Sir,
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first identified in Wuhan, China, in December 2019 as the aetiological agent of Coronavirus disease 2019 (COVID-19).1 , 2 Since then, the disease has spread rapidly worldwide and the World Health Organization (WHO) declared a pandemic on 11 March 2020.3 , 4 At the beginning of the outbreak, rapid development and implementation of reliable detection methods became an immediate priority for clinical laboratories worldwide, and reverse transcription polymerase chain reaction (RT-PCR) methods, including those provided by the WHO,5 , 6 have been implemented broadly. At the early stage of the outbreak, however, positive control material for RT-PCR assays (from positive patient samples, or viral culture) were not readily available. In such circumstances laboratories often turn to using synthetic controls (synDNA fragments or plasmids).7 , 8 These synthetic controls have their advantages, particularly in that the controls can be acquired as readily as PCR primers and probes. Yet, depending on how they are designed, precautions must be taken when handling such controls as trace amounts of this material can potentially cause contamination in the same way as that caused by PCR products. Here we report contamination of a SARS-CoV-2 probe that our evidence suggests occurred at the oligonucleotide manufacturer, and was due to the manufacturer synthesising full length control oligonucleotides (spanning from the forward to reverse primers) in parallel with our probe orders.
Two commonly used assays, E-gene and RdRP, reported by Corman et al.,5 were utilised by our laboratory in the early stages of the pandemic. To establish the assays, we were fortunate enough to have nucleotide stocks and positive control material provided to us from another local laboratory and the assays performed well in our hands. Needing new oligonucleotide stocks, we ordered primers and probes from supplier ‘X’ on 28 January 2020. The primers arrived on 31 January and were subjected to routine quality control checks. These included checking and recording oligonucleotide batch and reconstitution details, and master mix using new primers or probe was prepared and tested against previously checked reagents. The new primers passed quality control checks. The probes arrived on 11 February, however both were contaminated, providing positive results in the negative controls of both the E-gene and RdRP assays.
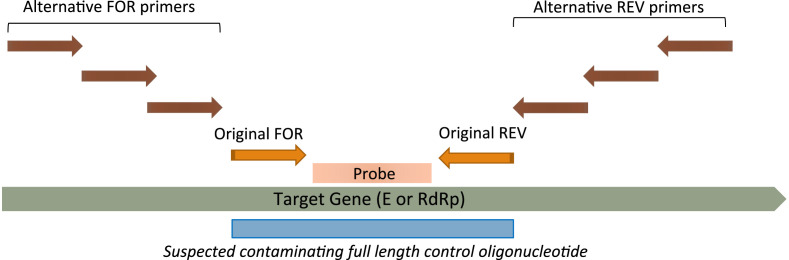
Suspecting that the probes had been contaminated by ‘full length control oligonucleotides’ (ordered by customers from elsewhere) we developed a series of alternative flanking primers for both assays (Fig. 1 , Table 1 ). Each of these alternative flanking primers was designed to gradually ‘step away’ from the original target region. Full-length synthetic controls typically would only contain sequences from the original target region (i.e., not any additional sequences sitting outside of the original primer pair), and so this type of contamination can be identified by testing alternative primer sets targeting regions further away from the original target site. Therefore, for our reagents, if the contamination was from synthetic controls, the original primers would produce false positive results while the flanking primers would generate negative results when testing non-template controls (NTCs). The flanking primers as well as the original primers from supplier X were tested against the supplier X probes. The experiments were replicated using probes sourced from another supplier, supplier ‘Y’. Two SARS-CoV-2 positive clinical samples and two NTCs were tested in each primer probe combination. This study was approved by the Children's Health Queensland Human Research Ethics Committee (HREC/LNR/19/QCHQ/49476).
Fig. 1.
Illustration of flanking primer designs for detection of contaminated probe. Note that six additional primers were designed for the E-gene but only four for the RdRP assay. FOR, forward primer; REV, reverse primer.
Table 1.
