Abstract
Background
Research investigating treatments and interventions for cognitive decline fail due to difficulties in accurately recognizing behavioral signatures in the presymptomatic stages of the disease. For this validation study, we took our previously constructed digital biomarker‐based prognostic models and focused on generalizability and robustness of the models.
Method
We validated prognostic models characterizing subjects using digital biomarkers in a longitudinal, multi‐site, 40‐month prospective study collecting data in memory clinics, general practitioner offices, and home environments.
Results
Our models were able to accurately discriminate between healthy subjects and individuals at risk to progress to dementia within 3 years. The model was also able to differentiate between people with or without amyloid neuropathology and classify fast and slow cognitive decliners with a very good diagnostic performance.
Conclusion
Digital biomarker prognostic models can be a useful tool to assist large‐scale population screening for the early detection of cognitive impairment and patient monitoring over time.
Keywords: Altoida Neuro Motor Index, Alzheimer's disease, artificial intelligence, augmented reality, cognitive aging, digital biomarker, machine learning, risk prediction
1. INTRODUCTION
Alzheimer's disease (AD) is the most common form of neurodegeneration, with an estimated 6 million North Americans and 46.8 million people worldwide living with the disease, costing health care and support systems approximately $1 trillion. AD's long prodromal phase renders the a priori identification of the individuals at risk complicated, yet roughly 50% of the people with incidental mild cognitive impairment (MCI) develop dementia within 3 years. 1 , 2 Intriguingly, other forms of dementia not associated to AD, such as vascular dementia, frontotemporal and Lewy Body dementia, are also characterized by an initial period of MCI negative to AD biomarkers (eg, increase amyloid beta [Aβ] load) often with multiple cognitive domain impairment (naMCI), leading to dementia within the same average 3‐year period. 2 What is still a matter of debate is who, among the subjects diagnosed with incidental MCI, will develop dementia and who will not? Several attempts were made to identify MCI subgroups, defined by impaired neuropsychological domains or presence of certain AD biomarkers that would provide this prediction, with interesting, but not definitive, results. 2 , 3
Among them, one of the most promising efforts has been proposed by the ABIDE (Alzheimer's Biomarkers in Daily Practice) project, whose ATN biomarker‐based prediction models combine the AD biomarkers for amyloid beta (Aβ) load (“A”), tauopathy (“T”), and atrophy neurodegeneration (“N”) in the context of the individual characteristics. 4 Once the large prospective, longitudinal validation study aimed to compare expected versus observed MCI conversion into AD dementia will be completed, a wealth of data will be available for building reliable predictive (or “prognostic”) models that will have profound implications for timely interventions and treatment planning for MCI patients, in accordance to the American Academy of Neurology guidelines. 5
Current innovation in how to build predictive models stems from the rapidly evolving field of machine‐learning (ML) and artificial neural networks (ANN) algorithms. 6 , 7 , 8 , 9 , 10 , 11 , 12 However, to date, only a few studies have been focusing on the validation of specific biomarker‐based predictive models using longitudinal clinical data with the aim to provide useful prognostic information of individual risk rather than group statistics. 9 , 10 , 11 Proper validation of predictive models based on ML‐ANN requires large and high‐quality databases obtained from longitudinal cohort studies, ideally planned to include different populations so as to reflect the heterogeneity observed in the real world. So far, most successful ML‐ANN studies have used biomarkers from a restricted number of publicly available databases with relatively homogeneous populations, namely Alzheimer's Disease Neuroimaging Initiative (ADNI) or Australian Imaging, Biomarker & Lifestyle Flagship Study of Aging (AIBL), 12 , 13 , 14 with few exceptions. 15 , 16 This is of relevance because important differences (up to three‐fold) in the conversion rate MCI‐to‐dementia have been reported depending if the study population was recruited in memory clinics or from the community. 17
The recent widespread availability of digital devices and applications running on smartphones and tablets assessing cognition and behavior has recently opened the possibility to add other biomarkers with potential prognostic value that could be easily used in clinical and community settings. In a first, proof‐of‐concept study, 18 we proposed a smartphone/tablet‐based digital biomarker implementing augmented reality tasks inspired by complex functional instrumental activities of daily living (Altoida iADL tasks), mostly related to spatial memory and navigation abilities. During a predefined task sequence, the app collected data from the smartphone/tablet built‐in sensors, profiling hands’ micromovements, screen touch frequency, reaction time, correct responses, walking bouts and speed, navigation trajectory, and many others, 19 presently defined as Neuro Motor Index (NMI). The study was held in two memory clinics, recruiting 215 subjects that were either cognitively normal (NC), with MCI or early dementia followed over the course of 60 months. 20 In that study the digital biomarker alone explained most of the variability for impaired executive function assessed with ADNI‐Exec score compared to other biomarkers, while prediction of MCI‐to‐dementia transition assessed using Cox proportional hazard model showed specificity and sensitivity >90%.
In the present article we report the outcome of a novel multisite, prospective cohort study that followed 548 subjects with either preclinical AD with no cognitive deficits or with MCI diagnosis, for up to 40 months (AltoidaML). 21 These subjects were recruited to be assessed in memory clinics, in community facilities, or at home. Data from each subject were obtained at baseline using smartphone/tablet augmented reality tasks whose NMI outcome was analyzed with a ML algorithm to predict the progression of individuals with MCI into dementia within 3 years. Taking our previous risk prediction models as a starting point, the aim of this extensive external validation study was to establish robust, generalizable prediction models using NMI digital biomarkers only.
HIGHLIGHTS
Digital biomarker prognostic models can accurately classify individuals at risk to progress to dementia within 3 years.
Digital biomarker prognostic models can accurately classify individuals with amyloid neuropathology.
Digital biomarker prognostic models can accurately classify individuals with slow or rapid cognitive decline.
Digital biomarker prognostic models show similar performance in both supervised and unsupervised environments.
Digital biomarker prognostic models can be a useful tool to assist large‐scale population screening.
RESEARCH IN CONTEXT
Systematic review: The authors reviewed the literature using traditional (eg, PubMed) sources and meeting abstracts and presentations, searching, without language restriction, for articles published up to April 1, 2020, on prognosis in people with mild cognitive impairment (MCI), at an individual level, on the basis of digital or biological biomarker evidence, using the terms “([mild cognitive impairment] AND [prognosis] OR [prognostic factor] OR [digital biomarker] OR [prediction model]).” We found a wealth of literature on the prognostic performance of biological biomarkers in individuals with MCI at the group level and very few that directly translate to the individual. On the other hand, we found no prospective studies on the prognostic performance of digital biomarkers, besides our own study that examined a complex instrumental activity of daily living marker on an individual level in people with MCI. For this validation study, we took our previously constructed digital biomarker‐based prognostic models and focused on the generalizability and robustness of our prediction models (external validation) during a 3‐year clinical trial at different clinical sites both in Europe and the United States.
