Summary
Convalescing coronavirus disease 2019 (COVID-19) patients mount robust T cell responses against SARS-CoV-2, suggesting an important role of T cells in viral clearance. To date, the phenotypes of SARS-CoV-2-specific T cells remain poorly defined. Using 38-parameter CyTOF, we phenotyped longitudinal specimens of SARS-CoV-2-specific CD4+ and CD8+ T cells from nine individuals who recovered from mild COVID-19. SARS-CoV-2-specific CD4+ T cells were exclusively Th1 cells and predominantly Tcm cells with phenotypic features of robust helper function. SARS-CoV-2-specific CD8+ T cells were predominantly Temra cells in a state of less terminal differentiation than most Temra cells. Subsets of SARS-CoV-2-specific T cells express CD127, can proliferate homeostatically, and can persist for over 2 months. Our results suggest that long-lived and robust T cell immunity is generated following natural SARS-CoV-2 infection and support an important role of SARS-CoV-2-specific T cells in host control of COVID-19.
Keywords: SARS-CoV-2, COVID-19, T cells, CD127, IL7, homeostatic proliferation, CyTOF, convalescent individuals, mild COVID-19
Graphical Abstract
Highlights
SARS-CoV-2-specific T cells are diverse, express CD127, and can proliferate
SARS-CoV-2-specific CD4+ T cells are IFNγ-producing, lymphoid-homing Tfh cells
SARS-CoV-2-specific CD8+ T cells are predominantly less-differentiated Temra cells
Homing features of SARS-CoV-2-specific CD4+ and CD8+ T cells differ
Combining CyTOF with single-cell detection of antigen-specific cells, Neidleman et al. provide an in-depth view of the phenotypic features of CD4+ and CD8+ T cells recognizing SARS-CoV-2 epitopes. These cells are different from T cells recognizing CMV, harbor diverse homing properties and effector functions, and include CD127-expressing cells.
Introduction
The first cases of coronavirus disease 2019 (COVID-19) were reported in December of 2019 in Wuhan, China, and soon thereafter its causative agent was identified as SARS-CoV-2, a betacoronavirus with 79% sequence identity to the SARS-CoV that had emerged in 2003.1 SARS-CoV-2 has proven to be much more transmissible than its SARS-CoV counterpart, quickly spreading around the world and causing what was declared a pandemic on March 11, 2020. By June of 2020, confirmed cases of COVID-19 surpassed 6 million, nearly 6% of which were fatal.2 The outcome of exposure to SARS-CoV-2 can range from asymptomatic infection to death, most often caused by respiratory failure because of acute respiratory distress syndrome (ARDS). Limited genetic diversity has been observed in circulating SARS-CoV-2 strains, and genetic variations do not seem to correlate with disease severity,3 suggesting that the variable COVID-19 outcomes are driven by variable host responses.
Many individuals exposed to SARS-CoV-2 are asymptomatic or exhibit only a mild course of disease, suggesting that natural immunity can effectively combat this virus. Although most studies of SARS-CoV-2 immunity have focused on the humoral immune response, emerging data suggest that T cell-mediated immunity is also likely to play an important role in eliminating the virus. Lymphopenia, characterized by reduced numbers of CD4+ and CD8+ T cells, is predictive of disease severity.3 In addition, levels of activated T cells increase at the time of SARS-CoV-2 clearance.4 Furthermore, the clonality of T cell receptor (TCR) sequences5 is higher in patients with mild than severe COVID-19, suggesting a role of antigen-specific T cell responses in symptom resolution. A beneficial role of T cells in combating COVID-19 would be in line with observations that CD4+ and CD8+ T cells are protective against the closely related SARS-CoV.6, 7, 8 However, the nature of the response is also important because Th1 responses appear to be protective against SARS-CoV, whereas Th2 responses are associated with immunopathology.9, 10, 11 Th17 responses have also been implicated in immunopathology during coronavirus infection.12
Only a limited number of studies have characterized the SARS-CoV-2-specific T cell response. These studies have focused on the breadth of the T cell response and detected T cells recognizing spike and non-spike epitopes.13, 14, 15, 16, 17 In some13,16 but not other17 studies, responses were also detected in individuals who had not been infected with SARS-CoV-2, presumably reflecting recognition of cross-reactive epitopes from other coronaviruses. These responses primarily involved non-spike open reading frames (ORFs).16 ELISA revealed that peptide-treated peripheral blood mononuclear cells (PBMCs) upregulated interferon γ (IFNγ) but not interleukin-4 (IL-4) or IL-17, suggesting a Th1 response.14,16 Limited phenotyping based on CD45RA and CCR7 suggested SARS-CoV-2-specific CD4+ T cells to be more of the T central memory (Tcm) phenotype, whereas SARS-CoV-2-specific CD8+ T cells were biased toward terminally differentiated effector (Temra) cells.14 These results are consistent with longitudinal analysis of two individuals with COVID-19 by bulk TCR sequencing, where clonal TCR sequences (assumed to be SARS-CoV-2 specific) were prevalent among Tcm cells for CD4+ T cells and Temra cells for CD8+ T cells.18 However, the phenotypic features of SARS-CoV-2-specific CD4+ and CD8+ T cells have not been investigated systematically, and little is actually known about the functional properties of these cells and their ability to persist long term in convalescent individuals.
In this study, we conducted an in-depth phenotypic analysis of SARS-CoV-2-specific CD4+ and CD8+ T cells circulating in the bloodstream of individuals who had recently recovered from COVID-19. This was achieved by combining detection of SARS-CoV-2-specific T cells together with CyTOF, a mass spectrometry-based single-cell phenotyping method that uses antibodies conjugated to metal lanthanides to quantify expression levels of surface and intracellular proteins.19 Because spectral overlap is not a limitation with CyTOF, large phenotyping panels of nearly 40 parameters can be implemented, allowing a high-resolution view of immune cells and use of high-dimensional analytical methods to monitor cellular remodeling.20,21 We report here that SARS-CoV-2-specific CD4+ and CD8+ T cells from convalescent individuals are diverse, exhibit features different from antigen-specific T cells against Cytomegalovirus (CMV), include cells with lymphoid and tissue homing potential, harbor phenotypic features of functional effector cells, and are long lived and capable of homeostatic proliferation.
Results
SARS-CoV-2-Specific T Cells from Convalescent Individuals Produce IFNγ
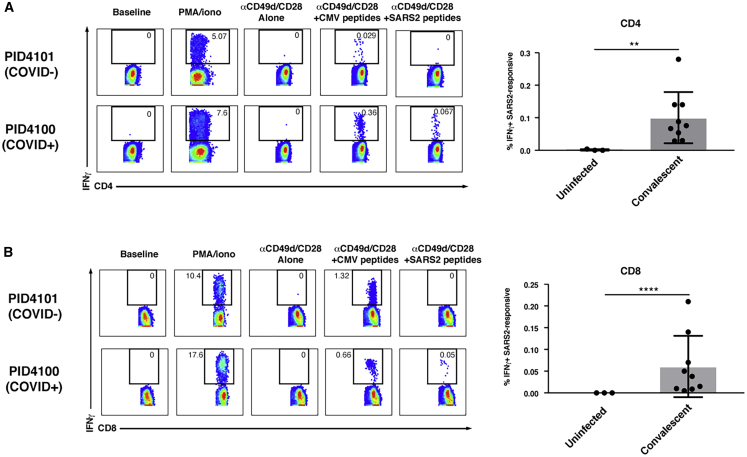
Nine convalescent and three uninfected participants (Table S1) were recruited from the University of California, San Francisco (UCSF) acute COVID-19 Host Immune Response Pathogenesis (CHIRP) study for blood donation. Blood specimens were obtained 20–47 days after the participants tested positive for SARS-CoV-2 by RT-PCR. PBMCs were purified from the freshly isolated blood specimens and then phenotyped immediately by CyTOF or stimulated for 6 h with overlapping peptides in the presence of co-stimulation to enable detection of antigen-specific T cells. Overlapping 15-mer peptides from spike, an immunodominant SARS-CoV-2 antigen,13,16 were used for characterization of the SARS-CoV-2-specific response, whereas overlapping peptides against pp65, an immunodominant CMV antigen,22 were used for comparison. To enable high-parameter phenotyping of antigen-specific cells, we modified a recently developed human T cell CyTOF panel21 so it would detect cells producing the cytokines IFNγ, IL-4, or IL-17 (Table S2). CD4+ and CD8+ T cells were identified by sequential gating on live, singlet CD3+ cells expressing the corresponding co-receptor (Figure S1). Spike-specific CD4+ and CD8+ T cells producing IFNγ were detected in convalescent but not uninfected individuals (Figure 1; Table S3), suggesting a robust spike-specific Th1 response. In contrast, spike-specific CD4+ T cells did not include cells of the Th2 and Th17 lineages because no IL-4- or IL-17-producing cells were detected following spike peptide stimulation, although such cells were detected following PMA/ionomycin stimulation (Figure S2).
Figure 1.
Antigen-Specific CD4+ and CD8+ T Cells against SARS-CoV-2 Spike Secrete IFNγ
(A and B) Shown on the left are pseudocolor plots of CyTOF datasets reflecting the percentage of CD4+ (A) or CD8+ (B) T cells producing IFNγ in response to the indicated treatment condition for one representative uninfected (COVID−) and recovered convalescent (COVID+) donor. Numbers correspond to the percentage of cells within the gates. The baseline condition corresponds to cells phenotyped by CyTOF immediately following isolation of PBMCs from freshly drawn blood, whereas for all other treatment conditions, cells were cultured for 6 h prior to phenotyping by CyTOF. PMA/ionomycin treatment was used as a positive control. Anti-CD49d/CD28 was used to provide co-stimulation during peptide treatment. Shown on the right are cumulative data from three uninfected individuals and nine recovered convalescent individuals (Table S1). Results are gated on live singlet CD4+ or CD8+ T cells. ∗∗p < 0.01, ∗∗∗∗p < 0.0001, as assessed using Student’s unpaired t test.
