Abstract
Rheumatoid arthritis (RA) is a systemic autoimmune disease characterized by chronic destructive joint disease. To date, the etiology and pathogenesis of RA have not been fully elucidated, but a large number of studies have indicated that hypoxia is an important feature of RA. Our study was designed to probe how hypoxia-induced exosome (exo) derived from synovial fibroblasts (SFs) affect RA. In this study, we found that hypoxic environment existed in synovial tissue of RA, and miR-424 expression was increased in RA, and exosome derived from synovial fibroblasts (SFs-exo) could significantly induce T cells differentiation, which Th17 cells increased and Treg cells decreased. Besides, SFs-exo affected the expression of related inflammatory cytokines. And, we also found that FOXP3 was a target gene of miR-424 and exo-miR-424 KD inhibited RA worsening. These results suggested that SFs-exo in hypoxia aggravates rheumatoid arthritis by regulating Treg/Th17 balance and thus may be a potential therapeutic target for RA.
Impact statement
A comparative study of osteoarthritis (OA) and RA mice was implemented to suggest that miR-424 expression was increased in RA, and exosome-miR-424 derived from synovial fibroblasts (SFs-exo) could significantly induce T cells differentiation in which Th17 cells increased and Treg cells decreased via targeting FOXP3. And thus, miR-424 may be a potential therapeutic target for RA.
Keywords: Hypoxia, exosome, rheumatoid arthritis, Th17/Treg cells
Introduction
Rheumatoid arthritis (RA) is a common clinical disease, which is widely distributed around the world. Statistically, the incidence of RA is 0.32% to 0.36% in China and 0.5–1% globally,1,2 and the incidence of RA in females is two to four times than that in males.3 The main features of RA are chronic, symmetrical, and progressive polyarthritis, which is manifested by chronic inflammation of synovial, cell infiltration, synovial pannus formation, invasion of cartilage and bone tissue, and destruction of joint structure, accompanied by joint systemic damage.4 RA belongs to autoimmune inflammatory diseases. RA lesions mainly exist in the synovial joints, such as hands, feet, and knee joints.5 Joint redness, swelling, heat and pain, and dysfunction are common symptoms in the early stage, and joints with varying degrees of stiffness may appear in the late stage, accompanied by bone destruction and atrophy of skeletal muscle, which can easily cause disability6 and seriously affect people’s quality of life. Besides, RA complications include pleurisy, pericarditis, and vasculitis, which increase the risk of cancer in severe cases,7 leading to death and bring heavy economic burden on society and families.8 The prevalence and harmfulness of RA make it urgent to explore its pathogenesis. Recent studies have found that Th17/Treg cells and cytokine profiles are induced by mononuclear cells from RA patients.9 As a type of autoimmune disease, RA progression requires the involvement of autoimmune CD4 + T cells. Th17 cells, the CD4 + T cells that secret IL-17 (IL-17A), contributes to the progression of RA by mediating inflammatory response.10–12 In contrast to Th17 cells, Tregs cells function in preserving immunological unresponsiveness to self-constituents and negative control of immune responses to attenuate the progression of RA.13 Moreover, the development of RA is associated with Th17/Treg imbalance.14
Exosomes are bioactive vesicles originating from late endosomes which were secreted by different types of cells to promote intercellular communication.15 It is reported that exosomes encapsulate various biomolecules such as protein markers (CD9, CD63 and CD81), single- and double-stranded DNA, various RNAs (mRNA, miRNA) and long non-coding RNA (lncRNA) from cells.16,17 Studies have shown that exosomes involve in the pathogenesis of RA and play important roles in antigen expression, inflammation, angiogenesis, and cell signaling.18 Exosomes transmit proteins and miRNAs with immunoregulatory properties between cells. Related exosomes have been examined in different joint diseases, suggesting that exosomes can act as potential biomarkers for diagnosis or assessment of RA activity.19,20 Recent studies proved that exosomal miR-155 inhibited the production of TNF-α, MMP-13 in SFs.21 In SFs of RA patients, miR146a expression can be up-regulated by TNF-α and IL-1β, thereby inhibiting pro-inflammatory cytokines secretion.22 In addition, miR-424 could regulate cells proliferation and the release of inflammatory factors.23 So, exosomes could be new targets on regulating the cell interaction related pathogenesis of RA.
Hypoxic microenvironment is an important pathological feature of RA.24 The high metabolic demand of inflammatory synovial tissue and rapid proliferation of synovial membrane together lead to hypoxic state in RA joints. As early as 1970, Lund-Olesen reported that the joint cavity of RA patients was hypoxic.25 Hypoxia promotes the pathological progression of RA by hypoxia-inducible factor-1 (HIF-1) to induce inflammation.26 However, whether hyperoxia could influence the Th17/Treg balance is still unknown.
In this research, we investigated the role of SFs-exo in the pathogenesis of RA. Herein, SFs-exo promoted T cell differentiation, where Th17 cells count was raised, but Treg cells count was dropped. Our study suggested that exosomal miR-424 inhibited inflammatory factors’ expression by targeting FOXP3. Our study provides new therapeutic clues and potential intervention targets for the treatment of RA.
Materials and methods
Rheumatoid arthritis and osteoarthritis model establishment
Balb/c mice (male, six to eight weeks) were purchased from Charles River (Beijing, China) to establish rheumatoid arthritis model as previously reported.27,28 Simply, 1 mg of inactivated Mycobacterium tuberculosis was suspended in 100 μL Freund’s complete adjuvant (FCA) and intradermally injected into the rat tail root. After iodoacetic acid was injected into the knee joint of mice, regional degeneration and necrosis of chondrocytes could be found one week later to establish osteoarthritis model. Early signs of arthritis appeared after two days. All animal experiments were conformed to the Guide for the Care and Use of Laboratory Animal (NIH publication, 8th edition, 2011), and approved by the Experimental Animal Care and Use Committee of Xi’an Jiaotong University. The mice were kept in a temperature-controlled room on a 12 h light-dark cycle with free food and water. All efforts were made to minimize animals’ suffering and to reduce the number of animals used.
Cell culture and transfection
CD4+ T cells and SFs were obtained from RA and osteoarthritis (OA) mice. Two hundred and ninety-three T cells were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). Two hundred and ninety-three T cells and CD4+ T cells were cultured in 1640 complete medium with 10% fetal bovine serum (FBS), 100 U penicillin, and 100 μg/mL streptomycin at 37°C in a humidified atmosphere incubator containing 5% CO2. SFs were cultured in Dulbecco’s modified Eagle medium (DMEM) complete medium containing 10% FBS, 100 U penicillin, and 100 μg/mL streptomycin. Besides, CD4+ T cells were incubated with SFs-derived exosomes for 2 h.
