Abstract
As the novel SARS-CoV-2 was detected in faeces, environmental researchers have been using centrifugal ultrafiltration, polyethylene glycol precipitation and aluminium hydroxide flocculation to describe its presence in wastewater samples. High recoveries (up to 65%) are described with electronegative filtration when using surrogate viruses, but few literature reports recovery efficiencies using accurate quantification of enveloped viruses. Considering that every single virus will have a different behaviour during viral concentration, it is recommended to use an enveloped virus, and if possible, a betacoronaviruses as murine hepatitis virus, as a surrogate. In this review, we show new data from a newly available technology that provides a quick ultrafiltration protocol for SARS-CoV-2. Wastewater surveillance is an efficient system for the evaluation of the relative prevalence of SARS-CoV-2 infections in a community, and there is the need of using reliable concentration methods for an accurate and sensitive quantification of the virus in water.
Keywords: Concentration methods, Enveloped virus, SARS-CoV-2, Recovery efficiency, Surrogate virus
Graphical abstract
Introduction
Many viruses that infect humans are excreted in large amounts through faeces, urine or skin desquamation, contributing to wastewater virome. Wastewater is a complex matrix that comprises a large variety of pathogenic and commensal viruses and provides important information about virus circulation, the introduction of emergent viruses and how they are transmitted among the population [1]. Waterborne viruses are generally nonenveloped and excreted in high numbers by infected individuals with or without disease, and in some cases long after the resolution of symptoms [2]. The study of excreted viruses is a very useful tool known as wastewater-based epidemiology, which has the potential to act as a complementary approach for current infectious disease surveillance systems and an early warning system for disease outbreaks [3].
The incidence of emerging microbes is a serious health concern worldwide. The increase of human–livestock contacts [4], population mobility and trade networks [5,6], climate change [7] or the wild meat trade and loss of animal habitats [8] has raised the risk of a global pandemics. Since 1980, nearly 90 novel human pathogen species have been discovered, more than 70 of those corresponded to novel human viruses, that compared to other pathogens have the potential to evolve more rapidly, being 80 of these associated with nonhuman reservoirs [9,10]. Influenza viruses (H1N1, H7N1 and H7N9), human immunodeficiency virus, Ebola virus, coronaviruses as SARS-CoV, MERS and the SARS-CoV-2 causing the COVID-19 pandemic have been the most significant.
SARS-CoV-2 was identified in China at the end of 2019 [11] and has become the first pandemic coronavirus (CoV). After the first case report of the presence of SARS-CoV-2 RNA in faeces [12], and because of the presence in the past of SARS-CoV-1 in faeces and sewage [13, 14, 15], the scientific community started to investigate if this virus could spread into the environment. Specific stability of SARS-CoV-2 has only been tested in aerosols and surfaces [16], but it is known that enveloped viruses are capable of retaining infectivity for days to months in aqueous environments [17, 18, 19]. On March 30th, SARS-CoV-2 was reported as detectable in wastewater 3 weeks before the first case was reported in the Netherlands [20]. On the following weeks, studies from Australia, China, Italy and Spain reported the presence of SARS-CoV-2 and concentrations in raw wastewater to be between 104 and 106 GC/L [21, ∗22, 23, 24].
One of the major challenges in SARS-CoV-2 research in wastewater samples is the lack of standardized protocols for its detection. From sample collection to virus concentration, there is still no consensus on the most efficient procedure. The way the sample is collected, or the virus is concentrated seems to be crucial to avoid false-negative results or inaccurate reported concentrations. Although viral titres in composite samples are being reported to be lower than in noncomposite ones, the persistent variability between noncomposite replicates suggest using an autosampler that collects a volume proportional to flow as the best sampling strategy. Also, the fact that different studies use different nucleic acid extraction and detection methods made difficult to establish comparisons among different studies.
After conducting an extensive revision on the most commonly used methods for concentrating viruses from wastewater samples in the last 2 years, Bofill-Mas and Rusiñol described that viral concentration methods had been mostly focused on combinations of flocculation/precipitation strategies [25]. Traditionally, viral environmental surveillance has considered principally RNA enteric viruses and also DNA viruses abundantly excreted in faeces, urine or skin desquamation as adenoviruses, polyomaviruses and papillomaviruses, which are all nonenveloped virus [2]. In fact, in 2015, Wigginton et al. [26] noticed that research should focus on the study of enveloped viruses in the urban water cycle as future pandemics could involve this type of viruses.
