Abstract
With the FDA approval of larotrectinib, NTRK fusion assessment has recently become a standard part of management for patients with locally advanced or metastatic cancers. Unlike somatic mutation assessment, the detection of NTRK fusions is not straightforward, and various assays exist at the DNA, RNA, and protein level. Here, we investigate the performance of immunohistochemistry and DNA-based next-generation sequencing to indirectly or directly detect NTRK fusions relative to an RNA-based next-generation sequencing approach in the largest cohort of NTRK fusion positive solid tumors to date. A retrospective analysis of 38,095 samples from 33,997 patients sequenced by a targeted DNA-based next-generation sequencing panel (MSK-IMPACT), 2189 of which were also examined by an RNA-based sequencing assay (MSK-Fusion), identified 87 patients with oncogenic NTRK1-3 fusions. All available institutional NTRK fusion positive cases were assessed by pan-Trk immunohistochemistry along with a cohort of control cases negative for NTRK fusions by next-generation sequencing. DNA-based sequencing showed an overall sensitivity and specificity of 81.1% and 99.9%, respectively, for the detection of NTRK fusions when compared to RNA-based sequencing. False negatives occurred when fusions involved breakpoints not covered by the assay. Immunohistochemistry showed overall sensitivity of 87.9% and specificity of 81.1%, with high sensitivity for NTRK1 (96%) and NTRK2 (100%) fusions and lower sensitivity for NTRK3 fusions (79%). Specificity was 100% for carcinomas of the colon, lung, thyroid, pancreas, and biliary tract. Decreased specificity was seen in breast and salivary gland carcinomas (82% and 52%, respectively), and positive staining was often seen in tumors with neural differentiation. Both sensitivity and specificity were poor in sarcomas. Selection of the appropriate assay for NTRK fusion detection therefore depends on tumor type and genes involved, as well as consideration of other factors such as available material, accessibility of various clinical assays, and whether comprehensive genomic testing is needed concurrently.
Introduction
Promising recent clinical trial data and the subsequent accelerated FDA approval of the selective Trk inhibitor, larotrectinib, for the treatment of solid tumors with an oncogenic NTRK fusion has created an immediate need for NTRK fusion testing in the oncology community. This need is reflected in the National Comprehensive Cancer Center management guidelines for several cancers [1]. Larotrectinib and entrectinib have shown a dramatic and durable activity against locally advanced and metastatic solid tumors with NTRK fusions [2, 3]. Notably, this remarkable activity was seen regardless of tumor site of origin, histologic classification, or NTRK fusion type [2].
Oncogenic fusions occur when one of the three neurotrophic tyrosine receptor kinase genes NTRK1, NTRK2, and NTRK3 (that encode TrkA, B, and C, respectively) fuses with any of a wide variety of fusion partners. Normally, physiologic binding of neurotrophin to the extracellular portion of the Trk receptor is required for receptor dimerization, transphosphorylation, and activation of downstream signaling cascades [4]. However, the oncogenic fusion of the C-terminal kinase domain with the N-terminal fusion partner causes constitutive activation of the Trk pathway and drives tumorigenesis [4–6]. While NTRK fusions were first described in colorectal carcinoma [7], the t(12;15)(p13; q25) translocation resulting in an ETV6-NTRK3 fusion was later found to be characteristic of infantile fibrosarcoma [8, 9], mesoblastic nephroma [10], and secretory carcinomas of the breast and salivary gland [11–13]. More recently, however, NTRK fusions have also been identified at low frequencies across a variety of more common malignancies [14–24].
Because there are three different NTRK genes, a variety of potential fusion partners, and a few possible breakpoints at which different exons of the NTRK tyrosine kinase would join the fusion partner, screening for NTRK fusions is somewhat complex. Assays commonly used to identify NTRK fusions include DNA-level sequencing using large targeted cancer gene panels with inclusion of NTRK introns, RNA level sequencing, and pan-Trk immunohistochemical staining to identify protein-level expression. Here we report the sensitivity, specificity, and pitfalls of various clinically validated assays used to identify NTRK fusions in a cohort of 87 NTRK fusion positive, as well as NTRK fusion negative cancers of different subtypes.
Materials and methods
Case selection
After approval from our institutional review board, a retrospective review of the MSK-IMPACT (DNA sequencing) and MSK-Fusion panel (RNA sequencing) results from January 1st, 2014 to March 30th, 2019 was performed. All 38,095 tumor samples from 33,997 patients were formalin-fixed paraffin embedded tissue, and all testing was performed in CLIA approved laboratories and reported clinically.
DNA sequencing
DNA sequencing was performed using MSK-IMPACT, a hybridization capture based next-generation sequencing assay that interrogates all exons (including those of NTRK1-3) and select introns of 468 genes to identify mutations, copy number changes, microsatellite instability status, and select structural variants in its current iteration (previous versions interrogated 341 and 410 genes) [25]. Sensitivity to detect NTRK1-3 fusions is increased by the inclusion of capture probes for introns 3 and 7 through 12 in NTRK1, intron 15 in NTRK2 and introns 4 and 5 in ETV6, the most common NTRK3 fusion partner. Kinase domain introns in NTRK3 are not captured due to a large size (introns 13, 14, and 15 where most of the fusion breakpoints occur span 193 kb), and additionally these intronic sequences are repetitive, rendering them not feasible to include. DNA-level structural variants involving NTRK1-3 detected by MSK-IMPACT often predicted previously described fusions with known partners. Novel structural variants, including intrachromosomal gene rearrangements, that were not previously known to result in a transcribed fusion were further characterized at the RNA level with an RNA-based next-generation sequencing assay. If a novel structural variant did not result in a transcribed fusion, the structural variant that was detected by MSK-IMPACT was deemed a false positive.