MiR-424 mimic and the corresponding negative control (miR-NC) were purchased from Thermo Fisher Scientific. MiR-424 inhibitor and the corresponding negative control (inhibitor-NC) were purchased from Ribo Bio (Guangzhou, China). These plasmids were transfected by using Lipofectamine 2000 (Invitrogen, CA, USA) when the cells reached 80% confluence on the manufacturer’s instruction.
HE staining
Synovial tissues were separated from OA and RA mice. The prepared synovial tissue slices were dewaxed and hydrated. After washed by water, synovial tissue slices were stained in hematoxylin solution for 5 min. Next, after differentiated by 1% hydrochloric alcohol for 15 s, the slices were washed with water. And then synovial tissue slices were stained by eosin solution for 1 min. Finally, synovial tissue slices were dehydrated, transparentized, and sealed by neutral gum. Photos of slices were taken by optical microscope (Olympus, Tokyo, Japan).
Immunohistochemistry staining
Slices were treated with 3% hydrogen peroxide for 10 min at room temperature. After blocking the sections by using goat serum at 25°C for 4 h, the sections were incubated with primary anti-HIF1α antibody (1:100) or anti-IL-17A antibody (1:100) at 4°C overnight. Then, the sections were washed with PBS for three times to remove the primary antibodies and were incubated with biotinylated secondary antibody (Bioworld, USA) for 2 h, and 3,3 N-diaminobenzidine tertrahydrochloride buffer (Beyotime, Shanghai, China) was used to perform the color reaction. The slices were observed under microscope (Olympus, Tokyo, Japan).
Immunofluorescence staining
SFs were treated by 4% paraformaldehyde for 30 min. After washing with PBS, the cells were permeabilized with 0.1% Triton X-100 for 10 min, and then blocked with BSA for 1 h. Next, SFs were incubated with the diluted primary antibody against HIF1α (1:100) for 1 h. After washing with PBS to remove the primary antibody, SFs were incubated with the secondary fluorescein anti-rabbit IgG (1:1000) at room temperature for 1 h. And DAPI was used to stain the nucleus for 5 min. Eventually, photos of SFs were taken by a fluorescence microscope (Olympus, Tokyo, Japan).
Flow cytometry
Flow cytometry was used to analyze the infusion of exosomes with the cells. The detection of PKH-67-labeled exosomes infusion with T cells was performed as previously described.29 Briefly, 20 μg of the PKH67-stained exosomes were washed five times to remove excess dye and then added to T cells in culture. Cells were harvested at 24 h, washed three times, and analyzed by flow cytometry using a BD FACSAria (BD Biosciences). For the detection of the cell marker, anti-FOXP3 and anti-IL-17 antibodies were used for flow cytometry were purchased from BD Biosciences-Pharmingen or eBioscience (San Diego, CA, USA). Cells were stained in FACS buffer with antibodies for 30 min on ice and washed with FACS buffer. For intracellular staining, cells were fixed and permeabilized and then stained with anti-FOXP3 or anti IL-17 antibody for 30 min on ice and washed. Samples were analyzed by Flow Cytometric Analysis (Beckman Coulter, CA, USA).
Luciferase reporter assay
The putative binding sites of miR‐424 on the FOXP3 3′ UTR were predicted by TargetScan 7.0. The sequences that bind to miR-424 were partly mutated and inserted into the reporter plasmid to identify the binding specificity. The miR-424 mimic and its control (Hanbio, Shanghai, China) were transfected into SFs. After 48 h of transfection, the relative luciferase activities were detected by the Dual-Glo Luciferase Assay System (Promega, Shanghai, China) approved by the manufacturer’s instructions.
Western blot
Samples were cracked in RIPA lysis buffer plus PMSF in low temperature, and BCA assay kit (Santa Cruz, California, USA) detected the total protein concentration. Prepared protein samples were separated in SDS-PAGE, and transferred into 0.22 μm PVDF membranes and incubated with prepared antibodies. Finally, enhanced chemiluminescence (ECL, ThermoFisher, MA, USA) visualized this membrane. The protein band analysis was conducted with ImageJ software. Antibodies against HIF1α and GAPDH were purchased from CST (Beverly, MA, USA). Antibodies against CD9, CD81, and TSG101 were purchased from Abcam (Cambridge, MA, USA).
Q-PCR
Total RNA of the samples was separated by Trizol reagent (Invitrogen, Shanghai, China) following the manufacturer’s protocols. Reverse transcriptase reactions were performed using HiScript® III 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing, China). qRT-PCR was done using quantitative PCR with ChamQTM Universal SYBR QPCR Master Mix (Vazyme, Nanjing, China). Eventually, the product was analyzed on a RT-PCR Detection System (Analytic, Jena, Germany). The quantitative measures were obtained using the ΔΔCT method and were normalized to GAPDH or U6 mRNA levels.
Exosomes extraction and identification
SFs were plated at a 15 cm cell culture dish. After cultured for 48 h, the culture medium from six dishes was collected. Then the medium was centrifuged (300g, 10 min, room temperature, then 2000g, 20 min, 4°C, then 12,000g, 40 min, 4°C). Supernatant was then centrifuged twice (100,000g, 1 h; 10 min, 4°C). The 100,000g pellet was further subjected to gradient centrifugation (20%–60% sucrose) (100,000g, 4°C, 16 h) for purification. Exosome were observed by using an electron microscope (Philips CM200F) operating at 200 keV. The size distribution and concentration of exosomes were analyzed by using a NanoSight NS300 system with a 405 nm laser. In brief, exosomes suspension diluted with sterile 1×PBS as the ratio of 1:100 was pushed slowly using a 1 mL syringe and illuminated with a laser. Their movements under Brownian motion were recorded and analyzed with the NTA analytical software (NanoSight, version 3.0). At least three videos were captured for each individual sample. The capture and analysis settings were manually set in accordance with the manufacturer’s instructions.
ELISA analysis
Expression levels of IL-10, TGF-β, IL-17, IL-22, IL-1β, and TNF-α in the cell culture medium, synovial fluid, and serum were measured using the commercialized ELISA Kit (Elabscience, TX, USA) according to the manufacturer’s instructions. The absorbance was examined at 450 nm using a micro plate reader (BIO-TEK, INC).
Statistical analysis
Statistical analysis were performed by using the Graphpad prism 8.0 software with one-way ANOVA and post hoc Dunnett’s T3 test to compare the differences among and between groups, respectively. All experiments were repeated three times, and the data are presented as the mean ± SEM. P < 0.05 was considered to be statistically significant.