This review provides a brief on what it is known about the efficiency of viral concentration methods for CoV as well as for other enveloped viruses and new data of a comparative study analysing three concentration methods, skimmed milk flocculation (SMF), a new quick technology for ultrafiltration and a centrifugal ultrafiltration (CeUF) protocol.
SARS-CoV-2 in wastewater studies
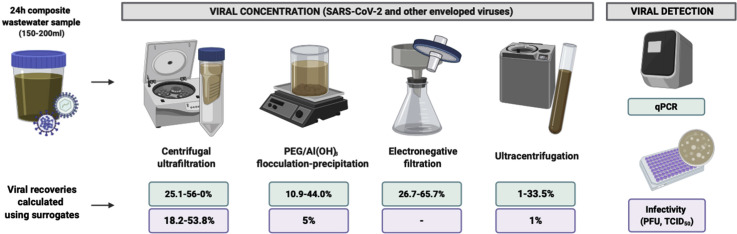
To date, the published SARS-CoV-2 surveillance studies use CeUF [20,21], methods including polyethylene glycol (PEG) or aluminium hydroxide (Al(OH)3) flocculation–precipitation [∗22, 23, 24] to concentrate SARS-CoV-2 from untreated wastewater. Figure 1 summarizes the methods used in recently published studies to concentrate SARS-CoV-2 from wastewater samples.
Figure 1.
Summary of the different strategies used in the published literature to concentrate SARS-CoV-2 from wastewater samples. PEG/Al(OH)3 flocculation- and precipitation-based methods [∗22, 23, 24] centrifugal ultrafiltration (CeUF) methods [20,21] and electronegative filtration [21] have been used to date in the published studies for SARS-CoV-2.
As wastewater becomes a surveillance tool for potential incidence regrowth, the interest to understand the performance of the concentration methods used increases as well as the interest towards those methods developed and validated for nonenveloped viruses testing. Culturing SARS-CoV-2 requires BSL-3 laboratories and specially trained personnel, thus the use of surrogate CoV (e.g. nonhuman infectious CoV strains, or other enveloped viruses) should be considered for methods development or as positive control at this stage of research.
La Rosa et al. [27] recently published a review on CoV in water environments, including data on occurrence, persistence and survival. Also, Carducci et al. [28] revised the current state of the art regarding CoV in water and highlighted the research gaps of the methods commonly used for sampling and concentration of enteric viruses, which need to adapt to enveloped viruses. Both reviews are focused in the four available studies on human CoV that use two-step methodologies based on a precentrifugation and ultrafiltration [18], glass wool filtration and PEG elution [29,30] and electropositive filter media columns and PEG precipitation [31]. Kitajima et al. [32] reviewed the state of the knowledge regarding the potential role of wastewater in the transmission of SARS-CoV-2.
The murine hepatitis virus (MHV), a surrogate for human CoV, has been used for persistence, survival and method comparison studies [17,18,33]. Ye et al. [18] compared, by means of MHV recoveries, three methodologies to concentrate enveloped viruses from wastewater samples, PEG precipitation and ultracentrifugation recovered approximately 5% of the spiked viruses, whereas with ultrafiltration protocol, the concentration was significantly higher (25%). The best performing method involved the removal of debris, prefiltering 250 mL of wastewater through a 0.22 μm PES membrane, followed by Centricon® Plus-70 10 kDa filtration. Recently, Ahmed et al. [33] have also evaluated six concentration strategies using MHV as a surrogate. The three filtration methods assayed provided the highest mean recoveries: when MgCl2 pretreatment was included, 65% of the MHV were recovered, when sample was directly filtered through 0.45-μm pore-size electronegative membranes, MHV recoveries were 60%, but when preacidifying the sample, the mean recovery decreased to 27%. Between the two CeUF methods tested, the Amicon® Ultra-15 30 kDa recovered 56% of the spiked surrogate and Centricon® Plus-70 10 kDa recovered 28%. Finally, by means of PEG precipitation and ultracentrifugation, MHV recoveries were 44% and 33%, respectively.
