
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
ETHICAL APPROVAL
The research protocols of the study were approved by the Ethics Committee of Jiangxi Provincial People’s Hospital Affiliated to Nanchang University.
FUNDING
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by grants from the National Natural Science Foundation of China [No. 81960432] and the Natural Science Foundation of Jiangxi Province, China [No. 20181BAB205051].
ORCID iD
Yuan-Feng Zeng https://orcid.org/0000-0003-4174-3575
SUPPLEMENTAL MATERIAL
Supplemental material for this article is available online.
REFERENCES
- 1.Recio-Boiles A, Waheed A, Cagir B. Cancer, colon. Treasure Island, FL: StatPearls, 2020 [Google Scholar]
- 2.Guye ML, Schoellhammer HF, Chiu LW, Kim J, Lai LL, Singh G. Designing liver resections and pushing the envelope with resections for hepatic colorectal metastases. Indian J Surg Oncol 2013; 4:349–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coppede F, Lopomo A, Spisni R, Migliore L. Genetic and epigenetic biomarkers for diagnosis, prognosis and treatment of colorectal cancer. World J Gastroenterol 2014; 20:943–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A 1976; 73:3852–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013; 495:333–8 [DOI] [PubMed] [Google Scholar]
- 6.Wang F, Nazarali AJ, Ji S. Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am J Cancer Res 2016; 6:1167–76 [PMC free article] [PubMed] [Google Scholar]
- 7.Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, Jiang C, Parast MM, Murry CE, Laurent LC, Salzman J. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol 2015; 16:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siede D, Rapti K, Gorska AA, Katus HA, Altmuller J, Boeckel JN, Meder B, Maack C, Volkers M, Muller OJ, Backs J, Dieterich C. Identification of circular RNAs with host gene-independent expression in human model systems for cardiac differentiation and disease. J Mol Cell Cardiol 2017; 109:48–56 [DOI] [PubMed] [Google Scholar]
- 9.He J, Xie Q, Xu H, Li J, Li Y. Circular RNAs and cancer. Cancer Lett 2017; 396:138–44 [DOI] [PubMed] [Google Scholar]
- 10.Chen S, Zhang L, Su Y, Zhang X. Screening potential biomarkers for colorectal cancer based on circular RNA chips. Oncol Rep 2018; 39:2499–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeng Y, Xu Y, Shu R, Sun L, Tian Y, Shi C, Zheng Z, Wang K, Luo H. Altered expression profiles of circular RNA in colorectal cancer tissues from patients with lung metastasis. Int J Mol Med 2017; 40:1818–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001; 25:402–8 [DOI] [PubMed] [Google Scholar]
- 13.Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep 2016; 6:30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao J, Li L, Wang Q, Han H, Zhan Q, Xu M. Circular RNA expression profile in early-stage lung adenocarcinoma patients. Cell Physiol Biochem 2017; 44:2138–46 [DOI] [PubMed] [Google Scholar]
- 15.Zhang J, Chang Y, Xu L, Qin L. Elevated expression of circular RNA circ_0008450 predicts dismal prognosis in hepatocellular carcinoma and regulates cell proliferation, apoptosis, and invasion via sponging miR-548p. J Cell Biochem 2019; 120:9487–94 [DOI] [PubMed] [Google Scholar]
- 16.Huang G, Zhu H, Shi Y, Wu W, Cai H, Chen X. cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/beta-catenin pathway. PloS One 2015; 10:e0131225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao D, Qi X, Zhang X, Fang K, Guo Z, Li L. hsa_circular RNA_0006528 as a competing endogenous RNA promotes human breast cancer progression by sponging miR-7-5p and activating the MAPK/ERK signaling pathway. Mol Carcinog 2019; 58:554–64 [DOI] [PubMed] [Google Scholar]
- 18.Liu L, Liu FB, Huang M, Xie K, Xie QS, Liu CH, Shen MJ, Huang Q. Circular RNA ciRS-7 promotes the proliferation and metastasis of pancreatic cancer by regulating miR-7-mediated EGFR/STAT3 signaling pathway. Hepatobiliary Pancreat Dis Int: 2019; 18:580–6 [DOI] [PubMed] [Google Scholar]
- 19.Zhang X, Wang S, Wang H, Cao J, Huang X, Chen Z, Xu P, Sun G, Xu J, Lv J, Xu Z. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer 2019; 18:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren S, Xin Z, Xu Y, Xu J, Wang G. Construction and analysis of circular RNA molecular regulatory networks in liver cancer. Cell Cycle 2017; 16:2204–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Wan B, Liu L, Zhou L, Zeng Q. Circular RNA circMTO1 suppresses bladder cancer metastasis by sponging miR-221 and inhibiting epithelial-to-mesenchymal transition. Biochem Biophys Res Commun 2019; 508:991–6 [DOI] [PubMed] [Google Scholar]
- 22.Hentze MW, Preiss T. Circular RNAs: splicing’s enigma variations. EMBO J 2013; 32:923–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013; 19:141–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bak RO, Hollensen AK, Mikkelsen JG. Managing microRNAs with vector-encoded decoy-type inhibitors. Mol Ther 2013; 21:1478–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haraguchi T, Ozaki Y, Iba H. Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res 2009; 37:e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lei B, Tian Z, Fan W, Ni B. Circular RNA: a novel biomarker and therapeutic target for human cancers. Int J Med Sci 2019; 16:292–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li XN, Wang ZJ, Ye CX, Zhao BC, Li ZL, Yang Y. RNA sequencing reveals the expression profiles of circular RNA and indicates that circDDX17 acts as a tumor suppressor in colorectal cancer. J Exp Clin Cancer Res 2018; 37:325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen LY, Zhi Z, Wang L, Zhao YY, Deng M, Liu YH, Qin Y, Tian MM, Liu Y, Shen T, Sun LN, Li JM. NSD2 circular RNA promotes metastasis of colorectal cancer by targeting miR-199b-5p-mediated DDR1 and JAG1 signalling. J Pathol 2019; 248:103–15 [DOI] [PubMed] [Google Scholar]
- 29.Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y, Yang S, Zeng Z, Liao W, Ding YQ, Liang L. Emerging roles of circular RNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget 2016; 7:26680–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhuo F, Lin H, Chen Z, Huang Z, Hu J. The expression profile and clinical significance of circular RNA0003906 in colorectal cancer. OncoTargets Ther 2017; 10:5187–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-ebm-10.1177_1535370220929965 for CircFADS2: A potential prognostic biomarker of colorectal cancer by Yi-Sheng Xiao, Hua-Zhang Tong, Xi-Hong Yuan, Chang-Hui Xiong, Xiao-Yang Xu and Yuan-Feng Zeng in Experimental Biology and Medicine
Supplemental material, sj-pdf-2-ebm-10.1177_1535370220929965 for CircFADS2: A potential prognostic biomarker of colorectal cancer by Yi-Sheng Xiao, Hua-Zhang Tong, Xi-Hong Yuan, Chang-Hui Xiong, Xiao-Yang Xu and Yuan-Feng Zeng in Experimental Biology and Medicine