Results
HIF1α expression is increased in RA
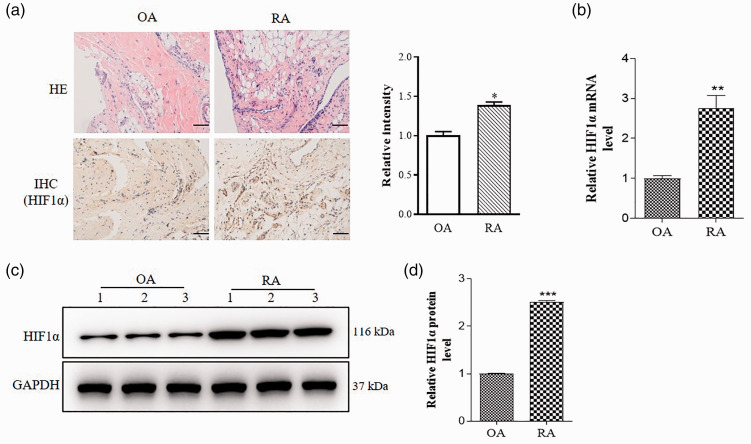
Hypoxia promotes the pathological progression of RA by hypoxia-inducible factor-1 (HIF-1).26 Given the important role of hypoxia in the pathogenesis of RA, we detected HIF1α expression in RA and OA. It is reported that HE staining could reveal the general morphological and structural characteristics of the tissue or cell components and lesions.30 HE and immunohistochemistry (IHC) staining results showed that HIF1α expression was higher in RA mice than OA mice (Figure 1(a)). Furthermore, HIF1α miRNA and protein expression were also significantly increased in RA compared to OA (Figure 1(b) to (d)).
Figure 1.
HIF1α expression is increased in RA. Synovial tissue was extracted from OA and RA model mice. (a) Representative images of HE staining and immunohistochemistry staining for HIF1α (brown color). Relative intensity of HIF1α was analyzed by Image J. Scale bar = 50 μm. (b) qRT-PCR was performed to examine relative values of HIF1α mRNA. (c) HIF1α protein expressions in synovial tissue of OA and RA mice were measured by Western blot. Each group is shown three representative blot images. (d) Relative values of HIF1α protein expressions were analyzed by Image J. Data are shown as mean ± S.E.M. *P < 0.05, **P < 0.01, ***P < 0.001, n = 6. (A color version of this figure is available in the online journal.)
Hypoxia induces exosomal miR-424 expression in SFs
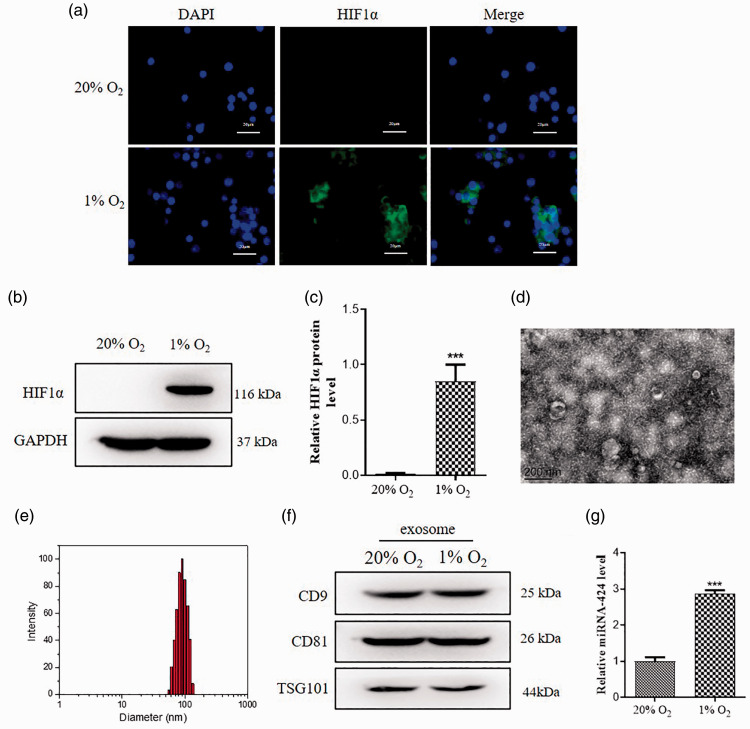
In order to further investigate the role of hypoxia in RA, we performed cell experiments using SFs. According to previous studies, we used 1% O2 to treat SFs to create a hypoxic microenvironment similar to RA.31 Immunofluorescence data indicated that 1% O2 obviously increased HIF1α expression compared to 20% O2 in SFs (Figure 2(a)). Additionally, Western blot also got similar results (Figure 2(b) and (c)). Recent researches demonstrate that exosomes exert a vital role in the pathological process of RA,20 so we extracted exosomes by repeated high-speed centrifugation from SFs to follow-up experiments. Transmission electron microscope (TEM) showed exosome double-layer capsule ultrastructure under the condition of 1% O2 (Figure 2(d)). Nanoparticle Tracking Analysis (NTA) indicated that the size of extracted particles is mostly around 100 nm, which is in line with the expected exosome characteristics (Figure 2(e)). Exosomes marker of CD9, CD81, and TSG101 expression showed no significant changes in 1% O2 group compared to 20% O2 group (Figure 2(f)). And we found that exosomal miR-424 level was higher in 1% O2 compared to 20% O2 (Figure 2(g)). These results indicated that hypoxia induces exosomal miR-424 expression in SFs.
Figure 2.
Hypoxia induces exosomal miR-424 expression in SFs. Synovial fibroblasts were treated with 20% O2 or 1% O2 to establish cell model in vitro. (a) Representative images of immunofluorescence staining for HIF1α. HIF1α is shown as green color and the nuclei are shown as blue color. Scale bar = 20 μm. (b) HIF1α protein expressions in OA and RA mice were measured by Western blot. (c) Relative values of HIF1α protein expressions was analyzed by Image J. (d) Representative TEM images of exosomes. Scale bar = 200 nm. (e) The average diameter of exosome size analyzed by NTA system. (f) Exosomes marker CD9 and CD81, TSG101 in the CAFs lysate and CAFs-derived exosomes were detected by Western blot. (g) Relative values of CD9 and CD81, TSG101 protein expressions was analyzed by Image J. Data are shown as mean ± S.E.M. **P < 0.01, ***P < 0.001, n = 6. (A color version of this figure is available in the online journal.)