Although some enveloped viruses could be adequate surrogates for betacoronavirus concentration, only 5 of 15 published studies on SARS-CoV-2 occurrence in wastewater have used whole process controls, some nonenveloped virus including RNA phages [20] and Mengo virus [22], and an enveloped virus as porcine epidemic diarrhoea virus (PEDV) [22]. The use of these controls proves that the protocol worked correctly and provide with an estimation of the recovery efficiency of the method for the control, although this could be different for the virus of interest. Highest recoveries were obtained with CeUF devices, like Centricon® Plus-70 30 kDa, reaching 73% of the seeded F-specific RNA phages [20]. Randazzo et al. [22] used a surrogate CoV to calculate recovery. It is remarkable that with the Al(OH)3 flocculation method, a similar recovery (11%) was obtained for the enveloped virus, PEDV, and the nonenveloped virus, Mengo virus. Different viruses, even those sharing physical properties, use to show a different recovery when concentrated by the same method. To observe, similar recovery values could have been a mere casualty or it could be that both viruses attached to flocs with similar efficiencies due to their negative charge when they are above the isoelectric point [34].
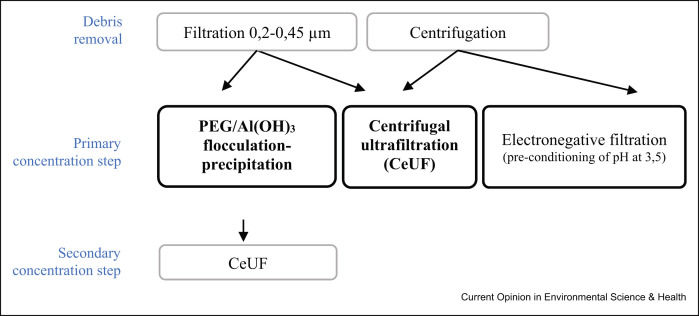
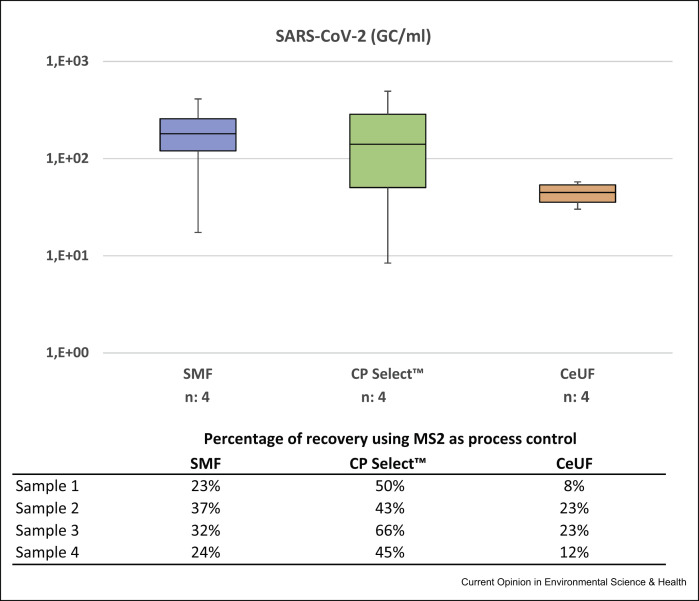
Preliminary data obtained by our research group in a study analysing different concentration methods for the detection of SARS-CoV-2 in wastewater from Catalonia (Spain), using MS2 as a process control, showed no statistically significant differences (p-value of the ANOVA test: 0.332) between the quantitative data (RT-qPCR) produced by the three viral concentration methods both for SARS-CoV-2 and for MS2. Four wastewater samples were concentrated using the SMF protocol [35] with an initial sample volume of 250 mL, the CeUF of 70 mL of the sample with Centricon® Plus-70 100 kDa (CeUF) [20] and a new and quick 80-mL ultrafiltration protocol using the automatic Concentrating Pipette (CP-Select™) from Innovaprep using 150 kDa ultrafiltration tips (www.innovaprep.com) (Figure 2 ). Debris was removed before the ultrafiltration by pelleting using centrifugation at 4750×g for 30 min. A volume of the three concentration methodologies, the equivalent of 2 mL of sewage was analysed at the qPCR.