RNA sequencing
Tumors were tested for RNA-level fusions using the MSK-Fusion panel, a custom RNA sequencing panel via a next generation sequencing platform that uses anchored multiplex PCR (via the Archer platform) as previously described [26, 27]. The custom panel covers fusions involving the kinase domains of the following genes: ALK, BRAF, EGFR, ERBB2, ERBB4, FGFR1, FGFR2, FGFR3, KIT, MET, NTRK1, NTRK2, NTRK3, RET, and ROS1. Tumors tested included cases where there was a high suspicion of fusion based on outside testing results or morphology (such as infantile fibrosarcoma, congenital mesoblastic nephroma, or secretory carcinomas of the breast or salivary gland), and cases involving select tumor types commonly enriched in MAPK driver alterations (lung adenocarcinoma, colon adenocarcinoma, pancreas adenocarcinoma) that were negative for MAPK driver alterations (i.e., BRAF p.V600E, KRAS/NRAS/EGFR hotspot) on MSK-IMPACT testing [28].
Definition of positive cases
Tumors positive for NTRK1-3 fusions were defined as those with either a transcribed in-frame fusion involving the kinase domain of NTRK detected by the RNA-based MSK-Fusion panel or, if RNA sequencing was not possible, a previously described NTRK fusion involving the kinase domain of the NTRK gene and a known fusion partner detected by the DNA-based sequencing panel MSK-IMPACT. Novel structural variants involving NTRK detected by MSK-IMPACT were always confirmed at the RNA level by MSK-Fusion. In our cohort, there were two exceptions to the above definition. There was one instance of a secretory carcinoma of the salivary gland with classic histologic features where fluorescence in situ hybridization using an ETV6 breakapart probe was used to define this patient as NTRK3 fusion positive. Additionally, a single melanoma case (cBioPortal ID: P-0009855) was excluded from the study. This case demonstrated a GON4L-NTRK1 structural rearrangement on both DNA-based and RNA-based sequencing, but the tumor also harbored an NRAS p. Q61R driver mutation and whole gene amplification of NTRK1, and the patient was clinically determined to be indeterminate for NTRK fusion.
Immunohistochemistry
Pan-Trk immunohistochemical staining using clone EPR17341 (Abcam, Cambridge, MA) was performed to assess for protein expression as previously described [29]. This rabbit recombinant monoclonal antibody, used in clinical labs and a number of recent studies [29–31], reacts with a conserved proprietary peptide in the C-terminal portion of TrkA, B, and C. Positive staining was defined as staining above background in at least 1% of tumor cells in any pattern including membranous, cytoplasmic, perinuclear, or nuclear. NTRK fusion positive cases that were negative for pan-Trk immunohistochemistry were re-stained with an increased concentration using a 1:25 antibody dilution if additional material was present.
Material was available for 66 cases positive for NTRK fusions as defined above. Negative control cases included 16 cases where a novel NTRK1-3 structural variant was detected by DNA sequencing yet reflex RNA sequencing was negative, 13 cases where NTRK1-3 amplification/copy number gain was detected by DNA sequencing yet RNA sequencing was negative, 241 cases of various tumor types negative for NTRK fusions by RNA sequencing, and 47 additional cases with strong MAPK pathway (BRAF p. V600E, KRAS/NRAS hotspot) drivers seen by DNA sequencing in the absence of NTRK1-3 structural rearrangements. The latter were included as previous studies have shown that NTRK1-3 fusions are mutually exclusive with the aforementioned mutations in the vast majority of cases [27].
Results
NTRK1-3 fusion prevalence
A total of 38,095 samples from 33,997 patients had MSK-IMPACT and/or MSK-Fusion testing at Memorial Sloan Kettering Cancer Center (MSKCC). The 87 tumors identified with oncogenic NTRK1-3 fusions and the tumor types are described in Table 1. In some of the tumor categories listed, which are classified by primary site, there are histological classifications that are enriched for NTRK fusions, and in these cases, histologic features can be suggestive of NTRK fusions (e.g., secretory carcinomas of the salivary gland and breast or infantile fibrosarcoma, see Supplementary Fig. 1). In the other categories, however, histological features have not been identified to be indicative of the presence of NTRK fusions. In carcinomas of the lung, pancreas, biliary tract, and appendix, NTRK fusions are seen in our cohort at a prevalence ranging from 0.3% to 0.5%. In thyroid carcinoma, NTRK fusions appear to be more prevalent, seen in 2.3% of tumors, with one explanation for the increased prevalence being that NTRK fusions are associated with radiation history for this tumor type [32]. NTRK fusions are seen in 0.36% of melanomas, including both cutaneous and mucosal primary sites, and are seen in 0.55% of gliomas and other primary CNS tumors. The positive tumors with the specific NTRK translocation identified and detailed molecular results are in Supplementary Table 1. In this study, NTRK fusions were mutually exclusive with strong activating MAPK pathway alterations. No KRAS, BRAF, NRAS, EGFR hotspot mutations nor kinase fusions were detected in the NTRK fusion cohort in Trk-inhibitor naive cancers.
Table 1.