SFs-exosomes promote T cell differentiation
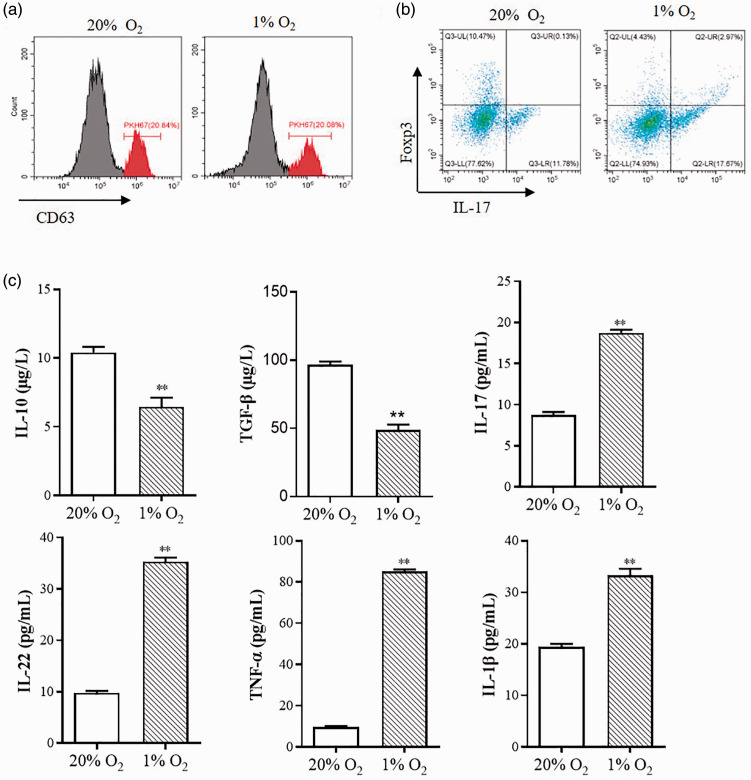
T cells, which are a major part of synovial infiltrating cells, especially CD4+ T cells, are considered to be closely related to RA.32 CD4+ T cells differentiate into Th1 cells and Th2 cells, Th17 cells, and Treg cells when they were activated. Different types of CD4+ T cells have different functions such as activating cellular and non-cellular immunity, direct cytolytic activity, and suppressing the immune response.21 Recent research shows flexible plasticity between Treg cells and Th17 cells.33 In this study, we found that SFs-exo could bind to the CD4+ T cells membrane, as suggested by the PKH-67-labeling assay (Figure 3(a)). And SFs-exo induced T-cell differentiation, in which Th17 cells count was raised, but Treg cells count was dropped (Figure 3(b)). We also detected the change of inflammatory cytokines induced by hypoxia. As shown in Figure 3(c), anti-inflammatory Treg-related cytokines IL-10 and TGF-β were decreased under the hypoxic condition, while the pro-inflammatory Th17-related cytokines IL-17, IL-22, IL-1β, and TNF-α increased under the hypoxic condition, indicating that hypoxic inducing inflammatory response and imbalance of Treg/Th17.
Figure 3.
SFs-exosomes promote T cell differentiation. Exosomes extracted from the synovial fibroblasts which were treated with 20% O2 or 1% O2 were used to incubated with T cells. (a) PKH67-labeled SFs-derived exosome were taken up by T cells. PKH67-labeled exosomes were incubated with T cells at 37°C for 12 h. T cells fused with exosomes were analyzed by flow cytometry. (b) IL-17 and FOXP3-positive cells were determined by flow cytometric analysis to indicate the T cells differentiation. (c) ELISA were used to measure the IL-10, IL-17, IL-22, IL-1β, IFN-γ, TNF-α, and TGF-β expression of exosomes-treated T cells. Data are shown as mean ± S.E.M. **P < 0.01, ***P < 0.001, n = 6. (A color version of this figure is available in the online journal.)
FOXP3 is a target gene of miR-424
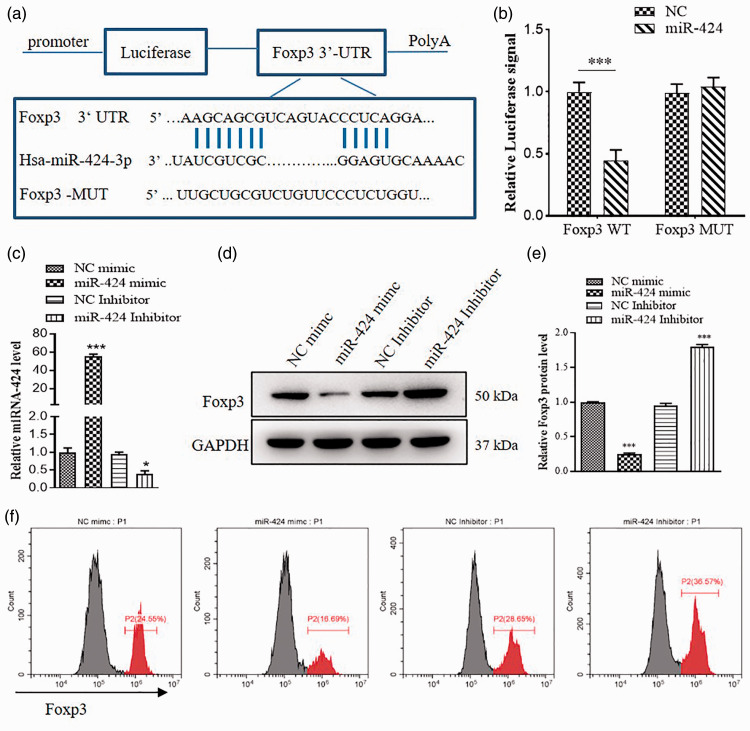
To further explore the molecular mechanism of miR-424 on RA, we used RNAhybrid to predict miR-424 targets, determined FOXP3 (Figure 4(a)). Transcription factor FOXP3, specifically expressed on Treg cells, is closely related to the growth and development and functional maintenance of on Treg cells.34 To identify the relationship of miR-424 and FOXP3, we designed luciferase reporter plasmids to detection. The luciferase activity significantly reduced following co-transfection with pMIR-FOXP3-3′-UTR-WT and miR-424, compared with co-transfection with pMIR-FOXP3-3′-UTR-Mut and miR-424 in 293 T cells (Figure 4(b)). This result demonstrated that miR-424 negatively regulates the FOXP3 expression via specifically binding to the 3′-UTR of FOXP3 (Figure 4(b)). miR-424 expression was increased by miR-424 mimic, and miR-424 inhibitor reduced miR-424 (Figure 4(c)). Furthermore, Western blot showed that miR-424 mimic significantly decreased FOXP3 expression (Figure 4(d) and (e)). And flow cytometry results were similar to Western blot (Figure 4(f)).
Figure 4.
FOXP3 is a target gene of miR-424. (a) The predicted FOXP3 binding sites with miR-424 &was shown. (b) Two hundred and ninety-three T cells were transfected with luciferase reporter plasmid containing WT or mutant form of FOXP3 and miR-424 mimic or miR-424 inhibitor, the luciferase signal were assessed 24 h later. (c) Relative values of miR-424 expression in NC mimic, miR-424 mimic, NC inhibitor and miR-424 inhibitor was measured by qRT-PCR. (d) FOXP3 expression in cells treated with miR-424 mimic and miR-424 inhibitor was examined by western blot. (e) Relative values of FOXP3 protein expression was analyzed by Image J. (f) Flow cytometric analysis was used to identify the FOXP3-positive cells (P2). Data are shown as mean ± S.E.M. *P < 0.05, **P < 0.01, ***P < 0.001, n = 6. (A color version of this figure is available in the online journal.)