Figure 2.
Barplots of the concentrations of naturally occurring SARS-CoV-2 in four sewage samples using three different concentration methods: SMF, InnovaPrep concentrating pipette with single-use ultrafiltration tips 150 kDa (CP Select™) and CeUF with Centricon Plus 70 100 kDa (CeUF).
Concentration of other enveloped viruses with pandemic potential in wastewater
On the lack of much data regarding CoV recovery efficiency when using commonly applied methods and until more data will be available, we should rely on what it is known for other enveloped viruses considering that every single virus will have a different behaviour during viral concentration. Alone or combined, the electropositive and electronegative filtration, CeUF, the organic flocculation and the PEG/Al(OH)3 precipitation methods have been used in different studies covering enveloped viruses’ detection in environmental water sources. Table 1 revises the concentration methods used until now for enveloped virus and summarises a selection of studies reporting recovery efficiencies.
Table 1.
Concentration methods and mean recoveries for enveloped viruses.
| Sample type | Family, genera and virus | Concentration method | Mean recovery ± SD (detection method) | Ref | |
|---|---|---|---|---|---|
| Wastewater | Coronaviridae, Betacoronavirus | MHV | Ultrafiltration (Centricon® Plus-70 100 kDa) | 25.1 ± 3.6% (PFU) | [18] |
| PEG/NaCl flocculation–precipitation | 5% (PFU) | ||||
| Ultracentrifugation | 1% (PFU) | ||||
| Ultrafiltration (Centricon® Plus-70 30 KDa) | 28.0 ± 9.10% (qPCR) | [33] | |||
| Ultrafiltration (Amicon® Ultra-15 30 KDa) | 56.0 ± 32.3% (qPCR) | ||||
| Electronegative filtration (preacidification) | 26.7 ± 15.3% (qPCR) | ||||
| Electronegative filtration (direct filtration) | 60.5 ± 22.2% (qPCR) | ||||
| Electronegative filtration (pretreated MgCl2) | 65.7 ± 23.0% (qPCR) | ||||
| PEG/NaCl flocculation–precipitation | 44.0 ± 27.7% (qPCR) | ||||
| Ultracentrifugation | 33.5 ± 12.1% (qPCR) | ||||
| SARS-CoV | Positive charged filter media + PEG elution | 1.02% (TCID50) | [31] | ||
| PEDV | Al(OH)3 flocculation–precipitation | Influent 10.90 ± 3.54% (qPCR) Effluent 3.29 ± 1.58% (qPCR) |
[22] | ||
| Cystoviridae, Cystovirus | Phi 6 | Ultrafiltration (Centricon® Plus-70 100 kDa) | 18.2 ± 9.5% (PFU) | [18] | |
| PEG/NaCl flocculation–precipitation | 5% (PFU) | ||||
| Ultracentrifugation | 1% (PFU) | ||||
| Orthomyxoviridae, Alphainfluenzavirus | Influenza A (H5N1) | Ultrafiltration (Centricon® Plus-70 30 KDa) | Influent 53.8% (qPCR) Effluent 42.7% (qPCR) |
[43] | |
| Surface water | Coronaviridae, Alphacoronavirus | TGEV | Glass wool (electropositive filtration) + 20% PEG elution | 51.3 ± 10.5% (qPCR) | [30] |
| Coronaviridae, Betacoronavirus | BCoV | Glass wool (electropositive filtration) + 10% PEG elution | Low turbidity 0.5 NTU: 25.8 ± 21.3% (qPCR) Medium turbidity 125 NTU: 9.2 ± 2.4% (qPCR) High turbidity 447 NTU: 19.5 ± 27.1% (qPCR) |
[29] | |
| Flaviviridae, Pestivirus | BVDV type 1 | Glass wool (electropositive filtration) + 10% PEG elution | Low turbidity 0.5 NTU: 12.9 ± 5.4% (qPCR) Medium turbidity 125 NTU: 12.9 ± 13.3% (qPCR) High turbidity 447 NTU: 21.1 ± 5.3% (qPCR) |
[29] | |
| BVDV | Skimmed milk flocculation | 15 ± 1.6% (qPCR) 0.7 ± 0.13% (TCID50) |
[35] | ||
| Orthomyxoviridae, Alphainfluenzavirus | Influenza A (H5N1) | Glass wool (electropositive filtration) + 10% PEG elution | River water 1% (TCID50) Rain water 3.63––13.79% (qPCR) Lake water 0.01–7.89% (qPCR) |
[44] | |
| Ultrafiltration (Hemoflow F80S) | Surface water 5.4% (qPCR) | [43] | |||
| Influenza A (H5N3) | Prefiltration + borosilicate glass membrane GF/F (electropositive filtration) | River water: 4.7 ± 0.05% (qPCR) Seawater: 16.7 ± 0.04% (qPCR) |
[45] | ||
| Electronegative filtration (SMWP membranes) | River water: 1.5 ± 0.01% (qPCR) Seawater: 5.00 (qPCR) |
||||
TGEV, transmissible gastroenteritis virus; BCoV, bovine coronavirus.