Prevalence of NTRK fusions in various tumor types
| Number of cases with NTRK fusions | Total patients for which molecular testing was performed | Percentage | |
|---|---|---|---|
| Salivary gland carcinoma | 13 | 256 | 5.08% |
| Thyroid carcinoma | 13 | 571 | 2.28% |
| Sarcoma | 13 | 1915 | 0.68% |
| Lung adenocarcinoma | 9 | 3993 | 0.23% |
| Colorectal carcinoma | 9 | 2929 | 0.31% |
| Glioma/neuroepithelial tumor | 8 | 1465 | 0.55% |
| Breast carcinoma | 6 | 4458 | 0.13% |
| Pancreatic adenocarcinoma | 5 | 1492 | 0.34% |
| Melanoma | 4 | 1125 | 0.36% |
| Inflammatory myofibroblastic tumor | 3 | 17 | 17.7% |
| Cholangiocarcinoma | 2 | 787 | 0.25% |
| Appendiceal adenocarcinoma | 1 | 208 | 0.48% |
| Neuroendocrine tumor | 1 | 322 | 0.31% |
DNA-based next-generation sequencing by MSK-IMPACT
Through March 30, 2019, tumors from 33,997 patients had been tested by MSK-IMPACT, and the ability of this assay for detection of NTRK fusion positive cases as defined in the Materials and Methods are shown in Table 2. NTRK1-3 structural variants with novel partners or breakpoints underwent subsequent RNA testing for confirmation of an NTRK fusion. There were 107 cases with NTRK structural variants identified by MSK-IMPACT, but 46 did not exhibit a transcribed NTRK fusion transcript when reflexed to RNA testing. In terms of sensitivity, IMPACT was not able to detect 14 NTRK fusions identified by the RNA-based MSK-Fusion panel because the breakpoints were likely outside the areas covered by MSK-IMPACT, as only selected introns in NTRK1, NTRK2, and ETV6 are covered (see methods). It is worth noting that for ETV6-NTRK3 fusions, all 29 were detected by IMPACT, while NTRK3 fusions with other partners were only detected in 1 out of 10 cases. The single case that was positive by IMPACT was because the breakpoint was within 133 bp of intron 15-exon 16 boundary. The NTRK fusions that were missed by MSK-IMPACT were four cases with EML4-NTRK3, two cases with SQSTM1-NTRK3, two cases with RBPMS-NTRK3, and one case each with TPM4-NTRK3, TRAF2-NTRK2, BCR-NTRK2, KANK1-NTRK2, STRN-NTRK2, and EPS15-NTRK1. However, this last case, a lung adenocarcinoma with the EPS15-NTRK1 fusion, had very low tumor content and no other alterations were seen by MSK-IMPACT, likely because the variant allele frequency was too low. It should be noted that an EPS15-NTRK1 fusion was picked up in a different lung adenocarcinoma case with adequate tumor content.
Table 2.
Sensitivity and specificity of DNA-based cancer gene panel next generation sequencing (MSK-IMPACT) for detecting NTRK fusions
| Sensitivity | Specificity | |
|---|---|---|
| NTRK1 | 96.8% (30/31) | |
| NTRK2 | 0% (0/4) | |
| NTRK3 | 76.9% (30/39) | |
| Total | 81.1% (60/74) | 99.86% (33877/33923) |
While 14 NTRK fusion positive cases were not detected by MSK-IMPACT, copy number analysis in a few situations provided clues that led to further testing. In two of the cases that were falsely negative by IMPACT, (a TRAF-NTRK2 and an EML4-NTRK3) an intragenic gain of NTRK kinase domain exons was seen, suggestive of a structural variant. Further analysis at the RNA level by MSK-Fusion confirmed the fusion. Although for the purposes of this study, these cases were called a false negative by MSK-IMPACT since a structural variant was not called by the pipeline, the clinical reports mentioned the intragenic gain, and the pathologist sent the case for follow-up testing with MSK-Fusion at the time of signout.
Immunohistochemical testing
A total of 66 fusion positive cases and 317 fusion negative cases were tested by pan-Trk immunohistochemistry. The negative cases included 13 cases with NTRK amplification as this has been reported to cause Trk overexpression [33], 16 cases with DNA-level NTRK structural variants that did not result in an expressed fusion transcript by MSK-Fusion, a mixture of common tumor types negative by MSK-Fusion that mirrored the tumor types where NTRK fusions were detected (carcinomas of the salivary gland, colon, thyroid, breast, lung, pancreas, and biliary tract), as well as tumor types with smooth muscle or neural differentiation (sarcomas, gliomas, neuroendocrine tumors, and small round blue cell tumors), as this has been reported to lead to false positive pan-Trk immunohistochemistry results [29]. All of the cases tested along with their molecular testing and immunohistochemical staining results are listed in Supplementary Tables 1 and 2. For pan-Trk immunohistochemistry, sensitivity was stratified by NTRK gene or tumor type, and the specificity was stratified by tumor type (Table 3).
Table 3.
