Abstract
The clinical translation of cationic alpha-helical antimicrobial peptides (AMPs) has been hindered by structural instability, proteolytic degradation, and in vivo toxicity from non-specific membrane lysis. Although analyses of hydrophobic content and charge distribution have informed the design of synthetic AMPs with increased potency and reduced in vitro hemolysis, non-specific membrane toxicity in vivo continues to impede AMP drug development. Here, we analyzed a 58-member library of stapled AMPs (StAMPs) based on Magainin-II, and applied the insights from structure-function-toxicity measurements to devise an algorithm for the design of stable, protease-resistant, potent, and nontoxic StAMP prototypes. We show that a lead double-stapled StAMP named Mag(i+4)1,15(A9K,B21A,N22K,S23K) can kill multidrug resistant Gram-negative pathogens, such as colistin-resistant Acinetobacter baumannii in a mouse peritonitis-sepsis model, without observed hemolysis or renal injury in murine toxicity studies. Inputting the amino acid sequences alone, we further generated membrane-selective StAMPs of pleurocidin, CAP18, and esculentin, highlighting the generalizability of our design platform.
INTRODUCTION
AMPs are a subclass of natural peptides expressed by diverse species to combat infections, predominantly of the skin or localized compartments1–3. Cationic AMPs kill bacteria by membrane lysis, a mechanism significantly less prone to inducing antibiotic resistance.1, 4 Indeed, AMPs have been proposed as a potential solution to the global threat of multidrug resistance5–7. However, linear AMPs are structurally unstable, proteolytically labile, and cause non-specific membrane toxicity that has precluded their clinical translation for internal use. Since their discovery8, 9, three decades of groundbreaking research have elucidated the properties of natural and non-natural AMPs4, but the insights have not yielded AMP-based antibiotic drugs.
One established strategy to overcome the proteolytic lability of AMPs involves the use of D-amino acids10–12. Analyses of the synthetic construct, V13KL, revealed qualitative design principles for locating positive charge and hydrophobic residues to increase antibiotic activity and mitigate the lysis of human red blood cells (RBCs), an in vitro surrogate for non-specific mammalian cell toxicity13. A key “specificity determinant” based on positioning a lysine residue in the middle of the nonpolar face of the D-amino acid sequence was identified13. However, D-amino acid-based peptides are expensive to synthesize, have unpredictable structures in solution, and require individualized optimization13, 14. To date, only one D-amino acid-based AMP (XMP.629) reached phase II clinical trials for the topical treatment of acne but failed to demonstrate clinical benefit15.
An alternative approach to reducing proteolytic lability of peptides in vivo involves enforcing α-helical structure by inserting all-hydrocarbon staples16–18. All-hydrocarbon stapling has been applied to maximize the utility of bioactive helices for targeting protein interactions16, 18–21, with the first clinical-grade anti-cancer stapled peptide in phase I and II trials22. Attempts to install staples into AMP sequences have yielded constructs with improved α-helicity and proteolytic resistance but variable antimicrobial activity and indiscriminate membrane lysis, reflective of a low therapeutic index23–28. Thus, stapling has generally been viewed as contributing to nonspecific AMP toxicity23, 25.
In our early work with stapled peptides for intracellular targeting, we discovered that select constructs achieved robust mammalian cell uptake by a pinocytotic mechanism18, whereas select amphipathic α-helical sequences caused nonspecific membrane lysis, a feature that we tested for and screened out20. Indeed, we recently identified excessive hydrophobicity and positive charge - natural features of AMPs - as inherent risk factors for generating stapled α-helical peptides with membrane-lytic properties19. Here, we set out to transform this liability into an advantage for developing AMP-based therapies. By analyzing staple-scanning and mutagenesis libraries of StAMPs, we uncovered structure-function-toxicity relationships that enabled us to develop a predictive algorithm for effective in silico design of StAMPs that are stable, active, and importantly, membrane-selective in vivo.