See also Figures S1 and S2.
SARS-CoV-2-Specific T Cells Are Phenotypically Diverse and Different from CMV-Specific T Cells
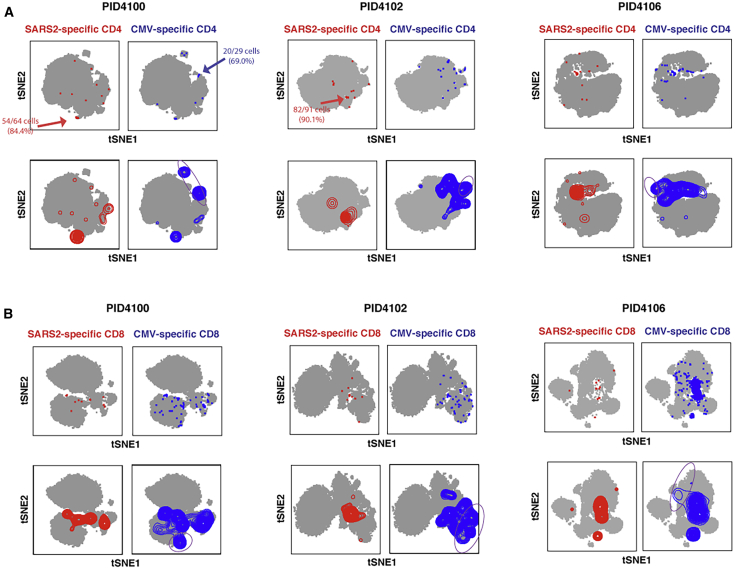
To obtain a global view of the phenotypic features of SARS-CoV-2-specific T cells, we visualized the data by t-Distributed Stochastic Neighbor Embedding (t-SNE).23 We gated on the IFNγ-producing spike-specific cells (Figure 1) and overlaid these on total T cells treated with co-stimulation alone. Because most of the convalescent donors were CMV+ (Tables S1 and S3), we compared the locations of the spike-specific cells and CMV-specific cells in three representative donors. Spike-specific CD4+ (Figures 2A and S3A) and CD8+ (Figures 2B and S3B) T cells were diverse in that they occupied multiple regions of the t-SNE. However, these cells were concentrated in one or two major regions of the t-SNE, suggesting that their phenotypes are biased toward particular subsets. The phenotypes of spike-specific T cells were not identical to those of CMV-specific T cells; in every donor, there were regions of the t-SNE occupied by CMV-specific T cells that were devoid of spike-specific cells (Figure 2, purple ovals), although overall the CD8+ T cells against the two viruses were more similar to each other than the CD4+ T cells. This was further confirmed by demonstrating that the distributions of subset clusters, as defined using the clustering algorithm DensVM24, were different among T cells specific for SARS-CoV-2 compared with those specific for CMV, particularly for CD4+ T cells (Figure S4). These results suggest that spike-specific T cells are not randomly distributed among T cell subsets and differ from CMV-specific T cells, a finding that is perhaps expected because CMV differs from SARS-CoV-2. To assess whether the spike-specific T cell response is representative of the overall SARS-CoV-2-specific T cell response, we stimulated parallel T cells from two convalescent individuals with the spike peptides or with a mix of overlapping 15-mer peptides against envelope (env) and nucleocapsid (NC), two other structural proteins of SARS-CoV-2. These results revealed the phenotypes of env/NC-specific cells to be similar to those of cells recognizing spike (Figure S5). Hereafter, we focus on the spike-specific cells and refer to them as “SARS-CoV-2-specific.”
Figure 2.
SARS-CoV-2 Spike-Specific CD4+ and CD8+ T Cells Are Diverse and Not Phenotypically Identical to Their CMV-Specific Counterparts
(A and B) Shown are t-SNE plots of CyTOF datasets reflecting CD4+ (A) or CD8+ (B) T cells from three representative COVID-19 convalescent donors who had also sustained a previous CMV infection. Cells shown in gray correspond to CD4+ or CD8+ T cells from specimens stimulated with anti-CD49d/CD28 in the absence of any peptides. The top pairs of plots show SARS-CoV-2 spike-specific (red) or CMV pp65-specific (blue) cells as individual dots, with some regions concentrated in antigen-specific cells indicated. The bottom pairs of plots show the same data but with antigen-specific cells shown as contours instead of dots to better visualize regions with the highest densities of antigen-specific cells. Purple ovals outline examples of regions harboring CMV-specific but not SARS-CoV-2-specific cells.
See also Figures S3–S6.
SARS-CoV-2-Specific Th1 Cells Exhibit Phenotypic Features Characteristic of Lymph Node Homing, Robust Helper Function, and Longevity
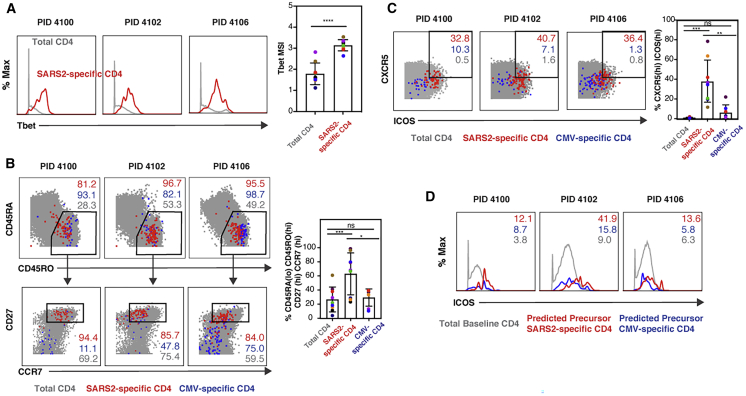
We then characterized the specific phenotypic features of SARS-CoV-2-specific CD4+ T cells. These cells expressed high levels of the transcription factor Tbet, which, together with their ability to induce IFNγ (Figure 3A), confirms their Th1 differentiation state. These cells, like their CMV-specific counterparts, almost exclusively expressed high levels of CD45RO and low levels of CD45RA, suggesting that they are memory cells. Interestingly, although most of the SARS-CoV-2-specific cells expressed CD27 and CCR7, the vast majority of CMV-specific cells did not (Figure 3B). This suggests that SARS-CoV-2-specific CD4+ T cells are mostly Tcm cells, memory cells that home to lymph nodes, where they can help B cells undergo affinity maturation. To evaluate the potential helper function of these cells, we assessed their expression of CXCR5 and ICOS, markers of circulating T follicular helper (cTfh) cells, cells that efficiently induce virus-specific memory B cells to differentiate into plasma cells and whose levels are associated with protective antibody responses.25 SARS-CoV-2-specific CD4+ T cells expressed higher levels of CXCR5 and ICOS than total and CMV-specific CD4+ T cells (Figure 3C). ICOS serves a critical role in Tfh function26 but is also an activation marker; therefore, one concern was that its levels were high on SARS-CoV-2-specific cells simply because these cells were responding to antigen stimulation. We believe that not to be the case because (1) ICOS−CXCR5− cells do not upregulate ICOS or CXCR5 during 6 h of in vitro stimulation,25,27 and (2) SARS-CoV-2-specific CD4+ T cells had higher ICOS expression than CMV-specific CD4+ T cells, which were stimulated similarly. To verify that SARS-CoV-2-specific cells at baseline express high levels of ICOS, we implemented predicted precursor as determined by SLIDE (PP-SLIDE),20,21 a bioinformatics pseudotime analysis approach that can predict the original phenotypes of cells before cellular perturbation. SARS-CoV-2- and CMV-specific CD4+ T cells were traced back to their predicted original states by matching their high-dimensional CyTOF profiles against the “atlas” of all CD4+ T cells phenotyped by CyTOF at baseline (prior to the 6 h of stimulation). The predicted original states of SARS-CoV-2 had high levels of ICOS, supporting the notion that these cells exhibit phenotypic features of cells with robust helper function (Figure 3D).
Figure 3.
SARS-CoV-2-Specific CD4+ Th1 Cells Are Tcm and cTfh Cells
(A) SARS-CoV2-specific CD4+ T cells are Th1 cells. Shown are the expression levels of Tbet, a transcription factor that directs Th1 differentiation, in total (gray) or SARS-CoV2-specific (red) CD4+ T cells from the blood of 3 representative convalescent individuals. Shown on the right are cumulative data from all 9 convalescent individuals analyzed in this study. ∗∗∗∗p < 0.0001 as assessed using Student’s paired t test.
(B) SARS-CoV-2-specific but not CMV-specific CD4+ T cells are predominantly Tcm cells. The phenotypes of total (gray), SARS-CoV-2-specific (red), and CMV-specific (blue) CD4+ T cells are shown as dot plots for 3 representative donors. Top: SARS-CoV-2-specific and CMV-specific CD4+ T cells are predominantly CD45RA−CD45RO+, characteristic of canonical memory cells. Bottom: most memory (CD45RA−CD45RO+) SARS-CoV-2-specific CD4+ T cells are CD27+CCR7+, characteristic of Tcm cells, whereas most CMV-specific memory CD4+ T cells are CD27−CCR7−, characteristic of Tem cells. The percentage of total, SARS-CoV-2-specific, and CMV-specific cells within the indicates gates are shown in gray, red, and blue, respectively. Shown on the right are cumulative data from all 9 convalescent individuals analyzed in this study. ∗p < 0.05, ∗∗∗p < 0.001, as assessed using Student’s unpaired t test.