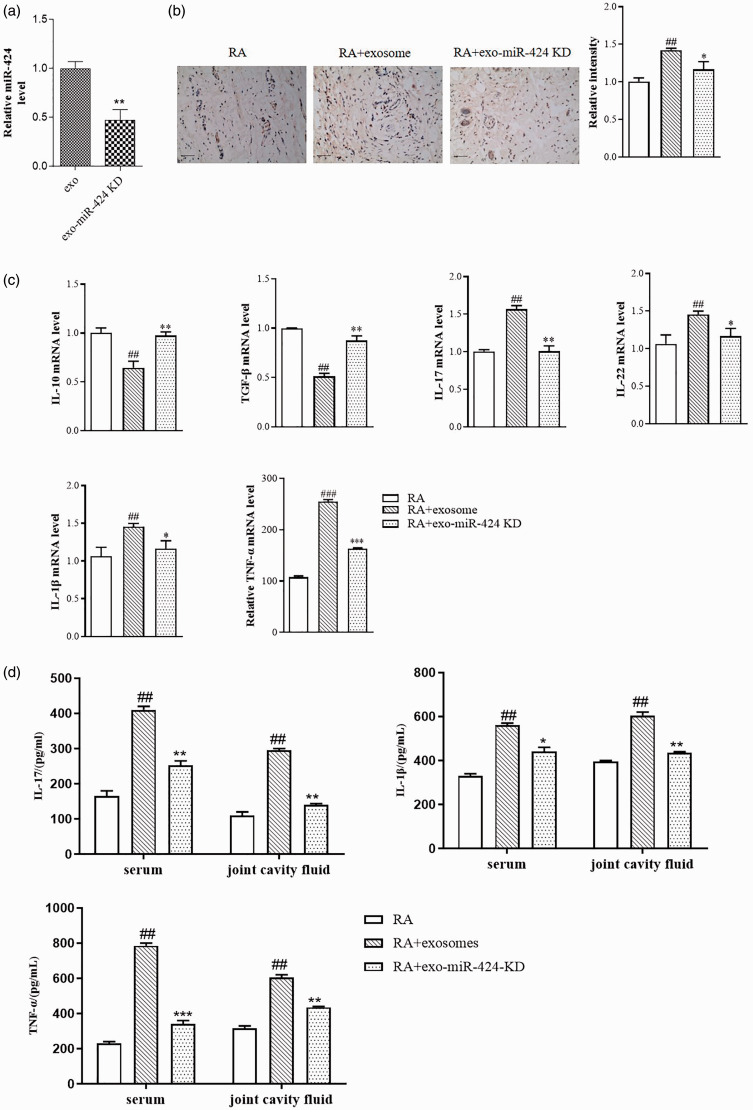
Exosomal miR-424 promotes the development of RA
We then verified the effect of exosomal miR-424 in RA mice. We extracted exosomes from SFs which were knocked down of miR-424 or not. The miR-424 level in exosomes is shown in Figure 5(a). IHC results of IL-17 showed that mice treated with SFs-exo showed significant increase of IL-17 expression, and knockdown of miR-424 could abolish the effect of SFs-exo (Figure 5(b)). We also detected the change of inflammatory cytokines in the synovial by qRT-PCR. As shown in Figure 5(c), anti-inflammatory Treg-related cytokines IL-10 and TGF-β were decreased in SFs-exo-treated mice, and the pro-inflammatory Th17-related cytokines IL-17, IL-22, IL-1β, and TNF-α increased in SFs-exo-treated mice, while knockdown of miR-424 could abolish the effect of SFs-exo. ELISA results showed that SFs-exo induced increase of IL-17, IL-1β, and TNF-α levels in the serum and joint cavity fluid, and exosomes extracted from miR-424 knockdown cells showed no obvious increase of the inflammatory factors compared with the RA group (Figure 5(d)). All the data suggested that exosomal miR-424 could enhance the inflammatory response of RA.
Figure 5.
Exosomal miR-424 promotes the development of RA. Exosome were extracted from SFs treated with miR-424 inhibitor or not, and then administrated to RA mice by tail injection. (a) The efficacy of miR-424 KD was analyzed by qRT-PCR. (b) Representative images of immunohistochemistry staining for IL-17 (brown color) in RA model. Relative intensity of IL-17 was analyzed by Image J. scale bar = 50 μm. (c) Expressions of IL-10, IL-17, IL-22, IL-1β, IFN-γ, TNF-α, and TGF-β in synovial tissue were determined by qRT-PCR. (d) Expressions of IL-17, IL-1β, and TNF-α in serum and joint cavity fluid were examined by ELISA. Data are shown as mean ± S.E.M. ##P < 0.01, ###P < 0.001, compared with the RA group; *P < 0.05, **P < 0.01, ***P < 0.001, compared with the RA+exosome group. n = 6. (A color version of this figure is available in the online journal.)
Discussion
In our research, HIF1α protein and mRNA expression were up-regulated in RA compared to OA, which indicated that there is hypoxic microenvironment in RA. Moreover, miR-424 is one part of miR-16 family, which involves in the proliferation and phenotypic differentiation of immune cells. We found that miR-424 from SFs-exo was dramatically increased under 1% O2. And then SFs-exo promotes T cell differentiation, in which Th17 cells count was raised, but Treg cells count was dropped. Our study data suggested that exosomal miR-424 inhibited inflammatory factors expression by targeting FOXP3, one of the key transcription factors that control the development and function of Treg cells. We also found that miR-424 KD could improve RA in vivo.
miRNA regulates gene expression by binding to 3′-UTR after transcription.35 Previous studies have shown that a class of miRNA has a key role in regulating CD4+ T cell differentiation. In our study, we found that miR-424 from SFs-exo was significantly upregulated under 1% O2. To date, at least five different subsets of CD4+ T cells have been identified, namely Th1, Th2, Th17, Treg, and Tfh cells.36 It has been reported that miR-146a inhibits Th1 cell differentiation by targeting STAT1,37 and miR-155 can downregulate the INF-γ on the surface of T cells by suppression of IFNγ regulator to promote the differentiation of T cells into Th1 cells.38 Furthermore, multiple miRNAs were found to be related to Th2 cell differentiation. Sawant et al. found that miR-21 downregulates Sprouty1 by over-expression on T cells, thereby stimulating Th2 differentiation.39 Th17 and Treg cells differentiation are also regulated by miRNAs. For example, miR-26a inhibits the generation of Th17 cells by targeting IL-6, which is positively correlated with FOXP3 expression and promotes the development of Treg cells, thus suppresses self-reactive inflammation.9 We found that SFs-exo promotes T cell differentiation, which Th17 cells were increased, Treg cells were decreased. miR-21 maintains the biological function of Treg cells by positively regulating FOXP3 expression.40 In RA patients, the low content of miR-21 inhibits the activity of STAT5 and limits Treg cells differentiation and development, resulting in an imbalance in the Th17/Treg ratio and the generation of chronic inflammation.41 In our study, we found that miR-424 exerts a role in the pathology Of RA.
miR-424 KD significantly inhibited pro-inflammatory factors’ expression in RA.miR-424 belongs to the miR-16 family, which plays a regulatory part in cell cycle and phenotype. A large number of studies have shown that miR-424 has a clinical prognostic effect and acts as a potential molecular therapeutic target. Studies have found that miR-424 has a significant change in different organs during the onset of infectious and immune diseases and also involves in regulating viral replication,41 the proliferation of lesion cells,42 and the proliferation and phenotypic differentiation of immune cells.43 Therefore, miR-424 regulates the occurrence and development of infectious diseases such as tuberculosis, AIDS, viral hepatitis, enteritis, psoriasis, and pemphigus, as well as immune diseases such as leukemia and lymphoma. Recovery of miR-424 expression reversed chemoresistance via activating T cell immune response through blocking the immune checkpoints of PD-L1 in vitro and in vivo.44 Our study data suggested that exosomal miR-424 increased inflammatory factors’ expression.