Sensitivity and specificity of pan-Trk immunohistochemistry for detecting NTRK fusions
| Sensitivity | ||
|---|---|---|
| NTRK1 | 96.2% (26/27) | |
| NTRK2 | 100% (5/5) | |
| NTRK3 | 79.4% (27/34) | |
| Sensitivity | Specificity | |
| Total | 87.9% (58/66) | 81.1% (257/317) |
| Colon | 87.5% (7/8) | 100% (25/25) |
| Lung | 87.5% (7/8) | 100% (24/24) |
| Thyroid | 81.8% (9/11) | 100% (27/27) |
| Salivary | 88.9% (8/9) | 52% (13/25) |
| Breast | 80% (4/5) | 82.1% (23/28) |
| Inflammatory myofibroblastic tumor | 100% (3/3) | 100% (5/5) |
| Sarcoma | 80% (8/10) | 74.4% (29/39) |
| Pancreas | (0/0)a | 100% (20/20) |
| Appendix | 100% (1/1) | 100% (1/1) |
| Cholangio | 100% (2/2) | 100% (19/19) |
| Glioma | 100% (6/6) | 20.8% (5/24) |
| Melanoma | 100% (3/3) | 100% (17/17) |
| Neuroendocrine | (0/0)a | 88.9% (8/9) |
| Small round cell tumorb | (0/0) | 45.8% (11/24) |
| Otherc | (0/0) | 100% (30/30) |
The NTRK fusion positive pancreatic adenocarcinoma and pancreatic neuroendocrine tumor cases were detected on cytology specimens, and no material remained available for immunohistochemical studies
Small round cell tumors included embryonal rhabdomyosarcoma (8), neuroblastoma (5), Ewing sarcoma (5), desmoplastic small round cell tumor (5), and NUT midline carcinoma (1)
Other tumor types included prostatic adenocarcinoma (9), high-grade serous ovarian carcinoma (7), endometrioid adenocarcinoma (4), ampullary carcinoma (2), esophageal adenocarcinoma (2), esophageal squamous cell carcinoma (1), uterine perivascular epithelioid cell tumor (1), histiocytosis (1), myoepithelial carcinoma (1), granular cell tumor (1), and cancer of unknown primary (1)
Immunohistochemistry sensitivity
The sensitivity of immunohistochemistry was lower for detecting NTRK3 fusions (79.4%) as compared to that in NTRK1 and NTRK2 fusions (combined 96.9%). In some NTRK3 fusion positive cases with pan-Trk expression, staining was weak and focal (in less than 5% of cells) (Fig. 1a, b). Therefore, any positive staining in at least 1% tumor cells was classified as positive. While sometimes focal, staining in ETV6-NTRK3 fusion positive cancers was often nuclear, which was not observed in false positive cases. When stratified by tumor type, sensitivity ranged from 80% in sarcomas and breast carcinomas to 100% in inflammatory myofibroblastic tumor, appendiceal adenocarcinoma, cholangiocarcinoma, glioma, and melanoma. It was hypothesized that for some of the tumor types, the reduced sensitivity could be due to the over-representation of NTRK3 fusions. Therefore, we compared the subgroup of cases where there could be a high suspicion of NTRK fusions due to histology (secretory carcinoma or infantile fibrosarcoma that could be alternatively confirmed by fluorescent in situ hybridization) and found that the sensitivity was 81.8% (9 of 11 cases) versus 89.1% (49 of 55 cases) in the remaining cases (p = 0.50, Chi-square test). The pattern of immunohistochemistry expression (cytoplasmic only, nuclear, peri-nuclear, or membranous) varied with the fusion partner as previously described (Fig. 1c–f) [29].
Fig. 1.
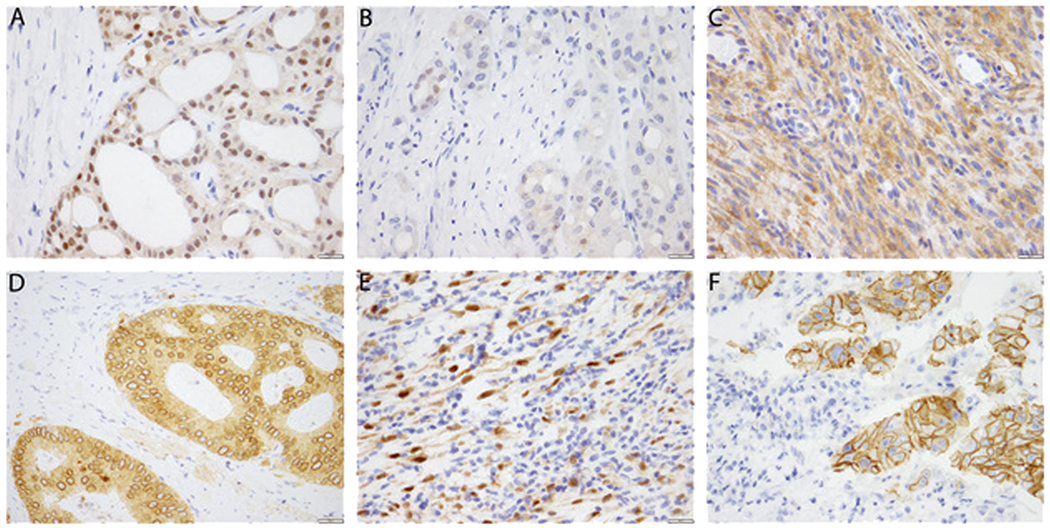
Patterns of pan-Trk immunohistochemistry expression in NTRK fusion positive cancers. a Strong cytoplasmic and nuclear staining in this secretory carcinoma of the salivary gland with canonical ETV6-NTRK3 fusion. b Staining is occasionally weak and focal, seen in approximately 1% of tumor cells, as in this secretory carcinoma of the salivary gland. c Cytoplasmic staining is seen in this Lipofibromatosis-like Neural Tumor with a TPM3-NTRK1 fusion. d Cytoplasmic and perinuclear staining is seen in this colonic adenocarcinoma with a LMNA-NTRK1 fusion. e Cytoplasmic and nuclear staining is seen in this inflammatory myofibroblastic tumor with an ETV6-NTRK3 fusion. f Membranous staining is seen in this intrahepatic cholangiocarcinoma with a PLEKHA6-NTRK1 fusion
Immunohistochemistry specificity
The overall specificity of pan-Trk immunohistochemistry in our cohort of MSK-Fusion negative cases was 81.1%. Some tumor types, most notably colon, lung, thyroid, pancreatobiliary, and melanoma showed 100% specificity in our cohort, while other selected tumor types showed a higher incidence of false positive staining (Table 3), which was always cytoplasmic (Fig. 2).
