Abstract
Humoral responses in coronavirus disease 2019 (COVID-19) are often of limited durability, as seen with other human coronavirus epidemics. To address the underlying etiology, we examined post mortem thoracic lymph nodes and spleens in acute SARS-CoV-2 infection and observed the absence of germinal centers and a striking reduction in Bcl-6+ germinal center B cells but preservation of AID+ B cells. Absence of germinal centers correlated with an early specific block in Bcl-6+ TFH cell differentiation together with an increase in T-bet+ TH1 cells and aberrant extra-follicular TNF-α accumulation. Parallel peripheral blood studies revealed loss of transitional and follicular B cells in severe disease and accumulation of SARS-CoV-2-specific “disease-related” B cell populations. These data identify defective Bcl-6+ TFH cell generation and dysregulated humoral immune induction early in COVID-19 disease, providing a mechanistic explanation for the limited durability of antibody responses in coronavirus infections, and suggest that achieving herd immunity through natural infection may be difficult.
Keywords: SARS-CoV-2, COVID-19, T follicular helper cells, germinal centers, double-negative B cells, extra-follicular B cells, plasmablasts, humoral immunity, TNF-α, cytokine dysregulation
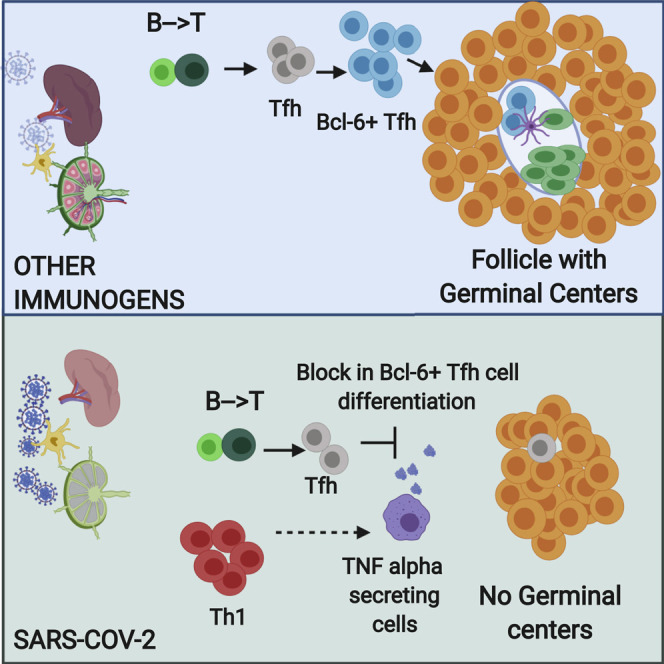
Graphical Abstract
Highlights
-
•
Germinal centers are lost in lymph nodes and spleens in acute COVID-19
-
•
Bcl-6+ GC B cells and Bcl-6+ T follicular helper cells are markedly diminished
-
•
Abundant TH1 cells and aberrant TNF-α production are seen in COVID-19 lymph nodes
-
•
SARS-CoV-2-specific activated B cells accumulate in the blood of patients
Shiv Pillai and colleagues show that in acute COVID-19, there is a striking loss of germinal centers in lymph nodes and spleens and depletion of Bcl-6+ B cells but preservation of AID+ B cells. A specific block in germinal center type Bcl-6+ T follicular helper cell differentiation may explain the loss of germinal centers and the accumulation of non-germinal-center-derived activated B cells. These data suggest an underlying basis for the lower quality and lack of durability of humoral immune responses observed during natural infection with SARS-CoV-2 and have significant implications for expectations of herd immunity.
Introduction
Adaptive immunity is initiated in secondary lymphoid organs and is influenced by the milieu generated by the initial activation of the innate immune system. Longitudinal studies on humoral immunity in coronavirus disease 2019 (COVID-19) as well as studies in convalescent subjects indicate that humoral immunity is often short lived and that most SARS-CoV-2 antibodies exhibit limited somatic hypermutation (Brouwer et al., 2020; Long et al., 2020; Robbiani et al., 2020). Understanding how the adaptive immune system is modulated in severe COVID-19 disease thus requires interrogation of secondary lymphoid organs in the acute phase of infection, where these responses are generated, but most studies to date have largely focused on peripheral blood samples.
SARS-CoV-2 infection results in a broad spectrum of clinical manifestations from asymptomatic to rapidly fatal, but the reasons for this heterogeneity are not known. Severely ill patients experience a life-threatening acute respiratory distress syndrome, and even in an advanced care setting, some patients sustain severe lung damage and succumb early (Zhu et al., 2020; Zhou et al., 2020). Virus is found in the lungs and airways early in infection, but not as the disease progresses (Schaefer et al., 2020). Damage-associated molecular patterns (DAMPs) released by infected pneumocytes likely combine with viral pathogen-associated molecular patterns (PAMPs) to activate innate immunity (Vardhana and Wolchok, 2020). The cytokine milieu thus generated would be predicted to influence the induction of lymphocyte activation by antigen conveyed directly in the lymph or by dendritic cells to draining lymph nodes. Viremia likely also leads to the initiation of immune responses in the spleen.
Many of the features of severe human coronavirus disease in COVID-19 and in severe acute respiratory syndrome (SARS) are strikingly similar. Progressive lymphopenia has been described in SARS-CoV-2 infection (Guan et al., 2020) and the degree of lymphopenia has been correlated with increases in circulating interleukin-6 (IL-6) and IL-8 (Zhang et al., 2020). Lymphopenia was also observed in SARS at the peak of active disease, which was also characterized by cytokine storm and acute respiratory distress (Perlman and Dandekar, 2005). Autopsy studies in SARS showed atrophy of lymphoid organs, including lymph nodes, spleen, and Peyer’s patches, and loss of germinal centers (Gu et al., 2005). Autopsy studies in COVID-19 have also identified splenic white pulp atrophy (Xu et al., 2020; Buja et al., 2020) and lymphocyte depletion in spleen and lymph nodes (Lax et al., 2020). However, numerous viral and non-viral infections do give rise to cytokine storm, acute respiratory distress, and lymphopenia (Tisoncik et al., 2012). Splenic white pulp atrophy has also been histo-pathologically demonstrated in Ebola and Marburg disease (Martines et al., 2015; Rippey et. al., 1984) and in H5N1 influenza (Gao et al., 2010; Lu et al., 2008). These data, taken together, suggest that many different viral and infectious triggers can contribute to a similar constellation of immunological phenomena that may drive pathology.
In persons with COVID-19, the magnitude and durability of antibody responses are greater in those with more severe disease (Ju et al., 2020; Amanat et al., 2020) but are often of low magnitude (Robbiani et al., 2020) and appear to lack durability (Long et al., 2020). This may be similar to SARS and Middle East respiratory syndrome (MERS), where humoral responses were generally not durable except in a subset of individuals (Mo et al., 2006; Zumla et al., 2015). Impaired infection-induced protective immunity has also been documented by repeated infections with the human coronaviruses CoV 229E, NL63, OC43, and HKU1 in patients with less severe respiratory tract infections (Galanti et al., 2019). Reinfection could be possibly attributed to viral strain subtypes, but the reason/s for the general lack of durable humoral immune responses to coronaviruses has never been established.
A better understanding of alterations to components of the humoral immune system, especially in secondary lymphoid organs, provides an opportunity to decipher why natural infections with coronaviruses often do not provide durable immunity. A granular analysis of B and T lymphocytes in draining lymph nodes and spleens of SARS or MERS patients was never reported, leaving the underlying basis for the lymphopenia and the general lack of durability of antibody responses in those diseases unresolved. Because COVID-19 disease most significantly affects the lungs, we undertook an analysis of thoracic lymph nodes, examining lymphoid architecture and lymphocyte populations using multi-color immunofluorescence, multispectral imaging, and cell-cell interaction analyses from the time of disease onset in persons with diverse disease outcomes. Given that viremia has been observed in this illness (Zheng et al., 2020; Lescure et al., 2020), we also interrogated spleens both in the acute- and late-disease settings and complemented these studies with examination of peripheral blood samples in a separate cohort, wherein convalescence could also be studied. Our results identify a striking absence of lymph node and splenic germinal centers and Bcl-6-expressing B cells, defective Bcl-6+ T follicular helper cell generation and differentiation, and dysregulated SARS-CoV-2-specific humoral immunity early in COVID-19 disease, providing a mechanistic explanation for the limited durability of humoral immunity and the less robust somatic hypermutation seen in this disease following natural infection.
Results
Absence of Germinal Centers and Loss of Bcl-6+ Germinal Center B Cells but Preservation of AID+ B Cells in Lymph Nodes Early in COVID-19 Disease
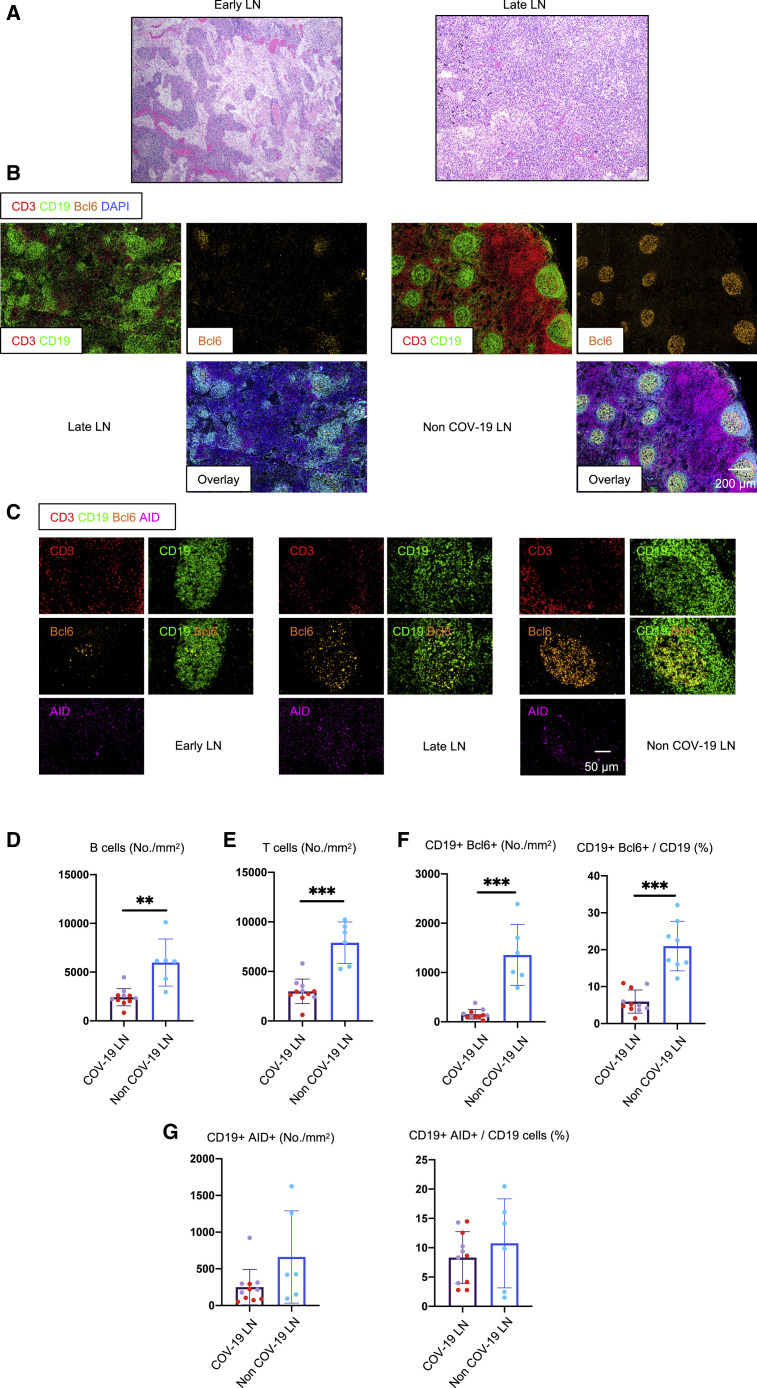
We have used a human-tissue-imaging platform with quantitative high-resolution automated slide-scanning microscopy, exploiting both regular and confocal approaches and multispectral imaging, in order to interrogate human lymphoid and non-lymphoid organs at the single-cell level. These approaches crucially preserve architecture over broad swaths of tissue. Thoracic lymph nodes in severely ill COVID-19 patients who succumbed in less than 8 days after admission (the group designated “early”; less than 10 days from the onset of respiratory symptoms; Table S1) displayed a lack of germinal centers, and these were also absent in those who succumbed later (15–36 days after admission, categorized as “late”; Figures 1A and 1B). Controls were thoracic lymph nodes from age-matched individuals who succumbed from non-COVID-19 causes (Table S2). Quantitation revealed dramatic early loss of both B and T cells, absolute numbers declining to about one-third of their non-COVID-19 controls, and this persisted in late disease (Figures 1D and 1E), though distinct T and B cell zones could always be clearly discerned. Human control lymph nodes contain germinal centers possibly because of ongoing adaptive immunity initiated by commensal antigens. The absence of germinal centers in the thoracic lymph nodes of acutely ill COVID-19 patients in whose lungs we have already described very high viral loads (Schaefer et al., 2020) was particularly surprising.