To further explore the function of miR-424 on RA, FOXP3 was predicted to contain an miR-424 3-UTR binding site and experimentally validated. FOXP3 is a member of the forkhead transcription factor family, encoded by the X chromosome gene, and plays an important transcriptional regulatory role in the development and function of Treg cells.45 Human FOXP3 gene mutations could lead to the occurrence of autoimmune diseases at an early stage, such as IPEX (immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome).46 Similarly, the lack of FOXP3 in mice can cause severe autoimmune disease.47 Studies have found that FOXP3 in combination with different transcription factors has different physiological functions to mediate the functional heterogeneity of different Treg subtypes. Treg cell specifically inhibits Th2 cell function via FOXP3 binding to IRF4, an important transcription factor in Th2 cells.48 Moreover, the interaction of FOXP3 and T-bet, a major transcription factor expressed in Th1 cells, can promote the inhibition of Treg cells on Th1 cells,49 and FOXP3 combined with signal transducers and activators of transcription (STAT3), the key factors of Th17 cells, helps Treg cells effectively inhibit Th17 cell function.50 In this study, FOXP3 is a target gene of miR-424, and miR-424 aggravated the development of RA via negatively regulating FOXP3 expression.
We also detected the change of inflammatory cytokines induced by hypoxia in vitro and by exosomal miR-424 in vivo. Change of the balance of Th17/Treg may result in the expression change of a serious of inflammatory cytokines.51 Th17 cell is a novel cell lineage of CD4+T that is characterized by the secretion of distinct IL-17 cytokines which promote inflammation.52 IL-22 is another important pro-inflammatory cytokines secreted by Th17 cells.53 In the present research, we investigate the IL-17 and IL-22 expression to identify the differentiation to Th17 cells and the level of inflammation. We found that hypoxia induced increase level of IL-17 and IL-22 in vitro, and exosomal miR-424 induced increased level of IL-17 and IL-22 in vivo. Treg cells secrete IL-10 and other inhibitory cytokines such as TGF-β, to suppress the immune response harmful to a host.54,55 In the present study, we found that hypoxia induced decreased level of IL-10 and TGF-β in vitro, and exosomal miR-424 induced decreased level of IL-10 and TGF-β in vivo. TNF-α and IL-1β are important inflammatory cytokines contributing to the progression of RA which could be used as the inflammatory marker of RA.56,57 It was reported that TNF-α promoted Th17 cell differentiation through IL-1β which was produced by monocytes in RA, and inhibition of IL-1β and TNF-α exhibited beneficial effects in preventing RA.58,59 We found that hypoxia induced increased level of IL-1β and TNF-α in vitro, and exosomal miR-424 induced increased level of IL-1β and TNF-α in vivo. All these data suggested that exosomal miR-424 could induce Th17 differentiation and inhibit Treg differentiation, with the increase of pro-inflammatory cytokines and decrease of anti-inflammatory cytokines.
In a word, our data indicated SFs-exo under hypoxia aggravate RA by regulating Treg/Th17 balance. And miR-424 plays main roles in the pathological process of RA by regulating FOXP3. In-depth research on the functions and molecular regulatory mechanisms of exosomes in disease microenvironment will provide new drug targets to overcome these immune-related diseases.
Acknowledgements
We would like to thank all the researchers and study participants for their contributions.
Authors’ contributions
WSF conceived and designed the experiments; DYJ wrote the manuscript; WLF designed the figures. WHQ and ZQ contributed to the writing and editing of the article. All authors and participants reviewed the paper and approved the final version of the article.
Data availability statement
All processed data and models used during the study are available from the corresponding author by request.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING
This study was supported by The Science and Technology Development Project of Kaifeng (Grant no. 2017123), Subject of Scientific Research on Traditional Chinese Medicine in Henan Province (Grant no. 2017ZY3044) and Medical Science and Technology Joint Project of Henan Province (Grant no. 2018020329).