Fig. 2.
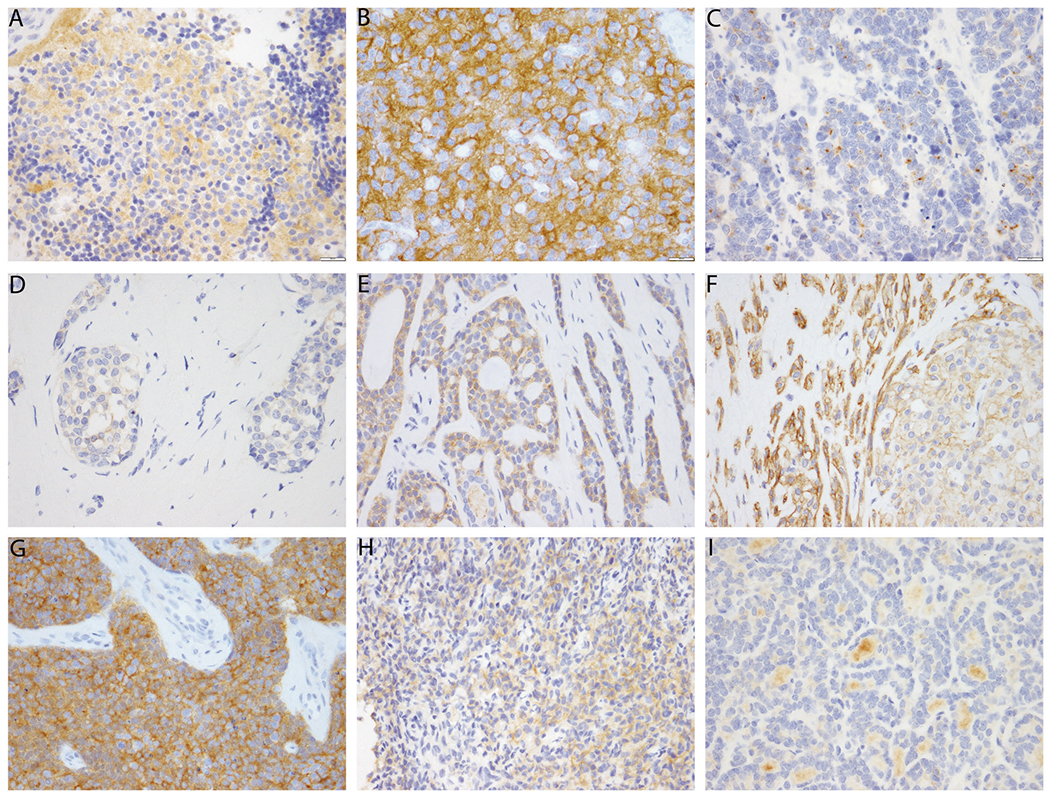
Pan-Trk immunohistochemistry expression in NTRK wild type carcinomas. a, b This pitfall can be seen in neural derived tumors such as neuroblastoma (a) and oligodendroglioma (b). c Occasionally focal cytoplasmic staining can be seen in carcinoma with neuroendocrine differentiation. d Weak cytoplasmic staining is seen in a minority of breast invasive ductal carcinomas. e Adenoid cystic carcinoma often shows moderate to strong cytoplasmic staining. f Cytoplasmic and membranous staining is seen in this atypical pleomorphic adenoma. The myoepithelial cells show particularly strong staining. g, h There is often staining in sarcomas, particularly those with neural or smooth muscle differentiation or those with other translocations, such as this desmoplastic small round cell tumor (g) and this sarcoma with BCOR-CCNB3 translocation (h). i In this papillary thyroid carcinoma, there is nonspecific staining of the colloid, but the tumor cells themselves show no staining, and the stain is therefore interpreted as negative
Since Trk is physiologically expressed in neural and muscle tissue, tumors derived from or involving such organ systems can show pan-Trk staining. Gliomas and neuroblastomas showed particularly poor specificity and strong background staining given the presence of neuropil (Fig. 2a, b). Occasionally focal cytoplasmic staining was also be seen in carcinomas with neuroendocrine differentiation (Fig. 2c). Interestingly, two carcinoma types showed a higher rate of false positive expression: breast carcinomas showed false positive pan-Trk expression in 18% of cases, and salivary gland tumors showed false positive staining in 48% of cases. All 5 false positive breast carcinomas demonstrated weak and focal cytoplasmic staining (Fig. 2d). In the salivary gland, adenoid cystic carcinoma showed positive cytoplasmic staining in 8 of 12 cases and was occasionally moderate to strong intensity (Fig. 2e). The staining pattern was cytoplasmic and often in the outer layer of tumor cells. Although the differential diagnosis should not overlap with secretory carcinoma because of differential mammaglobin expression [34], even discounting these cases, the specificity was still only 69% (9/13) in other salivary gland tumors (Fig. 2f).
In sarcomas, pan-Trk staining often showed faint cytoplasmic positivity in tumor cells, while in rarer cases, the staining can be quite striking. Pan-Trk staining is often positive in tumors within the small round blue cell category, including 5 of 5 desmoplastic small round cell tumors (Fig. 2g), 1 of 5 Ewing sarcomas, and 1 of 7 rhabdomyosarcomas. Pan-Trk staining was also positive in 3 of 5 sarcomas with BCOR translocations (Fig. 2h). Notably, all false positive cases demonstrated cytoplasmic or membranous expression only, and no nuclear staining was seen in these false positive cases. It should be noted that in some cases, the tumor cells themselves can be negative, but the high levels of background staining can make interpretation extremely difficult (Fig. 2i).