It has been reported that higher percentage of enveloped viruses adsorb to the solid fraction of wastewater compared with nonenveloped viruses [18] and it is believed that these suspended solids protect viruses from inactivation [19,36,37]. None of the published studies included the first step separated solids into the analysis, but most of them involved an initial step to remove wastewater solids and then focused on recovering the viruses from the liquid phase.
Despite the proposed viral concentration methods for SARS-CoV-2 or generally for CoV, extensively reviewed by others, the organic flocculation, has been also used for the concentration of viruses in water including enveloped viruses. The enveloped virus, bovine viral diarrhoea virus (BVDV), presented mean recoveries of 15% when tested with qPCR and 0.7% when tested for infectivity, but acid pH (for approximately 16 h) that is used in the SMF protocol seems to reduce the infectivity, as the log10 ratio RT-qPCR/infectivity for that virus was 2.03 [35,38,39]. The same observation has been described for PEG precipitation methods, which disrupt the lipid bilayers and thus are not optimal for recovering infective enveloped viruses [18,40].
When testing viral recovery methods, it is relevant to consider how recovery rates are calculated, and at this point, the quantification of viral stocks used for spiking is of relevance because different values may be obtained when the quantification is done directly from viral stocks used for spiking or when quantifying after adding viral stock into a similar matrix from which recovered viruses will be quantified. Different enzymatic inhibition could be observed depending on the matrix in which viruses are embedded. On the other hand, if recovery is calculated according to infectivity by means of plaque forming unit's quantification assays, viral aggregation phenomena could lead to an under quantification of viral stocks. Disaggregation protocols before spiking should be considered to correct this effect [41]. Finally, direct quantification of viral stocks without prepurification or enzymatic pretreatment may overestimate the real amount of infectious viruses as the presence of free RNA may be quantified in viral suspensions from cell cultures [42].
Future research directions and conclusions
Agents causing novel infections are often zoonotic, crossing from the natural host into the human population. Hence, a one-health surveillance approach of virus-infected animals as well as humans is required. Structural and biochemical differences between enveloped viruses suggest that the same methods would not exhibit the same recoveries between them. As there is a potential for new outbreaks, the molecular detection of SARS-CoV-2 RNA in wastewater and the correlation between its concentration and reported prevalence of COVID-19 may be a sensitive monitoring tool to evaluate the prevalence of the virus in a community, becoming a potential source of epidemiological data and public health risks information [20].
To face off novel outbreaks, important public health organizations such as the Centers for Disease Control and Prevention (CDC), the European Centre for Disease Prevention and Control (ECDC) or World Health Organisation, highlight the role of scientific research to combat infectious disease, especially those emerging or re-emerging disease that may reappear in a more threatening form. CDC establishes that detection and identification should be prioritized by expanding research on ecological and environmental factors, influencing disease emergence and transmission, meanwhile the ECDC highlights as a general surveillance objective, detect and monitor food- and waterborne and zoonotic outbreaks with respect to source, time, population and place to provide a rationale for public health actions. Consequently, one of the World Health Organisation actions is to provide an integrated global alert and response system for epidemics and other public health emergencies for an effective international coordinated response. More scientific research is needed to identify viral transmission routes, characterizing protocols and early detection strategies for a better understanding of the factors involved in disease emergence, prevention and elimination. To furnish health management models with wastewater surveillance data, more research should focus on optimizing and evaluating concentration methods able to recover enveloped potentially pandemic viruses or their surrogates from environmental samples.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the project AGL2017-86797-C2-1-R, PCI2019-103643, RTI2018-097346-B-I00 and AGL2017-86797-C2-1-R projects, all funded by the Spanish Ministry of Science, Innovation and Universities. S Bofill-Mas is a Serra Hunter fellow at the University of Barcelona. Graphical abstract was created with Biorender.com.