Interpretation: In the current study of 496 individuals with diagnosis of cognitively normal or MCI from multicenter cohorts in Europe and North America, we validated and updated, according to the TRIPOD (transparent reporting of a multivariable prediction model for individual prognosis or diagnosis) guidelines, multivariable, biomarker‐based models for the prediction of dementia on the basis of digital biomarkers only. We showed that the models had good generalizability and were well calibrated up to more than 40 months of follow‐up.
Future directions: A validated digital biomarker has significant implications on clinical practice and clinical decision‐making for people with mild cognitive impairment. We have shown generalizability and robustness of our predictions. Our models could facilitate a more timely and accurate diagnosis and prognosis of MCI, which is of high importance at the clinical practice. As a next step our models can allow clinical researchers to calculate trajectories of dementia progression within a given period of time and design therapeutic interventions that stabilize or reverse those trajectories, as a good starting point for dementia care.
2. METHODS
2.1. Database, study design, and participants
Data collected from two different studies (study A and B) that shared similar entry criteria, clinical scale, and AD biomarker characterization were considered for the analysis. Subjects between 55 and 90 years of age with cognitive impairments consistent with MCI and AD dementia diagnosis according to core criteria of National Institute on Aging‐Alzheimer's Association (NIA‐AA) revised guidelines 22 were included independently of their biomarker status. Clinical assessment included the Wechsler Memory Scale (adjusted for education), Mini‐Mental State Exam (MMSE), Clinical Dementia Rating (CDR), and Memory Box score.
AD biomarkers, consisting of Aβ and tau protein cerebrospinal fluid (CSF) levels, brain magnetic resonance imaging (MRI), and apolipoprotein E (APOE ɛ3/ɛ4) genotype, were collected at baseline, when also the Altoida iADL tasks were performed. Classification in the diagnostic clusters of MCI and dementia due to AD (aMCI and ADD) or MCI and dementia not associated with AD (naMCI and nADD) were performed based on the Aβ and tau protein CSF levels biomarker. Among the exclusion criteria, any significant neurologic disease, such as Parkinson's disease, Huntington's disease, normal pressure hydrocephalus, brain tumor, progressive supranuclear palsy, seizure disorder, subdural hematoma, multiple sclerosis, or history of significant head trauma followed by persistent neurologic defaults or known structural brain abnormalities. Every 12 month, each subject was assessed for their clinical and neuropsychological conditions with either MMSE or Montreal Cognitive Assessment (MOCA) tested in a clinical setting, when decision about the occurrence of a transition from MCI to early AD was made based on the diagnostic core criteria of NIA‐AA 22 for the MCI and early dementia condition, respectively. Clinical outcomes were ascertained by investigators blinded to the predictor variables during the various periodic visits. All studies were approved by the local Institutional Review Board (IRB). Upon enrollment, all participants gave written informed consent for participation and for reuse of the data.
Study A was a semi‐naturalistic observational study that included 215 subjects, ages 55 to 90 years, with preclinical AD, MCI, and early AD diagnosis recruited in two memory clinics, one in Greece and one in California. Subjects were tested every 12 months for a total duration of 60 months between 2009 and 2014 and results were published in 2015. 20 A summary of the study population is shown in Table 1. At baseline, cognitive deficits compatible with MCI diagnosis were found in 61 subjects.
TABLE 1.
Demographic and clinical characteristics of participants in the cohorts included in the Altoida database at baseline
| First study (Tarnasas et al. 2015) (n = 215) | Altoida ML study (NCT02843529) (n = 496) | |
|---|---|---|
| Follow‐up time, years | 4.5 (2.6) | 2.6 (1.6) |
| MCI subjects | 61 | 213 |
| Number of MCI subjects progressing to dementia | 37 (60%) | 100 (47%) |
| Number of MCI subjects with β‐amyloid biomarker progressing to AD dementia | 30 (49%) | 79 (37%) |
| MCI subjects progressing to other types of dementia | 7 (11%) | 21 (10%) |
| Average age, years (SD) | 72 (9) | 67 (8) |
| Female | 118 (55%) | 306 (62%) |
| Male | 97 (45%) | 190 (38%) |
| MMSE | 26 (2) | 27 (2) |
| Hippocampal volume, cm³ | 5.4 (1.6) | 6.2 (1.2) |
| CSF biomarkers, pg/ML | ||
| Amyloid beta (Aβ1‐42) | 925 (297) | 790 (378) |
| Total tau (t‐tau and p‐tau‐181/tau ratio) | 286 (191) | 270 (140) |
| Phosphorylated tau (p‐tau‐181/p‐tau231) | 27 (15) | 33 (18) |
Data are n (%) or mean (SD).
Abbreviations: AD, Alzheimer's diseaseCSF, cerebrospinal fluid; MCI, mild cognitive impairment; MMSE, Mini‐Mental State Examination; SD, standard deviation.
Study B (ClinicalTrials.gov Identifier: NCT02843529) was a semi‐naturalistic observational multicenter study performed in both memory clinics and primary care centers that enrolled 496 subjects with diagnosis of NC or MCI from 10 European memory clinics and primary care centers and two primary care community centers in the United States, collected between October 2017 and February 2020. More specifically, a total of seven European memory clinics were: Greek Alzheimer's Association and Related Disorders “Ag. Giannis” and “Ag. Eleni” memory clinics in Thessaloniki, Greece; the University of Roma La Sapienza memory clinic in Rome; IRCCS Centro San Giovanni di Dio Fatebenefratelli memory clinic in Brescia and Neuromed IRCCS memory clinic in Naples, Italy; Fundacion Clinic per a la Recerca Biomédica memory clinic in Barcelona, Spain; and University of Dublin, Trinity College, St James memory clinic in Dublin, Ireland. The three primary care centers from Europe were: BiHELab–Bioinformatics and Human Electrophysiology Lab and affiliated primary physicians’ network in Corfu, Greece and two offices from the Practice for Personalized Medicine of the Hirslanden Private Hospital in Switzerland (Zurich & Aarau). Finally, the two primary care community centers in the United States were Scripps Health at La Jolla, California and the Center for Brain Health—The University of Texas at Dallas.
The key inclusion criteria were: (1) 55–90 years of age; (2) fluency in English, French, Spanish, German or Italian; and (3) familiarity with digital devices, including currently possessing and actively using an iPad Pro or iPhone with an at‐home Wi‐Fi network for the remote assessments. Biomarkers for CSF, brain MRI, and APOE ɛ3/ɛ4 genotype were assessed at baseline together with the Altoida iADL test. A control group of 283 healthy individuals matched by age and that underwent the same procedure were provided by Global Brain Health Institute (GBHI) at Trinity College, Dublin. The summary of the study population is shown in Table 2. At baseline, cognitive deficits compatible with MCI diagnosis were found in 213 subjects, 170 from the memory clinics and primary care centers in Europe, and 43 from the community centers in the United States.