Immunohistochemistry specificity in cases with NTRK amplifications and structural variants
Of the 13 cases that showed NTRK amplification, two showed cytoplasmic pan-Trk staining. Both of these cases occurred in tumor types where physiological expression is often seen: one neuroblastoma (Fig. 2a) and one undifferentiated pleomorphic sarcoma. Of the 16 cases that showed NTRK structural variants that did not result in an expressed fusion transcript, three showed pan-Trk staining. All 3 of the cases occurred in tumor types where physiologic expression is seen, including 1 oligodendroglioma (Fig. 2b), 1 anaplastic astrocytoma, and 1 gastrointestinal stromal tumor. Overall, the specificity of pan-Trk staining in cases with NTRK amplifications and structural variants is not significantly different from the specificity in the entire cohort. In the cases that were interpreted as positive, the false positive staining can be attributed to neural differentiation or to the fact that nonspecific staining is often seen in sarcomas, and it is therefore not due to the genomic alterations involving NTRK.
Discussion
This study represents the largest cohort of NTRK fusion positive solid tumors stained with pan-Trk immunohistochemistry, and it is the first to detail the sensitivity and specificity of a targeted DNA-based next-generation sequencing panel for the detection of NTRK1-3 fusions. Pan-Trk immunohistochemistry demonstrated an overall sensitivity of 88%, with NTRK3 fusions comprising the vast majority of false negatives. Similarly, DNA-based sequencing had a sensitivity of 81%, with NTRK2 and NTRK3 fusions comprising the majority of false negatives. The specificity was over 99% for DNA-based sequencing, while the specificity for immunohistochemistry was largely dependent on the type of tumor tested.
The immunohistochemistry findings in the current study are in line with previous studies [23, 30, 31], with false positives occurring mainly in tumors with smooth muscle or neural differentiation and false negatives occurring in NTRK3 fusion positive cancers. A positive predictive value of 100% was seen when used for examining carcinomas of the colon, lung, thyroid, pancreas and biliary tract, and for melanomas. In these tumors, therefore, one could theoretically use immunohistochemistry to definitively rule in an NTRK fusion, but it cannot rule out an NTRK fusion due to less than 100% sensitivity. Although our study did not show immunostaining in NTRK amplified tumors that could not be explained by other factors, such as neural differentiation, previous studies have suggested the opposite [33]. Therefore, if a positive immunohistochemistry result is obtained in these tumors, further workup with other ancillary testing should be performed.
Specificity of immunohistochemistry was worse in breast and salivary gland carcinomas (82% and 52%, respectively), which is a novel finding to our knowledge. Therefore, in breast and salivary gland tumors, caution should be exercised, and other methods of fusion detection may be more cost effective and tissue conservative. Finally, in sarcomas, both sensitivity and specificity are poor. If a fusion is suspected, a DNA or RNA based assay should be considered. Considering these results, pan-Trk immunohistochemistry is a time and tissue efficient fusion screen in several specific primary tumor types in which NTRK1-3 fusions have been identified and are rare.
In this study, we also show that NTRK1-3 fusion testing is feasible with relatively high sensitivity and specificity with a targeted panel DNA based sequencing assay, MSK-IMPACT. The DNA-based next generation sequencing cancer gene panel MSK-IMPACT showed very high sensitivity for detecting NTRK1 and ETV6-NTRK3 fusions (97% and 100%, respectively) because the NTRK1 and ETV6 introns are covered by the panel. However, rarer NTRK2 and NTRK3 fusions involving partners other than ETV6 were missed. The challenge of detecting these latter fusions are not unique to MSK-IMPACT, as other large targeted DNA-based next-generation sequencing panels such as FoundationONE CDx do not cover these introns either. In some NTRK fusion positive cases without structural variants detected by MSK-IMPACT, intragenic gains of the kinase domain were seen by copy number analysis. This finding prompted further investigation by RNA testing, which was often positive for an NTRK fusion where the breakpoints were not covered by MSK-IMPACT. Although these intragenic gains are seen in a minority of cases, an algorithm in a bioinformatic pipeline to highlight such alterations could be beneficial to suggest further testing in targeted panel DNA-based assays to increase sensitivity, especially for NTRK3 fusions not involving ETV6.
In the current study, we use the detection of a transcribed NTRK fusion by RNA-based sequencing to define an NTRK fusion positive cancer, which reflects our clinical practice. RNA-based sequencing appears to be the optimal way to approach NTRK fusions because the splicing out of introns simplifies the technical requirements of adequate coverage and because detection of RNA-level fusions provides direct evidence of functional transcription. However, even though it provides many advantages, RNA-based testing is not perfect. Our MSK-Fusion assay is a custom design that examines the most likely breakpoints but does not interrogate every exon within the NTRK genes. It is therefore possible that an event may be missed that could be detected by a more comprehensive targeted panel or whole transcriptome sequencing. Further, it is possible that some of the cases that were called false positive by immunohistochemistry or DNA-based sequencing in this study could actually be a false negative by MSK-Fusion. Other significant drawbacks to RNA sequencing more generally include its cost, relative unavailability, and labile nature of RNA compared to other methodologies. Therefore, other assays described here clearly have a role to play. Overall, the development of a testing algorithm for identification of NTRK fusions requires consideration of accessibility of testing modalities and economic implications. Consideration of MAP kinase driver status can be used to triage specimens for further workup, given that in this cohort, the presence of a conventional MAP kinase driver was mutually exclusive with NTRK fusions. For sarcoma cases, immunohistochemistry should be eschewed for more specific testing methods, such as RNA-based sequencing, particularly if there is neural or smooth muscle differentiation, as this would not only be able to definitively determine NTRK fusion status but may identify other fusions characteristic of certain sarcomas.