(C) SARS-CoV-2-specific CD4+ T cells express high levels of CXCR5 and ICOS relative to total and CMV-specific CD4+ T cells. Numbers correspond to the percentages of SARS-CoV-2-specific (red), CMV-specific (blue), and total (gray) CD4+ T cells in the gates for 3 representative donors. Shown on the right are cumulative data from all 9 convalescent individuals analyzed in this study. ∗∗p < 0.01, ∗∗∗p < 0.001, as assessed using Student’s unpaired t test.
(D) ICOS is expressed at high levels on predicted precursors of IFNγ-producing SARS-CoV-2-specific CD4+ T cells. PP-SLIDE20,21 was conducted to predict the original phenotypic features of SARS-CoV-2-specific (red) and CMV-specific (blue) cells prior to IFNγ induction. The expression levels of ICOS on these cells were compared with those on total CD4+ T cells phenotyped by CyTOF immediately following PBMC isolation. Numbers correspond to mean signal intensity (MSI) of ICOS expression for the populations indicated at the bottom.
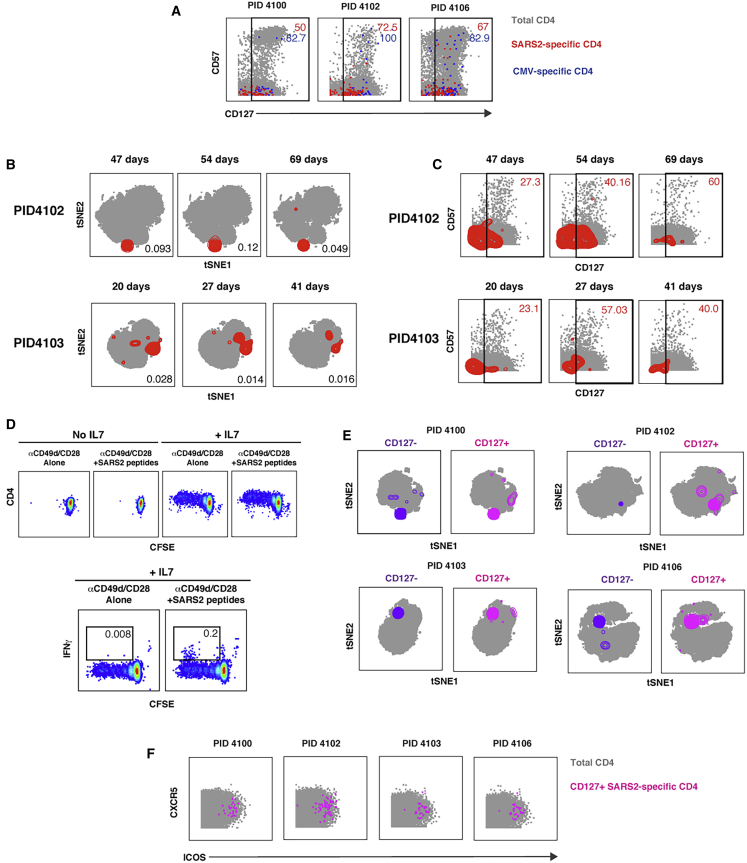
We next assessed whether SARS-CoV-2-specific CD4+ T cells exhibit features denoting longevity and an ability to proliferate. CD127, the α chain of the IL-7 receptor, is involved in cell survival and required for IL-7-driven homeostatic proliferation.28 We found that, among the nine convalescent donors, on average 58.5% ± 20.5% of SARS-CoV-2-specific CD4+ T cells expressed CD127. Although the vast majority of CMV-specific CD4+ T cells also expressed CD127, these cells differed from their SARS-CoV-2-specific counterparts in that a higher proportion additionally expressed high levels of the terminal differentiation marker CD57 (Figure 4A). To assess whether CD127+ SARS-CoV-2-specific CD4+ T cells are maintained over time, we conducted a phenotypic analysis of these cells in longitudinal specimens from two participants. SARS-CoV-2-specific CD4+ T cells exhibited stable phenotypes over time and were detected more than 2 months post-infection (Figure 4B). The proportions of CD127+ SARS-CoV-2-specific CD4+ T cells did not decrease over time and, in fact, tended to increase (Figure 4C).
Figure 4.
SARS-CoV-2-Specific CD4+ T Cells Express CD127 and Can Persist for Over 2 Months
(A) A subset of SARS-CoV-2-specific CD4+ T cells express CD127. The indicated cell populations were examined for expression levels of the terminal differentiation marker CD57 and the IL-7 receptor CD127. Numbers correspond to the percentages of the corresponding populations within the gates.
(B) SARS-CoV-2-specific CD4+ T cells can persist for over 2 months. Shown are t-SNE plots of CyTOF datasets of CD4+ T cells from two COVID-19 convalescent donors who were sampled longitudinally at the indicated time points post-infection (infection was defined as the time of testing positive for SARS-CoV-2). Cells shown in gray correspond to total CD4+ T cells from specimens stimulated with anti-CD49d/CD28 in the absence of any peptides. The locations of SARS-CoV-2-specific cells are shown as red contours. The percentage of CD4+ T cells that are SARS-CoV-2 specific is indicated at the bottom right of each plot.
(C) Persistent SARS-CoV-2-specific CD4+ T cells retain CD127 expression. Longitudinal specimens characterized in (B) were analyzed for expression levels of CD57 and CD127. SARS-CoV-2-specific CD4+ T cells are shown as red contours, whereas total CD4+ T cells are shown as gray dots. The percent of SARS-CoV-2-specific CD4+ T cells in the gates is shown at the top right of each plot. Note that the proportions of CD127+ SARS-CoV-2-specific CD4+ T cells do not decrease over time.
(D) SARS-CoV-2-specific CD4+ T cells can proliferate homeostatically in response to IL-7. PBMCs from convalescent donor PID4102 were labeled with CFSE and cultured for 5 days in the absence or presence of IL-7. Thereafter the cells were treated for 6 h with co-stimulation alone or in the presence of overlapping peptides from SARS-CoV-2 spike and then analyzed by flow cytometry. Homeostatic proliferation, as assessed by CFSE dye dilution, only occurred in the presence of IL-7 (top). A subpopulation of CFSElow cells produced IFNγ in response to peptide stimulation (bottom), demonstrating that cells driven to proliferate by IL-7 treatment included SARS-CoV-2-specific CD4+ T cells. Results are gated on live singlet CD3+CD4+CD8− cells and are representative of one of two donors.
(E) CD127+ SARS-CoV-2-specific CD4+ T cells are distributed similarly as their CD127- counterparts. Shown are t-SNE plots of CD127− (purple) and CD127+ (pink) SARS-CoV-2-specific CD4+ T cells overlaid on total CD4+ T cells treated with co-stimulation alone (gray). Note that, for each donor, CD127+ cells occupy similar regions of t-SNE space as CD127− cells.
(F) CD127+ SARS-CoV-2-specific CD4+ T cells express CXCR5 and ICOS.
To directly assess whether SARS-CoV-2-specific T cells were capable of homeostatic proliferation, we conducted an in vitro proliferation assay. PBMCs from convalescent donors were labeled with the proliferation dye CFSE and then cultured for 5 days in the absence or presence of IL-7. Treatment with IL-7 induced proliferation of the cells, as reflected by dilution of the CFSE dye in a subset of the cells (Figure 4D, top). After the 5 days of culture, the cells were subjected to intracellular cytokine staining analysis following treatment with co-stimulation alone or in the presence of SARS-CoV-2 spike peptides. SARS-CoV-2-specific CD4+ T cells were readily detected among CFSElow CD4+ T cells (Figure 4D, bottom), demonstrating that the cells that had proliferated homeostatically in response to IL-7 treatment included SARS-CoV-2-specific CD4+ T cells.
To assess the phenotypic features of CD127+ SARS-CoV-2-specific CD4+ T cells, we compared them with their CD127− counterparts. These two populations of cells occupied similar regions of t-SNE space (Figure 4E), suggesting that CD127+ and CD127− cells are phenotypically similar. Of note, CD127+ cells include cells expressing high levels of CXCR5 and ICOS (Figure 4F). Aside from CD127, CXCR4 was the only antigen that was expressed at significantly higher levels (p = 0.02 by pairwise test) on CD127+ versus CD127− SARS-CoV-2-specific CD4+ T cells; however, this difference was no longer significant when adjusted for multiple comparison using the Benjamini-Hochberg procedure (data not shown), consistent with the notion that CD127+ and CD127− SARS-CoV-2-specific CD4+ T cells exhibit similar phenotypes.
In summary, SARS-CoV-2-specific CD4+ T cells from convalescent individuals exhibit phenotypic features consistent with an ability to migrate into lymph node follicles, provide robust helper function for B cells, and to be long lived. A global view of all antigens differentially expressed in SARS-CoV-2-specific CD4+ T cells is presented in Figure S6.