TABLE 2.
Characteristics of the MCI population included in validation analyses
| Characteristic | EU population (n = 170) | U.S. population (n = 43) |
|---|---|---|
| Data collection period | 2017–2020 | 2017–2020 |
| Study design | Multicenter longitudinal cohort study | Multicenter longitudinal cohort study |
| Setting | Memory clinics and primary care | Primary care |
| Inclusion criteria | Memory complaints verified by study partner, abnormal memory functioning, MMSE of 24–30, Clinical Dementia Rating scale of 0·5, does not fulfill the criteria for dementia | Unspecified memory complaints, abnormal memory functioning, MMSE of 24–30, Clinical Dementia Rating scale of 0·5, does not fulfill the criteria for dementia |
| Participants who developed dementia | 74 (44%) | 16 (35%) |
| Follow‐up | Clinical follow‐up every 12 months | 6–12‐month interval |
| MRI measurements available | 149 (88%) | 40 (95%) |
| MRI quantification method | FreeSurfer version 5.3 | FreeSurfer version 5.3 |
| CSF samples with biomarkers measurements available | 124 (73%) | 17 (41%) |
| CSF platform | Innotest | Innotest |
Abbreviations: CSF, cerebrospinal fluid; MCI, mild cognitive impairment; MRI, magnetic resonance imaging.
An additional substudy was included aimed to test the smartphone version of the Altoida iADL when the subjects were at home rather than in the clinics. It consisted of 100 subjects that were provided with an iPhone 6 Plus to be used as their device for Altoida iADL home assessments. The study was approved by IRBs and is reported in accordance with the TRIPOD (transparent reporting of a multivariable prediction model for individual prognosis or diagnosis) guidelines. 23
2.2. Digital device and data generation
The Altoida iADL test is presented as a smartphone/tablet app providing augmented reality interactive tasks with varying difficulty levels inspired by iADL that require coordination of information by eliciting medium‐to‐high cognitive control, including spatial memory, prospective memory, executive function, and psychomotor processing speed. 20 Initially the user is asked to follow instructions and perform fine and gross motor tests on the touch screen to calibrate the device and assess the subject interface‐use competence; then other introductive information prompts the user to perform the navigation (NAV) task consisting of “hiding” three virtual objects (eg, star, heart, teddy bear) appearing on the screen while exploring the environment through the live camera by targeting conspicuous landmarks and clicking on the home button. After a few minutes, the subject is asked to recover the hidden virtual objects from the selected environmental landmarks and find them in a different order while performing a simple odd‐ball task used as distraction to increase the task difficulty. During the object‐finding task, a total of 731 different metrics are collected from the smartphone/tablet touch screen, gyroscope, and accelerometers assessing time and intensity of various activities engaged during the tasks. These measurements were organized in 109 key functional motor behaviors related to the tasks, including gait strides and speed, grip strength, purposeless screen touch, spatial distribution of the motility path complexity when finding the augmented reality objects, time‐to‐hide and time‐to‐find each object, location, and order errors in object finding, etc. (see Figure 1 and Table S1 in supporting information). These data are collectively integrated into the NMI that represent the overall outcome of the individual task performance. Results are visualized on a dashboard available to the study team and stored for statistical analysis. Instructions about how to perform the task were generally provided by trained clinical staff, but self‐teaching videos were also developed and used, in particular for the home testing.
FIGURE 1.
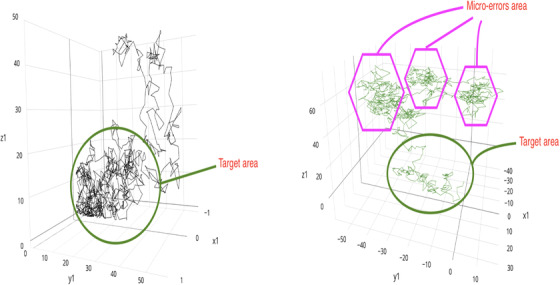
Visual representation of three motor features during augmented reality task. Visual representation of three features (out of 109 key functional motor behaviors) corresponding to the “distribution of the path complexity when finding the augmented reality objects” (see Table S1). LEFT PANEL, Typical digital phenotype of mild cognitive impairment (MCI) subjects that will remain stable over time (n = 113). RIGHT PANEL, Typical digital phenotype of MCI that would progress to dementia over time for a period of 3 years
Altoida iADL with NMI output were recognized as 513 g exempt Class 2 medical devices by the U.S. Food and Drug Administration (FDA) and Class 1 medical devices by CE in the EU. All data collected are encrypted in transit, sent to the cloud and stored on Altoida's Digital Biomarker Platform. This platform is compliant with the Health Insurance Portability and Accountability Act and the FDA Code of Federal Regulations (CFR) Title 21 Part 11.
From the smartphone/tablet neuro‐motor recordings and individual demographic data, 112 predictor variables were identified per subject for use in the prediction models. These predictor variables included: three demographic characteristics (age, years of education, and sex) and the 109 neuro‐motor parameters that generate the NMI (see Table S1). In previous studies these neuro‐motor parameters were seen to correlate to different cognitive subdomains tested with validated neuropsychological tests 19 , 20 (ie, perceptual motor function; motor coordination, visual perception, complex attention; processing speed, executive function; planning and decision making) as shown in the supplemental material (p 5‐9). No other biomarker was included in the model, being used only to define post hoc the disease progression.
2.3. Machine learning model
We used the XGBoost classification algorithm 24 as the prediction model, using binary logistic regression of conversion to dementia or not, as the learning objective. In addition, we used SMOTE (synthetic minority oversampling technique) 25 as the oversampling technique during model training to address class imbalance. To improve computation time, we applied hyperparameter optimization within each training set, using the data for MCI converting to dementia. For all other classification models, we use the thus‐found optimal model. For hyperparameter optimization, we split the data into multiple training, validation, and test sets using nested k‐fold cross‐validation 26 with five outer and five inner folds. Each outer fold returned one classifier after model building, validation, and hyperparameter optimization over the inner folds. Generalization error for this model was estimated by averaging test set scores over the five outer dataset splits. As a hyperparameter optimization strategy, we used a grid search, optimizing the learning rate, number of estimators, maximum tree depth, column subsampling, and row subsampling while optimizing for area under the receiving operating curve (ROC‐AUC). The optimal parameters, 0.001, 5000, 7, 0.35, and 0.95, respectively, were then used for all classifiers.