RESULTS
Molecular Basis of the Membrane Selectivity of StAMPs
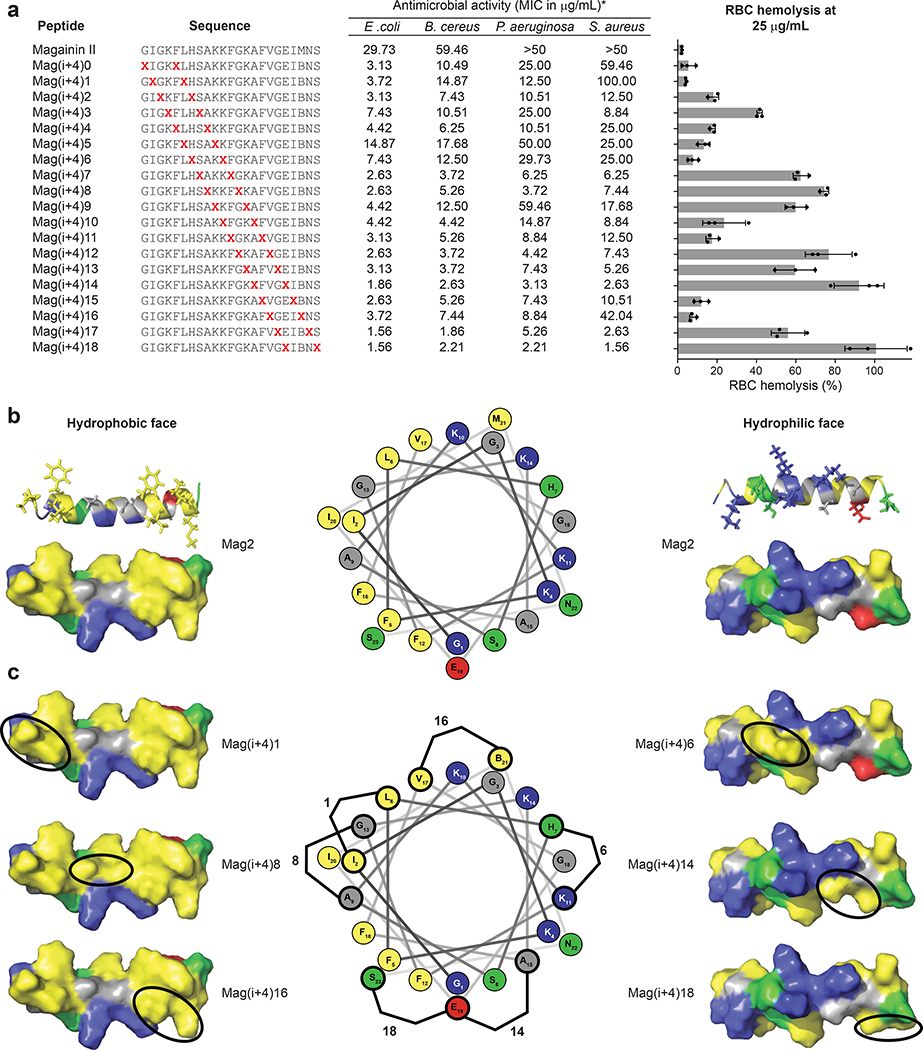
One of the most widely studied AMPs is a 23-amino acid peptide known as Magainin-II (Mag2)9, 29, 30. The capacity of Mag2 to lyse membranes is highly dependent on its α-helical structure31, making Mag2 an ideal template for stapling analyses. To search for intrinsic compositional features that could potentially discern between bacterial and mammalian membranes, we generated i, i+4 and i, i+7 staple-scanning libraries of Mag2, termed Mag(i+4) and Mag(i+7), and screened them for antimicrobial activity vs. human RBC hemolysis (Fig. 1a, Supplementary Fig. 1). While the Mag(i+7) library demonstrated minimum inhibitory concentrations (MICs) of 4–25 fold enhanced potency against Gram-positive (Bacillus cereus, Staphylococcus aureus) and Gram-negative (Escherichia coli, Pseudomonas aeruginosa) pathogens compared to Mag2, robust and indiscriminate hemolytic activity was also observed (Supplementary Fig. 1). In contrast, the MIC results for the Mag(i+4) library were more variable, with select StAMPs such as Mag(i+4)14 and Mag(i+4)18 displaying potent and broad spectrum antimicrobial activity, whereas others such as Mag(i+4)0 and Mag(i+4)1 relatively less potent (Fig. 1a). Also, there was notable variability among the constructs with respect to RBC hemolysis. Antibiotic potency did not uniformly correlate with the percentage of RBC hemolysis (for example, comparing Mag(i+4)7 with Mag(i+4)9), suggesting that the biophysical basis for the results was more complex. To investigate contributory factors that could explain the variable trends in antimicrobial activity and hemolysis of the i, i+4 stapled peptides, we measured and calculated a series of biophysical parameters; however, among the variables examined, including the percentage of α-helicity in solution, C18-HPLC retention time, isoelectric point, overall hydrophobicity, and three-dimensional hydrophobic moment32, none revealed a significant correlation (Supplementary Table 1, Supplementary Fig. 2). As an example, Mag(i+4)6 and Mag(i+4)18 had nearly identical hydrophobic content yet their hemolytic activities were at opposite ends of the spectrum (Fig. 1a, Supplementary Table 1).