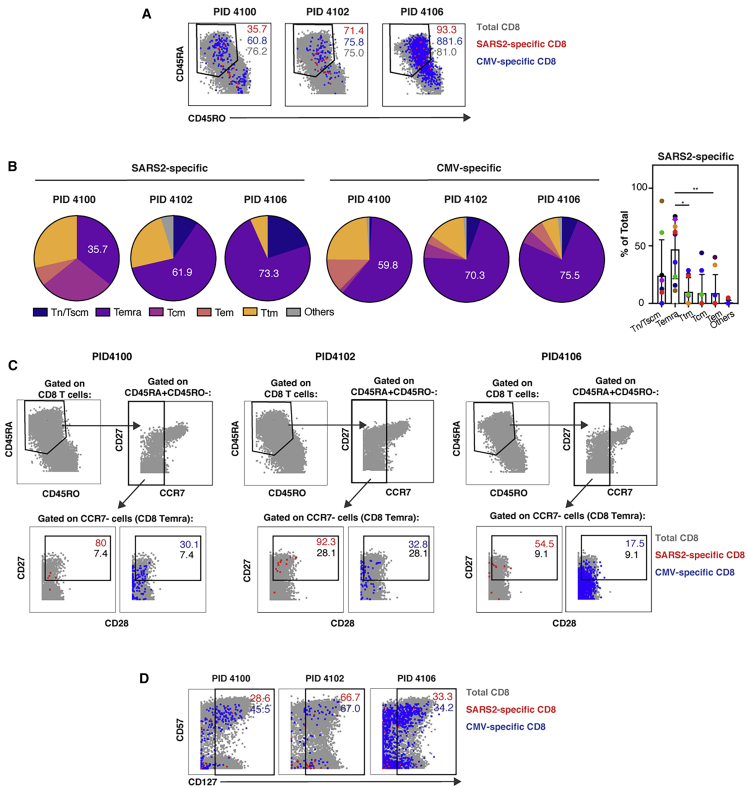
SARS-CoV-2-Specific CD8+ T Cells Exhibit Phenotypic Features of Less Differentiated CD8+ Temra Cells and Express CD127
We next characterized the phenotypic features of SARS-CoV-2-specific CD8+ T cells. These cells, in stark contrast to their CD4+ counterparts, included a prominent population of CD45RA-expressing cells (Figure 5A). CD45RA+ T cells include naive T (Tn) cells but also Temra and stem cell memory T (Tscm) cells. To determine the nature of the CD45RA-expressing CD8+ T cells, we assessed the proportions of CD8+ T cells that belonged to the Tn/Tscm (CD45RA+CD45RO−CCR7+) and Temra (CD45RA+CD45RO−CCR7−) subsets as well as the canonical memory subsets Tcm (CD45RA−CD45RO+CCR7+CD27+), T effector memory (Tem; CD45RA−CD45RO+CCR7−CD27−), and T transitional memory (Ttm; CD45RA−CD45RO+CCR7−CD27+). We then compared the subset distribution of SARS-CoV-2-specific cells with those specific for CMV. Consistent with prior studies, CMV-specific CD8+ T cells were predominantly Temra cells (Figure 5B).29 Temra cells were also the largest subset of SARS-CoV-2-specific CD8+ T cells (Figure 5B). Because CD8+ T cells recognizing influenza (flu) were also detected in a couple of convalescent individuals, we compared their features with those recognizing SARS-CoV-2. Flu-specific and SARS-CoV-2 CD8+ T cells occupied different regions of the t-SNE, suggesting that they are phenotypically distinct (Figure S7A). Although in one donor the flu-specific CD8+ T cells were not predominantly Temra cells, in the second they were (Figures S7B and S7C). The variability in the phenotypes of the flu-specific T cells may be due to different times of flu exposure in these convalescent individuals.
Figure 5.
SARS-CoV-2-Specific CD8+ T Cells Are Predominantly a Less Differentiated Subset of Temra Cells and Include Long-Lived CD127-Expressing Cells
(A) SARS-CoV2-specific CD8+ T cells include CD45RA+CD45RO− and CD45RA−CD45RO+ cells. The phenotypes of total (gray), SARS-CoV-2-specific (red), and CMV-specific (blue) CD8+ T cells are shown as dot plots for three representative donors.
(B) SARS-CoV-2-specific CD8+ T cells are predominantly Temra cells. The proportions of SARS-CoV-2-specific and CMV-specific CD8+ T cells belonging to each subset are depicted as pie graphs. Numbers correspond to the percentages of cells belonging to the Temra cell subset. Shown on the right are cumulative data for SARS-CoV-2-specific CD8+ T cells from all 9 convalescent individuals analyzed in this study. ∗p < 0.05, ∗*p < 0.01, as assessed using Student’s paired t test.
(C) Compared with CMV-specific and total CD8+ Temra cells, SARS-CoV-2-specific CD8+ Temra cells express high levels of CD27 and CD28. Top: Temra cells were defined as CD45RA+CD45RO− cells expressing low levels of CCR7. Bottom: CD8+ Temra cells were further assessed for expression levels of CD27 and CD28. Numbers correspond to the percentage of total (gray), SARS-CoV-2-specific (red), and CMV-specific (blue) cells within the gate.
(D) A subset of SARS-CoV-2-specific CD8+ T cells express CD127. The indicated cell populations were examined for expression levels of CD57 and CD127.
See also Figure S7.
To better define the features of SARS-CoV-2-specific CD8+ Temra cells and compare them with their CMV-specific counterparts, we gated on these cells and assessed for expression levels of CD27 and CD28. These two co-stimulatory receptors have been used to distinguish the most terminally differentiated Temra cells (CD27−CD28−) from less differentiated ones (CD27+CD28+).30 The vast majority of total CD8+ Temra cells in all donors expressed low levels of CD27, consistent with prior reports,29 whereas the majority of SARS-CoV-2-specific CD8+ Temra cells were CD27+ (Figure 5C), suggesting that SARS-CoV-2-specific CD8+ Temra cells were less terminally differentiated than typical Temra cells. These less differentiated CD8+ Temra cells were also present among CMV-specific cells but in markedly lower proportions (Figure 5C). Although CD28 staining was weak, it nonetheless suggested that CD28 levels were higher among SARS-CoV-2-specific CD8+ T cells than among total or CMV-specific CD8+ T cells. These results suggest that, although a large proportion of SARS-CoV-2-specific CD8+ T cells are Temra cells, they exhibit features characteristic of a less terminally differentiated state possibly capable of expansion.
Finally, to determine whether SARS-CoV-2-specific CD8+ T cells, like their CD4+ counterparts, may be capable of homeostatic proliferation, we assessed for expression levels of CD127. Although a lower proportion of SARS-CoV-2-specific CD8+ T cells expressed CD127 relative to their CD4+ counterparts, when averaged over the nine convalescent donors, these cells still accounted for an average of 58.0% ± 25.5% of the population. These CD127+ cells were similar to their CMV counterparts in that they included CD57− and CD57+ cells (Figure 5D). These results suggest that a subset of SARS-CoV-2-specific CD8+ T cells express CD127 and may therefore be long lived. A global view of all antigens differentially expressed in SARS-CoV-2-specific CD8+ T cells is presented in Figure S6.
Discussion
In this study, we define phenotypic features of the T cell response against SARS-CoV-2 in nine convalescent individuals who recovered from COVID-19 after only experiencing mild symptoms. SARS-CoV-2-specific responses were readily detected in CD4+ and CD8+ T cells from all nine individuals. The phenotypes of SARS-CoV-2-specific CD4+ T cells were markedly different from those of their CD8+ counterparts and included long-lived cells capable of homeostatic proliferation.
Recent studies measuring the levels of cytokines in supernatants of convalescent COVID-19 PBMCs treated with SARS-CoV-2 peptides revealed upregulation of IFNγ, suggesting a Th1 CD4+ T cell response.14,16 However, in those studies, the cellular source of IFNγ was not determined. We confirmed the existence of SARS-CoV-2-specific CD4+ Th1 cells by demonstrating that Tbet-expressing CD4+ T cells from convalescent individuals secrete IFNγ in response to SARS-CoV-2 peptide treatment. These results suggest that, similar to what has been observed for SARS-CoV,9 a Th1 response directed against SARS-CoV-2 is associated with effective resolution of infection and symptoms. In comparison, the lack of any detectable SARS-CoV-2-specific CD4+ T cells producing IL4 or IL-17 suggests that antigen-specific Th2 or Th17 responses are not key for recovery from COVID-19. In fact, Th2 and Th17 responses have been suggested to be detrimental for recovery from highly pathogenic coronavirus infections—Th2 responses because of their association with lung immunopathology in the context of SARS-CoV10,11 and Th17 responses because of the close association of ARDS (exacerbated by Th17 responses) and IL-6 (a Th17-associated cytokine) with severe COVID-19.12,31 Interestingly, although we did not detect any IL-6 production by SARS-CoV-2-specific CD4+ T cells (whereas IL-6 induction was readily detectable upon LPS stimulation of monocytes; data not shown), IL-6+ CD4+ T cells have been reported to be elevated during severe COVID-19.32 Future studies are warranted to determine whether SARS-CoV-2 CD4+ T cells from severely ill patients, particularly those in the ICU or those who succumb to disease, produce IL-4, IL-17, or IL-6.