We validated our results using both internal‐external and external validation. 27 The Study A and Study B datasets consisted of data from 711 patients in total, with 109 of them diagnosed with AD and 27 with other types of dementia as of the last study time point. For each patient Pi (1 ≤i ≤N), the Study A and B dataset included Li separate examinations by a physician and N was the total number of patients in the dataset. Then, each NMI examination of the ith patient could be defined as a multidimensional vector, where di was the date of the examination and ci was the clinical state of the patient (normal, MCI, or dementia) as measured during that examination. Generally speaking, Study A was used as a training and internal‐external validation dataset, whereas Study B was used as external validation to test the ability of the trained machine learning model to predict the progression of AD on an independent patient population. Because the Study B data were only used to test the machine learning models and not for training, we used a fixed set of hyper‐parameters for the XGBoost algorithm with a time variable that represents the number of months into the future that the machine learning model should make a prediction. The probabilities calculated by the machine learning algorithm based on these examinations were averaged to generate predicted probabilities for patient Pi at time t. When comparing the model's predictions against the actual diagnoses in Study B, the predicted probability was compared to the examination date documenting a new diagnosis in Study B. An illustration of the method is at Figure 2.
FIGURE 2.
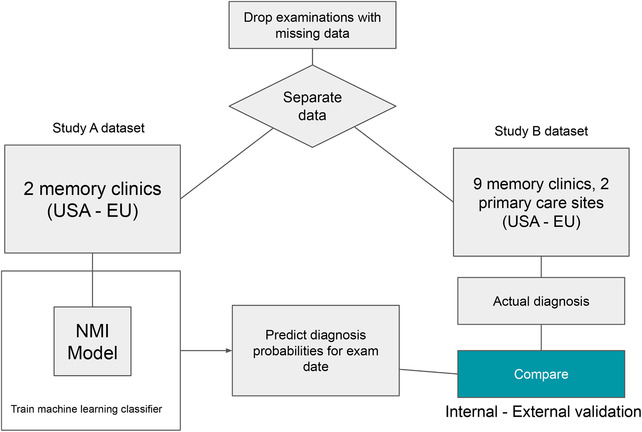
Flowchart showing methodology for training and evaluating machine learning models
2.4. Outcomes for machine learning
The primary outcome in the training set was the conversion from MCI‐to‐dementia assessed using clinical measurements (ie, either MMSE or MOCA, and CDR) to any stage of cognitive impairment at any time. In a secondary analysis, the same outcome was considered for those MCI subjects that would progress into dementia due to AD (aMCI) or not associated with AD (naMCI).
2.5. Statistical analysis
To identify the size of the dataset necessary for obtaining a clinically relevant analysis, we used the information available from NC and MCI participants in our previous study. 20 Using Clinical Dementia Rating scale sum of boxes (CDR‐SB) as an outcome at 40 months, a minimum of 200 individuals per class was needed to demonstrate difference in the longitudinal disease progression, that is, stability over time or significant changes toward dementia, in keeping with the recent literature. 28 To assess real‐world performance, the NMI models were trained on the entire Study A dataset and then evaluated on data derived from Study B and compared to actual diagnoses, asking whether NMI can predict their later diagnoses. The training dataset from Study A was covering 60 months with an average of 5.45 examinations per patient, totaling 1171.75 examinations. The actual examination dates in Study B vary from patient to patient, but they generally cover a 40‐month period containing a total of 2961.12 examinations, or an average of 5.97 examinations per patient.
For all analyses, except the cognitive domain comparison, we used all 112 predictors without further feature selection. For the cognitive domain comparison, we trained multiple prediction models using only the predictors corresponding to each cognitive domain. For all analyses, predictors with missing data were handled automatically by the XGBoost algorithm and we applied no manual imputation.
We validated our results using both internal‐external and external validation as proposed by Steyerberg and Harrell (2016). 27 For internal‐external validation, we used five‐fold cross‐validation using all applicable subjects from Study A and a leave‐maximum‐one‐center‐out scheme. Because not all recruitment centers had all study population classes represented (healthy, MCI, MCI due to AD), our test sets consisted of either a single center or of two centers selected such that each test set contained all population classes. Each data collection site was only used once in a test set. Each of the test sets was left out once for the validation of a model based on all remaining data. For external validation we used the Study B dataset.
Model performance was primarily measured using the ROC‐AUC, with a mean and standard deviation performance based on averages over the testing folds. In some cases, the precision‐sensitivity curve was also used to offer information about the reliability of the prediction, particularly for imbalanced data. Representative model accuracy, sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), prognostic summary index (PSI), positive likelihood ratio (LR+), and negative likelihood ratio (LR–) are presented regarding the point on the ROC curve corresponding to Youden's index.
3. RESULTS
We included all 496 participants in the external validation dataset, with mean age 67 years (standard deviation [SD] 8), mean MMSE of 27 (SD 2), and of whom 306 (63%) were female (Table 2). During a mean of 3.3 years (SD 2) of follow‐up, 100 (47%) of the MCI participants progressed to dementia, whereas 113 remained relatively stable over time. Characteristics of the MCI participants are shown at Table 1.
3.1. Prediction of conversion from MCI to dementia
The external validation performance of NMI for our primary outcome was significantly better than chance. Clinically defined MCI subjects converting to dementia, independently of their Aβ biomarker value, when contrasted with those MCI subjects that remained stable over the 3‐year period were predicted by NMI with a ROC‐AUC of 91% after cross‐validation, as shown in Figure 2, left panel, as supported by a good discriminative metric profile described in Table 3, column 1.
TABLE 3.
Performance metrics of the NMI model
| Column A | Column B | Column C | Column D | |||||
|---|---|---|---|---|---|---|---|---|
| Statistic | Mean | SD | Mean | SD | Mean | SD | Mean | SD |
| ROC‐AUC | 0.91 | 0.03 | 0.92 | 0.03 | 0.94 | 0.01 | 0.91 | 0.01 |
| Accuracy | 0.85 | 0.05 | 0.86 | 0.02 | 0.88 | 0.02 | 0.89 | 0.03 |
| Precision | 0.90 | 0.04 | 0.83 | 0.04 | 0.94 | 0.07 | 0.96 | 0.02 |
| F1‐score | 0.85 | 0.06 | 0.83 | 0.03 | 0.88 | 0.05 | 0.93 | 0.02 |
| Sensitivity | 0.81 | 0.08 | 0.84 | 0.04 | 0.84 | 0.11 | 0.91 | 0.05 |
| Specificity | 0.90 | 0.04 | 0.88 | 0.04 | 0.95 | 0.03 | 0.82 | 0.09 |
| NPV | 0.82 | 0.06 | 0.89 | 0.02 | 0.88 | 0.07 | 0.71 | 0.11 |
| PSI | 0.72 | 0.10 | 0.72 | 0.05 | 0.82 | 0.06 | 0.67 | 0.10 |
| LR+ | 10.98 | 5.92 | 7.90 | 3.52 | 10.20 | 5.42 | 6.40 | 2.50 |
| LR‐ | 0.22 | 0.09 | 0.18 | 0.04 | 0.19 | 0.04 | 0.11 | 0.05 |
Abbreviations: AUC, area under the curve; LR‐, negative likelihood ratio; LR+, positive likelihood ratio; NMI, Neuro Motor Index; NPV, Negative Predictive Value; PSI, Prognostic Summary Index; ROC, receiver operating characteristic.