Figure 1. Antimicrobial and hemolytic activities of an i, i+4-stapled Mag2 library.
a, The antimicrobial activity (MIC in μg/mL) and percentage of hemolysis at 25 μg/mL were determined for a library of i, i+4 stapled peptides based on the Mag2 sequence. The MIC is a geometric mean of four independent experiments. The percentage hemolysis data are mean ± s.d. of three independent experiments (shown as dots) and the bar plot reflects a periodicity of lytic activity that relates to staple position. X, S5 stapling amino acid; B, norleucine (substituted for methionine to maximize the efficiency of ruthenium-catalyzed olefin metathesis). b, Surface and ribbon views of the hydrophobic (left) and hydrophilic (right) faces of the amphipathic Mag2 α-helix (Protein Data Bank ID: 2MAG). The hydrophobic face contains two patches of highly hydrophobic residues (I, F, L, V, I, M; yellow) separated by a small patch of low hydrophobicity (A, G; grey). Cationic, hydrophilic and anionic residues are represented in blue, green and red, respectively, on the structures and helical wheel. c, When the hydrocarbon staple (represented by a black oval) is located within a patch of high hydrophobicity, such as in Mag(i+4)1 and Mag(i+4)16, or on the hydrophilic face of the α-helix, such as in Mag(i+4)6, the hydrophobic landscape remains similar to that of Mag2, resulting in little to no increase in hemolytic activity. However, when the staple connects the highly hydrophobic patches into a continuous region of high hydrophobicity, as in Mag(i+4)8, or expands the borders of the hydrophobic face, as in Mag(i+4)14 and Mag(i+4)18, there is a notable increase in RBC hemolysis. Staple locations for each StAMP are indicated on the helical wheel.
An important mechanistic clue came from inspection of the RBC hemolysis results, which revealed a periodicity to the recorded values that tracked with the sequential placement of the staple along the length of the peptide helix (Fig. 1a). To visualize the consequences of moving the staple along the AMP structure, we examined and compared the variety of StAMP surfaces, as generated in Pymol based on the NMR structure of Mag2 in dodecylphosphocholine micelles (Protein Data Bank ID: 2MAG)31. The hydrophobic surface of Mag2 is composed of two highly hydrophobic patches at the N-terminal (I2, G3, F5, L6; yellow) and C-terminal (F12, F16, V17, I20, M21; yellow) regions that are joined by a smaller patch of lower hydrophobicity (A9, G13; grey) (Fig. 1b). When the staple was positioned within one of the high hydrophobicity patches, such as in Mag(i+4)1 and Mag(i+4)16, there was little to no increase in hemolytic activity compared to Mag2 (Fig. 1c); however, when the patch of low hydrophobicity was replaced with the staple moiety, as in Mag(i+4)8, the entire hydrophobic face became even more hydrophobic, and hemolytic activity was markedly increased. In the case of Mag(i+4)6, substitution of H7 and K11 with the hydrophobic staple increased its calculated hydrophobicity (Supplementary Table 1) but had little to no effect on baseline hemolysis. Inspection of the Mag(i+4)6 structure demonstrated the localization of the staple on the hydrophilic face of the Mag2 helix surrounded by positively-charged residues, thus causing no change in the landscape of the hydrophobic surface (Fig. 1c). We further noted that insertion of the staple at the amphipathic boundary, as in Mag(i+4)14 and Mag(i+4)18, extended the hydrophobic surface onto the hydrophilic face resulting in dramatic increases in hemolytic activity (Fig. 1a, c). Thus, we observed that staple placements that expand the confluence of highly hydrophobic patches on the hydrophobic face of the amphipathic Mag2 α-helix triggered hemolytic activity, whereas increasing overall hydrophobicity without expanding the hydrophobic landscape of the hydrophobic interaction surface had no such effect.