One important function of CD4+ T cells is to help B cells undergo affinity maturation, a process that typically occurs in lymph nodes draining the site of infection. We found that most SARS-CoV-2-specific CD4+ T cells were Tcm cells, cells poised to enter lymph nodes because they express the lymph-node-homing chemokine receptor CCR7. Furthermore, we found that most SARS-CoV-2-specific CD4+ T cells expressed CXCR5, a chemokine receptor that directs Tfh into the germinal centers of lymph nodes, where B cells mature.33,34 CXCR5 has been used as a marker of cTfh cells, the blood counterpart of lymphoid Tfh cells.35 Some cTfh cells also express high levels of ICOS, a co-stimulatory molecule that plays a critical role in Tfh function, including helping with development of high-affinity B cells.26 We found that SARS-CoV-2-specific CD4+ T cells expressed especially high levels of ICOS, suggesting an active helper function of B cells. These findings suggest that SARS-CoV-2-specific cTfh cells play a key role in effective immunity and are in line with the observation that the frequency of total cTfh cells increases at the time of SARS-CoV-2 clearance4 and detection of robust Tfh cell responses following exposure of rhesus macaques to SARS-CoV-2.36 Interestingly, the SARS-CoV-2-specific cTfh cells we identified exhibit features similar to a population of cTfh cells shown to be important in generation of vaccine-induced antibodies against the flu.25 Both express high levels of CXCR5 and ICOS. Both also appear to be Th1 cells: ours as defined by expression of Tbet and IFNγ and the flu-responsive ones as defined by expression of CXCR3, a chemokine receptor preferentially expressed on Th1 cells.37,38 Interestingly, the flu-responsive cTfh cells were shown to effectively help memory but not naive B cells25. This, together with the observation that the frequencies of Th1 cTfh cells were associated with flu-specific antibody titers only in individuals with pre-existing antibodies against other flu strains,25 suggests that these cells may be most effective at helping pre-existing cross-reactive memory B cells. Whether this is also true in the context of COVID-19 is not clear, but given that 40%–60% of unexposed individuals have T cells that react against SARS-CoV-2 peptides, likely because of cross-reactivity with endemic coronaviruses,16 it is possible that the SARS-CoV-2-specific CD4+ T cells we identified are acting on previously generated cross-reactive memory B cells. We speculate that the convalescent individuals we analyzed had previously been exposed to endemic coronaviruses and that this prior exposure, along with conditions favoring a Th1 response in these individuals, provided the necessary conditions for eliciting a robust and effective response against SARS-CoV-2. Testing this intriguing hypothesis will require serological and immune cell analyses of a large collection of specimens collected before and after exposure to SARS-CoV-2.
In contrast to their CD4+ counterparts, SARS-CoV-2-specific CD8+ T cells were predominantly Temra cells, antigen-experienced cells that re-express the naive cell marker CD45RA. Because of lack of CCR7 expression, CD8+ Temra cells do not home efficiently to lymph nodes and, instead, predominantly reside in the blood, spleen, and lungs.39 These cells have been shown to be protective against a variety of viral pathogens, including CMV, the flu, Epstein-Barr virus (EBV), and Human Immunodeficiency Virus (HIV).40, 41, 42, 43 Exceptionally high numbers of CMV-specific Temra cells have been observed in CMV-infected individuals,44 and this subset also constitutes a major proportion of dengue-specific CD8+ T cells in individuals that have recovered from dengue infection.45,46 Although generally thought to be terminally differentiated, some CD8+ Temra cells can be long-lived. The most terminally differentiated CD8+ Temra cells express low levels of CD27 and CD28, and these cells are thought to have differentiated from a more pluripotent state of Temra cells expressing CD27 and CD28.30 Interestingly, SARS-CoV-2-specific CD8+ Temra cells were heavily biased toward the CD27+CD28+ subset compared with total as well as CMV-specific CD8+ Temra cells. These phenotypic features are similar to the previously described EBV-specific CD8+ Temra cells, a subset that has been demonstrated to be apoptosis resistant because of expression of bcl2, to have long telomeres relative to other memory subsets (suggesting high replicative potential), and to be capable cytotoxic function.42 Natural killer (NK)-like cytotoxic functions have also been observed in dengue-specific CD8+ Temra cells.45 We postulate that, based on their phenotypic similarities to EBV-specific CD8+ Temra cells, SARS-CoV-2-specific CD8+ Temra cells are cytotoxic and long-lived, although this will need to be confirmed in follow-up studies.
Consistent with the notion of long-lived cells, we found that a substantial fraction of CD4+ and CD8+ T cells specific for SARS-CoV-2 expressed CD127. This surface protein corresponds to the α chain of the IL-7 receptor, and together with the common γ chain (CD132) shared by the IL-2 and IL-15 receptors, enables T cell proliferation in response to IL-7. Signaling by IL-7 is involved in many key aspects of T cell survival and proliferation and acts by inducing JAK/STAT signaling, which results in increased expression of the Bcl2 anti-apoptotic gene family.28,47 Consistent with the notion of persisting CD127-expressing SARS-CoV-2-specific T cells is our observation that these cells could still be detected 69 days post-infection and that they can proliferate in response to IL-7. Collectively, these results suggest that SARS-CoV-2-specific T cell responses have the potential to be long lived, something that has been found to be true for SARS-CoV, where memory T cells persist for up to 11 years post-infection.48,49 Having long-lived protective T cells against SARS-CoV-2 may be particularly important because no neutralizing antibodies were detected in 18% of convalescent individuals ∼30 days after symptom onset, suggesting that the antibody response may be short lived, at least in some individuals.50 Of note, a clinical trial (NCT04379076) has been initiated to treat hospitalized lymphopenic COVID-19 patients with IL-7 to restore T cell counts.51 Whether such treatment will boost the numbers of SARS-CoV-2-specific T cells remains to be determined but is conceivable, given that CD127 is expressed on these cells.
In summary, our phenotypic analysis reveals long-lived lymph-node-homing cTfh CD4+ T cells and CD27+CD28+ CD8+ Temra cells as the major subsets of SARS-CoV-2-specific T cells that persist following recovery from mild COVID-19. The system we developed here to conduct high-parameter phenotypic analysis of SARS-CoV-2-specific T cells can be used in future studies to better understand what constitutes an effective versus an immunopathological T cell response against the virus and to assess the features of vaccine-induced SARS-CoV-2-specific T cell responses. Because our study was limited to analyzing T cells from nine individuals who recovered efficiently from COVID-19, it did not allow us to determine exactly why some individuals fight off the infection whereas others succumb to it. Nonetheless, it revealed common features of effective immunity against SARS-CoV-2 and suggests that mobilization of a Th1 response plays a key role in viral clearance and establishment of long-lived CD4+ and CD8+ T cells. Vaccination strategies aimed at conferring long-term COVID-19 immunity should strongly consider approaches that include elicitation of long-lived CD4+ and CD8+ T cells against this pandemic virus.
Limitations of Study
This study has several limitations. This study only examined SARS-CoV-2-specific T cell responses from 9 convalescent individuals that had recovered from mild COVID-19. Follow-up studies should be conducted in larger cohorts to validate the findings reported here. This study did not characterize T cell responses in asymptomatic individuals and those that had severe COVID-19 (e.g., individuals hospitalized in the ICU) and focused on the spike-specific T cell response. Future studies comparing longitudinal T cell responses against different SARS-CoV-2 antigens in asymptomatic, mildly symptomatic, and severely symptomatic individuals are warranted.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| HLADR | Thermofisher | Cat#Q22158 |
| RORγt | Fisher Scientific | Cat#5013565 |
| CD49d (α4) | Fluidigm | Cat#3141004B |
| CTLA4 | Fisher Scientific | Cat#5012919 |
| NFAT | Fluidigm | Cat#3143023A |
| CCR5 | Fluidigm | Cat#3144007A |
| CD20 | Biolegend | Cat#302343 |
| CD95 | Fisher Scientific | Cat#MAB326100 |
| CD7 | Fluidigm | Cat#3147006B |
| ICOS | Fluidigm | Cat#3148019B |
| Tbet | Fisher Scientific | Cat#5013190 |
| IL4 | Biolegend | Cat#500829 |
| CD2 | Fluidigm | Cat#3151003B |
| IL17 | Biolegend | Cat#512331 |
| CD62L | Fluidigm | Cat#3153004B |
| TIGIT | Fludigm | Cat#3154016B |
| CCR6 | BD Biosciences | Cat#559560 |
| IL6 | Biolegend | Cat#501115 |
| CD8 | Biolegend | Cat#301053 |
| CD19 | Biolegend | Cat#302247 |
| CD14 | Biolegend | Cat#301843 |
| OX40 | Fluidigm | Cat#3158012B |
| CCR7 | Fluidigm | Cat#3159003A |
| CD28 | Fluidigm | Cat#3160003B |
| CD45RO | Biolegend | Cat#304239 |
| CD69 | Fluidigm | Cat#3162001B |
| CRTH2 | Fluidigm | Cat#3163003B |
| PD-1 | Biolegend | Cat#329941 |
| CD127 | Fluidigm | Cat#3165008B |
| CXCR5 | BD Biosciences | Cat#552032 |
| CD27 | Fluidigm | Cat#3167006B |
| IFNγ | Fluidigm | Cat#3168005B |
| CD45RA | Fluidigm | Cat#3169008B |
| CD3 | Fluidigm | Cat#3170001B |
| CD57 | Biolegend | Cat#359602 |
| CD38 | Fluidigm | Cat#3172007B |
| CD4 | Fluidigm | Cat#3174004B |
| CXCR4 | Fluidigm | Cat#3175001B |
| CD25 | Biolegend | Cat#356102 |
| CD161 | Biolegend | Cat#339919 |
| CD3 (APC/Cyanine) | Biolegend | Cat#344817 |
| CD4 (PE/Dazzle™ 594) | Biolegend | Cat#300547 |
| CD8 (Alexa Fluor® 700) | Biolegend | Cat#344723 |
| IFNγ (PE) | BD Biosciences | Cat#554701 |
| Purified Mouse anti-Human CD49d | BD Biosciences | Cat#340976 |
| Purified Mouse anti-Human CD28 | BD Biosciences | Cat#340975 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Lymphoprep™ | STEMCELL Technologies | Cat#07801 |
| Cisplatin | Sigma-Aldrich | Cat#P4394 |
| Paraformaldehyde | Electron Microscopy Sciences | Cat#15710 |
| Metal Contaminant-Free PBS | Rockland | Cat#MB-008 |
| Brefeldin A Solution | Thermofisher | Cat#00-4506-51 s |
| PepMix SARS-CoV-2 Peptide (Spike Glycoprotein) | JPT Peptide Technologies | Cat#PM-WCPV-S |
| PepMix SARS-CoV-2 Peptide (VEMP) | JPT Peptide Technologies | Cat#PM-WCPV-VEMP |
| PepMix SARS-CoV-2 Peptide (NCAP) | JPT Peptide Technologies | Cat#PM-WCPV-NCAP |
| CMV pp65 Peptide Pool | NIH AIDS Reagent Program | Cat#11549 |
| Influenza Virus Control Peptide Pool | Anaspec | Cat#AS-62340 |
| PMA | Sigma-Aldrich | Cat#1585 |
| Ionomycin calcium salt | Sigma-Aldrich | Cat#I0634 |
| Normal Mouse Serum | ThermoFisher | Cat#10410 |
| Normal Rat Serum | ThermoFisher | Cat#10710C |
| Human Serum From Male AB Plasma | Sigma-Aldrich | Cat#H4511 |
| Intracellular Fixation & Permeabilization Buffer | ThermoFisher | Cat#88-8823-88 |
| Permeabilization Buffer | ThermoFisher | Cat#00-8333-56 |
| Iridium Interchelator Solution | Fluidigm | Cat#201192B |
| MaxPar® cell staining buffer | Fluidigm | Cat#201068 |
| Cell acquisition solution | Fluidigm | Cat#201240 |
| EQ Four Element Calibration Beads | Fluidigm | Cat#201078 |
| Recombinant Human IL-7 Protein | R&D Systems | Cat#207-IL-005 |
| Zombie Violet™ Fixable Viability Kit | Biolegend | Cat#423113 |
| CFSE | ThermoFisher | Cat#65-0850-84 |
| Critical Commercial Assays | ||
| MaxPar® X8 Antibody Labeling Kit | Fluidigm | Cat#201169B |
| Cell-ID™ 20-Plex Pd Barcoding Kit | Fluidigm | Cat#201060 |
| Deposited Data | ||
| Raw CyTOF datasets | This paper | https://doi.org/10.7272/Q6RJ4GPP |
| Software and Algorithms | ||
| FlowJo | BD Biosciences | https://www.flowjo.com/ |
| Cytobank | Cytobank | https://www.cytobank.org/ |
| DensVM | Becher et al.24 | https://github.com/JinmiaoChenLab/cytofkit |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Nadia Roan (nadia.roan@ucsf.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The raw CyTOF datasets generated from this study are available for download through the public repository Dryad via the following link: https://doi.org/10.7272/Q6RJ4GPP.