This review comes from a themed issue on Environmental Health: COVID-19
Edited by Avelino Núñez-Delgado
References
- 1.Martínez-Puchol S., Rusiñol M., Fernández-Cassi X., Timoneda N., Itarte M., Andrés C., Antón A., Abril J.F., Girones R., Bofill-Mas S. Characterisation of the sewage virome: comparison of NGS tools and occurrence of significant pathogens. Sci Total Environ. 2020:713. doi: 10.1016/j.scitotenv.2020.136604. [DOI] [PubMed] [Google Scholar]
- 2.Rusiñol M., Girones R. In: Global water pathogens project. Rose J.B., Jiménez-Cisneros B., editors. Michigan State University, E. Lansing, MI, UNESCO; 2017. Summary of excreted and waterborne viruses. [Google Scholar]
- 3.Sims N., Kasprzyk-Hordern B. Future perspectives of wastewater-based epidemiology: monitoring infectious disease spread and resistance to the community level. Environ Int. 2020;139:105689. doi: 10.1016/j.envint.2020.105689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klous G., Huss A., Heederik D.J.J., Coutinho R.A. Human–livestock contacts and their relationship to transmission of zoonotic pathogens, a systematic review of literature. One Heal. 2016;2:65–76. doi: 10.1016/j.onehlt.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karesh W.B., Dobson A., Lloyd-Smith J.O., Lubroth J., Dixon M.A., Bennett M., Aldrich S., Harrington T., Formenty P., Loh E.H., et al. Zoonoses 1 Ecology of zoonoses: natural and unnatural histories. Lancet. 2012:380. doi: 10.1016/S0140-6736(12)61678-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friend Milton, Milton F. vol. 1285. Geological Survey, Circular; U.S: 2006. Zoonoses and travel; p. 400. (Disease emergence and resurgence: the wildlife-human connection). [Google Scholar]
- 7.Cann K.F., Thomas D.R., Salmon R.L., Wyn-Jones A.P., Kay D. Extreme water-related weather events and waterborne disease. Epidemiol Infect. 2013;141:671–686. doi: 10.1017/S0950268812001653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cantlay J.C., Ingram D.J., Meredith A.L. A review of zoonotic infection risks associated with the wild meat trade in Malaysia. EcoHealth. 2017;14:361–388. doi: 10.1007/s10393-017-1229-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morse S.S., Mazet J.A.K., Woolhouse M., Parrish C.R., Carroll D., Karesh W.B., Zambrana-Torrelio C., Lipkin I., Daszak P. Zoonoses 3 Prediction and prevention of the next pandemic zoonosis. Lancet. 2012:380. doi: 10.1016/S0140-6736(12)61684-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keusch G.T., Pappaioanou M., Gonzalez M.C., Scott K.A., Tsai P. Intitute of Medicine and National Research Council; 2009. Sustaining global surveillance and response to emerging zoonotic diseases. [PubMed] [Google Scholar]
- 11.WHO: WHO Timeline – COVID-19. www.who.int.2020,
- 12.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., et al. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X.-W., Li J.-W.J.-S.J.-F., Guo T.-K., Zhen B., Kong Q.-X., Yi B., Li Z., Song N., Jin M., Xiao W.-J., et al. Concentration and detection of SARS coronavirus in sewage from xiao tang Shan hospital and the 309th hospital. J Virol Methods. 2005;128:156–161. doi: 10.1016/j.jviromet.2005.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng P.K.C., Wong D.A., Tong L.K.L., Ip S.-M., Lo A.C.T., Lau C.-S., Yeung E.Y.H., Lim W.W.L. Viral shedding patterns of coronavirus in patients with probable severe acute respiratory syndrome. Lancet. 2004;363:1699–1700. doi: 10.1016/S0140-6736(04)16255-7. (London, England) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hung I.F.N., Cheng V.C.C., Wu A.K.L., Tang B.S.F., Chan K.H., Chu C.M., Wong M.M.L., Hui W.T., Poon L.L.M., Tse D.M.W., et al. Viral loads in clinical specimens and SARS manifestations. Emerg Infect Dis. 2004;10:1550–1557. doi: 10.3201/eid1009.040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., Tamin A., Harcourt J.L., Thornburg N.J., Gerber S.I., et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med. 2020;382 doi: 10.1056/NEJMc2004973. 0–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casanova L., Rutala W., Weber D., Sobsey M. Survival of surrogate coronaviruses in water. Water Res. 2009;43 doi: 10.1016/j.watres.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ Sci Technol. 2016;50:5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- 19.Gundy P.M., Gerba C.P., Pepper I.L. Survival of coronaviruses in water and wastewater. Food Environ Virol. 2009;1:10–14. [Google Scholar]