List of oligonucleotides used in this study
| Name | Oligonucleotides | Nucleotide positiona | Notes |
|---|---|---|---|
| Sarbeco_E-F1 | 5ʹACAGGTACGTTAATAGTTAATAGCGT | 26237–26262 | E-Gene Original primer5 |
| Sarbeco_E-R2 | 5ʹATATTGCAGCAGTACGCACACA | 26328–26349 | E-Gene Original primer5 |
| Sarbeco_E-probe | 5ʹACACTAGCCATCCTTACTGCGCTTCG | 26300–26325 | E-Gene Original probe5 |
| Sarbeco_E-altF2 | 5ʹCTTATGTACTCATTCGTTTCGGAAGA | 26210–26235 | E-Gene Flanking primer |
| Sarbeco_E-altF3 | 5ʹGTAAGCACAAGCTGATGAGTACGA | 26185–26208 | E-Gene Flanking primer |
| Sarbeco_E-altF4 | 5ʹ GACGACGACTACTAGCGTGCCTT | 26161–26183 | E-Gene Flanking primer |
| Sarbeco_E-altR2 | 5ʹ GAAGGTTTTACAAGACTCACGTTAACA | 26350–26376 | E-Gene Flanking primer |
| Sarbeco_E-altR3 | 5ʹ GAAGAATTCAGATTTTTAACACGAGAGTAAA | 26385–26415 | E-Gene Flanking primer |
| Sarbeco_E-altR4 | 5ʹ GTTCGTTTAGACCAGAAGATCAGGAA | 26421–26446 | E-Gene Flanking primer |
| RdRP_SARSr-F2 | 5ʹ GTGARATGGTCATGTGTGGCGG | 15399–15420 | RdRP Gene Original primer5 |
| RdRP_SARSr-R1 | 5ʹ CARATGTTAAASACACTATTAGCATA | 15473–15498 | RdRP Gene Original primer5 |
| RdRP_SARSr-P2 | 5ʹ CAGGTGGAACCTCATCAGGAGATGC | 15438–15462 | RdRP Gene Original probe5 |
| RdRP_SARSr-altF3 | 5ʹ GTTTCTATAGATTAGCTAATGAGTGTGCTCAA | 15360–15391 | RdRP Gene Flanking primer |
| RdRP_SARSr-altF4 | 5ʹ CTTGTTCTTGCTCGCAAACATACAA | 15314–15338 | RdRP Gene Flanking primer |
| RdRP_SARSr-altR2 | 5ʹ GCATTAACATTGGCCGTGACA | 15505–15525 | RdRP Gene Flanking primer |
| RdRP_SARSr-altR3 | 5ʹ TCGGCAATTTTGTTACCATCAGTAGATA | 15531–15558 | RdRP Gene Flanking primer |
Nucleotide position used reference genome Genbank ID: MN938384.
All results are shown in Table 2 . In brief, the known positive samples were positive by all oligonucleotide combinations. Notably, the NTCs were only positive in the supplier X probes using the original supplier X primers, and not in any other NTC, including the original supplier X primers with the supplier Y probes. Of concern were the cycle threshold (Ct) values for the supplier X probe NTCs for the RdRP assay which were very low at ∼26 cycles (Table 2), indicating very high levels of contamination, whereas the NTC Ct values for the E assay were ∼36 cycles. These results show the supplier X probes were contaminated with nucleic acid fragments consistent with the size, but not larger than, the expected PCR products for the E and RdRP assays. We have since contacted supplier X and they have now implemented new quality control measures to address this issue.
Table 2.