ORCID iD
Yanjie Ding https://orcid.org/0000-0002-0515-8427
References
- 1.Mateen S, Shahzad S, Ahmad S, Naeem SS, Khalid S, Akhtar K, Rizvi W, Moin S. Cinnamaldehyde and eugenol attenuates collagen induced arthritis via reduction of free radicals and pro-inflammatory cytokines. Phytomedicine 2019; 53:70–8 [DOI] [PubMed] [Google Scholar]
- 2.Helmick CG, Felson DT, Lawrence RC, Gabriel S, Hirsch R, Kwoh CK, Liang MH, Kremers HM, Mayes MD, Merkel PA, Pillemer SR, Reveille JD, Stone JH. National arthritis data W. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Part I. Arthritis Rheum 2008; 58:15–25 [DOI] [PubMed] [Google Scholar]
- 3.Harrison P, Pointon JJ, Chapman K, Roddam A, Wordsworth BP. Interleukin-1 promoter region polymorphism role in rheumatoid arthritis: a meta-analysis of IL-1B-511A/G variant reveals association with rheumatoid arthritis. Rheumatology (Oxford) 2008; 47:1768–70 [DOI] [PubMed] [Google Scholar]
- 4.Aletaha D, Smolen JS. Diagnosis and management of rheumatoid arthritis: a review. JAMA 2018; 320:1360–72 [DOI] [PubMed] [Google Scholar]
- 5.Ye X, Zhao K, Wu C, Hu P, Fu H. Associations between genetic variants in immunoregulatory genes and risk of non-Hodgkin lymphoma in a Chinese population. Oncotarget 2017; 8:10450–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stuart S, Maher BH, Sutherland H, Benton M, Rodriguez A, Lea RA, Haupt LM, Griffiths LR. Genetic variation in cytokine-related genes and migraine susceptibility. Twin Res Hum Genet 2013; 16:1079–86 [DOI] [PubMed] [Google Scholar]
- 7.Liu Q, Tian J, Xu Y, Li C, Meng X, Fu F. Protective effect of RA on myocardial Infarction-Induced cardiac fibrosis via AT1R/p38 MAPK pathway signaling and modulation of the ACE2/ACE ratio. J Agric Food Chem 2016; 64:6716–22 [DOI] [PubMed] [Google Scholar]
- 8.Keen HI, Emery P. How should we manage early rheumatoid arthritis? From imaging to intervention. Curr Opin Rheumatol 2005; 17:280–5 [DOI] [PubMed] [Google Scholar]
- 9.Zhang R, Tian A, Wang J, Shen X, Qi G, Tang Y. miR26a modulates Th17/T reg balance in the EAE model of multiple sclerosis by targeting IL6. Neuromol Med 2015; 17:24–34 [DOI] [PubMed] [Google Scholar]
- 10.Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, McClanahan T, Kastelein RA, Cua DJ. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med 2005; 201:233–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, Weaver CT. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol 2005; 6:1123–32 [DOI] [PubMed] [Google Scholar]
- 12.Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YH, Wang Y, Hood L, Zhu Z, Tian Q, Dong C. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol 2005; 6:1133–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boissier MC, Assier E, Biton J, Denys A, Falgarone G, Bessis N. Regulatory T cells (Treg) in rheumatoid arthritis. Joint Bone Spine 2009; 76:10–4 [DOI] [PubMed] [Google Scholar]
- 14.Niu Q, Cai B, Huang ZC, Shi YY, Wang LL. Disturbed Th17/Treg balance in patients with rheumatoid arthritis. Rheumatol Int 2012; 32:2731–6 [DOI] [PubMed] [Google Scholar]
- 15.Tan L, Wu H, Liu Y, Zhao M, Li D, Lu Q. Recent advances of exosomes in immune modulation and autoimmune diseases. Autoimmunity 2016; 49:357–65 [DOI] [PubMed] [Google Scholar]
- 16.Kowal J, Tkach M, Thery C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol 2014; 29:116–25 [DOI] [PubMed] [Google Scholar]
- 17.Thakur BK, Zhang H, Becker A, Matei I, Huang Y, Costa-Silva B, Zheng Y, Hoshino A, Brazier H, Xiang J, Williams C, Rodriguez-Barrueco R, Silva JM, Zhang W, Hearn S, Elemento O, Paknejad N, Manova-Todorova K, Welte K, Bromberg J, Peinado H, Lyden D. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res 2014; 24:766–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shao H, Im H, Castro CM, Breakefield X, Weissleder R, Lee H. New technologies for analysis of extracellular vesicles. Chem Rev 2018; 118:1917–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song J, Kim D, Han J, Kim Y, Lee M, Jin EJ. PBMC and exosome-derived hotair is a critical regulator and potent marker for rheumatoid arthritis. Clin Exp Med 2015; 15:121–6 [DOI] [PubMed] [Google Scholar]
- 20.Zakeri Z, Salmaninejad A, Hosseini N, Shahbakhsh Y, Fadaee E, Shahrzad MK, Fadaei S. MicroRNA and exosome: key players in rheumatoid arthritis. J Cell Biochem 2019; [DOI] [PubMed] [Google Scholar]
- 21.Alexander M, Hu R, Runtsch MC, Kagele DA, Mosbruger TL, Tolmachova T, Seabra MC, Round JL, Ward DM, O’Connell RM. Exosome-delivered microRNAs modulate the inflammatory response to endotoxin. Nat Commun 2015; 6:7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakasa T, Miyaki S, Okubo A, Hashimoto M, Nishida K, Ochi M, Asahara H. Expression of microRNA-146 in rheumatoid arthritis synovial tissue. Arthritis Rheum 2008; 58:1284–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee A, Papangeli I, Park Y, Jeong HN, Choi J, Kang H, Jo HN, Kim J, Chun HJ. A PPARgamma-dependent miR-424/503-CD40 axis regulates inflammation mediated angiogenesis. Sci Rep 2017; 7:2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fearon U, Canavan M, Biniecka M, Veale DJ. Hypoxia, mitochondrial dysfunction and synovial invasiveness in rheumatoid arthritis. Nat Rev Rheumatol 2016; 12:385–97 [DOI] [PubMed] [Google Scholar]
- 25.Lund-Olesen K. Oxygen tension in synovial fluids. Arthritis Rheum 1970; 13:769–76 [DOI] [PubMed] [Google Scholar]
- 26.Hu F, Mu R, Zhu J, Shi L, Li Y, Liu X, Shao W, Li G, Li M, Su Y, Cohen PL, Qiu X, Li Z. Hypoxia and hypoxia-inducible factor-1alpha provoke toll-like receptor signalling-induced inflammation in rheumatoid arthritis. Ann Rheum Dis 2014; 73:928–36 [DOI] [PubMed] [Google Scholar]
- 27.Bevaart L, Vervoordeldonk MJ, Tak PP. Evaluation of therapeutic targets in animal models of arthritis: how does it relate to rheumatoid arthritis? Arthritis Rheum 2010; 62:2192–205 [DOI] [PubMed] [Google Scholar]
- 28.Yang X, Zhao Y, Jia X, Wang C, Wu Y, Zhang L, Chang Y, Wei W. CP-25 combined with MTX/LEF ameliorates the progression of adjuvant-induced arthritis by the inhibition on GRK2 translocation. Biomed Pharmacother 2019; 110:834–43 [DOI] [PubMed] [Google Scholar]
- 29.Xiao H, Lasser C, Shelke GV, Wang J, Radinger M, Lunavat TR, Malmhall C, Lin LH, Li J, Li L, Lotvall J. Mast cell exosomes promote lung adenocarcinoma cell proliferation – role of KIT-stem cell factor signaling. Cell Commun Signal 2014; 12:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han L, Song N, Hu X, Zhu A, Wei X, Liu J, Yuan S, Mao W, Chen X. Inhibition of RELM-beta prevents hypoxia-induced overproliferation of human pulmonary artery smooth muscle cells by reversing PLC-mediated KCNK3 decline. Life Sci 2020; 246:117419. [DOI] [PubMed] [Google Scholar]