- 20.Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environ Sci Technol Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- 21.Ahmed W., Angel N., Edson J., Bibby K., Bivins A., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181:115942. doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study includes data on recovery of enveloped surrogates and detection process controls within a surveillance study of the prevalence of SARS-CoV-2 in wastewater.
- 23.La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-COV-2 in untreated wastewaters in Italy. Sci Total Environ. 2020;736:139652. doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang D., Ling H., Huang X., Li J., Li W., Yi C., Zhang T., Jiang Y., He Y., Deng S., et al. Potential spreading risks and disinfection challenges of medical wastewater by the presence of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) viral RNA in septic tanks of fangcang hospital. Sci Total Environ. 2020;741:140445. doi: 10.1016/j.scitotenv.2020.140445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bofill-Mas S., Rusiñol M. Recent trends on methods for the concentration of viruses from water samples. Curr Opin Environ Sci Heal. 2020;16:7–13. [Google Scholar]
- 26.Wigginton K.R., Ye Y., Ellenberg R.M. Emerging investigators series: the source and fate of pandemic viruses in the urban water cycle. Environ Sci Water Res Technol. 2015;1:735–746. [Google Scholar]
- 27.La Rosa G., Bonadonna L., Lucentini L., Kenmoe S., Suffredini E. Coronavirus in water environments: occurrence, persistence and concentration methods - a scoping review. Water Res. 2020;179:115899. doi: 10.1016/j.watres.2020.115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carducci A., Federigi I., Liu D., Thompson J.R., Verani M. Making waves: coronavirus detection, presence and persistence in the water environment: state of the art and knowledge needs for public health. Water Res. 2020;179:115907. doi: 10.1016/j.watres.2020.115907. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors reviewed specifically human coronavirus prevalence and persistance in the environment.
- 29.Abd-Elmaksoud S., Spencer S.K., Gerba C.P., Tamimi A.H., Jokela W.E., Borchardt M.A. Simultaneous concentration of bovine viruses and agricultural zoonotic bacteria from water using sodocalcic glass wool filters. Food Env Virol. 2014;6:253–259. doi: 10.1007/s12560-014-9159-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blanco A., Abid I., Al-Otaibi N., Pérez-Rodríguez F., Fuentes C., Guix S., Pintó R.M., Bosch A. Glass wool concentration optimization for the detection of enveloped and non-enveloped waterborne viruses. Food Environ Virol. 2019;11:184–192. doi: 10.1007/s12560-019-09378-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X.-W., Li J.-S., Jin M., Zhen B., Kong Q.-X., Song N., Xiao W.-J., Yin J., Wei W., Wang G.-J., et al. Study on the resistance of severe acute respiratory syndrome-associated coronavirus. J Virol Methods. 2005;126:171–177. doi: 10.1016/j.jviromet.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors performed an extensive review on the state of the art regarding SARS-CoV-2 research in wastewater, including summary tables of SARS-CoV-2 RT-qPCR and RT-PCR assays, detection of the virus in human specimens, water and aerosols, survival of coronaviruses in wastewater and QMRA parameters.