E-gene and RdRP RT-PCR results with different oligonucleotide combinations
| Assay and oligonucleotides | Notes | SARS-CoV-2 positive clinical sample 1 (Ct) | SARS-CoV-2 positive clinical sample 2 (Ct) | NTC 1 (Ct) | NTC 2 (Ct) |
|---|---|---|---|---|---|
| E-gene | |||||
| F1/R2 + probe Y | Original primer set with probe from supplier Y | 26.37 | 20.42 | ND | ND |
| F1/R2 + probe X | Original primer set with probe from supplier X | 29.04 | 22.59 | 35.44 | 37.05 |
| F1/altR2 + probe X | Alternative primer combinations with probe from supplier X | 29.51 | 23.15 | ND | ND |
| F1/altR3 + probe X | 29.3 | 23.25 | ND | ND | |
| F1/altR4 + probe X | 30.14 | 23.71 | ND | ND | |
| altF2/R2 + Probe X | 28.85 | 22.94 | ND | ND | |
| altF2/altR2 + Probe X | 29.49 | 23.5 | ND | ND | |
| altF2/altR3 + Probe X | 30.44 | 23.93 | ND | ND | |
| altF2/altR4 + Probe X | 29.86 | 23.76 | ND | ND | |
| altF3/R2 + Probe X | 29.32 | 23.2 | ND | ND | |
| altF3/altR2 + Probe X | 29.87 | 23.84 | ND | ND | |
| altF3/altR3 + Probe X | 30.51 | 24.24 | ND | ND | |
| altF3/altR4 + Probe X | 30.22 | 24.24 | ND | ND | |
| altF4/R2 + Probe X | 29.9 | 23.8 | ND | ND | |
| altF4/altR2 + Probe X | 30.85 | 24.65 | ND | ND | |
| altF4/altR3 + Probe X | 30.48 | 24.29 | ND | ND | |
| altF4/altR4 + Probe X | 30.75 | 24.72 | ND | ND | |
| RdRP | |||||
| F2/R1 + probe Y | Original primer set with probe from supplier Y | 29.48 | 23.97 | ND | ND |
| F2/R1 + probe X | Original primer set with probe from supplier X | 26 | 24.31 | 26.24 | 26.11 |
| F2/altR2 + probe X | Alternative primer combinations with probe from supplier X | ND | 28.74 | ND | ND |
| F2/altR3 + probe X | 29.27 | 23.63 | ND | ND | |
| altF3/R1 + probe X | ND | 26.14 | ND | ND | |
| altF3/altR2 + probe X | 28.49 | 22.97 | ND | ND | |
| altF3/altR3 + probe X | 29.06 | 23.46 | ND | ND | |
| altF4/R1 + probe X | 31.29 | 25.72 | ND | ND | |
| altF4/altR2 + probe X | 28.64 | 23.05 | ND | ND | |
| altF4/altR3 + probe X | 29.31 | 23.36 | ND | ND | |
alt, alternative; Ct, cycle threshold; F, forward primer; ND, not detected; R, reverse primer.
Overall, our study highlights the potential for contamination of oligonucleotide probes at the manufacturer and is due to customers ordering ‘full length control oligonucleotides’. This is alarming in the context of reagent shortages and delays associated with the pandemic, and would have left our laboratory in a precarious position had we not also ordered probes from supplier Y. We affirm that synthetic controls can be useful as positive control material for rare or emergent diseases but should be manufactured and used carefully. Oligonucleotide suppliers should consider how to better handle such requests.
Acknowledgements
We thank Pathology Queensland staff for their assistance with provision of samples for this study.
Conflicts of interest and sources of funding
PH reports financial support from Sandoz, MSD, Shionogi and Pfizer outside the submitted work. DW reports financial support from SpeeDx Pty Ltd outside the submitted work. The authors state that there are no conflicts of interest to disclose.
References
- 1.Guo Y.R., Cao Q.D., Hong Z.S., et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Mil Med Res. 2020;7:11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lv M., Luo X., Estill J., et al. Coronavirus disease (COVID-19): a scoping review. Euro Surveill. 2020;25:2000125. doi: 10.2807/1560-7917.ES.2020.25.15.2000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rabi F.A., Al Zoubi M.S., Kasasbeh G.A., et al. SARS-CoV-2 and coronavirus disease 2019: what we know so far. Pathogens. 2020;9:231. doi: 10.3390/pathogens9030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organisation (WHO) 2020. WHO Director-General’s opening remarks at the media briefing on COVID-19.https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 Cited 2 Jun 2020. [Google Scholar]
- 5.Corman V.M., Landt O., Kaiser M., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organisation (WHO) 2020. Coronavirus disease (COVID-19) pandemic.https://www.who.int/emergencies/diseases/novel-coronavirus-2019 [Google Scholar]
- 7.Smith G., Smith I., Harrower B., et al. A simple method for preparing synthetic controls for conventional and real-time PCR for the identification of endemic and exotic disease agents. J Virol Methods. 2006;135:229–234. doi: 10.1016/j.jviromet.2006.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conte J., Potoczniak M.J., Tobe S.S. Using synthetic oligonucleotides as standards in probe-based qPCR. Biotechniques. 2018;64:177–179. doi: 10.2144/btn-2018-2000. [DOI] [PubMed] [Google Scholar]