Prediction of conversion to dementia of MCI subjects, independently of their Aβ biomarker value, was tested against a mixed population characterized by a stable clinical profile over time for 3 years. To this aim, MCI subjects that did not convert to dementia (n = 113) and healthy control subjects that remained stable over 3 years (n = 283) were included in the models and contrasted with MCI subjects that converted to dementia (n = 100). The performance of the model was characterized by ROC‐AUC of 0.92 (SD 0.3), as shown in Figure 3, left panel, and by a good model metric profile described in Table 3, column 2. Overall, these results are consistent with a good predictive value of the models.
FIGURE 3.
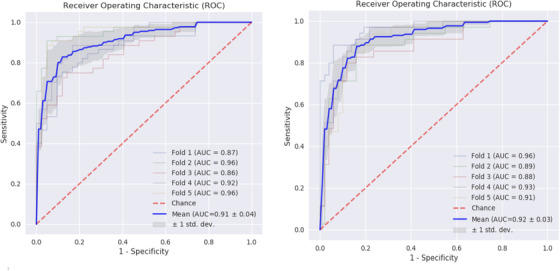
Receiver operating characteristic (ROC) curves for internal cross‐validated results of predicting mild cognitive impairment (MCI) to dementia. Internally five‐fold cross‐validated ROC‐area under the curve (AUC) curves AUC‐ROC analysis, in identifying MCI subjects that progress to dementia in a 3‐year period. LEFT PANEL, Assessment versus MCI subjects that will remain stable over time (n = 113). RIGHT PANEL, Assessment versus the combined population of MCI and healthy control subjects that will remain stable over time for a period of 3 years
3.2. Prediction of conversion from aMCI to AD dementia
We also assessed a prediction model to distinguish between aMCI, defined by positive Aβ and tau biomarker at baseline, which would convert to AD dementia, versus a pooled population characterized by a stable response over 3 years, consisting of those aMCI that do not convert to dementia and healthy control but positive Aβ biomarker subjects. To test this prediction model, we used 450 subjects in total from which 67 healthy controls with Aβ biomarker measured and 167 aMCI subjects, 79 of those converting to dementia. All these subjects were assessed in a clinical setting. After cross‐validation, the estimated generalization error of the prediction model gave a ROC‐AUC of 0.94 (SD 0.01) and good model performance metrics shown in Figure 4, left panel and Table 3, column 3. These data are consistent with a good predictive value of the model.
FIGURE 4.
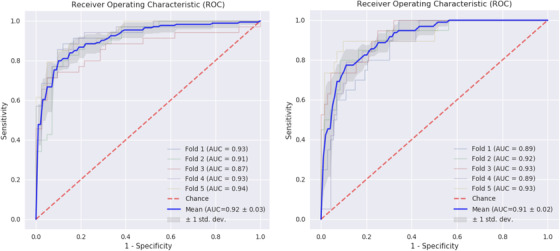
Receiver operating characteristic (ROC) curves for internal cross‐validated results of predicting amnestic mild cognitive impairment (aMCI) to Alzheimer's disease (AD) dementia. Internally five‐fold cross‐validated ROC curves predicting the conversion of subjects with aMCI to dementia due to AD versus a stable pooled population of non‐converters (LEFT PANEL) and predicting aMCI subject undergoing rapid conversion (<24 month) versus late conversion (RIGHT PANEL)
Moreover, within the group aMCI subjects likely to convert to AD, the predictive model was trained to discriminate between rapid progressors that convert into AD within 18 months and slow decliners that convert into AD afterward. The generalization error of this model, namely the rate of decline, was estimated at a ROC‐AUC of 91% (SD 0.01). Performance metrics of the model are shown in Figure 4, right panel and Table 3, column 4. Overall, these results indicate the possibility to identify at baseline subpopulations of aMCI subjects that will progress to AD within 3 years and also those with a faster rate than other patients with positive Aβ and/or tau biomarkers.
External validation of the predictive model was further implemented for the classification of MCI subjects converting to AD (aMCI) as defined by positivity of Aβ biomarker at baseline (AD neuropathology) versus all MCI subjects converting to other types of dementia (naMCI) characterized by Aβ negative biomarker. The model was trained using 46 naMCI (n = 46) and aMCI (n = 167) subjects. The estimated generalization error gave a ROC‐AUC of 90% (0.01 SD) with sensitivity of 0.90 (SD 0.06), specificity of 0.81 (SD 0.10), precision 0.95 (SD 0.03), accuracy (0.88 [SD 0.04], NPV 0.70 [SD 0.12], and PSI 0.65 [SD 0.11]; see Figure S4 and Table S4 in supporting information).
3.3. Testing heterogeneity: predictive performance when the task is assessed in the community, memory clinic, or at home
Regarding the unsupervised assessment, aMCI subjects and age‐matched healthy controls agreed to volunteer for unsupervised, home‐based tests, and for yearly follow‐up assessment. The predictive ML model was trained with data from aMCI subjects (n = 45) and control subjects (n = 55). The estimated generalization error of this predictor gave a ROC‐AUC of 0.93 (SD 0.2) with sensitivity of 0.82 (SD 0.07), specificity of 0.91 (SD 0.8), precision 0.89 (SD 0.10), accuracy 0.87 (SD 0.04), F1 0.84 (0.04), NPV 0.88 (SD 0.04), and PSI 0.76 (SD 0.09) (supplemental material p 14).
Furthermore, an internal‐external validation process was performed to account for any center‐specific effects, that is, memory clinic, primary care, and unsupervised assessment (home environment). For this analysis, we left a maximum of two settings out at a time to cross‐validate the ML model developed in other settings, aimed to deliver our primary outcome (conversion to dementia), and secondary outcome (conversion to AD dementia). The obtained performance metrics are described in Table 4.
TABLE 4.