A Design Algorithm for Bacterial-Selective StAMPs
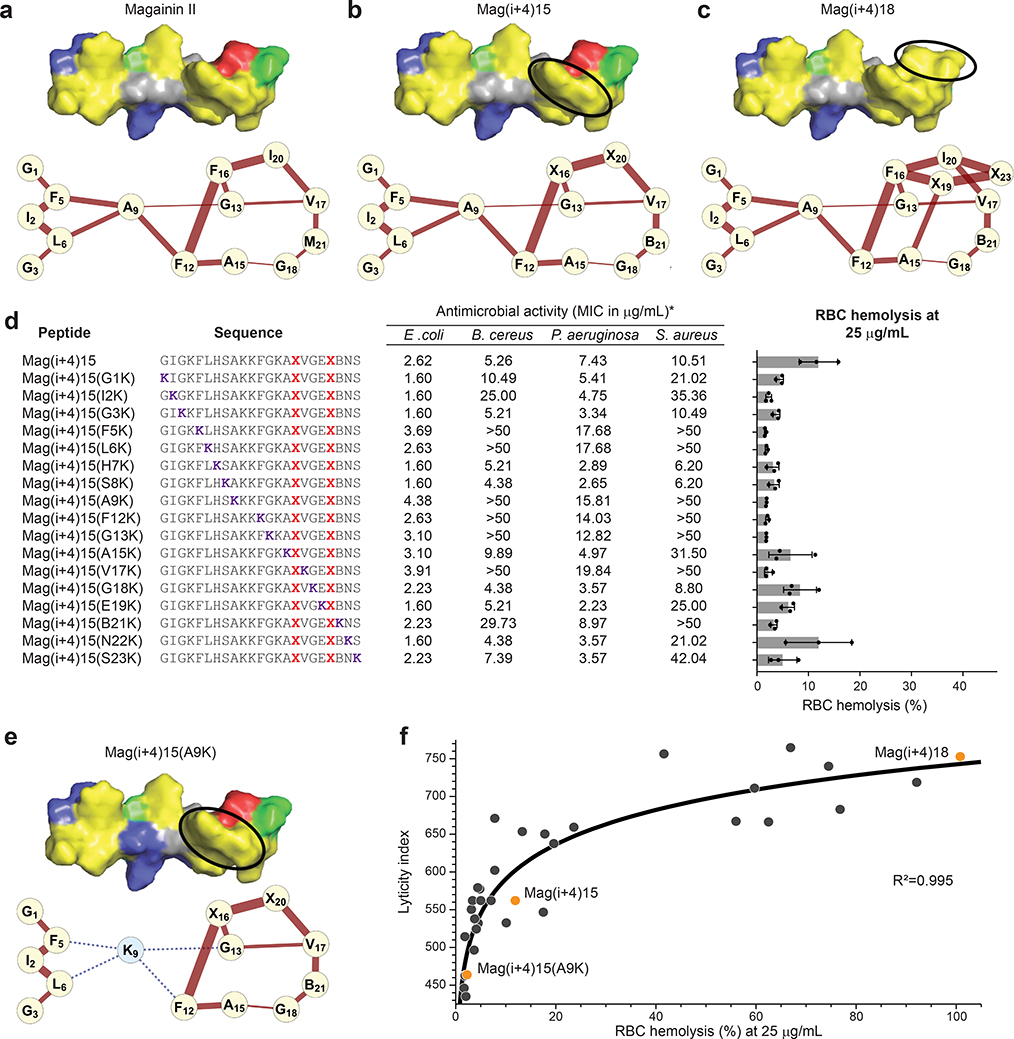
We next aimed to transform the conceptual advance that emerged from our empiric observations of differentially stapled AMPs into an algorithm that could be broadly applied to design optimal StAMPs for therapeutic development. Building on the two-dimensional approach of representing hydrophobic interactions on peptide surfaces as α-helical nets12, 13, we created a model to systematically visualize and quantitate the hydrophobic interactions along the z-axis of a StAMP α-helix. We applied a network analysis of the hydrophobic residues based on i, i+3 and i, i+4 interaction proximity to establish “hydrophobicity network maps” (HNMs) that demonstrate both the connectivity and strength of hydrophobic interactions on the three-dimensional α-helical surface (Fig. 2a–c, Supplementary Fig. 3). The HNM of Mag(i+4)15 demonstrated that when a staple is installed within a hydrophobic patch, the number of hydrophobic interactions is not altered compared to Mag2 (Fig. 2a–b), but the high hydrophobicity of the staple could increase the strength of hydrophobic interactions within the existing network, as represented by the relative thickness of the interaction line(s) (Fig. 2b). An HNM of Mag(i+4)18, the most hemolytic StAMP in the Mag(i+4) library, demonstrated that by substituting E18 and S23 with the all-hydrocarbon staple, the surface area of the hydrophobic patch increased, as reflected by the higher number of linkages within the network, along with the strength of the interactions, which are represented by the thicker network linkages (Fig. 2c). While the mild changes in Mag(i+4)15 caused little hemolytic effect, the prominent expansion of the hydrophobic landscape in Mag(i+4)18 resulted in marked RBC hemolysis (Fig. 1a).