Our study has several limitations. The site of the current study was a major site for the phase I larotrectinib clinical trial [2], and a major referral center in general. We thus have referral bias that may result in a slight over-representation of NTRK fusions in advanced cancers. We also did not investigate the specificity of pan-Trk immunohistochemistry in every single tumor type. Tumor types without any NTRK1-3 fusion positive cases (e.g., renal cell carcinoma) were not retrospectively investigated with pan-Trk immunohistochemistry due to the extremely low likelihood of finding an NTRK fusion in clinical practice.
In conclusion, newer methods of screening for NTRK fusions include immunohistochemistry and DNA-based next-generation sequencing. Each assay has advantages and disadvantages and works best in selected circumstances. Thus, selection of the appropriate assay depends on the tumor type, differentiation, and suspected fusion, as well as whether comprehensive genomic testing is needed concurrently.
Supplementary Material
Acknowledgments
Conflict of interest This study was funded by the National Cancer Institute (NCI) under the MSK Cancer Center Support Grant/Core Grant (P30 CA008748) and a research grant from Bayer AG. ER is supported by internal Memorial Sloan Kettering funding and Memorial Sloan Kettering Clinical Scholars grant 2T32CA009512-29A1. DMH is in a consulting or advisory role for Chugai Pharma, CytomX Therapeutics, Boehringer Ingelheim, AstraZenica, Pfizer, Bayer, and Genentech, and has received research funding from AstraZenica, Puma Biotechnology, Loxo, and Bayer. AD reports honoraria from Ignyta/Genentech/Roche, Loxo/Bayer/Lilly, Takeda/Ariad/Millenium, TP Therapeutics, AstraZeneca, Pfizer, Blueprint Medicines, Helsinn, Beigene, BergenBio, Hengrui Therapeutics, Exelixis, Tyra Biosciences, Verastem, MORE Health, Merck, Puma, Medscape, OncLive, PeerVoice, Physicians Education Resources, Targeted Oncology, and Research to Practice, research funding from Pfizer, Exelixis, GlaxoSmithKlein, Teva, Taiho, PharmaMar, and Foundation Medicine, and royalties from Wolters Kluwer. JFH has received honoraria from Axiom Healthcare Strategies, Cor2Ed, and Medscape, as well as research funding from Bayer. The remaining authors declare that they have no conflict of interest.
Footnotes
Supplementary information The online version of this article (https://doi.org/10.1038/s41379-019-0324-7) contains supplementary material, which is available to authorized users.
References
- 1.Food and Drug Administration. FDA approves an oncology drug that targets a key genetic driver of cancer, rather than a specific type of tumor. FDA news release; 2018. https://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm626710.htm. Accessed 26 May 2019. [Google Scholar]
- 2.Drilon A, Laetsch TW, Kummar S, DuBois SG, Lassen UN, Demetri GD, et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N Engl J Med. 2018;378:731–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drilon A, Siena S, Ou SI, Patel M, Ahn MJ, Lee J, et al. Safety and antitumor activity of the multitargeted Pan-TRK, ROS1, and ALK inhibitor entrectinib: combined results from two phase I trials (ALKA-372-001 and STARTRK-1). Cancer Disco. 2017;7:400–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaishnavi A, Le AT, Doebele RC. TRKing down an old oncogene in a new era of targeted therapy. Cancer Disco. 2015;5:25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kheder ES, Hong DS. Emerging targeted therapy for tumors with NTRK fusion proteins. Clin Cancer Res. 2018;24:5807–14. [DOI] [PubMed] [Google Scholar]
- 6.Cocco E, Scaltriti M, Drilon A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat Rev Clin Oncol. 2018;15:731–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin-Zanca D, Hughes SH, Barbacid M. A human oncogene formed by the fusion of truncated tropomyosin and protein tyrosine kinase sequences. Nature. 1986;319:743–8. [DOI] [PubMed] [Google Scholar]
- 8.Knezevich SR, McFadden DE, Tao W, Lim JF, Sorensen PH. A novel ETV6-NTRK3 gene fusion in congenital fibrosarcoma. Nat Genet. 1998;18:184–7. [DOI] [PubMed] [Google Scholar]
- 9.Bourgeois JM, Knezevich SR, Mathers JA, Sorensen PH. Molecular detection of the ETV6-NTRK3 gene fusion differentiates congenital fibrosarcoma from other childhood spindle cell tumors. Am J Surg Pathol. 2000;24:937–46. [DOI] [PubMed] [Google Scholar]
- 10.Anderson J, Gibson S, Sebire NJ. Expression of ETV6-NTRK in classical, cellular and mixed subtypes of congenital mesoblastic nephroma. Histopathology. 2006;48:748–53. [DOI] [PubMed] [Google Scholar]
- 11.Vasudev P, Onuma K. Secretory breast carcinoma: unique, triple-negative carcinoma with a favorable prognosis and characteristic molecular expression. Arch Pathol Lab Med. 2011; 135:1606–10. [DOI] [PubMed] [Google Scholar]