Performance metrics for internal‐external validation of center‐specific effects
| Internal‐external validation (dementia) | Internal‐external validation (AD) | |
|---|---|---|
| ROC‐AUC | 0.87 (0.05) | 0.85 (0.12) |
| Accuracy | 0.81 (0.06) | 0.86 (0.04) |
| Sensitivity | 0.80 (0.05) | 0.90 (0.06) |
| Specificity | 0.83 (0.13) | 0.81 (0.10) |
| PPV | 0.85 (0.09) | 0.85 (0.12) |
| NPV | 0.80 (0.04) | 0.84 (0.15) |
| PSI | 0.64 (0.11) | 0.65 (0.11) |
| LR+ | 8.15 (5.58) | 6.05 (2.56) |
| LR– | 0.25 (0.05) | 0.12 (0.07) |
Notes: Data are ROC‐AUC (std.) Leave‐maximum‐two‐sites‐out internal‐external validation metrics for memory clinics, primary care, and unsupervised, at home assessment settings. Representative accuracy, sensitivity, specificity, PPV, NPV, PSI, LR+, and LR– are based on the point on the receiver operating characteristic curve corresponding to Youden's index. Different thresholds can be taken on the basis of the requirements of the diagnostic test.
Abbreviations: AD, Alzheimer's disease; AUC, area under the curve; LR‐, negative likelihood ratio; LR+, positive likelihood ratio; NPV, Negative Predictive Value; PSI, Prognostic Summary Index.
3.4. Linking cognitive functions with NMI parameter performance in predicting dementia
To assess the predictive power of the most frequently used neuro‐motor predictors, we trained multiple prediction models each using only the predictor variables corresponding to the most frequently used everyday function and/or cognitive domains in performing the Altoida iADL tasks. The bar charts created show a stronger discriminative capacity (ROC‐AUC >0.80) for four cognitive domains, that is, perceptual motor coordination, complex attention, cognitive processing speed, and planning. Interestingly, the same cognitive dimensions were involved in both the primary outcome (prediction of conversion to dementia) and the secondary outcome (prediction of conversion to AD dementia; Figure 5).
FIGURE 5.
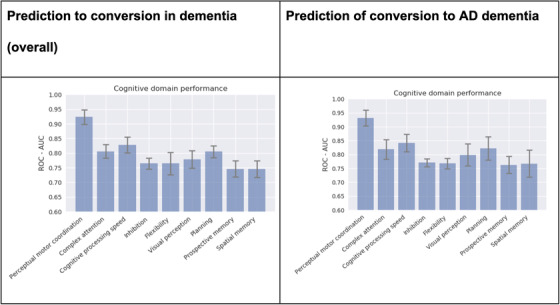
Individual performance of cognitive domains. Individual performance in each cognitive domain engaged by the Neuro Motor Index neuro‐motor parameters most critically involved in the Altoida iADL task performance that predict the conversion into either dementia (LEFT PANEL) or Alzheimer's disease dementia (RIGHT PANEL). The standard deviation is computed over five cross‐validation folds
4. DISCUSSION
In this study, we provided robust evidence that an agmented reality (AR) digital biomarker (NMI) obtained by profiling performance in tasks inspired to activities of daily living using a smartphone/tablet could deliver prognosis for dementia in individuals with MCI over a period of 3 years when performed at baseline, that is, the time of MCI diagnosis.
This result was achieved implementing ML analytics to a database consisting of a relatively heterogeneous set of data obtained in subjects with diagnosis of MCI, of relevance for the generalization value of the model. The data were collected from subjects of both sexes in two semi‐naturalistic observational studies executed in different periods during the last decade, recruited from both memory clinics and primary care settings in various European countries and the United States as well as at home, who spoke six different languages. This diversification in the training set was planned to help the models to generalize, potentially reducing the risk of cohort bias.
The ML model consisted of three demographic variables (age, sex, and years of education) and 109 neuro‐motor parameters collected from the signal generated by built‐in sensors of the smartphone/tablets used to perform the NMI tasks inspired by complex instrumental activities of daily living. 20 The model performance for predicting the main clinical outcome (ie, changes in MMSE/MOCA and CRD that define the dementia condition) had an excellent performance (ROC‐AUC 0.91) in those subjects with clinical MCI that convert to dementia compared to subjects that remain stable over 3 years. A similar performance (ROC‐AUC 0.92) was obtained in the subgroup of aMCI subjects characterized by positive Aβ and tau biomarkers at baseline that convert into dementia due to AD within 3 years. Intriguingly, also the subgroup of MCI subjects converting to AD (aMCI) as defined by positivity of Aβ biomarker only at baseline (AD neuropathology) versus naMCI, that is, those individuals with negative Aβ biomarkers that progress into non‐AD dementia, could be identified when tested with NMI at baseline. This result suggests a potential for extending the predictive capacity of NMI outside the strictly defined AD pathogenetic mechanisms, probably due to commonalities in the early regional dysfunction of brain circuits that characterize those MCI conditions evolving toward dementia.
ML modeling was performed on a series of neuro‐motor parameters collected from the smartphone‐tablet sensors during the Altoida iADL tasks that were previously related to a series of cognitive domains. 18 , 19 , 20 These parameters, all included in the NMI score, when assessed against the cognitive domains, indicated a principal role for perceptual motor coordination, complex attention, cognitive processing speed, and planning (as indicated by ROC‐AUC 0.81‐0.95). These impaired cognitive functions were similarly involved in predicting the conversion into dementia for subjects with clinically defined MCI, as well as the specific conversion in AD dementia in aMCI subjects with positive Aβ biomarker at baseline, suggesting possible commonalities on defective neural substrates already engaged at the MCI stage. Accordingly, metabolic positron emission tomography (PET) imaging studies consistently describe cortical hypometabolism, in the inferior and superior temporal gyri and inferior parietal lobule, and subcortical hypometabolism, in the primarily posterior cingulate cortex and the hippocampus, of MCI subjects that progress to dementia. 29 , 30 These regions are involved in visuo‐spatial motor coordination processing, complex attention, decision making, and procedural memory, all critically involved in iADL performance. Intriguingly, the data collected with the NMI Altoida iADL tasks and the neuroimaging profiling are in keeping with recent findings on the “Motoric Cognitive Risk Syndrome” 31 and the “Gait and Cognition Syndrome” 32 that also occur in MCI subjects that develop dementia. The present data also corroborate the predictive power of impairments in spatial memory and planning detected during walking or navigation tasks previously published. 33
Another result of the present study was the application of the ML model to discriminate between aMCI subjects that rapidly (ie, within 18 months from diagnosis) converted to dementia due to AD against those that show a slow decline and that convert after 36 months. The predictive performance was good (AUC 0.91). The fact of being able to differentiate between rapid and slow decliners at baseline may have important implications for the definition of the study population in clinical trials for novel disease‐modifying treatment. The model was also applied to identify those naMCI subjects that convert to non‐AD dementia within 3 years, with a very good performance (AUC 0.92).