- Ahmed W., Bertsch P., Bivins A., Bibby K., Farkas K., Gathercole A., Haramoto E., Gyawali P., Korajkic A., Mcminn B.R., et al. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study makes a comparison between four SARS-CoV-2 concentration methods using MHV surrogate to calculate enveloped virus recoveries.
- 34.Michen B., Graule T. Isoelectric points of viruses. J Appl Microbiol. 2010;109:388–397. doi: 10.1111/j.1365-2672.2010.04663.x. [DOI] [PubMed] [Google Scholar]
- Gonzales-Gustavson E., Cárdenas-Youngs Y., Calvo M., da Silva M.F.M., Hundesa A., Amorós I., Moreno Y., Moreno-Mesonero L., Rosell R., Ganges L., et al. Characterization of the efficiency and uncertainty of skimmed milk flocculation for the simultaneous concentration and quantification of water-borne viruses, bacteria and protozoa. J Microbiol Methods. 2017;134:46–53. doi: 10.1016/j.mimet.2017.01.006. [DOI] [PubMed] [Google Scholar]; The study includes an enveloped virus in the characterization of the Skimmed Milk Flocculation. It includes qPCR and infectivity results and characterises the variability and uncertainty of the whole method.
- 36.Barrett M., Fitzhenry K., O'Flaherty V., Dore W., Keaveney S., Cormican M., Rowan N., Clifford E. Detection, fate and inactivation of pathogenic norovirus employing settlement and UV treatment in wastewater treatment facilities. Sci Total Environ. 2016;568:1026–1036. doi: 10.1016/j.scitotenv.2016.06.067. [DOI] [PubMed] [Google Scholar]
- 37.Schaar H., Sommer R., Schürhagl R., Yillia P., Kreuzinger N. Microorganism inactivation by an ozonation step optimized for micropollutant removal from tertiary effluent. Water Sci Technol. 2013;68:311–318. doi: 10.2166/wst.2013.212. [DOI] [PubMed] [Google Scholar]
- 38.Ye K., Dhiman H.K., Suhan J., Schultz J.S. Effect of pH on infectivity and morphology of ecotropic moloney murine leukemia virus. Biotechnol Prog. 2003;19:538–543. doi: 10.1021/bp0200705. [DOI] [PubMed] [Google Scholar]
- 39.Costello D.A., Whittaker G.R., Daniel S. Variations in pH sensitivity, acid stability, and fusogenicity of three influenza virus H3 subtypes. J Virol. 2015;89:350–360. doi: 10.1128/JVI.01927-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boni L., Stewart T., Alderfer J., Hui S., Lt B. Lipid-polyethylene glycol interactions: II. Formation of defects in bilayers. J Membr Biol. 1981;62 doi: 10.1007/BF01870201. TP S, JL A, Sw H. [DOI] [PubMed] [Google Scholar]
- 41.Cotter C.A., Earl P.L., Wyatt L.S., Moss B. Preparation of cell cultures and vaccinia virus stocks. Curr Protein Pept Sci. 2017;89:5.12.1–5.12.18. doi: 10.1002/cpps.34. [DOI] [PubMed] [Google Scholar]
- 42.Leibowitz J., Kaufman G., Liu P. Coronaviruses: propagation, quantification, storage, and construction of recombinant mouse hepatitis virus. Curr Protoc Microbiol. 2011 doi: 10.1002/9780471729259.mc15e01s21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heijnen L., Medema G. Surveillance of Influenza A and the pandemic influenza A (H1N1) 2009 in sewage and surface water in The Netherlands. J Water Health. 2011;9 doi: 10.2166/wh.2011.019. [DOI] [PubMed] [Google Scholar]
- 44.Deboosere N., Horm S.V., Pinon A., Gachet J., Coldefy C., Buchy P., Le Vialette M. Development and validation of a concentration method for the detection of influenza A viruses from large volumes of surface water. Appl Environ Microbiol. 2011;77:3802–3808. doi: 10.1128/AEM.02484-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rönnqvist M., Ziegler T., von Bonsdorff C.H., Maunula L. Detection method for avian influenza viruses in water. Food Environ Virol. 2012;4:26–33. doi: 10.1007/s12560-011-9075-4. [DOI] [PubMed] [Google Scholar]