Figure 1.
Early Loss of Germinal Centers and Bcl-6-Expressing B Cells in COVID-19 Thoracic Lymph Nodes
(A) Hematoxylin-eosin staining of lymph nodes from early (left) and late (right) COVID-19 patients.
(B) Low-power images of CD3 (red), CD19 (green), Bcl-6 (orange), and DAPI (blue) staining in a lymph node from a late COVID-19 patient (left) and a non-COVID-19 thoracic lymph node (right).
(C) Representative multi-color immunofluorescence images of CD3 (red), CD19 (green), Bcl-6 (orange), and AID (purple) staining in lymph nodes from early (left) and late (middle) COVID-19 patients and a non-COVID-19 lymph node (right).
(D and E) Absolute numbers of CD19+ B cells (D) and CD3+ T cells (E) in lymph nodes from COVID-19 patients (purple, n = 11) and non-COVID-19 patients (blue, n = 6). COVID-19 samples include early (purple, n = 5) and late (red, n = 6) COVID-19 patients.
(F and G) Absolute numbers and relative proportion of Bcl6+ B cells (F) and AID+ B cells (G) in the pool of CD19+ B cells in lymph nodes from COVID-19 patients (purple, n = 11) and non-COVID-19 patients (blue, n = 6).
COV-19, COVID-19; LN, lymph node. Mann-Whitney U test was used to calculate p value. Error bars represent mean ± SEM. ∗∗p < 0.01; ∗∗∗p < 0.001. See also Figure S1 and Tables S1 and S2.
Bcl-6-expressing germinal center B cells were also markedly reduced in COVID-19, but there was a preservation of AID-expressing B cells, although these were diffusely distributed compared to controls (Figures 1C, 1F, and 1G).
Marked Reduction in Bcl-6+ Germinal Center B Cells in COVID-19 Spleens
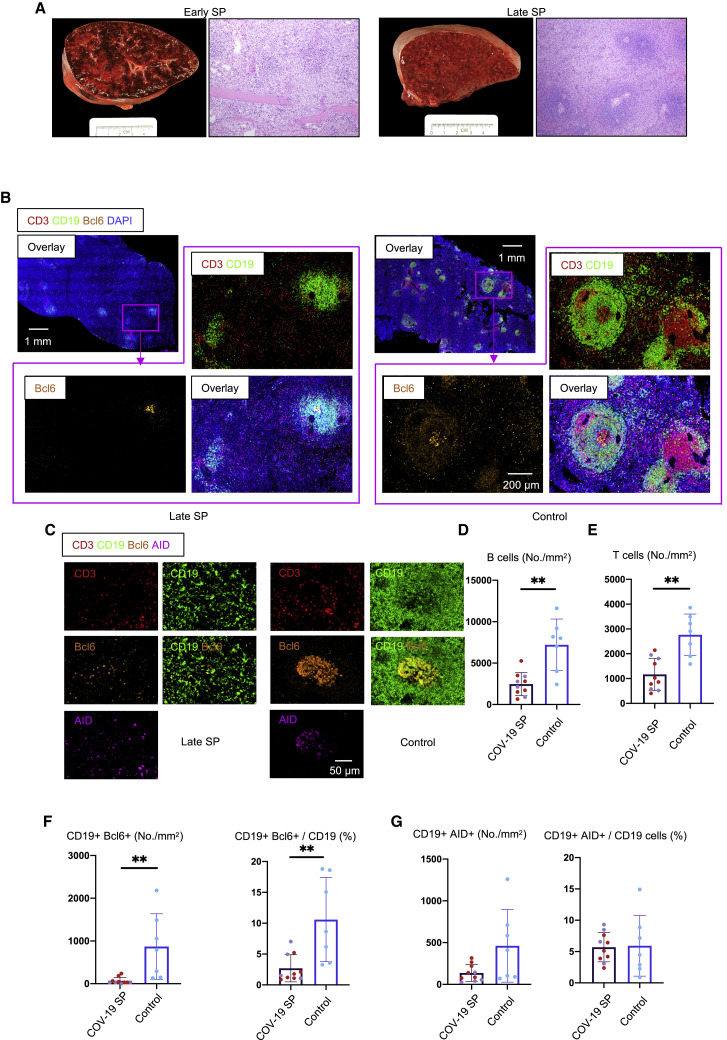
Analysis of spleens in this same group of COVID-19 patients also revealed a preponderance of red pulp and paucity of white pulp (Figures 2A and 2B), a marked reduction in B and T cell numbers (Figures 2B, 2D, and 2E), and a marked reduction in Bcl-6+ germinal center B cells (Figures 2C and 2F). There was, however, very clear and quantitative preservation of AID+ B cells in both early and late splenic tissue (Figures 2C and 2G). Importantly, follicular dendritic cells (FDCs) were present in both lymph nodes and spleen in these patients, indicating that the lack of these cells was not contributing to the lack of germinal center B cells (Figure S1 ). Together, these data indicate that, early in severe COVID-19 disease, even within 10 days of the onset of respiratory symptoms, there is severe attrition in B and T cell numbers and a striking reduction in Bcl-6+ B cells in lymph nodes and the spleen and the loss of germinal centers. Interestingly, AID+ B cells are preserved, indicating that activated helper T cells are still likely to be in frequent contact with antigen-specific B cells.
Figure 2.
White Pulp Attrition, Early Loss of Germinal Centers, and Bcl-6-Expressing B Cells in COVID-19 Spleens
(A) Cross-sectional view of whole spleen and hematoxylin-eosin staining from early (left) and late (right) COVID-19 patients.
(B) Low-power images of CD3 (red), CD19 (green), Bcl-6 (orange), and DAPI (blue) staining in a spleen from a late COVID-19 patient (left) and a control (right).
(C) Representative multi-color immunofluorescence image of CD3 (red), CD19 (green), Bcl-6 (orange), and AID (purple) staining in spleens from a late COVID-19 patient (left) and a control (right).
(D and E) Absolute numbers of CD19+ B cells (D) and CD3+ T cells (E) in spleens from early (purple, n = 4) and late (red, n = 6) COVID-19 patients and controls (blue, n = 7).
(F and G) Absolute numbers and relative proportion of Bcl-6+ B cells (F) and AID+ B cells (G) in spleens from early (purple, n = 4) and late (red, n = 6) COVID-19 patients and controls (blue, n = 7).
SP, spleen. Multiple comparisons are controlled for by Kruskal-Wallis test. Error bars represent mean ± SEM. ∗p < 0.05. See also Tables S2 and S3.
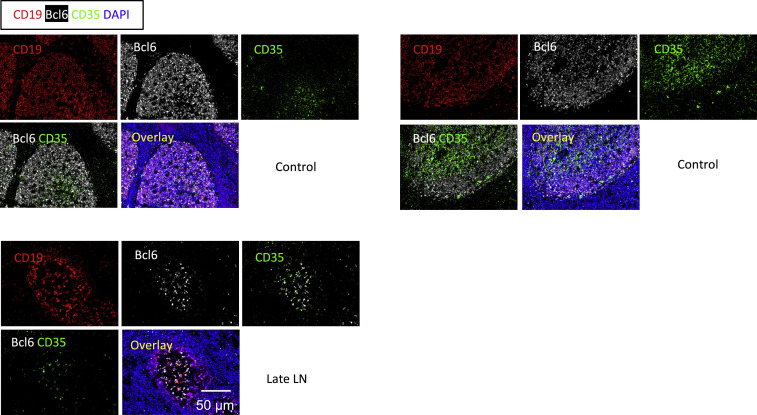
Figure S1.
FDCs Are Not Lost in COVID-19 Lymph Nodes, Related to Figure 1
Representative multi-color immunofluorescence images of CD19 (red), Bcl6 (white), CD35 (green) and DAPI (blue) staining in lymph nodes from late COVID-19 patient (lower panel) and controls (upper panels).
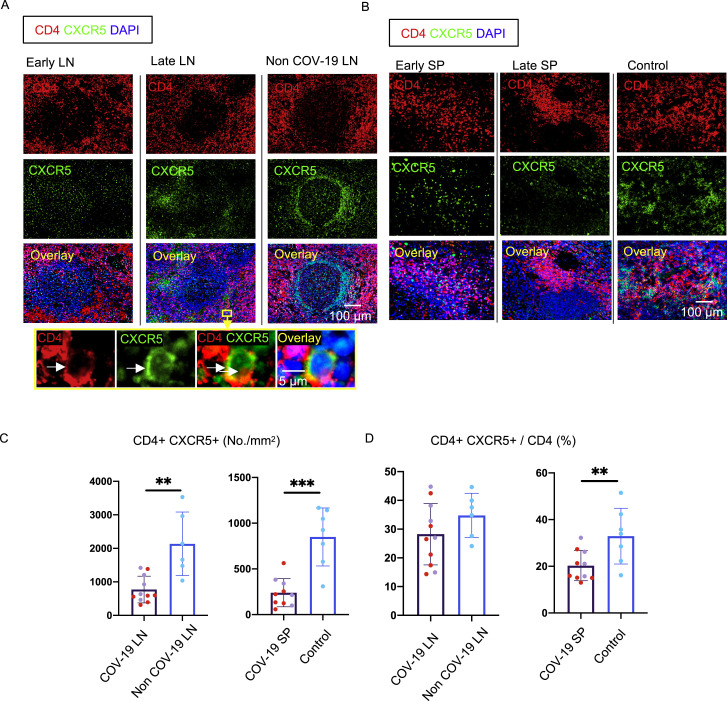
COVID-19-Related Reduction in CD4+CXCR5+Bcl-6+ Germinal Center T Follicular Helper Cells
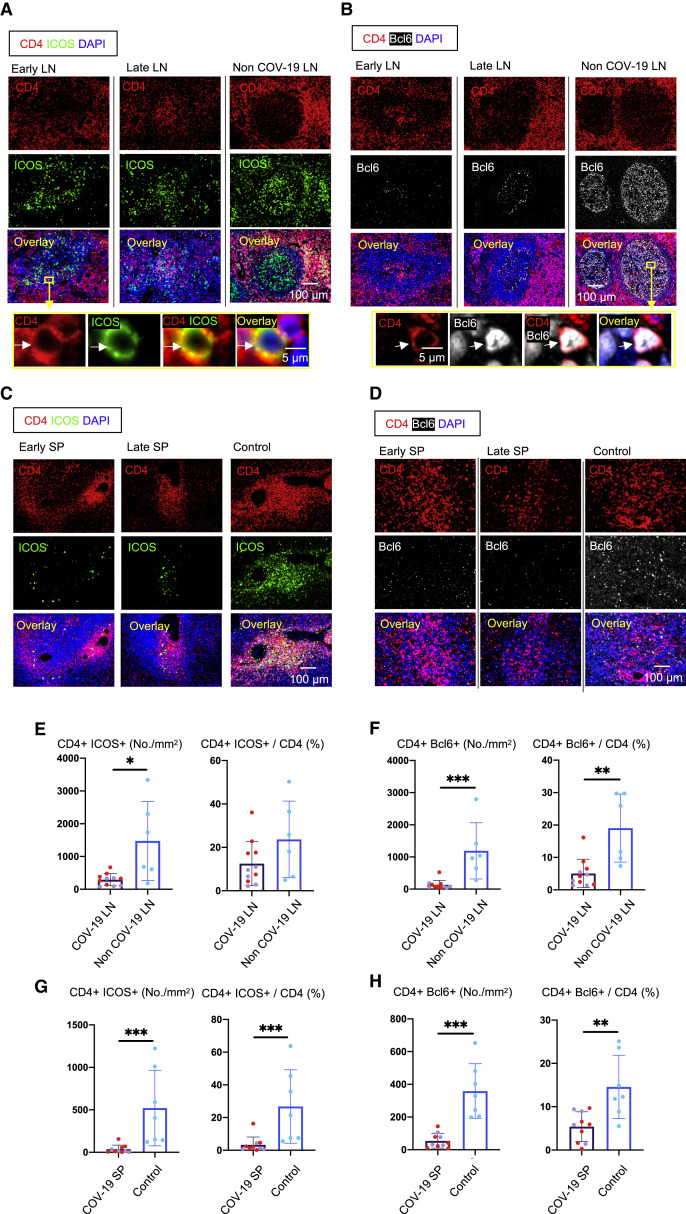
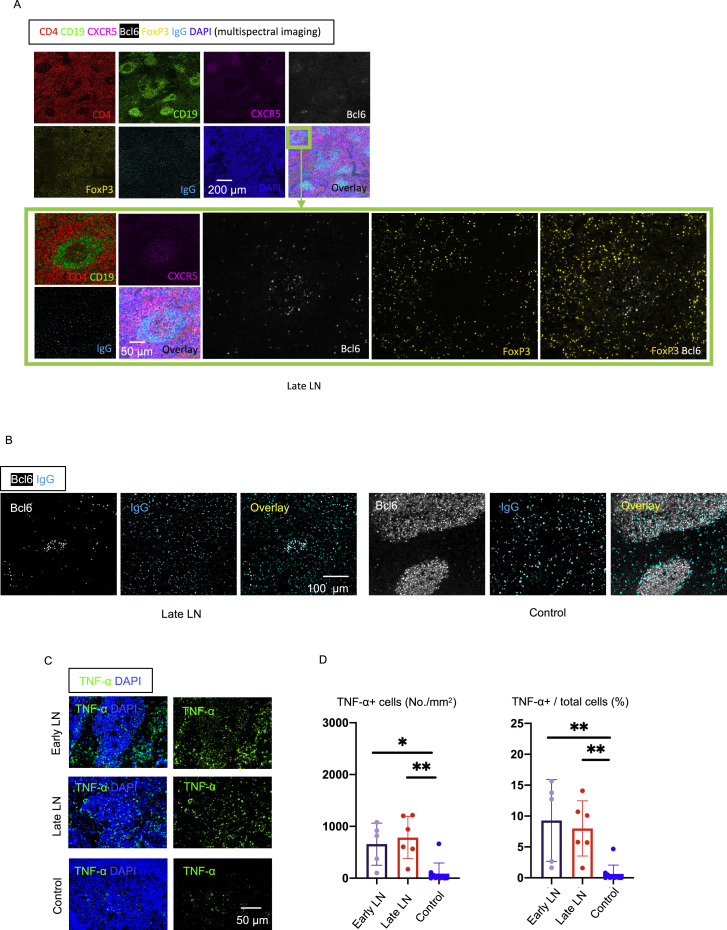
To better understand the absence of germinal centers in COVID-19, we explored the possibility that the tissue milieu might contribute to defective T follicular helper cell differentiation. In both the lymph nodes and spleen, in early as well as late disease, CD4+ ICOS+ TFH cells were diminished (Figures 3A, 3C, 3E, and 3G) and CD4+ CXCR5+ TFH cells were present but reduced in numbers (Figure S2 ), but the decrease in CD4+ Bcl-6+ germinal center type TFH (GC-TFH) cells was striking (Figures 3B, 3D, 3F, and 3H). Tissue quantitation confirmed significant differences for both early and late disease compared to controls. Because these changes were seen both in thoracic lymph nodes and in the spleen, these data are consistent with the view that circulating factors in severely ill COVID-19 patients may impair GC-TFH cell differentiation and thus abrogate the generation of germinal centers. Although, in principle, phenotypically defined CD4+ Bcl-6+ T cells could include both TFH cells and T follicular regulatory cells, we stained cells simultaneously with CD4, CXCR5, FOXP3, and Bcl-6 among other markers and used multispectral imaging to establish that, although there were FOXP3+ T reg cells present, there was no overlap in Bcl-6 and FoxP3 expression in COVID-19 secondary lymphoid organs, indicating that there are very few if any T follicular regulatory cells in COVID-19 (Figure S3 A). Although there is a developmental role for tumor necrosis factor alpha (TNF-α) in primary lymphoid follicular development (Pasparakis et al., 1996; Körner et al., 1997), germinal center loss has been described in the context of cytokine storm in mouse models, reversed by TNF-α blockade (Ryg-Cornejo et al., 2016; Popescu et al., 2019), and also linked genetically to an abundance of TNF-α (Popescu et al., 2019). We therefore also examined activated secondary lymphoid tissues from controls and COVID-19 lymph nodes for TNF-α expression. In this case, we used tonsils from non-COVID-infected patients as a control for activated lymphoid tissue. Although TNF-α is expressed at low levels in the follicle in controls, in COVID-19, it is expressed very abundantly both inside and outside the follicle (Figures S3C and S3D). These data indicate that the differentiation of activated CD4+ T cells into GC-type Bcl-6+ TFH cells is specifically blocked in COVID-19. Given the information obtained from the above animal models (Popescu et al., 2019; Ryg-Cornejo et al., 2016), it is possible that the aberrant and exuberant synthesis of TNF-α at the site of TFH differentiation in COVID-19 lymph nodes may contribute to the lack of germinal centers and the impaired quality and durability of the antibody response to SARS-CoV-2 in this disease.