One of the remarkable aspects of these results is that the predictive capacity of the digital biomarker NML when considered alone (not in conjunction with other biomarkers) and associated to three basic demographic parameters (ie, age, education, and sex) is similar or moderately better than those so far published and based on multiple biomarkers. For example, ABIDE, a comprehensive risk prediction model based on the ATN biomarker construct, included 2116 subjects from different North‐American and European cohorts, delivered the highest ROC‐AUC performance of 0.74, 14 which is in the low range of ROC‐AUC we obtained with NMI digital biomarker. Another model using support vector machine with linear kernel on the dataset of the Dementia Competence Network, a German multicenter cohort observational study performed on 115 subjects with MCI due to AD with annual follow‐up up to 3 years, showed that by combining up to four AD biomarkers in the same model, the ROC‐AUC predicting aMCI conversion to AD dementia was 0.82, significantly superior to the 0.77 AUC value obtained using the single best biomarker, that is, the hippocampal volume. 15
In recognition of the fact that intervention strategies are progressively using high‐frequency, unsupervised, home based assessments 29 results from the substudy on our 100 subjects suggest that unsupervised NMI obtained at home using a smartphone/tablet could also be used to predict the risk of progression to AD within the aMCI population (ROC‐AUC 0.93). While raising possible ethical problems about informing subjects of their risks to developing dementia, this evidence, if confirmed, has great potential for implementation in clinical trials and for optimizing the available treatment in a cost‐effective way. In addition, a recent meta‐analysis of data collected from tasks performed at different times of the day in unsupervised environment showed a preference for later‐in‐the‐day testing periods, when cognition and/or functionality could be different from the morning time according to the literature. 34 , 35 , 36
The present study has certain limitations. Among them, the predictions of our models were calibrated every 12 months (ie, assessing concordance between predicted and observed clinical outcome). This sample frequency was chosen mostly for feasibility, because in this case, higher frequency assessments, for example, every 3 months, would constitute a burden for the participant and be more expensive. Another limitation is that we did not include APOE ɛ3/ɛ4 genotype or polygenic risk score information in our models. Because genotyping is currently rarely used in clinical practice and we ultimately wanted to focus on the digital biomarker platform alone, in the present study we didn't combine the genotyping information with NMI. Finally, the time of testing at home was not controlled in the unsupervised environment, so it is unclear if time‐of‐day effects should have been added as predictors in the current study.
In conclusion, we have shown the generalizability and robustness of the NMI digital biomarker‐based ML models for prediction of progression to dementia in individuals with MCI, in particular for those progressing to AD. The ease of handling, the high diagnostic accuracy, the assessment's brevity, and the noninvasiveness renders NMI an ideal tool for participant profiling, cutting down enrolment periods and costs for cumbersome screenings, aside from risk prediction and disease monitoring. The unique capability to distinguish individuals in the MCI group that progress to dementia, as well as fast and slow aMCI cognitive decliners, is of utmost importance in the definition of the study population in clinical trials and for its recruitment, eventually reducing the risks of false positives.
While many digital biomarker efforts are in progress, particularly in neurodegenerative disorders, NMI sets itself apart through its high sensitivity and the rapidity of the assessment. In this light, it could serve as a starting point for precision medicine plans for AD.
FUNDING AND DISCLOSURE
The AltoidaML Study was an IRB‐approved multi‐site 40‐month randomized prospective clinical trial conducted by Altoida, Inc. on behalf of the European Institute of Innovation & Technology (EIT), the Alzheimer's Association USA through the Global Brain Health Institute (GBHI), and Takeda Pharmaceutical Company Ltd. The European Institute of Innovation & Technology (EIT), the Global Brain Health Institute (GBHI), and Takeda Pharmaceutical Company Ltd. provided partial support for data analyses. The funders of this study were not involved in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author and co‐authors had full access to all the data in the study and had final responsibility for the integrity of the data, analyses developed in the present study, and the decision to submit for publication.
CONFLICTS OF INTEREST
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this article.
The authors declare the following financial interests/personal relationships which may be considered potential competing interests:
Dr. Buegler, Dr. Meier, Dr. Harms, and Dr. Tarnanas report personal fees from Altoida Inc., outside the submitted work; Dr. Pich reports personal fees from Takeda Pharmaceuticals Ltd., outside the submitted work; in addition Dr. Tarnanas reports gludes human subjects and followrants from EIT Health, grants from Alzheimer's Association US – Global Brain Health Institute, grants from Takeda Pharmaceuticals Ltd., outside the submitted work; finally, Dr. Tarnanas has a patent “Apparatus, method, and program for determining a cognitive state of a user of a mobile device” pending.
DATA SHARING
The corresponding author can provide the dataset used or documentation on the analysis performed upon reasonable request.
Supporting information
Supplementary information
Buegler M, Harms RL, Balasa M, et al. Digital biomarker‐based individualized prognosis for people at risk of dementia. Alzheimer's Dement. 2020;12:e12073 10.1002/dad2.12073
The work includes human subjects and follows the Declaration of Helsinki guidelines. Informed consent was obtained for experimentation. The privacy rights of human subjects were always observed.