Figure 2. Hydrophobic network mapping predicts mammalian cell lysis.
a, The HNM of Mag2 depicts the i+3 and i+4 relationships of hydrophobic residues along the z-axis of an α-helix. The thickness of each connection reflects the combined hydrophobicity of the connected amino acids. b, The HNM of Mag(i+4)15 reveals that when a staple is installed within an established patch of high hydrophobicity, the number of hydrophobic interactions remains the same and the strength of the interactions can increase (e.g. X16-X20, X20-V17). c, When a staple is installed outside of a hydrophobic patch, as in Mag(i+4)18, the HNM demonstrates that the number of hydrophobic connections and the strength of these interactions are increased. d, The comparative antimicrobial and hemolytic activities of a Mag(i+4)15 lysine-scanning library demonstrate that site-specific lysine substitution can attenuate hemolytic activity while maintaining antimicrobial potency against Gram-negative pathogens. The MIC is a geometric mean of three independent experiments. Percentage hemolysis data are mean ± s.d. of three independent experiments (shown as dots). X, S5 stapling amino acid; B, norleucine. e, The HNM of Mag(i+4)15(A9K) reveals that A9K mutagenesis severs the continuous hydrophobic network, thereby reducing the number of highly hydrophobic connections. f, A plot of the calculated lyticity index versus the percentage of RBC hemolysis distributes StAMPs from the Mag(i+4) staple-scanning and Mag(i+4)15 lysine-scanning libraries (n=36 distinct StAMPs) along a logarithmic selectivity continuum, with an R2 value of 0.995 as calculated by Pearson correlation.
Comprehensive HNM analysis of the Mag(i+4) library revealed that StAMPs with high hemolytic activity had the largest and strongest network of hydrophobic interactions (Supplementary Fig. 4). This trend motivated us to investigate how pruning these interactions by sequential insertion of a lysine along the length of a StAMP helix influenced antimicrobial and hemolytic activities. Inspired by the impact of single-site specificity determinants13, we performed a lysine scan analysis, choosing Mag(i+4)15 as the template owing to its robust antimicrobial activity in the context of mild hemolytic activity (Fig. 1a). As anticipated, when a lysine was substituted at the hydrophilic face such that overall charge positivity of the interface increased, antimicrobial potency against Gram-positive and Gram-negative pathogens improved and a slight decrease in hemolytic activity was observed (Fig. 2d). When a lysine substitution on the hydrophobic face disrupted the hydrophobic network (Supplementary Fig. 5), we noted a series of functional consequences. First, reductions in hemolytic activity correlated with a corresponding decrease in the number and strength of hydrophobic interactions, as calculated by the HNM algorithm (Fig. 2d, Supplementary Fig. 5). For example, the Mag(i+4)15(A9K) HNM showed how A9K mutation severed the link between the N- and C-terminal hydrophobic patches, resulting in two discontinuous hydrophobic patch networks and negligible hemolysis (Fig. 2d–e), consistent with the reported influence of a single-site specificity determinant12–14. Second, activity against Gram-negative pathogens was variably and only modestly affected, depending on the relative hydrophobicity of the amino acid replaced by lysine (Fig. 2d, Supplementary Fig. 5). Finally, activity against Gram-positive bacteria was notably reduced, especially when lysine substitutions eliminated amino acids that conferred hydrophobic interactions to a hydrophobic network (Fig. 2d, Supplementary Fig. 5). This result, also observed in the context of V13KL13, suggests that lysing Gram-positive bacteria requires a more robust hydrophobic interaction network compared to Gram-negative bacteria, providing an opportunity to both tailor the spectrum of StAMP activity to resistant Gram-negative bacteria and expand the window of selectivity for bacterial versus mammalian cell membranes.