Figure 3.
Loss of Germinal Center Type Bcl-6+ T Follicular Helper Cells in COVID-19 Lymph Nodes and Spleens
(A) Representative multi-color immunofluorescence image of CD4 (red), ICOS (green), and DAPI (blue) staining in lymph nodes from early (left) and late (middle) COVID-19 patients and a non-COVID-19 control (right). Arrows indicate CD4+ ICOS+ TFH cells.
(B) Representative multi-color immunofluorescence images of CD4 (red), Bcl-6 (white), and DAPI (blue) staining in lymph nodes from early (left) and late (middle) COVID-19 patients and a non-COVID-19 control (right). Arrows indicate CD4+ Bcl6+ GC-type TFH cells.
(C) Representative multi-color immunofluorescence images of CD4 (red), ICOS (green), and DAPI (blue) staining in spleens from early (left) and late (middle) COVID-19 patients and a control (right).
(D) Representative multi-color immunofluorescence images of CD4 (red), Bcl-6 (white), and DAPI (blue) staining in spleens from early (left) and late (middle) COVID-19 patients and a control (right).
(E and F) Absolute numbers and relative proportions of CD4+ ICOS+ TFH cells (E) and CD4+ Bcl-6+ GC-type TFH cells (F) in lymph nodes from COVID-19 patients (purple, n = 11) and non-COVID-19 patients (blue, n = 6). COVID-19 samples include early (purple, n = 5) and late (red, n = 6) COVID-19 patients.
(G and H) Relative proportions of CD4+ ICOS+ TFH cells (G) and CD4+ Bcl-6+ GC-type TFH (H) in spleens from COVID-19 patients (purple, n = 10) and controls (blue, n = 7). COVID-19 samples include early (purple, n = 4) and late (red, n = 6) COVID-19 patients.
Mann-Whitney U test used to calculate p value. Error bars represent mean ± SEM. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. See also Figures S2, S3, and S4 and Tables S1, S2, and S3.
Figure S2.
Decrease in CXCR5+ T Follicular Helper Cells in COVID-19 Lymph Nodes and Spleens, Related to Figure 3
(A) Representative multi-color immunofluorescence images of CD4 (red), CXCR5 (green) and DAPI (blue) staining in lymph nodes from early (left) and late (middle) COVID-19 patients and a non-COVID-19 lymph node (right). Arrows indicate CD4+ CXCR5+ TFH cells. (B) Representative multi-color immunofluorescence images of CD4 (red), CXCR5 (green) and DAPI (blue) staining in spleens from early (left) and late (middle) COVID-19 patients and a control (right). (C and D) Absolute numbers (C) and relative proportions (D) of CD4+ CXCR5+ TFH cells in lymph nodes and spleens from COVID-19 patients and non-COVID-19 lymph nodes (blue) (n = 6) and control spleens (blue) (n = 7). COVID-19 samples include early (purple; lymph node; n = 5) (spleen; n = 4) and late (red dots) (n = 6) COVID-19 patients. Mann-Whitney U test used to calculate p value. Error bars represent mean ± SEM. ∗∗p < 0.01; ∗∗∗p < 0.001.
Figure S3.
Increased T reg Cells but No Differentiation into TFR Cells in COVID-19 Lymph Nodes, Related to Figure 3
(A) Representative multi-spectral 7 color immunofluorescence images showing CD4 (red), CD19 (green), CXCR5 (purple), Bcl6 (white), FoxP3 (yellow), IgG (light blue) and DAPI (blue) staining of lymph nodes from late COVID-19 patients. Images in the green box show high-power images. No FoxP3+/Bcl6+ cells were seen (white staining with no yellow overlap) in follicles.
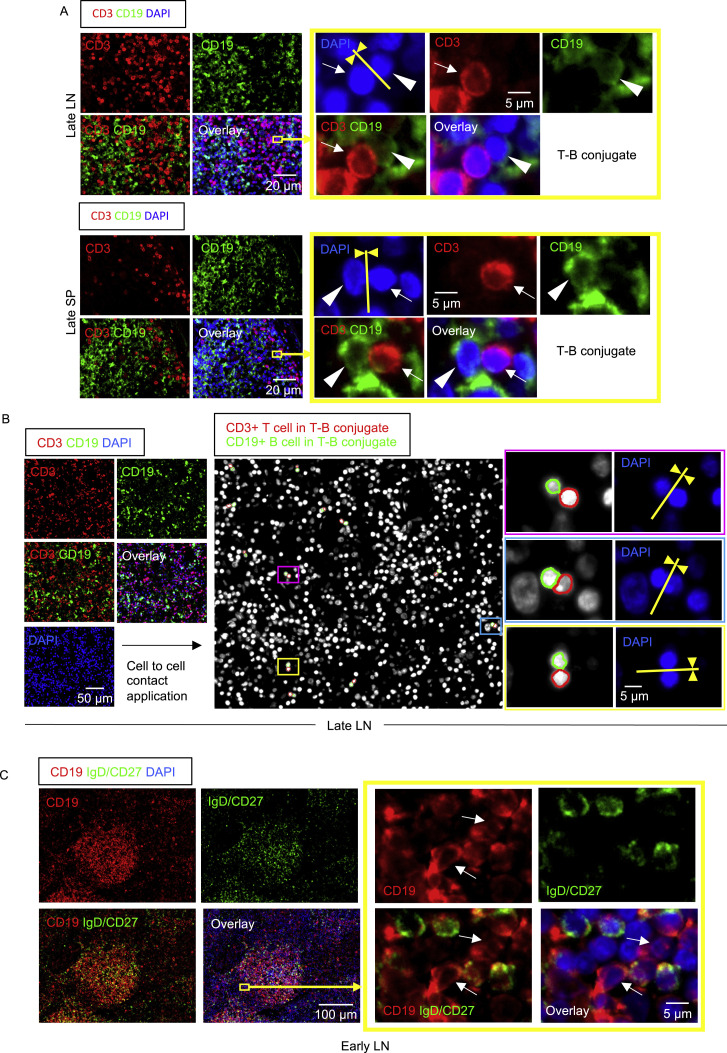
(B) IgG+ plasmablasts are found in follicular and extra-follicular areas in COVID-19 lymph nodes. Related to Figure 3. Representative multi-color immunofluorescence images of Bcl6 (white) and IgG (light blue) in lymph nodes of a late COVID-19 patient (left) and a control (right). IgG+ cells were abundant in follicular and extrafollicular areas in both COVID-19 lymph nodes and controls.
(C and D) Large increase in TNF-α production at both follicular and extra-follicular sites in COVID-19 lymph nodes, while controls have low levels of TNF-α localized to follicles. Related to Figure 3.
(C) Representative multi-color immunofluorescence images of TNF-α (green) and DAPI (blue) staining in lymph nodes from early and late COVID-19 patients and a control. (D) Absolute numbers (left) and percentages (right) of TNF-α+ cells in lymph nodes from early (purple) (n = 5) and late (red; n = 6) COVID-19 patients and controls (blue; n = 10). Multiple comparisons were controlled for by Kruskal-Wallis test. Error bars represent mean ± SEM. ∗p < 0.05; ∗∗p < 0.01.