Experimental Model and Subject Details
Human Subjects
Blood was obtained from eight individuals who had confirmed SARS-CoV-2 as assessed by RT-PCR, one household member of a confirmed SARS-CoV-2-positive individual who was not tested but exhibited COVID-19 symptoms (PID4107) and was therefore assumed to be infected, and three healthy controls. All participants were recruited as part of the UCSF acute COVID-19 Host Immune Pathogenesis (CHIRP) study. The timing of specimen collection relative to symptom onset, when SARS-CoV-2 infection was confirmed, and the sex and ages of the participants are presented in Table S1. Two individuals were additionally sampled at three longitudinal study visits. This study was approved by the University of California, San Francisco and informed consent was obtained from all subjects (IRB # 20-30588).
Method Details
Preparation of specimens for CyTOF
PBMCs were isolated from blood using Lymphoprep™ (StemCell Technologies) within 2 hours of blood collection. Six million cells were then immediately treated with cisplatin (Sigma-Aldrich) as a Live/Dead marker and fixed with paraformaldehyde (PFA) as previously described.21 Briefly, 6x106 cells were resuspended at room temperature in 2 mL PBS (Rockland) with 2 mM EDTA (Corning). Next, 2 mL of PBS containing 2 mM EDTA and 25 μM cisplatin (Sigma-Aldrich) were added to the cells. The cells were quickly mixed and incubated at room temperature for 60 s, after which 10 mL of CyFACS (metal contaminant-free PBS (Rockland) supplemented with 0.1% BSA and 0.1% sodium azide (Sigma-Aldrich)) was added to quench the reaction. The cells were then centrifuged and resuspended in 2% PFA in CyFACS, and incubated for 10 minutes at room temperature. The cells were then washed twice in CyFACS, after which they were resuspended in 100 μL of CyFACS containing 10% DMSO. These fixed cells were stored at −80°C until analysis by CyTOF.
For identification of antigen-specific T cells, unless otherwise indicated, 6 million freshly-isolated PBMCs were stimulated for 6 hours in RP10 media (RPMI 1640 medium (Corning) supplemented with 10% fetal bovine serum (FBS, VWR), 1% penicillin (GIBCO), and 1% streptomycin (GIBCO)) in the presence of 3 μg/ml Brefeldin A Solution (eBioscience) to enable detection of intracellular cytokines. For detection of antigen-specific T cells, 0.5 μg/ml anti-CD49d clone L25 and 0.5 μg/ml anti-CD28 clone L293 (both from BD Biosciences) were added as a source of co-stimulation, in the presence of 0.5 μm PepMix SARS-CoV-2 Peptide (Spike Glycoprotein) (JPT Peptide Technologies), human CMV pp65 Peptide Pool (NIH AIDS Reagent Program), or Influenza Virus Control Peptide Pool (Anaspec). As a positive control for cytokine detection, cells were treated with 16 nM PMA (Sigma-Aldrich) and 1 μm ionomycin (Sigma-Aldrich). Treated cells were then cisplatin-treated and PFA-fixed as described above. For longitudinal comparison of SARS-CoV-2-specific T cells, cryopreserved PBMCs from 3 time points each from participants PID4102 and PID4103 were simultaneously thawed, and then cultured overnight in RP10 media. Peptide stimulation was conducted for 6 hours as described above, using 0.5 μm PepMix SARS-CoV-2 Peptides for the following: spike glycoprotein, or a mix of envelope (env) and nucleocapsid (NC) (JPT Peptide Technologies), prior to cisplatin treatment and PFA fixation. Due to logistical reasons, PBMCs from participants PID4107, PID4108, PID4109, and PID4110 were also cryopreserved prior to peptide stimulation and CyTOF analysis.
CyTOF staining and data acquisition
Staining of cells for analysis by CyTOF was conducted similar to recently described methods.20,52,53 Briefly, cisplatin-treated cells were thawed and washed in Nunc 96 DeepWell™ polystyrene plates (Thermo Fisher) with CyFACS buffer at a concentration of 6x106 cells / 800 μL in each well. Cells were then pelleted and blocked with mouse (Thermo Fisher), rat (Thermo Fisher), and human AB (Sigma-Aldrich) sera for 15 minutes at 4°C. The samples were then washed twice in CyFACS, pelleted, and stained in a 100 μL cocktail of surface antibodies (Table S2) for 45 minutes at 4°C. The samples were then washed three times with CyFACS and fixed overnight at 4°C in 100 μL of freshly prepared 2% PFA in PBS (Rockland). Samples were then washed twice with Intracellular Fixation & Permeabilization Buffer (eBioscience) and incubated in this buffer for 45 minutes at 4°C. Next, samples were washed twice in Permeabilization Buffer (eBioscience). The samples were then blocked for 15 minutes at 4°C in 100 μL of mouse and rat sera diluted in Permeabilization Buffer, washed once with Permeabilization buffer, and incubated for 45 minutes at 4°C in a 100 μL cocktail of intracellular antibodies (Table S2) diluted in Permeabilization Buffer. The cells were then washed with CyFACS and stained for 20 minutes at room temperature with 250 nM of Cell-ID™ Intercalator-IR (Fluidigm). Finally, the cells were washed twice with CyFACS buffer, once with MaxPar® cell staining buffer (Fluidigm), and once with Cell acquisition solution (CAS, Fluidigm), and then resuspended in EQ Four Element Calibration Beads (Fluidigm) diluted in CAS. Sample concentration was adjusted to acquire at a rate of 200 - 350 events/sec using a wide-bore (WB) injector on a CyTOF2 instrument (Fluidigm) at the UCSF Parnassus flow core facility.
CFSE Proliferation Assay
Cryopreserved PBMCs from two convalescent donors (4102 and 4106) were resuspended at 2x107 cells/ml in PBS containing 0.1% FBS. The cells were then loaded for 10 minutes in the dark with 1 μM of CFSE (eBioscience). Labeling was stopped by adding 5 volumes of RP10. The cells were then incubated at 4°C for an additional 5 minutes. After 3 washes in RP10, the cells were cultured for 5 days in RP10 in the absence or presence of 10 ng/ml human IL-7 (R&D System). Cells were then stimulated for 6 hours with 0.5 μg/ml anti-CD49d clone L25 and 0.5 μg/ml anti-CD28 clone L293 (BD Biosciences), in the absence or presence of 0.5 μm PepMix SARS-CoV-2 Peptide (Spike Glycoprotein). Stimulation was conducted in the presence of Brefeldin A (3 μg/ml, eBioscience) to enable intracellular detection of induced IFNγ. Following the stimulation, cells were stained with APC/Cyanine7 anti-human CD3 (Clone SK7, Biolegend), PE/Dazzle™ 594 anti-human CD4 (Clone RPA-T4, Biolegend), Alexa Fluor 700 anti-human CD8 (Clone SK1, Biolegend), PE anti-IFNγ (Clone B27, BD Biosciences), and Zombie Violet Pacific Blue (Biolegend) as a Live/Dead discriminator. Stained cells were fixed and analyzed by FACS on an LSRII (BD Biosciences). Flowjo (BD Biosciences) was used for analysis. Live, singlet CD3+CD4+CD8- cells were assessed for proliferation by monitoring the loss of CFSE signal, and for the presence of SARS-CoV-2-specific cells by induction of IFNγ.