To broaden the utility of our HNMs beyond generating visual representations, we sought to assign numerical values to HNMs for high throughput analyses of candidate StAMP sequences (Supplementary Fig. 3). For example, establishing a threshold value for nonspecific lyticity could serve as a first-line filter to weed out nonspecifically disruptive sequences within a given StAMP library. Since the number and strength of the hydrophobic interactions that comprise the hydrophobic α-helical surface dictates hemolytic activity, we examined whether the sum of i, i+3 and i, i+4 hydrophobic relationships would correlate with the degree of hemolysis. We calculated a lyticity index (LI) (see Online Methods) and then plotted the LIs generated for the Mag(i+4) staple-scanning and Mag(i+4)15 lysine-scanning libraries against the percentage of hemolysis values (Fig. 2f), revealing a strong logarithmic correlation (R2=0.995) between the LI value and the observed percentage of hemolysis. An LI threshold of 600 captured all Mag(i+4) StAMPs with <10% hemolysis at the supratherapeutic dosing level of 25 μg/mL.
Mechanisms of Membrane Lysis by StAMPs
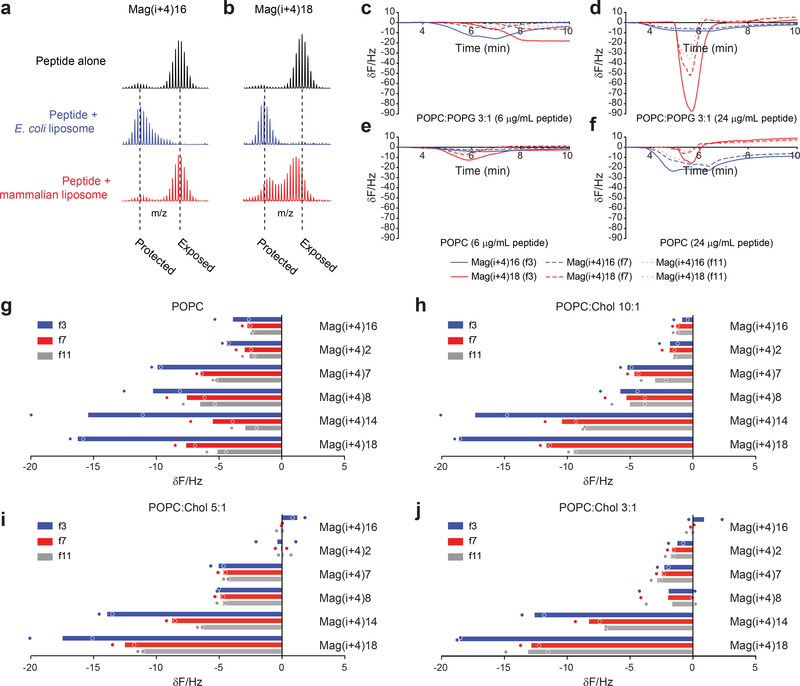
The consensus mechanism of Mag2-induced membrane lysis involves peptide folding into an active α-helical conformation upon exposure to negatively charged bacterial membranes and insertion as a transmembrane peptide-lipid complex that forms a toroidal pore1, 33, 34. Given the variation in bacterial and mammalian membrane lytic behavior among Mag(i+4) StAMPs, we sought to understand the mechanisms underlying the selectivity of exemplary StAMPs, such as Mag(i+4)16 and Mag(i+4)18, respectively. We hypothesized that differences in membrane tropism for StAMPs of different composition could explain their spectrum of membrane specificity. We evaluated this concept by studying the hydrogen-deuterium exchange mass spectrometry (HX-MS) profiles of StAMPs upon exposure to anionic and zwitterionic membranes that model the bacterial (e.g. E. coli) and mammalian contexts, respectively. HX-MS probes peptide structure by measuring the deuterium incorporation of exposed backbone amide hydrogen atoms35, 36. When Mag(i+4)16 was mixed with anionic liposomes composed of a 4:1 ratio of POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine):POPG (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol), marked protection from deuterium exchange was observed, as compared to the peptide in aqueous solution alone (Fig. 3a). Strikingly, when Mag(i+4)16 was instead mixed with zwitterionic liposomes composed of a 10:1 ratio of POPC:Cholesterol, no protection was observed. These data suggest that Mag(i+4)16 selectively interacts with anionic liposomes, consistent with the observed antimicrobial activity of Mag(i+4)16 in the absence of hemolysis. In contrast, Mag(i+4)18 was protected from deuterium exchange upon exposure to anionic liposomes and showed partial protection in the presence of zwitterionic liposomes (Fig. 3b), consistent with its indiscriminate lytic activity. Taken together, these data indicate that the capacity to selectively engage negatively charged membranes underlies the specificity of particular StAMPs for bacterial membranes.