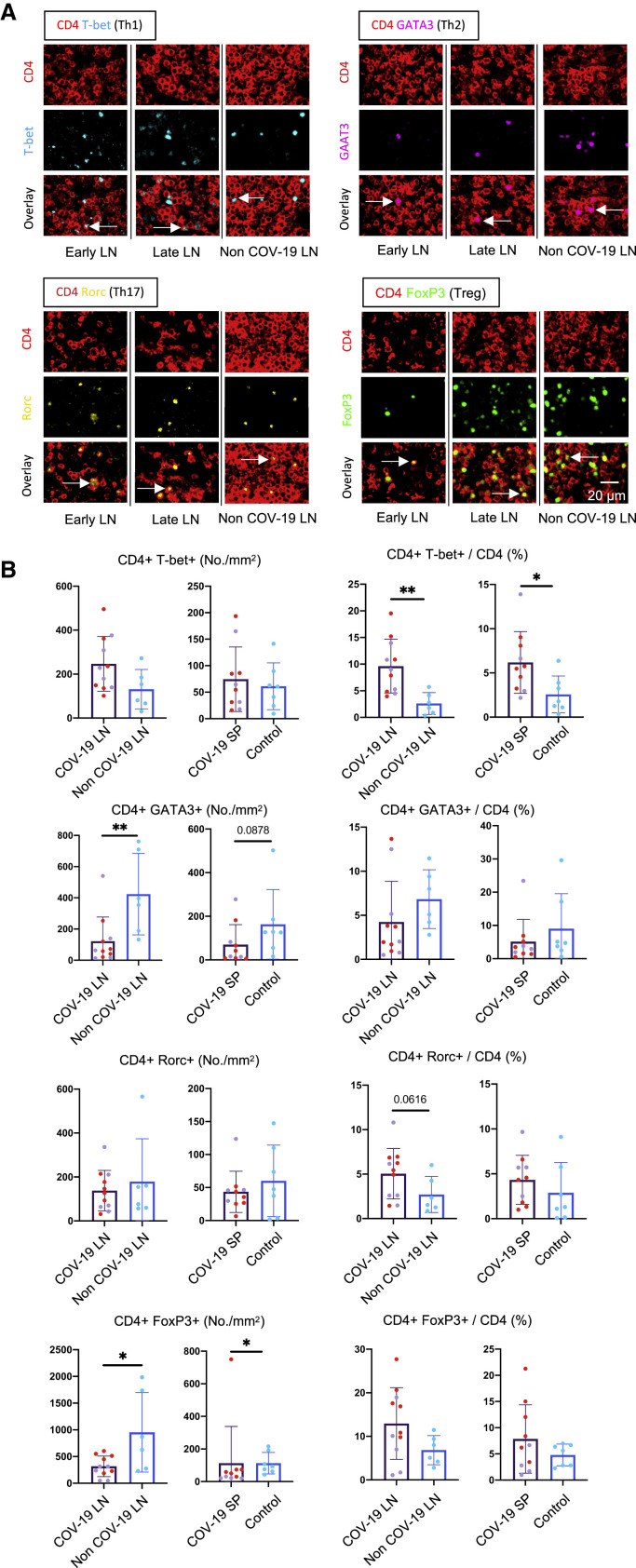
Increased Frequency of Secondary Lymphoid Organ TH1 Cells in Severe COVID-19
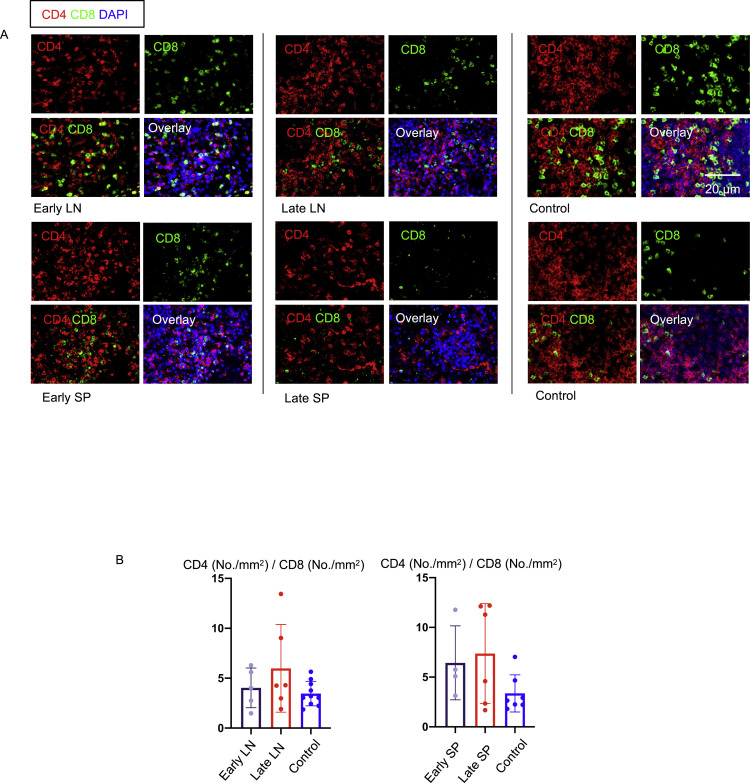
We hypothesized that the reduction in GC-TFH cell numbers likely reflects a block in differentiation and next sought to determine whether this reduction was specific to this particular CD4+ T cell subset. We quantitated CD4+ T cell subsets in the lymph nodes and spleens using the well-established transcription factors T-bet, GATA-3, RORγt, and FOXP3 as key markers. In contrast to the reduced GC-TFH cell numbers, TH1 cells were consistently increased early and late in both the lymph nodes and spleen, whereas an increase in TH17 cells was more variable (Figure 4 ). In contrast, a consistent reduction in TH2 cells was observed (Figure 4). Late in the disease, FOXP3+ T reg cells made up a large part of the population of CD4+ T cells. Overall, there was an increase in secondary lymphoid organ CD4+ T cells relative to CD8+ T cells in COVID-19 secondary lymphoid organs, though this was variable (Figure S4 ). These data indicate that the defect in GC-type TFH cell differentiation is specific and suggest that this defect may be indirectly linked to the strong TH1 response seen in this disease.
Figure 4.
TH1 Cells Are Expanded in Comparison to Other CD4+ T Cell Subsets in COVID-19 Thoracic Lymph Nodes and Spleens
(A) Representative multi-color staining showing TH1, TH2, TH17, and T reg cells in lymph nodes form early (left) and late (middle) COVID-19 patients and a non-COVID-19 control (right; TH1: CD4+ [red] T-bet+ [light blue]; TH2: CD4+ [red] GATA3+ [purple]; TH17: CD4+ [red] RORγ+ [yellow]; T reg cell: CD4+ [red] FOXP3+ [green]).
(B) Absolute numbers and relative proportions of TH1, TH2, TH17, and T reg cells in lymph nodes and spleens (purple) from early (purple, lymph nodes: n = 5; spleens: n = 4) and late (red, n = 6) COVID-19 patients and controls (blue, lymph nodes: n = 6; spleens: n = 7).
Mann-Whitney U test was used to calculate p value. Error bars represent mean ± SEM. ∗p < 0.05; ∗∗p < 0.01. See also Figures S5, S6, and S7 and Tables S1, S2, and S3.
Figure S4.
Increase in CD4+/CD8+ T Cell Ratio in Lymph Nodes and Spleens in COVID-19, Related to Figure 4
(A) Representative multi-color immunofluorescence images of CD4 (red), CD8 (green) and DAPI (blue) staining in lymph nodes (top 2 rows of images) and spleens (bottom 2 rows of pictures) from early (left) and late (middle) COVID-19 patients and controls (right). (B) Relative ratios of CD4 and CD8 T cells (No./mm2) in lymph nodes (left) and spleens (right) from early (purple) (n = 5/4) and late (red) (n = 6) COVID-19 patients and controls (blue) (n = 10/7).
Follicular and Extra-follicular T-B Conjugates and Activated IgD−CD27− Double-Negative B Cells in COVID-19 Lymph Nodes and Spleens
The preservation in COVID-19 of AID+ B cells and the relatively large proportions of CD4+ CXCR5+ TFH cells (that do not express Bcl-6) and TH1 cells, both known to express CD40L, led us to hypothesize that, even though there were no germinal centers, there may be frequent T-B conjugates in COVID-19 within follicles as well as in extra-follicular locations. The absence of germinal centers (most germinal center B cells are immunoglobulin D [IgD]−CD27−Bcl-6+AID+CXCR5+ CD19+ cells) offered the opportunity to directly ask whether many of the scattered activated AID+ B cells inside and outside follicles in COVID-19 secondary lymphoid organs were also IgD−CD27− “double-negative” B cells. These cells are often observed in chronic infectious contexts, including in viral infections as well as in autoimmunity (Portugal et al., 2017; Jenks et al., 2019). They are considered to be “disease-related” cells and are generally described as “extra-follicular,” implying they are not derived from the germinal center reaction but are frequently class switched and have the hallmarks of having been induced in a T-dependent manner without germinal-center-based selection. At extra-follicular and follicular sites, these double-negative B cells may be less capable of inducing the differentiation of the Bcl-6+GC-TFH cells that are required to induce germinal centers.
By applying computational tools to systematically quantitate the area of cytoplasmic overlap between sets of two cells of two different cell types, and using pre-determined cutoffs to assess true cell-cell interaction, we quantified the degree and intimacy of plasma membrane contacts between cells and observed the presence of numerous T-B conjugates in COVID-19 lymph nodes and spleens (Figure S5 ). The presence of IgD−CD27− double-negative cells both within and outside follicles in COVID-19 lymph nodes and spleens was clearly evident (Figure S5). IgG-expressing class-switched plasmablasts were prominent within both the follicular and extra-follicular areas (Figure S3B). These data indicate that, in the absence of germinal center formation in COVID-19, an extra-follicular type of class-switched B cell response, more typical of disease rather than of long-lasting protection, predominates in secondary lymphoid organs.
Figure S5.
Follicular and Extra-follicular T-B Conjugates and IgD−CD27− B Cells in COVID-19 Lymph Nodes, Related to Figure 4
(A) Immunofluorescence staining of CD3 (red), CD19 (green) and DAPI (blue) in a lymph node (top) and a spleen (bottom) in a late COVID-19 patient. Arrows and arrowheads indicate CD3+ T cells and CD19+ B cells respectively. T cell and B cells formed close and extensive intercellular plasma membrane contacts as highlighted in the yellow box. (B) Immunofluorescence staining (left panels) and visualization of T-B conjugates (middle and right panels) in a lymph node from a late COVID-19 patient using the StrataQuest cell-to-cell contact application. Masks of the nuclei based on DAPI staining establish the inner boundary of the cytoplasm and the software “looks” outward toward the plasma membrane boundary. An overlap of at least 3 pixels of adjacent cell markers was required to establish each “contact” criterion. Details are in the STAR Methods section. Nuclei circled in red and green respectively depict CD3+ T cells and CD19+ B cells in T-B conjugates. Each box (purple, light blue and yellow) highlights a T-B conjugate. (C) Representative multi-color immunofluorescence image of CD19 (red), IgD and CD27 (both in green) and DAPI (blue) staining in a lymph node from an early COVID-19 patient. IgD-/CD27- double negative B cells (red staining with no green overlap) are abundant inside the follicle and also outside. Boxed area depicts some of these cells outside the follicle. White arrows show IgD-/CD27- double negative B cells.
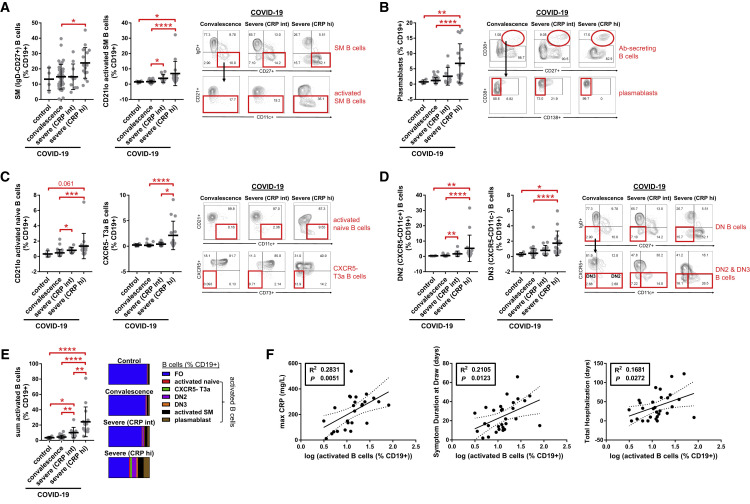
Decreases in Early Transitional and Follicular B Cells in Severe COVID-19 Patients Correlate with Systemic Inflammation
Although the interrogation of secondary lymphoid organs from autopsies helps provide snapshots regarding the sites of the induction of the immune response, we sought to obtain information from patients at different stages of the disease, including convalescence and ongoing severe illness, by the use of extended flow cytometry panels by examining peripheral blood from patients. Given the observed accumulation of activated non-germinal-center-derived B cells, such as IgD−CD27− double-negative B cells, in COVID-19 lymphoid organs, we examined whether similar activated B cell populations with binding specificity for SARS-CoV-2 could be found in the circulation in persons with disparate disease severity.
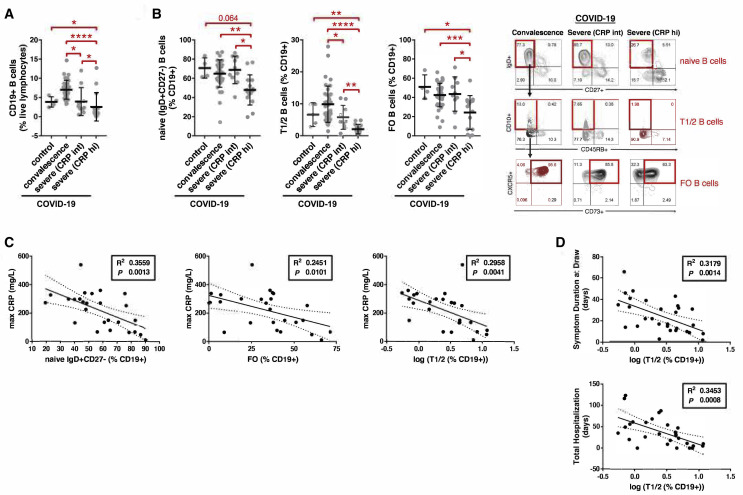
Patients were divided into gradations of severity that included asymptomatic convalescence and active severe illness requiring hospitalization that was further subdivided by intermediate (CRP < 200 mg/L; “CRP int”) and high (CRP > 200 mg/L; “CRP hi”) levels of maximum inflammatory marker elevation (Table S4). Studies on freshly isolated PBMCs revealed that the proportions and absolute numbers of total CD19+ B cells in addition to naive (IgD+CD27−), early transitional T1 and T2 (IgD+CD27−CD10+CD45RB−), and CXCR5+ follicular (IgD+CD27−CD10−CD73+; Farmer et al., 2019) B cell subsets were markedly reduced in severely ill COVID-19 patients with high CRP levels as compared to convalescent patients and healthy controls (Figures 5 A, 5B, S6 , and S7). The relative loss of these naive B cells correlated with the maximum level of CRP elevation (Figure 5C) and with the clinical parameters of symptom duration at the time of blood draw and total days of hospital admission for loss of early transitional B cells, specifically (Figure 5D). These data suggest a direct association between these B cell phenotypic changes in blood and patient clinical morbidity from COVID-19. They indicate that, in the milieu of severe COVID-19, B cells either develop very inefficiently in the bone marrow or are possibly generated but acquire an activated phenotype and are lost. This results in a marked reduction in early transitional and follicular B cells and likely further compounds defects in humoral immunity.