Quantification and Statical Analysis
CyTOF data export and PP-SLIDE analysis
The CyTOF data were exported as FCS files, and samples were de-barcoded according to manufacturer’s instructions (Fluidigm). Events corresponding to antigen-specific cells were identified by gating on live, singlet intact CD3+CD19- CD4+ or CD8+ T cells expressing IFNγ, and exported as the population of antigen-specific cells. Data export was conducted using FlowJo (BD Biosciences) and Cytobank software. t-SNE analyses were performed using the Cytobank software with default settings. All cellular markers not used in the upstream gating strategy were included in generating the t-SNE plots. Non-cellular markers (e.g., live/dead stain) and the cytokines IFNγ, IL4, and IL17 were excluded for the generation of t-SNE plots. Dot plots were generated using both Cytobank and FlowJo.
The clustering method density-based clustering aided by support vector machine (DensVM)24 was implemented in R to cluster, for each of the CD4+ or CD8+ T cell populations, total atlas cells, SARS-CoV-2-specific cells, or CMV-specific cells. DensVM builds on the method used by ACCENSE54 for categorizing subpopulations, and calculates density by transforming the t-SNE data using the Gaussian kernel transformation.
PP-SLIDE analysis to define the predicted original states of antigen-specific IFNγ-producing cells followed our previously described method.20,53 Each responding IFNγ+ cell from the stimulated sample was matched against every cell in the baseline unstimulated atlas phenotyped immediately after sample procurement, and then k-nearest neighbor (kNN) calculations were used to identify the phenotypically most similar cell in the atlas. The identified atlas cells harbor the predicted phenotypes of the original antigen-specific cells prior to IFNγ induction.
The statistical analyses used in the experiments are indicated within the Figure Legends. In all instances, the “n” refers to the number of different donors.
Acknowledgments
This work was supported by the Van Auken Private Foundation and David Henke; the Program for Breakthrough Biomedical Research, which is partly funded by the Sandler Foundation; and philanthropic funds donated to the Gladstone Institutes by The Roddenberry Foundation and individual donors devoted to COVID-19 research. We acknowledge NIH DRC Center grant P30 DK063720 and S10 1S10OD018040-01 for use of the CyTOF instrument. We also thank Stanley Tamaki, Tomoko Kakegawa Peech, and Caudia Bispo for CyTOF assistance at the Parnassus Flow Core; Nicole Lazarus and Eugene Butcher for the Act1 antibody; Françoise Chanut for editorial assistance; and Robin Givens for administrative assistance.
Author Contributions
J.N., J.F., G.X., and A.F.G. conducted experiments. J.N., X.L., M.M., T.M., A.K., E.G., and N.R.R. conducted data analysis. X.L. and M.M. wrote scripts. J.N., J.F., G.X., E.G., and N.R.R. designed the experiments. S.L. established the CHIRP cohort. G.G., E.S.S., and S.L. conducted CHIRP participant interviews, enrollment, and specimen collection. W.C.G. supervised data analysis and edited the manuscript. J.V., E.G., S.L., and N.R.R. conceived ideas for the study. N.R.R. wrote the manuscript. All authors read and approved the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: August 19, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.xcrm.2020.100081.
Contributor Information
Sulggi Lee, Email: sulggi.lee@ucsf.edu.
Nadia R. Roan, Email: nadia.roan@ucsf.edu.
Supplemental Information
References
- 1.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johns Hopkins Coronavirus Resource Center 2020. https://coronavirus.jhu.edu/map.html
- 3.Zhang X., Tan Y., Ling Y., Lu G., Liu F., Yi Z., Jia X., Wu M., Shi B., Xu S. Viral and host factors related to the clinical outcome of COVID-19. Nature. 2020;583:437–440. doi: 10.1038/s41586-020-2355-0. [DOI] [PubMed] [Google Scholar]
- 4.Thevarajan I., Nguyen T.H.O., Koutsakos M., Druce J., Caly L., van de Sandt C.E., Jia X., Nicholson S., Catton M., Cowie B. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat. Med. 2020;26:453–455. doi: 10.1038/s41591-020-0819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang L., Shi Y., Gong B., Jiang L., Liu X., Yang J., Tang J., You C., Jiang Q., Long B. Blood single cell immune profiling reveals the interferon-MAPK pathway mediated adaptive immune response for COVID-19. medRxiv. 2020 doi: 10.1101/2020.03.15.20033472. [DOI] [Google Scholar]
- 6.Roberts A., Deming D., Paddock C.D., Cheng A., Yount B., Vogel L., Herman B.D., Sheahan T., Heise M., Genrich G.L. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 2007;3:e5. doi: 10.1371/journal.ppat.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li C.K., Wu H., Yan H., Ma S., Wang L., Zhang M., Tang X., Temperton N.J., Weiss R.A., Brenchley J.M. T cell responses to whole SARS coronavirus in humans. J. Immunol. 2008;181:5490–5500. doi: 10.4049/jimmunol.181.8.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao J., Zhao J., Perlman S. T cell responses are required for protection from clinical disease and for virus clearance in severe acute respiratory syndrome coronavirus-infected mice. J. Virol. 2010;84:9318–9325. doi: 10.1128/JVI.01049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janice Oh H.L., Ken-En Gan S., Bertoletti A., Tan Y.J. Understanding the T cell immune response in SARS coronavirus infection. Emerg. Microbes Infect. 2012;1:e23. doi: 10.1038/emi.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deming D., Sheahan T., Heise M., Yount B., Davis N., Sims A., Suthar M., Harkema J., Whitmore A., Pickles R. Vaccine efficacy in senescent mice challenged with recombinant SARS-CoV bearing epidemic and zoonotic spike variants. PLoS Med. 2006;3:e525. doi: 10.1371/journal.pmed.0030525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yasui F., Kai C., Kitabatake M., Inoue S., Yoneda M., Yokochi S., Kase R., Sekiguchi S., Morita K., Hishima T. Prior immunization with severe acute respiratory syndrome (SARS)-associated coronavirus (SARS-CoV) nucleocapsid protein causes severe pneumonia in mice infected with SARS-CoV. J. Immunol. 2008;181:6337–6348. doi: 10.4049/jimmunol.181.9.6337. [DOI] [PubMed] [Google Scholar]
- 12.Hotez P.J., Bottazzi M.E., Corry D.B. The potential role of Th17 immune responses in coronavirus immunopathology and vaccine-induced immune enhancement. Microbes Infect. 2020;22:165–167. doi: 10.1016/j.micinf.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F.S.H., Dingeldey M., Kruse B., Fauchere F. Presence of SARS-CoV-2-reactive T cells in COVID-19 patients and healthy donors. medRxiv. 2020 doi: 10.1101/2020.04.17.20061440v1. [DOI] [PubMed] [Google Scholar]
- 14.Weiskopf D., Schmitz K.S., Raadsen M.P., Grifoni A., Okba N.M.A., Endeman H., van der Akker J.P.C., Molenkamp R., Koopmans M.P.G., van Gorp E.C.M. Phenotype of SARS-CoV-2-specific T-cells in COVID-19 patients with acute respiratory distress syndrome. medRxiv. 2020 doi: 10.1101/2020.04.11.20062349v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ni L., Ye F., Cheng M.L., Feng Y., Deng Y.Q., Zhao H., Wei P., Ge J., Gou M., Li X. Detection of SARS-CoV-2-Specific Humoral and Cellular Immunity in COVID-19 Convalescent Individuals. Immunity. 2020;52:971–977.e3. doi: 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng Y., Mentzer A.J., Liu G., Yao X., Yin Z., Dong D., Dejnirattisai W., Rostron T., Supasa P., Liu C. Broad and strong memory CD4 (+) and CD8 (+) T cells induced by SARS-CoV-2 in UK convalescent COVID-19 patients. bioRxiv. 2020 doi: 10.1101/2020.06.05.134551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minervina A.A., Komech E.A., Titov A., Koraichi M.B., Rosati E., Mamedov I.Z., Franke A., Efimov G.A., Chudakov D.M., Mora T. Longitudinal high-throughput TCR repertoire proling reveals the dynamics of T cell memory formation after mild COVID-19 infection. bioRxiv. 2020 doi: 10.1101/2020.05.18.100545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bendall S.C., Simonds E.F., Qiu P., Amir A.D., Krutzik P.O., Finck R., Bruggner R.V., Melamed R., Trejo A., Ornatsky O.I. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cavrois M., Banerjee T., Mukherjee G., Raman N., Hussien R., Rodriguez B.A., Vasquez J., Spitzer M.H., Lazarus N.H., Jones J.J. Mass Cytometric Analysis of HIV Entry, Replication, and Remodeling in Tissue CD4+ T Cells. Cell Rep. 2017;20:984–998. doi: 10.1016/j.celrep.2017.06.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma T., Luo X., George A.F., Mukherjee G., Sen N., Spitzer T.L., Giudice L.C., Greene W.C., Roan N.R. HIV efficiently infects T cells from the endometrium and remodels them to promote systemic viral spread. eLife. 2020;9:e55487. doi: 10.7554/eLife.55487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sylwester A.W., Mitchell B.L., Edgar J.B., Taormina C., Pelte C., Ruchti F., Sleath P.R., Grabstein K.H., Hosken N.A., Kern F. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J. Exp. Med. 2005;202:673–685. doi: 10.1084/jem.20050882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Maaten L., Hinton G. Visualizing Data using t-SNE. J. Mach. Learn. Res. 2008;9:2579–2605. [Google Scholar]