- 12.Skalova A, Vanecek T, Sima R, Laco J, Weinreb I, Perez-Ordonez B, et al. Mammary analogue secretory carcinoma of salivary glands, containing the ETV6-NTRK3 fusion gene: a hitherto undescribed salivary gland tumor entity. Am J Surg Pathol. 2010; 34:599–608. [DOI] [PubMed] [Google Scholar]
- 13.Tognon C, Knezevich SR, Huntsman D, Roskelley CD, Melnyk N, Mathers JA, et al. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell. 2002;2:367–76. [DOI] [PubMed] [Google Scholar]
- 14.Wang L, Busam KJ, Benayed R, Cimera R, Wang J, Denley R, et al. Identification of NTRK3 fusions in childhood melanocytic neoplasms. J Mol Diagn. 2017;19:387–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lezcano C, Shoushtari AN, Ariyan C, Hollmann TJ, Busam KJ. Primary and metastatic melanoma with NTRK fusions. Am J Surg Pathol. 2018;42:1052–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alassiri AH, Ali RH, Shen Y, Lum A, Strahlendorf C, Deyell R, et al. ETV6-NTRK3 is expressed in a subset of ALK-negative inflammatory myofibroblastic tumors. Am J Surg Pathol. 2016; 40:1051–61. [DOI] [PubMed] [Google Scholar]
- 17.Xu T, Wang H, Huang X, Li W, Huang Q, Yan Y, et al. Gene fusion in malignant glioma: an emerging target for next-generation personalized treatment. Transl Oncol. 2018;11:609–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farago AF, Taylor MS, Doebele RC, Zhu VW, Kummar S, Spira AI, et al. Clinicopathologic features of non-small-cell lung cancer harboring an NTRK gene fusion. JCO Precis Oncol. 2018;2:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pietrantonio F, Di Nicolantonio F, Schrock AB, Lee J, Tejpar S, Sartore-Bianchi A, et al. ALK, ROS1, and NTRK rearrangements in metastatic colorectal cancer. J Natl Cancer Inst. 2017;109:12. [DOI] [PubMed] [Google Scholar]
- 20.Brzezianska E, Karbownik M, Migdalska-Sek M, Pastuszak-Lewandoska D, Wloch J, Lewinski A. Molecular analysis of the RET and NTRK1 gene rearrangements in papillary thyroid carcinoma in the Polish population. Mutat Res. 2006;599:26–35. [DOI] [PubMed] [Google Scholar]
- 21.Greco A, Miranda C, Pierotti MA. Rearrangements of NTRK1 gene in papillary thyroid carcinoma. Mol Cell Endocrinol. 2010; 321:44–9. [DOI] [PubMed] [Google Scholar]
- 22.Ricarte-Filho JC, Li S, Garcia-Rendueles ME, Montero-Conde C, Voza F, Knauf JA, et al. Identification of kinase fusion oncogenes in post-Chernobyl radiation-induced thyroid cancers. J Clin Invest. 2013;123:4935–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiang S, Cotzia P, Hyman DM, Drilon A, Tap WD, Zhang L, et al. NTRK fusions define a novel uterine sarcoma subtype with features of fibrosarcoma. Am J Surg Pathol. 2018;42:791–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferguson SD, Zhou S, Huse JT, de Groot JF, Xiu J, Subramaniam DS, et al. Targetable gene fusions associate with the IDH wild-type astrocytic lineage in adult gliomas. J Neuropathol Exp Neurol. 2018;77:437–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017; 23:703–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng Z, Liebers M, Zhelyazkova B, Cao Y, Panditi D, Lynch KD, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med. 2014;20:1479–84. [DOI] [PubMed] [Google Scholar]
- 27.Cocco E, Benhamida J, Middha S, Zehir A, Mullaney K, Shia J, et al. Colorectal carcinomas containing hypermethylated MLH1 promoter and wild-type BRAF/KRAS are enriched for targetable kinase fusions. Cancer Res. 2019;79:1047–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benayed R, Offin M, Mullaney K, Sukhadia P, Rios K, Desmeules P, et al. High yield of RNA sequencing for targetable kinase fusions in lung adenocarcinomas with no driver alteration detected by DNA sequencing and low tumor mutation burden. Clin Cancer Res. 2019. 10.1158/1078-0432.CCR-19-0225. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hechtman JF, Benayed R, Hyman DM, Drilon A, Zehir A, Frosina D, et al. Pan-Trk immunohistochemistry is an efficient and reliable screen for the detection of NTRK fusions. Am J Surg Pathol. 2017;41:1547–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gatalica Z, Xiu J, Swensen J, Vranic S. Molecular characterization of cancers with NTRK gene fusions. Mod Pathol. 2018; 32:147–53. [DOI] [PubMed] [Google Scholar]
- 31.Rudzinski ER, Lockwood CM, Stohr BA, Vargas SO, Sheridan R, Black JO, et al. Pan-Trk immunohistochemistry identifies NTRK rearrangements in pediatric mesenchymal tumors. Am J Surg Pathol. 2018;42:927–35. [DOI] [PubMed] [Google Scholar]
- 32.Leeman-Neill RJ, Kelly LM, Liu P, Brenner AV, Little MP, Bogdanova TI, et al. ETV6-NTRK3 is a common chromosomal rearrangement in radiation-associated thyroid cancer. Cancer. 2014;120:799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee SJ, Kim NKD, Lee SH, Kim ST, Park SH, Park JO, et al. NTRK gene amplification in patients with metastatic cancer. Precis Future Med. 2017;1:129–37. [Google Scholar]
- 34.Zhu S, Schuerch C, Hunt J. Review and updates of immunohistochemistry in selected salivary gland and head and neck tumors. Arch Pathol Lab Med. 2015;139:55–66. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