Figure 5.
Decreased Early Transitional and Follicular B Cells in the Peripheral Blood of Patients with Severe COVID-19
(A and B) Quantitation of (A) total CD19+ B cells and (B) naive, early transitional (T1/2), and follicular (FO) B cell subsets in the peripheral blood of patients with COVID-19 at states of convalescence (n = 39), severe illness with an intermediate maximum CRP level during hospitalization (<200 mg/L; severe [CRP int]; n = 10), and severe illness with a high maximum CRP level during hospitalization (>200 mg/L; severe [CRP hi]; n = 15) as defined by the clinical criteria listed in Table S4 and compared to healthy controls (n = 4). Quantitation is shown by B cell level for each individual patient with mean, standard deviation, and significance by one-way ANOVA of log % B cell value indicated (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001). Representative COVID-19 contour plots shown (B) with healthy control contour plots and full B cell flow cytometry gating strategy outlined (Figure S6).
(C and D) Association of peripheral blood B cell frequency with maximum CRP level (C) and the clinical parameters of symptom duration at blood draw and total length of hospital stay (D) in all symptomatic COVID-19 patients (n = 29) with moderate to severe illness as defined by the clinical criteria listed in Table S4. Correlation is shown by linear regression with individual patients, 95% confidence bands, R2, and p values shown.
See also Figures S6 and S7 and Table S4.
Figure S6.
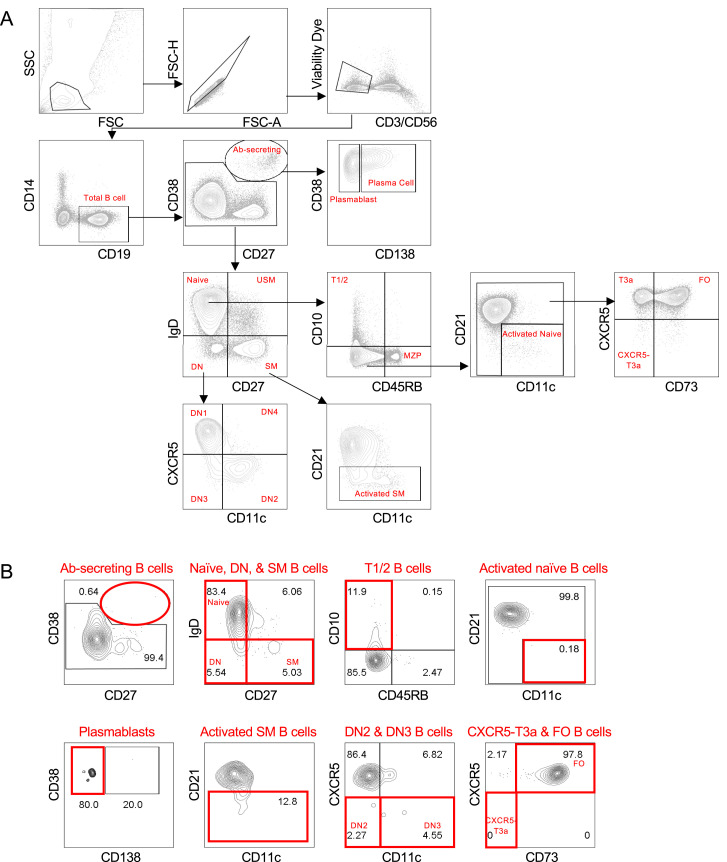
Analysis of Circulating B Cell Subsets by Flow Cytometry, Related to Figures 5, 6, and 7
(A) Gating strategy for COVID-19 PBMCs. (B) Representative flow plots of relevant B cell subsets from PBMCs derived from a healthy donor. Antibody-secreting (Ab-secreting), double negative (DN), follicular (FO), marginal zone precursor (MZP), switched memory (SM), transitional (T), unswitched memory (USM).
Figure S7.
Quantitation of B Cells, Related to Figures 5 and 6
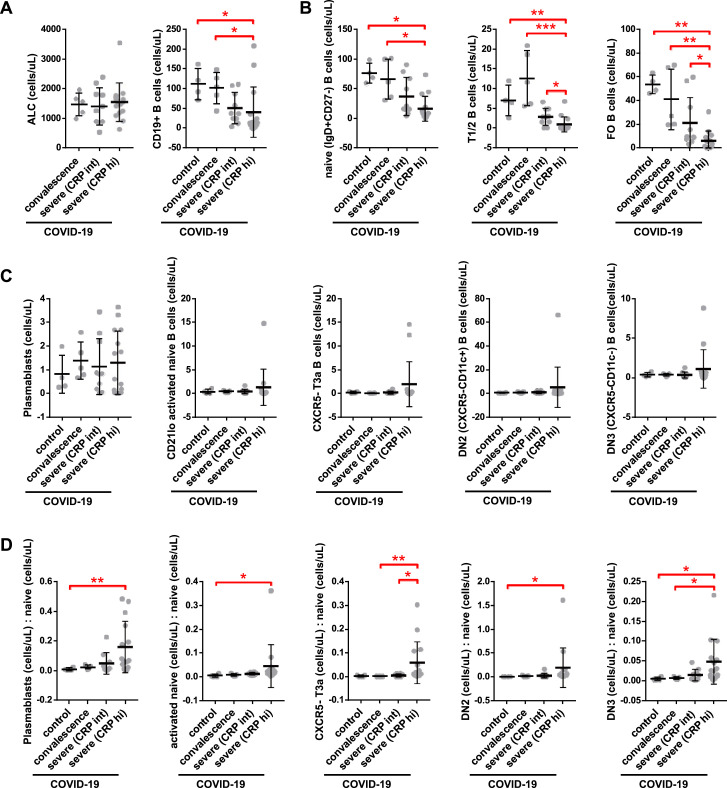
Quantitation of absolute B cell counts was undertaken in all COVID-19 patients in whom a proximal absolute lymphocyte count (ALC) was available in the electronic medical record at states of convalescence (n = 5), severe illness with an intermediate maximum CRP level during hospitalization (< 200 mg/L; severe (CRP int); n = 10), and severe illness with a high maximum CRP level during hospitalization (> 200 mg/L; severe (CRP hi); n = 15) as defined by the clinical criteria listed in Table S4. Data are compared to healthy controls (n = 4), assuming a healthy control ALC of 2.9 (K/uL). Quantitation shown by B cell level for each individual patient with mean, standard deviation, and significance by one-way ANOVA of log % B cell value indicated (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001) for (A) total CD19+ B cells, (B) naive, early transitional (T1/2), and follicular (FO) B cell subsets, (C) activated and double negative (DN) B cell subsets as indicated, and (D) activated and DN B cell subsets as a ratio to the available precursor population (naive, IgD+CD27-).
Increased Proportions of Activated Naive B Cells, IgD−CD27−CXCR5− B Cells, and Plasmablasts in Severely Ill COVID-19 Patients Correlate with Systemic Inflammation and Are Specific for SARS-CoV-2
Within the CD27+ memory B cell compartment, total switched memory (IgD−) B cells, including activated (CD21lo) switched memory B cells, and plasmablasts were significantly elevated (as a proportion of all B cells) in severely ill COVID-19 patients with high CRP levels as compared to convalescent COVID-19 patients and healthy controls (Figures 6A and 6B), though in terms of absolute numbers, these relative increases were blunted by the greater degree of B cell lymphopenia (Figure S7 ). Within the IgD+CD27− compartment, severely ill patients with COVID-19 and high CRP levels, specifically, showed an increase in a number of disease-related presumed non-germinal-center-derived activated B cells as compared to convalescent patients. These include activated naive B cells (IgD+CD27−CD21loCD11chi; Kaminski et al., 2012) as well as atypical late transitional B cells (IgD+CD27−CD10−CD73−CXCR5−; Figure 6C). Additionally, IgD−CD27−CXCR5− B cells that include populations described by Kaminski et al. based on the expression of CXCR5 and CD11c were expanded in severely ill COVID-19 patients with high CRP levels, specifically, as compared to convalescent patients and healthy controls (Figure 6D). The sum accumulation of these activated B cells was markedly higher in the severely ill COVID-19 patients with high versus intermediate CRP levels and also in comparison to convalescent patients and healthy controls (Figure 6E). Paralleling the trends observed with the loss of early transitional B cells, the gain in circulating activated B cells correlated with higher CRP levels and increased patient morbidity as measured by symptom duration and length of inpatient hospitalization (Figure 6F).
Figure 6.
Increased Proportions of Activated B Cell Subsets in the Peripheral Blood of Patients with Severe COVID-19
(A–E) Proportions of (A) switched memory (SM) B cells, (B) plasmablasts, (C) activated naive and CXCR5-late transitional (T3a) B cells, (D) IgD−CD27− double-negative (DN) B cells, and (E) total (sum) activated B cells in the peripheral blood of patients with COVID-19 at states of convalescence (n = 39), severe illness with an intermediate maximum CRP level during hospitalization (<200 mg/L; severe CRP int; n = 10), and severe illness with a high maximum CRP level during hospitalization (>200 mg/L; severe CRP hi; n = 15), as defined by the clinical criteria listed in Table S4 and compared to healthy controls (n = 4). Quantitation is shown by B cell level for each individual patient with mean, standard deviation, and significance by one-way ANOVA of log % B cell value indicated (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001). Representative COVID-19 contour plots are shown with healthy control contour plots and full B cell flow cytometry gating strategy outlined (Figure S6).
(F) Association of activated B cell frequency in peripheral blood with maximum CRP level, symptom duration at blood draw, and total length of hospital stay in all symptomatic COVID-19 patients (n = 29) with moderate to severe illness as defined by the clinical criteria listed in Table S4. Correlation is shown by linear regression with individual patients, 95% confidence bands, R2, and p values shown.
See also Figures S6 and S7 and Table S4.
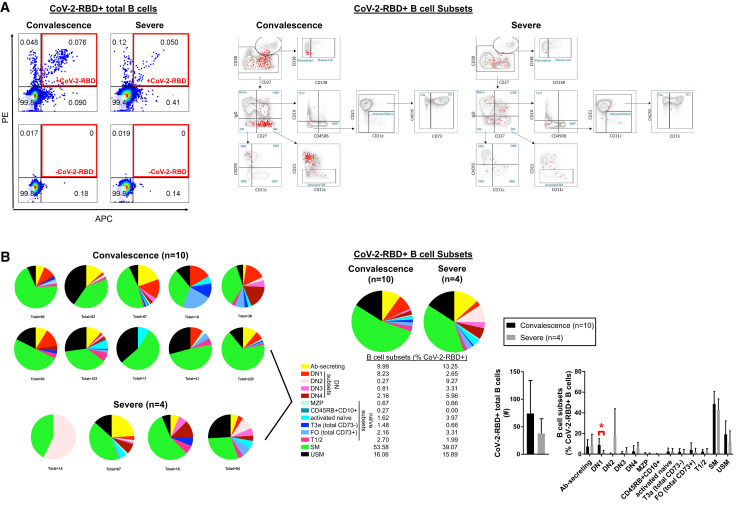
Although recent studies have also documented circulating activated B cell populations in COVID-19 patients (Mathew et al., 2020; Kuri-Cervantes et al., 2020; Woodruff et al., 2020), the antigen specificity of these cells was not evaluated. We tested whether the activated B cell populations we had identified were specific for the SARS-CoV-2 spike receptor binding domain (RBD) using recombinant RBD labeled separately with allophycocyanin (APC) and phycoerythrin (PE) fluorophores. Cells that stained with both labeled probes were considered authentic RBD-specific B cells (Figure 7 A). This probe contains a very small fraction of all the potential antibody epitopes in SARS-CoV-2 but is highly specific for this particular virus (Premkumar et al., 2020). All the activated and mainly disease-related populations of relevance in COVID-19 contain SARS-CoV-2-specific cells (Figures 7A and 7B), establishing that they were expanded as a result of an adaptive immune response to this virus. In particular, the presence of antigen-specific double-negative B cells, switched memory B cells, and plasmablasts is consistent with an extra-germinal center/extra-follicular type class-switched antibody response to SARS-CoV-2. These data establish that the aberrant non-germinal-center-type-activated B cells that accumulate in tissues also accumulate in the blood of severely ill and convalescent COVID-19 patients, and these include virus-specific B cells. Given that they bear the hallmarks of not being from germinal centers, they are therefore unlikely to provide optimal or durable humoral immunity.
Figure 7.
Activated B Cell Subsets in the Peripheral Blood of Patients with COVID-19 Are Specific for SARS-CoV-2-RBD
(A) Representative dot plots showing positive SARS-CoV-2-RBD staining in total CD19+ B cells (left; boxed in red) and B cell subsets (right; colored in red).
(B) All CD19+ B cells binding to SARS-CoV-2-RBD shown for COVID-19 patients at states of convalescence (n = 10) and severe illness (n = 4) with summation of total % CoV-2-RBD reactivity by indicated B cell subset and quantitated as mean ± SD with significance by Student’s t test of log % B cell value indicated (∗p < 0.05). DN2 and DN3 double-negative B cells are CXCR5 low, although DN1 and DN4 B cells are CXCR5 high.