- 31.Kinderlerer AR, Steinberg R, Johns M, Harten SK, Lidington EA, Haskard DO, Maxwell PH, Mason JC. Statin-induced expression of CD59 on vascular endothelium in hypoxia: a potential mechanism for the anti-inflammatory actions of statins in rheumatoid arthritis. Arthritis Res Ther 2006; 8:R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warrington KJ, Takemura S, Goronzy JJ, Weyand CC. CD4+, CD28-T cells in rheumatoid arthritis patients combine features of the innate and adaptive immune systems. Arthritis Rheum 2001; 44:13–20 [DOI] [PubMed] [Google Scholar]
- 33.Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity 2009; 30:646–55 [DOI] [PubMed] [Google Scholar]
- 34.Ali S, Leonard SA, Kukoly CA, Metzger WJ, Wooles WR, McGinty JF, Tanaka M, Sandrasagra A, Nyce JW. Absorption, distribution, metabolism, and excretion of a respirable antisense oligonucleotide for asthma. Am J Respir Crit Care Med 2001; 163:989–93 [DOI] [PubMed] [Google Scholar]
- 35.Felekkis K, Touvana E, Stefanou C, Deltas C. microRNAs: a newly described class of encoded molecules that play a role in health and disease. Hippokratia 2010; 14:236–40 [PMC free article] [PubMed] [Google Scholar]
- 36.Li P, Spolski R, Liao W, Leonard WJ. Complex interactions of transcription factors in mediating cytokine biology in T cells. Immunol Rev 2014; 261:141–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu LF, Boldin MP, Chaudhry A, Lin LL, Taganov KD, Hanada T, Yoshimura A, Baltimore D, Rudensky AY. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell 2010; 142:914–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huffaker TB, Hu R, Runtsch MC, Bake E, Chen X, Zhao J, Round JL, Baltimore D, O’Connell RM. Epistasis between microRNAs 155 and 146a during T cell-mediated antitumor immunity. Cell Rep 2012; 2:1697–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sawant DV, Wu H, Kaplan MH, Dent AL. The Bcl6 target gene microRNA-21 promotes Th2 differentiation by a T cell intrinsic pathway. Mol Immunol 2013; 54:435–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rouas R, Fayyad-Kazan H, El Zein N, Lewalle P, Rothe F, Simion A, Akl H, Mourtada M, El Rifai M, Burny A, Romero P, Martiat P, Badran B. Human natural treg microRNA signature: role of microRNA-31 and microRNA-21 in FOXP3 expression. Eur J Immunol 2009; 39:1608–18 [DOI] [PubMed] [Google Scholar]
- 41.Zhang S, Ouyang X, Jiang X, Gu D, Lin Y, Kong SK, Xie W. Dysregulated serum MicroRNA expression profile and potential biomarkers in hepatitis C virus-infected patients. Int J Med Sci 2015; 12:590–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim J, Kang Y, Kojima Y, Lighthouse JK, Hu X, Aldred MA, McLean DL, Park H, Comhair SA, Greif DM, Erzurum SC, Chun HJ. An endothelial apelin-FGF link mediated by miR-424 and miR-503 is disrupted in pulmonary arterial hypertension. Nat Med 2013; 19:74–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steinhilber J, Bonin M, Walter M, Fend F, Bonzheim I, Quintanilla-Martinez L. Next-generation sequencing identifies deregulation of microRNAs involved in both innate and adaptive immune response in ALK+ ALCL. PLoS One 2015; 10:e0117780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu S, Tao Z, Hai B, Liang H, Shi Y, Wang T, Song W, Chen Y, OuYang J, Chen J, Kong F, Dong Y, Jiang SW, Li W, Wang P, Yuan Z, Wan X, Wang C, Li W, Zhang X, Chen K. miR-424(322) reverses chemoresistance via T-cell immune response activation by blocking the PD-L1 immune checkpoint. Nat Commun 2016; 7:11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Di Nunzio S, Cecconi M, Passerini L, McMurchy AN, Baron U, Turbachova I, Vignola S, Valencic E, Tommasini A, Junker A, Cazzola G, Olek S, Levings MK, Perroni L, Roncarolo MG, Bacchetta R. Wild-type FOXP3 is selectively active in CD4+CD25(hi) regulatory T cells of healthy female carriers of different FOXP3 mutations. Blood 2009; 114:4138–41 [DOI] [PubMed] [Google Scholar]
- 46.Godfrey VL, Wilkinson JE, Russell LB. X-linked lymphoreticular disease in the scurfy (sf) mutant mouse. Am J Pathol 1991; 138:1379–87 [PMC free article] [PubMed] [Google Scholar]
- 47.Bennett CL, Christie J, Ramsdell F, Brunkow ME, Ferguson PJ, Whitesell L, Kelly TE, Saulsbury FT, Chance PF, Ochs HD. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet 2001; 27:20–1 [DOI] [PubMed] [Google Scholar]
- 48.Zheng Y, Chaudhry A, Kas A, deRoos P, Kim JM, Chu TT, Corcoran L, Treuting P, Klein U, Rudensky AY. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature 2009; 458:351–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koch MA, Tucker-Heard G, Perdue NR, Killebrew JR, Urdahl KB, Campbell DJ. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol 2009; 10:595–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chaudhry A, Rudra D, Treuting P, Samstein RM, Liang Y, Kas A, Rudensky AY. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science 2009; 326:986–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao M, Liu LX, Wu FL, Zhang X, Li YY, Shi T, Li DZ, Han TT. The Changes of Th17/Treg and related cytokines: IL-17, IL-23, IL-10, and TGF-beta in respiratory syncytial virus bronchiolitis rat model. Iran J Allergy Asthma Immunol 2017; 16:386–95 [PubMed] [Google Scholar]
- 52.Wakashin H, Hirose K, Maezawa Y, Kagami S, Suto A, Watanabe N, Saito Y, Hatano M, Tokuhisa T, Iwakura Y, Puccetti P, Iwamoto I, Nakajima H. IL-23 and Th17 cells enhance Th2-cell-mediated eosinophilic airway inflammation in mice. Am J Respir Crit Care Med 2008; 178:1023–32 [DOI] [PubMed] [Google Scholar]
- 53.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 cells. Annu Rev Immunol 2009; 27:485–517 [DOI] [PubMed] [Google Scholar]
- 54.Ohkura N, Kitagawa Y, Sakaguchi S. Development and maintenance of regulatory T cells. Immunity 2013; 38:414–23 [DOI] [PubMed] [Google Scholar]
- 55.Liston A, Gray DH. Homeostatic control of regulatory T cell diversity. Nat Rev Immunol 2014; 14:154–65 [DOI] [PubMed] [Google Scholar]
- 56.Szekanecz Z, Koch AE. Macrophages and their products in rheumatoid arthritis. Curr Opin Rheumatol 2007; 19:289–95 [DOI] [PubMed] [Google Scholar]
- 57.Li X, Tian F, Wang F. Rheumatoid arthritis-associated microRNA-155 targets SOCS1 and upregulates TNF-alpha and IL-1beta in PBMCs. Int J Mol Sci 2013; 14:23910–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng Y, Sun L, Jiang T, Zhang D, He D, Nie H. TNFalpha promotes Th17 cell differentiation through IL-6 and IL-1beta produced by monocytes in rheumatoid arthritis. J Immunol Res 2014; 2014:385352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tian J, Chen JW, Gao JS, Li L, Xie X. Resveratrol inhibits TNF-alpha-induced IL-1beta, MMP-3 production in human rheumatoid arthritis fibroblast-like synoviocytes via modulation of PI3kinase/Akt pathway. Rheumatol Int 2013; 33:1829–35 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All processed data and models used during the study are available from the corresponding author by request.