- 24.Becher B., Schlitzer A., Chen J., Mair F., Sumatoh H.R., Teng K.W., Low D., Ruedl C., Riccardi-Castagnoli P., Poidinger M. High-dimensional analysis of the murine myeloid cell system. Nat. Immunol. 2014;15:1181–1189. doi: 10.1038/ni.3006. [DOI] [PubMed] [Google Scholar]
- 25.Bentebibel S.E., Lopez S., Obermoser G., Schmitt N., Mueller C., Harrod C., Flano E., Mejias A., Albrecht R.A., Blankenship D. Induction of ICOS+CXCR3+CXCR5+ TH cells correlates with antibody responses to influenza vaccination. Sci. Transl. Med. 2013;5:176ra32. doi: 10.1126/scitranslmed.3005191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hutloff A., Dittrich A.M., Beier K.C., Eljaschewitsch B., Kraft R., Anagnostopoulos I., Kroczek R.A. ICOS is an inducible T-cell co-stimulator structurally and functionally related to CD28. Nature. 1999;397:263–266. doi: 10.1038/16717. [DOI] [PubMed] [Google Scholar]
- 27.Morita R., Schmitt N., Bentebibel S.E., Ranganathan R., Bourdery L., Zurawski G., Foucat E., Dullaers M., Oh S., Sabzghabaei N. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity. 2011;34:108–121. doi: 10.1016/j.immuni.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kondrack R.M., Harbertson J., Tan J.T., McBreen M.E., Surh C.D., Bradley L.M. Interleukin 7 regulates the survival and generation of memory CD4 cells. J. Exp. Med. 2003;198:1797–1806. doi: 10.1084/jem.20030735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Derhovanessian E., Maier A.B., Hähnel K., Beck R., de Craen A.J.M., Slagboom E.P., Westendorp R.G.J., Pawelec G. Infection with cytomegalovirus but not herpes simplex virus induces the accumulation of late-differentiated CD4+ and CD8+ T-cells in humans. J. Gen. Virol. 2011;92:2746–2756. doi: 10.1099/vir.0.036004-0. [DOI] [PubMed] [Google Scholar]
- 30.Koch S., Larbi A., Derhovanessian E., Ozcelik D., Naumova E., Pawelec G. Multiparameter flow cytometric analysis of CD4 and CD8 T cell subsets in young and old people. Immun. Ageing. 2008;5:6. doi: 10.1186/1742-4933-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pacha O., Sallman M.A., Evans S.E. COVID-19: a case for inhibiting IL-17? Nat. Rev. Immunol. 2020;20:345–346. doi: 10.1038/s41577-020-0328-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Y., Fu B., Zheng X., Wang D., Zhao C., Qi Y., Sun R., Tian Z., Xu X., Wei H. Pathogenic T cells and inflammatory monocytes incite inflammatory storm in severe COVID-19 patients. Natl. Sci. Rev. 2020:nwaa041. doi: 10.1093/nsr/nwaa041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Breitfeld D., Ohl L., Kremmer E., Ellwart J., Sallusto F., Lipp M., Förster R. Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. J. Exp. Med. 2000;192:1545–1552. doi: 10.1084/jem.192.11.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaerli P., Willimann K., Lang A.B., Lipp M., Loetscher P., Moser B. CXC chemokine receptor 5 expression defines follicular homing T cells with B cell helper function. J. Exp. Med. 2000;192:1553–1562. doi: 10.1084/jem.192.11.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chevalier N., Jarrossay D., Ho E., Avery D.T., Ma C.S., Yu D., Sallusto F., Tangye S.G., Mackay C.R. CXCR5 expressing human central memory CD4 T cells and their relevance for humoral immune responses. J. Immunol. 2011;186:5556–5568. doi: 10.4049/jimmunol.1002828. [DOI] [PubMed] [Google Scholar]
- 36.Elzaldi S.R., Lakshmanappa Y.S., Roh J.W., Schmidt B.A., Carroll T.D., Weaver K.D., Smith J.C., Deere J.D., Dutra J., Stone M. SARS-CoV-2 infection induces germinal center responses with robust stimulation of CD4 T follicular helper cells in rhesus macaques. bioRxiv. 2020 doi: 10.1101/2020.07.07.191007. [DOI] [Google Scholar]
- 37.Sallusto F., Lenig D., Mackay C.R., Lanzavecchia A. Flexible programs of chemokine receptor expression on human polarized T helper 1 and 2 lymphocytes. J. Exp. Med. 1998;187:875–883. doi: 10.1084/jem.187.6.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonecchi R., Bianchi G., Bordignon P.P., D’Ambrosio D., Lang R., Borsatti A., Sozzani S., Allavena P., Gray P.A., Mantovani A., Sinigaglia F. Differential expression of chemokine receptors and chemotactic responsiveness of type 1 T helper cells (Th1s) and Th2s. J. Exp. Med. 1998;187:129–134. doi: 10.1084/jem.187.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thome J.J., Yudanin N., Ohmura Y., Kubota M., Grinshpun B., Sathaliyawala T., Kato T., Lerner H., Shen Y., Farber D.L. Spatial map of human T cell compartmentalization and maintenance over decades of life. Cell. 2014;159:814–828. doi: 10.1016/j.cell.2014.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lilleri D., Fornara C., Revello M.G., Gerna G. Human cytomegalovirus-specific memory CD8+ and CD4+ T cell differentiation after primary infection. J. Infect. Dis. 2008;198:536–543. doi: 10.1086/590118. [DOI] [PubMed] [Google Scholar]
- 41.Sridhar S., Begom S., Bermingham A., Hoschler K., Adamson W., Carman W., Bean T., Barclay W., Deeks J.J., Lalvani A. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat. Med. 2013;19:1305–1312. doi: 10.1038/nm.3350. [DOI] [PubMed] [Google Scholar]
- 42.Dunne P.J., Faint J.M., Gudgeon N.H., Fletcher J.M., Plunkett F.J., Soares M.V., Hislop A.D., Annels N.E., Rickinson A.B., Salmon M., Akbar A.N. Epstein-Barr virus-specific CD8(+) T cells that re-express CD45RA are apoptosis-resistant memory cells that retain replicative potential. Blood. 2002;100:933–940. doi: 10.1182/blood-2002-01-0160. [DOI] [PubMed] [Google Scholar]
- 43.Northfield J.W., Loo C.P., Barbour J.D., Spotts G., Hecht F.M., Klenerman P., Nixon D.F., Michaëlsson J. Human immunodeficiency virus type 1 (HIV-1)-specific CD8+ T(EMRA) cells in early infection are linked to control of HIV-1 viremia and predict the subsequent viral load set point. J. Virol. 2007;81:5759–5765. doi: 10.1128/JVI.00045-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van den Berg S.P.H., Pardieck I.N., Lanfermeijer J., Sauce D., Klenerman P., van Baarle D., Arens R. The hallmarks of CMV-specific CD8 T-cell differentiation. Med. Microbiol. Immunol. (Berl.) 2019;208:365–373. doi: 10.1007/s00430-019-00608-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tian Y., Babor M., Lane J., Seumois G., Liang S., Goonawardhana N.D.S., De Silva A.D., Phillips E.J., Mallal S.A., da Silva Antunes R. Dengue-specific CD8+ T cell subsets display specialized transcriptomic and TCR profiles. J. Clin. Invest. 2019;129:1727–1741. doi: 10.1172/JCI123726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chng M.H.Y., Lim M.Q., Rouers A., Becht E., Lee B., MacAry P.A., Lye D.C., Leo Y.S., Chen J., Fink K. Large-Scale HLA Tetramer Tracking of T Cells during Dengue Infection Reveals Broad Acute Activation and Differentiation into Two Memory Cell Fates. Immunity. 2019;51:1119–1135.e5. doi: 10.1016/j.immuni.2019.10.007. [DOI] [PubMed] [Google Scholar]
- 47.Kaech S.M., Tan J.T., Wherry E.J., Konieczny B.T., Surh C.D., Ahmed R. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat. Immunol. 2003;4:1191–1198. doi: 10.1038/ni1009. [DOI] [PubMed] [Google Scholar]
- 48.Tang F., Quan Y., Xin Z.T., Wrammert J., Ma M.J., Lv H., Wang T.B., Yang H., Richardus J.H., Liu W., Cao W.C. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J. Immunol. 2011;186:7264–7268. doi: 10.4049/jimmunol.0903490. [DOI] [PubMed] [Google Scholar]
- 49.Ng O.W., Chia A., Tan A.T., Jadi R.S., Leong H.N., Bertoletti A., Tan Y.J. Memory T cell responses targeting the SARS coronavirus persist up to 11 years post-infection. Vaccine. 2016;34:2008–2014. doi: 10.1016/j.vaccine.2016.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S. Convergent Antibody Responses to SARS-CoV-2 Infection in Convalescent Individuals. bioRxiv. 2020 doi: 10.1101/2020.05.13.092619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gill, V., and Buchanan, R. (2020). Coronavirus: Immune clue sparks treatment hope. BBC News, May 22, 2020. https://www.bbc.com/news/health-52754280.
- 52.Trapecar M., Khan S., Roan N.R., Chen T.H., Telwatte S., Deswal M., Pao M., Somsouk M., Deeks S.G., Hunt P.W. An Optimized and Validated Method for Isolation and Characterization of Lymphocytes from HIV+ Human Gut Biopsies. AIDS Res. Hum. Retroviruses. 2017;33(S1):S31–S39. doi: 10.1089/aid.2017.0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma T., Luo X., George A., Mukherjee G., Sen N., Spitzer T., Giudice L.C., Greene W.C., Roan N.R. HIV efficiently infects T cells from the endometrium and remodels them to promote systemic viral spread. eLife. 2020;9:e55487. doi: 10.7554/eLife.55487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shekhar K., Brodin P., Davis M.M., Chakraborty A.K. Automatic Classification of Cellular Expression by Nonlinear Stochastic Embedding (ACCENSE) Proc. Natl. Acad. Sci. USA. 2014;111:202–207. doi: 10.1073/pnas.1321405111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw CyTOF datasets generated from this study are available for download through the public repository Dryad via the following link: https://doi.org/10.7272/Q6RJ4GPP.