Ab-secreting, antibody secreting; DN, double-negative; MZP, marginal zone precursor; SM, switched memory; T, transitional; USM, unswitched memory. Ab-secreting cells included both plasmablasts and plasma cell, though the latter were very rare. See also Figure S6 and Table S4.
Discussion
Long-lasting B cell memory and the highest affinity pathogen-specific antibodies are derived within germinal centers in secondary lymphoid organs. Germinal centers are anatomically structured to facilitate the selection of high-affinity B cell with long lifespans (Victora and Nussenzweig, 2012). Longevity of such responses exceeds decades for some infectious diseases and, when a substantial portion of a population is infected, can contribute to herd immunity. In contrast, antibody responses to SARS-CoV-2 appear to be similar to other human coronaviruses in being short lived in a large fraction of individuals (Long et al., 2020). Understanding the reasons for this decline in responses requires architectural studies of lymphoid organs from patients coupled with cell-cell interaction analyses, as well as approaches to reliably identify, quantify, and physically locate the diverse immune cell types that contribute to antibody induction. Using quantitative multi-color immunofluorescence combined with multispectral imaging and cell-cell interaction analyses of autopsy specimens as well as analyses of peripheral blood samples in parallel cohorts with acute SARS-CoV-2 infection, we show evidence for dysregulated humoral immune induction early in COVID-19, including a striking absence of germinal centers in the earliest stages of infection, defective Bcl6+ TFH cell generation, and aberrant lymphoid TNF-α production.
The absence of Bcl-6+ T follicular helper cells (and the consequent absence of germinal centers) in COVID-19 secondary lymphoid organs provides an explanation for a phenomenon anecdotally observed in autopsies of many different severe viral infections. These findings also provide a mechanistic basis for the recent descriptions of non-durable humoral immune responses, impaired humoral immunity, and the low levels of somatic hypermutation in antibodies from convalescent COVID-19 patients (Long et al., 2020; Robbiani et al., 2020; Brouwer et al., 2020). The alteration of the cytokine milieu in secondary lymphoid organs in this disease likely reflects a continuum across the spectrum of disease. Although T-dependent B cell activation, class switching, and some low-level somatic hypermutation do occur in COVID-19, the germinal center reaction is sub-optimal or totally absent, and this will likely, in due course of time, be reflected in less-durable class-switched antibody responses similar to those seen in SARS and MERS. These findings have some bearing on concepts such as herd immunity and immunity passports following natural infection with SARS-CoV-2. They strongly underscore the need and relevance of vaccination approaches to the prevention of COVID-19.
Severe infections by many different human viruses result in high levels of circulating cytokines and peripheral lymphopenia, but few studies have examined secondary lymphoid organs where immune responses are generated. Autopsy studies have revealed lymphoid depletion of the spleen and lymph nodes in Ebola, Marburg, and in H5N1 (Martines et al., 2015; Rippey et al., 1984; Lu et al., 2008; Gao et al., 2010). In SARS, lymphoid depletion and loss of germinal centers was also reported (Gu et al., 2005). Autopsy studies in these severe viral infections had not systematically examined lymphoid populations in secondary lymphoid organs. More detailed mechanistic studies of tissues have been undertaken in murine model systems. In a mouse model of severe malaria, germinal center responses were defective, and this was linked to a defect in T follicular helper cell differentiation (Ryg-Cornejo et al., 2016) that could be reversed by TNF-α blockade. Ehrlichia muris infection in mice has also been associated with the loss of germinal centers, and TNF-α blockade restored germinal centers as did the genetic deletion of TNF-α (Popescu et al., 2019). A mouse immunization model that involved prior generation of specific CD4+ T cell memory prior to infection with lymphocytic choriomeningitis virus generated a severe cytokine storm, splenic shrinkage, loss of germinal centers, and bone marrow hypocellularity, suggesting that lymphopenia and lymphoid organ abnormalities may be attributed to immune mechanisms rather than being a direct consequence of viral cytolysis (Penaloza-MacMaster et al., 2015). These studies in mice, taken together, suggest that significant elevation of secreted cytokines and chemokines seen in the context of protozoan, bacterial, and viral infections can cause the loss of germinal centers.
The studies of Ryg-Cornejo et al. in murine malaria and our studies in COVID-19 suggest that the observed cytokine and chemokine dysregulation may block germinal center type T follicular helper cell differentiation. The contribution of TNF-α to follicular development and germinal center formation (Pasparakis et al., 1996; Körner et al., 1997) as well as to the loss of germinal centers is complex and seemingly contradictory (Ryg-Cornejo et al., 2016; Popescu et al., 2019). Local cytokine concentrations at the site of T follicular helper cell differentiation likely have important consequences for the germinal center reaction. The differentiation of CD4+Bcl-6−CXCR5+ pre-germinal center TFH cells into CD4+Bcl-6+CXCR5+ GC-TFH cells likely occurs extra-follicularly at the T-B interface (Kerfoot et al., 2011; Kitano et al., 2011; Crotty, 2014; Vinuesa et al., 2016). Based on our findings, we suspect that the very high local levels of TNF-α and possibly other cytokines at this location in COVID-19 lymph nodes, possibly induced downstream of TH1 cell activation, block the final step in T follicular helper cell differentiation. In the murine malaria model, IFN-γ blockade also restored TFH cells and germinal centers, consistent with TH1 cells being upstream of the induction of TNF-α. Given that Bcl-6+ B cells, Bcl-6+ TFH cells, and Bcl-6+ T follicular regulatory cells are all extremely sparse or absent in COVID-19 secondary lymphoid organs, the formal possibility that excessive TNF signaling (or excessive signaling by some combination of cytokines in the extra-follicular area) negatively impacts the expression of BCL6 either transcriptionally or post-transcriptionally also needs to be investigated. Such deeper mechanistic studies can best be pursued in murine models.
Because of our focus on the loss of germinal centers, we have concentrated on TNF-α because of its known ability, when produced in excess, to contribute to impaired TFH cell differentiation and germinal center loss. Many other cytokines are induced in COVID-19 and probably contribute to some aspects of the phenotypes that we describe here. IL-6, for example, though it has pleiotropic effects, is known to suppress lymphopoiesis and induce myelopoiesis (Maeda et al., 2009), and it might thus contribute to the B lymphopenia that we document here.
Altered extra-follicular B cell activation could potentially also contribute to a defect in T follicular helper cell differentiation observed in SARS-CoV-2 infection. After the initial activation of naive CD4+ T cells by dendritic cells presenting the relevant major histocompatibility complex (MHC) class II molecule and peptide, along with co-stimulation, these T cells activate antigen-specific B cells that present the same MHC-peptide complex, and extra-follicular B cell foci are generated. It is in this vicinity that pre-germinal center T follicular helper cells are first generated, and we have shown in humans (Maehara et al., 2018), as others have in mice (Roco et al., 2019), that most isotype switching actually occurs at this location. In COVID-19, it is likely that some antibody generation occurs extra-follicularly, though we have identified IgG-expressing plasmablasts extra-follicularly as well as in the follicles bereft of germinal centers. Activated B cells expressing ICOSL provide additional differentiation signals to activated CD4+ T cells to acquire high levels of CXCR5, induce the expression of Bcl-6, and migrate into the follicles as fully differentiated germinal center type T follicular helper cells, which set up the germinal center reaction. Perhaps because cytokine alterations lead to the formation of dysfunctional B cells and plasmablasts outside the follicle, activated B cells in COVID-19 may be less capable of inducing cognate T cells to differentiate into germinal center type T follicular helper cells.
It is possible that our investigations may throw some light on the mechanisms of disease progression in severe COVID-19, although a deeper understanding will likely await the acquisition of more knowledge and the development of suitable animal models. The evasion of the anti-viral aspects of innate immunity and the overly aggressive activation of inflammation by the virus likely results in an altered milieu that contributes both to the relative attenuation of CD8+ T cell immunity and prevents the generation of Bcl-6+ T follicular helper cells. As a result of the latter, the early development of high-affinity antibodies that could contribute to some attenuation of viral spread may be compromised. Whether the increased CD4+/CD8+ T cell ratio we observe in lymph nodes and spleens of COVID-19 patients reflects selective activation of CD4+ T cells or a preferential depletion of CD8+ T cells is unclear. Why certain individuals are more prone to initial more aggressive and destructive immune responses also remains a very difficult question to address at this time. Based on our analyses, severe disease does not appear to be due to a paucity of regulatory T cells in secondary lymphoid organs at the earlier stages of the disease, though our approaches do not interrogate T reg cell function. The striking relative accumulation of FOXP3+ regulatory T cells that we observe late in disease could possibly reflect a homeostatic mechanism for the resolution of infection. More likely, it reflects the reduced overall numbers of recirculating T cells entering lymphoid organs and the recurring activation of naive CD4+ T cells into activated subsets that egress these lymphoid organs while lymphoid organ-resident regulatory T cells perhaps do not leave these sites and therefore appear to accumulate in lymph nodes and the spleen.
In summary, even in acutely ill COVID-19 patients, at a time when they have extremely high viral loads and abundant virus has been demonstrated in their lungs, there is a striking absence of germinal centers associated with a marked reduction of germinal center B cells but preservation of AID-expressing B cells. Thus, though there is robust T-cell-mediated activation of B cells, germinal centers do not form. The robust activation of non-germinal center type B cell responses does not give rise to long-lived memory or high-affinity B cells. The underlying basis for the loss of germinal centers is best explained by the striking failure of differentiation of Bcl-6+ T follicular helper cells, likely because of dramatic changes in the extra-follicular cytokine milieu driven by TH1 cells and the aberrant local production of TNF-α in lymphoid organs. We believe these findings will be relevant to a range of human viral and non-viral diseases in which there is a cytokine storm, a better understanding of which will require a granular function-focused analysis of the architecture and composition of lymph nodes and spleens. We predict that a broadly applicable common mechanistic basis similar to what we describe here will be elucidated in future studies on MERS, H1N1, Ebola, Marburg, and other viral diseases.
Limitations of Study
Our studies on secondary lymphoid organs focused by necessity on very ill patients, and although changes in the disease may represent a continuum, it is quite possible that patients with less-severe disease may induce better germinal center responses. Of necessity, the numbers for thoracic lymph node and spleen were relatively small. We used a single approach to study lymphocyte populations in the lymph nodes and the spleen, and the complementary use of an orthogonal approach would have been ideal.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-CD3 | BD Biosciences | Clone UCHT1 |
| Anti-CD56 | BD Biosciences | Clone NCAM16.2 |
| Anti-CD19 | BioLegend | Clone SJ25C1; RRID: AB_2564193 |
| Anti-CD27 | BD Biosciences | Clone L128; RRID: AB_2744349 |
| Anti-IgD | BioLegend | Clone IA6-2; AB_2740295 |
| Anti-CD38 | BioLegend | Clone HB-7; AB_2563875 |
| Anti-CD10 | BioLegend | Clone HI10a; RRID: AB_2565878 |
| Anti-CD45RB | BioLegend | Clone MEM-55; RRID: AB_2687314 |
| Anti-CD21 | BD Biosciences | Clone B-ly4 |
| Anti-CD73 | BioLegend | Clone AD2; RRID: AB_11204424 |
| Anti-CD138 | BioLegend | Clone MI15; RRID: AB_2562658 |
| Anti-CD11c | BioLegend | Clone Bu15; RRID: AB_2561503 |
| Anti-CXCR5 | BioLegend | Clone J252D4; RRID:AB_2561813 |
| Anti CD3 | DAKO | Clone A045229-2; RRID:AB_2335677 |
| Anti-CD4 | Abcam | Clone EPR685; RRID:AB_2750883 |
| Anti-CD19 | Biocare Medical | Clone SKU310; RRID:AB_2861287 |
| Anti-Bcl-6 | Biocare Medical | Clone LN22; RRID:AB_10890175 |
| Anti-AID | Invitrogen | Clone ZA001; RRID:AB_2533403 |
| Anti-T-bet | Abcam | Clone ab150440; RRID:AB_150440 |
| Anti-GATA-3 | Biocare Medical | Clone CM405A; RRID:AB_10895444 |
| Anti-ICOS | Cell Signaling Technology | Clone 89601; RRID:AB_2800142 |
| Anti-Rorc | Abcam | Clone ab212496 |
| Anti-CXCR5 | R&D Systems | Clone MAB190; RRID:AB_2292654 |
| Anti-FOXP3 | Cell Signaling Technology | Clone 98377; RRID:AB_2747370 |
| Ant-CD8 | Abcam | Clone ab85972; RRID:AB_10674324 |
| Anti-IgD | DAKO | Clone AA093; RRID:AB_578485 |
| Anti-CD27 | Abcam | Clone ab131254; RRID:AB_11155136 |
| Anti-IgG | Abcam | Clone ab109489; RRID:AB_10863040 |
| Anti-TNF-α | Abcam | Clone ab6671; RRID:AB_305641 |
| Anti-CD35 | Abcam | Clone ab25; RRID:AB_448530 |
| Chemicals and Recombinant Proteins | ||
| Prolong Diamond Antifade Mountant with DAPI | Invitrogen | CAT#P36962 |
| Opal Manual Multiplex IHC Kit | Perkin Elmer | CAT#NEK811001KT |
| FcR Blocking Reagent | Miltenyi Biotec | CAT#130-059-901 |
| Rainbow Calibration Particles | ThermoFisher | CAT#A34305 |
| LIVE/DEAD Fixable Blue Dead Cell Stain Kit | ThermoFisher | CAT# L23105 |
| 4% Paraformaldehyde solution | Santa Cruz Biotechnology | CAT#sc-281692 |
| Streptavidin-APC conjugate | Life Technologies | CAT#S32362 |
| Streptavidin-PE conjugate | Life Technologies | CAT#S21388 |
| Freestyle Expression Medium | Thermo Scientific | CAT#123388018 |
| Dulbecco’s Phosphate buffer saline | Corning | CAT#21031 |
| Fetal Bovine Serum (HyClone) | Fisher Scientific | CAT#SH3007003 |
| TALON Metal Affinity Resin | Takara | CAT#635652 |
| Experimental Models: Cell Lines | ||
| Human Expi293F | Thermo Fisher | CAT#A14527 |
| Recombinant DNA | ||
| pVRC-SARS-CoV-2-RBD | This study | N/A |
| Software and Algorithms | ||
| FlowJo | FlowJo | https://www.flowjo.com; RRID:SCR_008520 |
| FACSDIVA | BD Biosciences | https://www.bdbiosciences.com/en-us/instruments/research-instruments/research-software/flow-cytometry-acquisition/facsdiva-software; RRID:SCR_001456 |
| Prism 8 | GraphPad Software | www.graphpad.com/scientific-software/prism/; RRID:SCR_002798 |
| TissueQuest | TissueGnostics | https://www.tissuegnostics.com/products/software/tissuequest; RRID:SCR_014822 |
| StrataQuest | TissueGnostics | https://www.tissuegnostics.com/products/software/strataquest |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Shiv Pillai pillai@helix.mgh.harvard.edu
Materials Availability
The SARS-CoV-2 RBD protein is made freely available through Dr. Aaron Schmidt who may be reached through the Lead Contact or directly.