REFERENCES
- 1. Vos SJB, Verhey F, Frölic L, et al. Prevalence and prognosis of Alzheimer's disease at the mild cognitive impairment stage. Brain. 2015;138(Pt 5):1327‐1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Michaud TL, Su D, Siahpush M, Murman DL. The risk of incident mild cognitive impairment and progression to dementia considering mild cognitive impairment subtypes. Dement Geriatr Cogn Dis Extra. 2017;7(1):15‐29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fleisher AS, Sowell BB, Taylor C, et al. Clinical predictors of progression to Alzheimer disease in amnestic mild cognitive impairment. Neurology. 2007;68(19):1588‐1595. [DOI] [PubMed] [Google Scholar]
- 4. van Maurik IS, Zwan MD, Tijms BM, et al. Interpreting biomarker results in individual patients with mild cognitive impairment in the Alzheimer's biomarkers in daily practice (ABIDE) project. JAMA Neurol. 2017;74(12):1481‐1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Petersen RC, Lopez O, Armstrong MJ, et al. Practice guideline update summary: mild cognitive impairment. Report of the Guideline Development, Dissemination, and Implementation Subcommittee of the American Academy of Neurology. Neurology. 2018;90(3):126‐135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bhagwat N, Viviano JD, Voineskos AN, Chakravarty MM, Alzheimer's Disease Neuroimaging Initiative . Modeling and prediction of clinical symptom trajectories in Alzheimer's disease using longitudinal data. PLoS Comput Biol. 2018;14(9):e1006376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lorenzi M, Filippone M, Frisoni GB, Alexander DC, Ourselin S, Alzheimer's Disease Neuroimaging Initiative . Probabilistic disease progression modeling to characterize diagnostic uncertainty: application to staging and prediction in Alzheimer's disease. Neuroimage. 2019;190:56‐68. [DOI] [PubMed] [Google Scholar]
- 8. Davatzikos C, Bhatt P, Shaw LM, Batmanghelich KN, Trojanowski JQ. Prediction of MCI to AD conversion, via MRI, CSF biomarkers, and pattern classification. Neurobiol Aging. 2011;32(12):2322 e19‐27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Moradi E, Pepe A, Gaser C, Huttunen H, Tohka J, Alzheimer's Disease Neuroimaging Initiative . Machine learning framework for early MRI‐based Alzheimer's conversion prediction in MCI subjects. Neuroimage. 2015;104:398‐412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang T, Qiu RG, Yu M. Predictive modeling of the progression of Alzheimer's disease with recurrent neural networks. Sci Rep. 2018;8(1):9161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mathotaarachchi S, Pascoal TA, Shin M, et al. Identifying incipient dementia individuals using machine learning and amyloid imaging. Neurobiol Aging. 2017;59:80‐90. [DOI] [PubMed] [Google Scholar]
- 12. Fisher CK, Smith AM, Walsh JR. Machine learning for comprehensive forecasting of Alzheimer's disease progression. Sci Rep. 2019;9(1):13622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Frisoni GB, Boccardi M, Barkhof F, et al. Strategic roadmap for an early diagnosis of Alzheimer's disease based on biomarkers. Lancet Neurol. 2017;16(8):661‐676. [DOI] [PubMed] [Google Scholar]
- 14. van Maurik IS, Vos SJ, Bos I, et al. Biomarker‐based prognosis for people with mild cognitive impairment (ABIDE): a modelling study. Lancet Neurol. 2019;18(11):1034‐1044. [DOI] [PubMed] [Google Scholar]
- 15. Frölich L, Peters O, Lewczuk P, et al. Incremental value of biomarker combinations to predict progression of mild cognitive impairment to Alzheimer's dementia. Alzheimers Res Ther. 2017;9(1):84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. De Carli F, Nobili F, Pagani M, et al. Accuracy and generalization capability of an automatic method for the detection of typical brain hypometabolism in prodromal Alzheimer disease. Eur J Nucl Med Mol Imaging. 2019;46(2):334‐347. [DOI] [PubMed] [Google Scholar]
- 17. Chen Y, Denny KG, Harvey D, et al. Progression from normal cognition to mild cognitive impairment in a diverse clinic‐based and community‐based elderly cohort. Alzheimers Dement. 2017;13(4):399‐405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tarnanas I, Tsolaki M, Nef T, Müri RM, Mosimann UP. Can a novel computerized cognitive screening test provide additional information for early detection of Alzheimer's disease?. Alzheimers Dement. 2014;10(6):790‐798. [DOI] [PubMed] [Google Scholar]
- 19. Tarnanas I, Papagiannopoulos S, Kazis D, Wiederhold M, Widerhold B, Tsolaki M. Reliability of a novel serious game using dual‐task gait profiles to early characterize aMCI. Front Aging Neurosci. 2015;7:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tarnanas I, Tsolaki A, Wiederhold M, Wiederhold B, Tsolaki M. Five‐year biomarker progression variability for Alzheimer's disease dementia prediction: can a complex instrumental activities of daily living marker fill in the gaps?. Alzheimers Dement (Amst). 2015;1(4):521‐532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. ClinicalTrials.gov , Evaluation of a Computerized Complex Instrumental Activities of Daily Living Marker (Altoida™) (AltoidaML). Bethesda (MD): National Library of Medicine (US), 2016. [Google Scholar]
- 22. Jack CR, Jr , Albert MS, Knopman DS, et al. Introduction to the recommendations from the National Institute on Aging‐Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7(3):257‐262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Collins GS, Reitsma JB, Altman DG, Moons KGM, members of the TRIPOD group . Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. Eur Urol. 2015;67(6):1142‐1151. [DOI] [PubMed] [Google Scholar]
- 24. Chen TG, Carlos XG, Boost: A Scalable Tree Boosting System. 2016:785‐794.
- 25. Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP. SMOTE: synthetic minority over‐sampling technique. J Artificial Intelligence Res. 2002:321‐357. [Google Scholar]
- 26. Cawley GCT, Talbot NLC. On over‐fitting in model selection and subsequent selection bias in performance evaluation. J Mach Learn Res. 2010;11:2079‐2107. [Google Scholar]
- 27. Steyerberg EW, Harrell FE, Jr . Prediction models need appropriate internal, internal‐external, and external validation. J Clin Epidemiol. 2016;69:245‐247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Grill JD, Nuño MM, Gillen DL, Alzheimer's Disease Neuroimaging Initiative . Which MCI patients should be included in prodromal Alzheimer disease clinical trials?. Alzheimer Dis Assoc Disord. 2019;33(2):104‐112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mosconi L, Tsui WH, Herholz K, et al. Multicenter standardized 18F‐FDG PET diagnosis of mild cognitive impairment, Alzheimer's disease, and other dementias. J Nucl Med. 2008;49(3):390‐398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Greene SJ, Killiany RJ, Alzheimer's Disease Neuroimaging Initiative . Subregions of the inferior parietal lobule are affected in the progression to Alzheimer's disease. Neurobiol Aging. 2010;31(8):1304‐1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Verghese J, Annweiler C, Ayers E, et al. Motoric cognitive risk syndrome: multicountry prevalence and dementia risk. Neurology. 2014;83(8):718‐726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Montero‐Odasso M, Speechley M, Muir‐Hunter SW, et al. Motor and cognitive trajectories before dementia: results from gait and brain study. J Am Geriatr Soc. 2018;66(9):1676‐1683. [DOI] [PubMed] [Google Scholar]
- 33. Howett D, Castegnaro A, Krzywicka K, et al. Differentiation of mild cognitive impairment using an entorhinal cortex‐based test of virtual reality navigation. Brain. 2019;142(6):1751‐1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yaffe K, Falvey CM, Hoang T. Connections between sleep and cognition in older adults. Lancet Neurol. 2014;13(10):1017‐1028. [DOI] [PubMed] [Google Scholar]
- 35. André C, Tomadesso C, de Flores R, et al. Brain and cognitive correlates of sleep fragmentation in elderly subjects with and without cognitive deficits. Alzheimers Dement (Amst). 2019;11:142‐150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen R, Jankovic F, Marinsek N, et al. “Developing measures of cognitive impairment in the real world from consumergrade multimodal sensor streams”, presented at Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information