Data and Code Availability
The published article includes all data generated or analyzed during this study, and summarized in the accompanying tables, figures and Supplemental Information.
Experimental Model and Subject Details
Human Subjects
Tissue analysis Cohort
Thoracic lymph nodes and spleens samples from COVID-19 patients were obtained through the Brigham and Women’s hospital Department of Pathology.
Sample Size Estimation: Power calculations and sample size estimation were not performed before the initiation of this study which was based primarily on the availability of COVID-19 positive autopsies for analysis between March and May 2020. All cases were retrieved from the Anatomic Pathology files of Brigham and Women’s Hospital and included 17 patients (information on age and gender in Tables S1 and S2) including 11 with laboratory confirmed COVID-19 who underwent autopsy in 2020.
Sample allocation to experimental groups: All patients had tested positive for SARS-CoV-2 by RT-PCR of nasopharyngeal swabs in a laboratory during hospital admission. All cases were divided into two groups; early (less than ten days from respiratory symptoms onsets to death, hospitalization of up to 8 days), and late (hospitalized for 15-36 days prior to death). Thoracic lymph nodes were obtained from six age-matched non-COVID-19 patients who underwent autopsies at the Brigham and Women’s Hospital in the same time window. In addition, seven spleens from automobile accident victims were obtained from the Ragon Institute Tissue Core. These were histologically normal. Information on age and gender is in Table S3. Ten discarded control tonsils from Massachusetts General Hospital not matched for age and gender were also used for initial titration and validation of certain antibodies and as controls for TNF-α and CD8 staining studies,
Peripheral Blood Cohort
Peripheral blood samples were drawn from both Outpatients and Inpatients with COVID-19 at Massachusetts General Hospital and fresh blood was analyzed for flow cytometry using a multi-color panel.
Sample Size estimation: Power calculations were not undertaken pior to the initiation of these studies. However, the primary end-point was the sum of activated B cells (as a % CD19+ B cells in peripheral blood). Using a two sided Student’s t test to compare log values of convalescent and severe (CRP hi) COVID-19 patient samples, with groups of 10, we had more than 90% power to detect an effect of size of 1.20 between groups based on simulation studies using 10,000 Monte Carlo samples with a type 1 error rate of 5%.
Allocation to Experimental Groups: Data is presented on B cell populations from 68 patients, including moderately ill, severely ill and convalescent patients. Convalescence was defined as a clinically asymptomatic state on the date of blood draw, either from a baseline asymptomatic state or recuperated from moderate clinical symptoms of COVID- 19. Moderate disease was defined as active clinical symptoms of COVID-19 on the date of blood draw that did not necessitate a hospital admission. Severe disease was defined as active clinical symptoms of COVID-19 on the date of blood draw that did necessitate a hospital admission. Severe disease was further subdivided by maximum CRP level during hospital admission as CRP < 200 mg/L classified as ‘CRP intermediate (int)’ and CRP > 200 mg/L as ‘CRP high (hi).’
Study Approval
This study was performed with the approval of the Institutional Review Boards at the Massachusetts General Hospital and the Brigham and Women’s Hospital.
Method Details
Multi-color immunofluorescence staining
Tissue samples were fixed in formalin, embedded in paraffin, and sectioned. These specimens were incubated with the following antibodies: anti-CD3 (clone: A045229-2; DAKO), anti-CD4 (clone: EPR6855;Abcam), anti-CD19 (clone: SKU310; Biocare Medical), anti-Bcl6 (clone: LN22; Biocare Medical), anti-AID (clone: ZA001; Invitrogen), anti-T-bet (clone: ab150440; Abcam), GATA3 (clone: CM405A; Biocare), ICOS (clone: 89601; Cell Signaling Technology), Rorc (clone: ab212496; Abcam), CXCR5 (clone: MAB190; R&D Systems), Foxp3 (clone: 98377; Cell Signaling Technology), anti-CD8 (clone: ab85792; Abcam), anti-IgD (clone: AA093; DAKO), anti-CD27 (clone: ab131254; Abcam), anti-IgG (clone: ab109489; Abcam), anti-TNF-α (clone: ab6671; Abcam), and anti-CD35 (clone: ab25; Abcam) followed by incubation with a secondary antibody using an Opal Multiplex Kit (Perkin Elmer). The samples were mounted with ProLong Diamond Antifade mountant containing DAPI (Invitrogen).
Microscopy and Quantitative Image Analysis
Images of the tissue specimens were acquired using the TissueFAXS platform (TissueGnostics). For quantitative analysis, the entire area of the tissue was acquired as a digital grayscale image in five channels with filter settings for FITC, Cy3, Cy5 and AF75 in addition to DAPI. Cells of a given phenotype were identified and quantitated using the TissueQuest software (TissueGnostics), with cut-off values determined relative to the positive controls. This microscopy-based multicolor tissue cytometry software permits multicolor analysis of single cells within tissue sections similar to flow cytometry. In addition, multispectral images (seven-colors staining) were unmixed using spectral libraries built from images of single stained tissues for each reagent using the StrataQuest (TissueGnostics) software. StrataQuest software was also used to quantify cell-to-cell contact. In the StrataQuest cell-to-cell contact application, masks of the nuclei based on DAPI staining establish the inner boundary of the cytoplasm and the software “looks” outward toward the plasma membrane boundary. Overlap of at least 3 pixels of adjacent cell markers is required to establish a “contact” criterion. Although the software has been developed and validated more recently, the principle of the method and the algorithms used have been described in detail elsewhere (Ecker and Steiner, 2004).
Flow cytometry
1 Million fresh PBMCs were stained within 2 hours of isolation. Prior to antibody staining, Fc receptors were blocked using human FcR blocking reagent (Miltenyi) at a concentration of 1:50 at 4°C for 10 minutes. Cells were surface stained at 4°C, protected from light, using optimized concentrations of fluorochrome-conjugated primary antibodies for 30 minutes as well as live/dead fixable blue stain (Thermo Fisher) at a concentration of 1:20 using the following antibody panels (clone, manufacturer).
B cell panel: CD3 (UCHT1, BD Biosciences), CD56 (NCAM16.2, BD Biosciences), CD19 (SJ25C1, BioLegend), CD27 (L128, BD Biosciences), IgD (IA6-2, BD Biosciences), CD38 (HB-7, BioLegend), CD10 (HI10a, BioLegend), CD21 (B-ly4, BD Biosciences), CD73 (AD2, BioLegend), CD45RB (MEM-55, BioLegend), CD138 (MI15, BioLegend), CD11c (Bu15, BioLegend), and CXCR5 (J252D4, BioLegend).
For the RBD-specific characterization, 6-7 million fresh PBMCs were stained using fluorescently labeled SARS-CoV-2 RBD in addition to the B cell panel antibodies listed above.
Cells were then washed in PBS and fixed using 4% paraformaldehyde for 30 minutes at at 4°C. Flow cytometry was performed on a BD Symphony (BD Biosciences, San Jose, CA) and rainbow tracking beads (8 peaks calibration beads, Fisher) were used to ensure consistent signals between flow cytometry batches. FCS files were analyzed, and B cell subsets were quantified using FlowJo software (version 10).
RBD Expression and Purification
SARS-2 RBD (GenBank: MN975262.1) was cloned into pVRC vector containing an HRV 3C-cleavable C-terminal SBP-His8X tag and sequence confirmed by Genwiz. The construct was transiently transfected into mammalian Expi293F suspension cells for recombinant expression. 5 days post-transfection, supernatants were harvested and clarified by low-speed centrifugation. The RBD was purified by immobilized metal affinity chromatography (IMAC) using Cobalt-TALON resin (Takara) followed by size exclusion chromatography on Superdex 200 Increase 10/300 GL (GE Healthcare) in PBS. Purity was assessed by SDS-PAGE analysis. The fluorescent PE-SA (Invitrogen, Cat#12-4317-877) and APC-SA (Invitrogen; Cat#S32362) labels were added to the purified SBP-tagged RBD proteins through iterative complex formation, as previously described (Weaver et al., 2016). The fluorescent SA conjugates were added to SBP-RBD in five increments to sequentially form the complexes. In this case the final molar ratio of probe to streptavidin valency was 1:1 (one SA-fluorophore can bind two SBP tags). After each stepwise addition of the fluorescent label, the mixture was incubated for 20 minutes and set rotating at 4C within an opaque 1.5 mL Eppendorf spin tube. Using this method, we generated fluorescent probes at a final concentration of 0.1ug/ul. An example of SBP-RBD (31,000 g/mol) labeled with PE-SA (300,000 g/mol) is described for a total 10 assays (0.5 ug labeled protein per assay): First, 5uL of SBP-RBD at 1ug/ul was diluted in 20.8uL PBS. Fluorescent PE-SA at 1ug/ul was then added in five increments, with an incremental volume of 4.8 uL for a final volume of 50uL.
Quantification and Statistical Analysis
Flow cytometry, clinical correlations and tissue studies. GraphPad Prism version 8 was used for statistical analysis, curve fitting and linear regression. A two-tailed Mann-Whitney U test was used to calculate p values for continuous, non-parametric variables. For comparing more than one population, Kruskal-Wallis testing was used with Dunn’s multiple comparison testing. A p value of < 0.05 was considered significant.
Consortia
The members of the Mass CPR Specimen Working Group are Betelihem A. Abayneh, Patrick Allen, Diane Antille, Katrina Armstrong, Alejandro Balazs, Max Barbash, Siobhan Boyce, Joan Braley, Karen Branch, Katherine Broderick, George Daley, Ashley Ellman, Liz Fedirko, Keith Flaherty, Jeanne Flannery, Pamela Forde, Elise Gettings, David Golan, Amanda Griffin, Sheila Grimmel, Kathleen Grinke, Kathryn Hall, Meg Healey, Howard Heller, Deborah Henault, Grace Holland, Chantal Kayitesi, Evan C. Lam, Vlasta LaValle, Yuting Lu, Sara Luthern, Jordan Marchewska, Brittni Martino, Ilan Millstrom, Noah Miranda, Christian Nambu, Susan Nelson, Marjorie Noone, Claire O’Callaghan, Christine Ommerborn, Lois Chris Pacheco, Nicole Phan, Falisha A. Porto, Alexandra Reissis, Francis Ruzicka, Edward Ryan, Katheleen Selleck, Arlene Sharpe, Christianne Sharr, Sue Slaugenhaupt, Kimberly Smith Sheppard, Elizabeth Suschana, Vivine Wilson, and Daniel Worrall.
Acknowledgments
We thank Doug Kwon and Brooke Spencer of the Ragon Institute for access to Ragon Institute Tissue core specimens and Andrew Lichtman of BWH Pathology for helpful advice. This work was supported by NIH U19 AI110495 to S.P., NIH R01 AI146779 to A.G.S., and NIH R01AI137057 and DP2DA042422 to D.L. B.M.H. was supported by NIGMS T32 GM007753 and T.M.C. was supported by T32 AI007245. Funding for these studies from the Massachusetts Consortium of Pathogen Readiness, the Mark and Lisa Schwartz Foundation, and Enid Schwartz is also acknowledged. The graphical abstract was prepared using Biorender.
Author Contributions
Conceptualization, S.P., R.F.P., N.K., J.R.F., H.-H.K., J. Boucau, B.D.W., and X.G.Y.; Methodology, H.A.-C., N.K., J.R.F., T.J.D., M.T.W., H.-H.K., and J. Boucau; Investigation, N.K., H.-H.K., J. Boucau, A.P.-T., K.L., M.O., J. Bals, Y.C.B., N.B., J.C., F.C., K.E., J. Fallon, K.K.F., P.G.-B., C.A.H., C.J., P.K., M.K., X.L., H.L., J.L., N.L.L., A.R.M., Y.R., K.S., L.S., S.S., N.S., W.S., X.S., H.J.T., A.L.Z., and R.F.P.; Resources, R.F.P., B.D.W., X.G,Y., J.L., V.S.M., D.L., A.G.S., B.M.H., J. Feldman, T.M.C., J. Bals, G.A., and M.L.; Original Draft, S.P., N.K., J.R.F., J. Boucau, and H.-H.K.; Review/Editing, S.P., B.D.W., N.K., J.R.F., H.-H.K., J. Boucau, and H.A.-C.; Supervision, S.P. and X.G.Y.
Declaration of Interests
S.P. is on the SAB of Abpro Inc. and Pulsar Biopharma. G.A. is founder of Seromyx Systems Inc.
Published: August 19, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2020.08.025.
Supplemental Information
References
- Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buja L.M., Wolf D.A., Zhao B., Akkanti B., McDonald M., Lelenwa L., Reilly N., Ottaviani G., Elghetany M.T., Trujillo D.O. The emerging spectrum of cardiopulmonary pathology of the coronavirus disease 2019 (COVID-19): report of 3 autopsies from Houston, Texas, and review of autopsy findings from other United States cities. Cardiovasc. Pathol. 2020;48:107233. doi: 10.1016/j.carpath.2020.107233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41:529–542. doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker R.C., Steiner G.E. Microscopy-based multicolor tissue cytometry at the single-cell level. Cytometry A. 2004;59:182–190. doi: 10.1002/cyto.a.20052. [DOI] [PubMed] [Google Scholar]
- Farmer J.R., Allard-Chamard H., Sun N., Ahmad M., Bertocchi A., Mahajan V.S., Aicher T., Arnold J., Benson M.D., Morningstar J. Induction of metabolic quiescence defines the transitional to follicular B cell switch. Sci. Signal. 2019;12:eaaw5573. doi: 10.1126/scisignal.aaw5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanti M., Birger R., Ud-Dean M., Filip I., Morita H., Comito D., Anthony S., Freyer G.A., Ibrahim S., Lane B. Longitudinal active sampling for respiratory viral infections across age groups. Influenza Other Respir. Viruses. 2019;13:226–232. doi: 10.1111/irv.12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R., Dong L., Dong J., Wen L., Zhang Y., Yu H., Feng Z., Chen M., Tan Y., Mo Z. A systematic molecular pathology study of a laboratory confirmed H5N1 human case. PLoS ONE. 2010;5:e13315. doi: 10.1371/journal.pone.0013315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J., Gong E., Zhang B., Zheng J., Gao Z., Zhong Y., Zou W., Zhan J., Wang S., Xie Z. Multiple organ infection and the pathogenesis of SARS. J. Exp. Med. 2005;202:415–424. doi: 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., China Medical Treatment Expert Group for Covid-19 Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenks S.A., Cashman K.S., Woodruff M.C., Lee F.E., Sanz I. Extrafollicular responses in humans and SLE. Immunol. Rev. 2019;288:136–148. doi: 10.1111/imr.12741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Kaminski D.A., Wei C., Rosenberg A.F., Lee F.E., Sanz I. Multiparameter flow cytometry and bioanalytics for B cell profiling in systemic lupus erythematosus. Methods Mol. Biol. 2012;900:109–134. doi: 10.1007/978-1-60761-720-4_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerfoot S.M., Yaari G., Patel J.R., Johnson K.L., Gonzalez D.G., Kleinstein S.H., Haberman A.M. Germinal center B cell and T follicular helper cell development initiates in the interfollicular zone. Immunity. 2011;34:947–960. doi: 10.1016/j.immuni.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano M., Moriyama S., Ando Y., Hikida M., Mori Y., Kurosaki T., Okada T. Bcl6 protein expression shapes pre-germinal center B cell dynamics and follicular helper T cell heterogeneity. Immunity. 2011;34:961–972. doi: 10.1016/j.immuni.2011.03.025. [DOI] [PubMed] [Google Scholar]
- Körner H., Cook M., Riminton D.S., Lemckert F.A., Hoek R.M., Ledermann B., Köntgen F., Fazekas de St Groth B., Sedgwick J.D. Distinct roles for lymphotoxin-alpha and tumor necrosis factor in organogenesis and spatial organization of lymphoid tissue. Eur. J. Immunol. 1997;27:2600–2609. doi: 10.1002/eji.1830271020. [DOI] [PubMed] [Google Scholar]
- Kuri-Cervantes L., Pampena M.B., Meng W., Rosenfeld A.M., Ittner C.A.G., Weisman A.R., Agyekum R.S., Mathew D., Baxter A.E., Vella L.A. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci. Immunol. 2020;5:eabd7114. doi: 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lax S.F., Skok K., Zechner P., Kessler H.H., Kaufmann N., Koelblinger C., Vander K., Bargfrieder U., Trauner M. Pulmonary arterial thrombosis in COVID-19 with fatal outcome: results from a prospective, single-center, clinicopathologic case series. Ann. Intern. Med. 2020 doi: 10.7326/M20-2566. Published online May 14, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescure F.X., Bouadma L., Nguyen D., Parisey M., Wicky P.H., Behillil S., Gaymard A., Bouscambert-Duchamp M., Donati F., Le Hingrat Q. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect. Dis. 2020;20:697–706. doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q.X., Liu B.Z., Deng H.J., Wu G.C., Deng K., Chen Y.K., Liao P., Qiu J.F., Lin Y., Cai X.F. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- Lu M., Xie Z.G., Gao Z.C., Wang C., Li N., Li M., Shao H.Q., Wang Y.P., Gao Z.F. [Histopathologic study of avian influenza H5N1 infection in humans] Zhonghua Bing Li Xue Za Zhi. 2008;37:145–149. [PubMed] [Google Scholar]
- Maeda K., Malykhin A., Teague-Weber B.N., Sun X.H., Farris A.D., Coggeshall K.M. Interleukin-6 aborts lymphopoiesis and elevates production of myeloid cells in systemic lupus erythematosus-prone B6.Sle1.Yaa animals. Blood. 2009;113:4534–4540. doi: 10.1182/blood-2008-12-192559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehara T., Mattoo H., Mahajan V.S., Murphy S.J., Yuen G.J., Ishiguro N., Ohta M., Moriyama M., Saeki T., Yamamoto H. The expansion in lymphoid organs of IL-4+ BATF+ T follicular helper cells is linked to IgG4 class switching in vivo. Life Sci. Alliance. 2018;1:e201800050. doi: 10.26508/lsa.201800050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martines R.B., Ng D.L., Greer P.W., Rollin P.E., Zaki S.R. Tissue and cellular tropism, pathology and pathogenesis of Ebola and Marburg viruses. J. Pathol. 2015;235:153–174. doi: 10.1002/path.4456. [DOI] [PubMed] [Google Scholar]
- Mathew D., Giles J.R., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E., Alanio C., Kuri-Cervantes L., Pampena M.B., D’Andrea K. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020 doi: 10.1126/science.abc8511. Published online July 15, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo H., Zeng G., Ren X., Li H., Ke C., Tan Y., Cai C., Lai K., Chen R., Chan-Yeung M., Zhong N. Longitudinal profile of antibodies against SARS-coronavirus in SARS patients and their clinical significance. Respirology. 2006;11:49–53. doi: 10.1111/j.1440-1843.2006.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasparakis M., Alexopoulou L., Episkopou V., Kollias G. Immune and inflammatory responses in TNF alpha-deficient mice: a critical requirement for TNF alpha in the formation of primary B cell follicles, follicular dendritic cell networks and germinal centers, and in the maturation of the humoral immune response. J. Exp. Med. 1996;184:1397–1411. doi: 10.1084/jem.184.4.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penaloza-MacMaster P., Barber D.L., Wherry E.J., Provine N.M., Teigler J.E., Parenteau L., Blackmore S., Borducchi E.N., Larocca R.A., Yates K.B. Vaccine-elicited CD4 T cells induce immunopathology after chronic LCMV infection. Science. 2015;347:278–282. doi: 10.1126/science.aaa2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S., Dandekar A.A. Immunopathogenesis of coronavirus infections: implications for SARS. Nat. Rev. Immunol. 2005;5:917–927. doi: 10.1038/nri1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu M., Cabrera-Martinez B., Winslow G.M. TNF-α contributes to lymphoid tissue disorganization and germinal center B cell suppression during intracellular bacterial infection. J. Immunol. 2019;203:2415–2424. doi: 10.4049/jimmunol.1900484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portugal S., Obeng-Adjei N., Moir S., Crompton P.D., Pierce S.K. Atypical memory B cells in human chronic infectious diseases: an interim report. Cell. Immunol. 2017;321:18–25. doi: 10.1016/j.cellimm.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premkumar L., Segovia-Chumbez B., Jadi R., Martinez D.R., Raut R., Markmann A., Cornaby C., Bartelt L., Weiss S., Park Y. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci. Immunol. 2020;5:eabc8413. doi: 10.1126/sciimmunol.abc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippey J.J., Schepers N.J., Gear J.H. The pathology of Marburg virus disease. S. Afr. Med. J. 1984;66:50–54. [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roco J.A., Mesin L., Binder S.C., Nefzger C., Gonzalez-Figueroa P., Canete P.F., Ellyard J., Shen Q., Robert P.A., Cappello J. Class-switch recombination occurs infrequently in germinal centers. Immunity. 2019;51:337–350.e7. doi: 10.1016/j.immuni.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryg-Cornejo V., Ioannidis L.J., Ly A., Chiu C.Y., Tellier J., Hill D.L., Preston S.P., Pellegrini M., Yu D., Nutt S.L. Severe malaria infections impair germinal center responses by inhibiting T follicular helper cell differentiation. Cell Rep. 2016;14:68–81. doi: 10.1016/j.celrep.2015.12.006. [DOI] [PubMed] [Google Scholar]
- Schaefer I.M., Padera R.F., Solomon I.H., Kanjilal S., Hammer M.M., Hornick J.L., Sholl L.M. In situ detection of SARS-CoV-2 in lungs and airways of patients with COVID-19. Mod. Pathol. 2020;19:1–11. doi: 10.1038/s41379-020-0595-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisoncik J.R., Korth M.J., Simmons C.P., Farrar J., Martin T.R., Katze M.G. Into the eye of the cytokine storm. Microbiol. Mol. Biol. Rev. 2012;76:16–32. doi: 10.1128/MMBR.05015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardhana S.A., Wolchok J.D. The many faces of the anti-COVID immune response. J Exp Med. 2020;217:e20200678. doi: 10.1084/jem.20200678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora G.D., Nussenzweig M.C. Germinal centers. Annu. Rev. Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- Vinuesa C.G., Linterman M.A., Yu D., MacLennan I.C. Follicular helper T cells. Annu. Rev. Immunol. 2016;34:335–368. doi: 10.1146/annurev-immunol-041015-055605. [DOI] [PubMed] [Google Scholar]
- Weaver G.C., Villar R.F., Kanekiyo M., Nabel G.J., Mascola J.R., Lingwood D. In vitro reconstitution of B cell receptor-antigen interactions to evaluate potential vaccine candidates. Nat. Protoc. 2016;11:193–213. doi: 10.1038/nprot.2016.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff M., Ramonell R., Cashman K., Nguyen D., Ley A., Kyu S., Saini A., Haddad N., Chen W., Howell J.C. Critically ill SARS-CoV-2 patients display lupus-like hallmarks of extrafollicular B cell activation. medRxiv. 2020 doi: 10.1101/2020.04.29.20083717. [DOI] [Google Scholar]
- Xu X., Chang X.N., Pan H.X., Su H., Huang B., Yang M., Luo D.J., Weng M.X., Ma L., Nie X. [Pathological changes of the spleen in ten patients with new coronavirus disease 2019 (COVID-19) by postmortem needle autopsy] Zhonghua Bing Li Xue Za Zhi. 2020;49:576–582. doi: 10.3760/cma.j.cn112151-20200401-00278. [DOI] [PubMed] [Google Scholar]
- Zhang X., Tan Y., Ling Y., Lu G., Liu F., Yi Z., Jia X., Wu M., Shi B., Xu S. Viral and host factors related to the clinical outcome of COVID-19. Nature. 2020;583:437–440. doi: 10.1038/s41586-020-2355-0. [DOI] [PubMed] [Google Scholar]
- Zheng S., Fan J., Yu F., Feng B., Lou B., Zou Q., Xie G., Lin S., Wang R., Yang X. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369:m1443. doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., China Novel Coronavirus Investigating and Research Team A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A., Hui D.S., Perlman S. Middle East respiratory syndrome. Lancet. 2015;386:995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all data generated or analyzed during this study, and summarized in the accompanying tables, figures and Supplemental Information.