Figure 3. Transmembrane insertion mechanism of bacterial-selective StAMPs.
a-b, StAMP-membrane interaction, as reflected by a decrease in deuterium exchange compared to the aqueous control condition, is anionic liposome (POPC:POPG)-selective for Mag(i+4)16 (a), but promiscuous for Mag(i+4)18 (b), which is at least partially protected by both anionic and zwitterionic (POPC:Cholesterol) liposomes. HX-MS experiments were performed three times independently with similar results. c-f, QCM sensorgrams demonstrate the changes in resonant frequency of SLBs mimicking bacterial membranes (POPC:POPG) upon exposure to 6 (c) and 24 (d) μg/mL of Mag(i+4)16 (blue) and Mag(i+4)18 (red) peptides, and mammalian membranes (POPC) upon exposure to 6 (e) and 24 (f) μg/mL of Mag(i+4)16 and Mag(i+4)18 peptides, across the third, seventh, and eleventh harmonics, which probe the outer leaflet lipid headgroup-water interface (solid line), middle of the bilayer (dashed line), and the inner leaflet headgroup (dotted line), respectively. For each condition, an exemplary sensorgram is shown for experiments performed independently two times with similar results. g-j, Measured changes in resonant frequency across the third (blue), seventh (red), and eleventh (black) harmonics in response to the indicated StAMP treatments (8 μg/mL) of SLBs composed of POPC (g) and increasing concentrations of cholesterol (h-j). Data are mean of two independent experiments (shown as dots).
To link the functional selectivities of StAMPs with peptide orientation within membranes, we applied quartz crystal microbalance (QCM) analyses to Mag(i+4)16 and Mag(i+4)18. We analyzed the responses of supported phospholipid bilayers (SLBs) to applied peptides across the spectrum of harmonics of the sensor crystal37–39. Changes in the recorded frequency correspond to induced alterations in membrane mass upon peptide interaction with the phospholipid bilayer, either as a result of peptide addition or lipid loss. The distribution of these changes across distinct harmonics with differing penetrating power informs the variable depths of the SLB, providing an indication of peptide orientation37. We used POPC:POPG (3:1; anionic) and POPC (zwitterionic) SLBs to simulate bacterial and mammalian membranes, respectively. Consistent with the HX-MS results, prominent changes in resonant frequency were observed for both StAMPs when applied to POPC:POPG SLBs at low and high concentrations (Fig. 3c–d). For the nonselective Mag(i+4)18, the magnitude of changes increased with distance from the chip surface in each condition (Fig. 3c–f), indicating a surface-bound orientation consistent with the carpet-like mechanism previously reported for certain AMPs37. In contrast, Mag(i+4)16 induced only very modest changes in resonant frequency at 6 μg/mL in the context of zwitterionic POPC SLBs, indicating little to no membrane interaction (Fig. 3e). At 24 μg/mL, Mag(i+4)16 displayed similar changes across all harmonics with a slightly larger mass increase in the third harmonic, indicating that at this high concentration the peptide was predominantly interacting in a transmembrane orientation and with some residual surface association (Fig. 3e–f). In the context of the anionic POPC:POPG SLBs, Mag(i+4)16 interacted in a similar manner at 6 μg/mL (Fig. 3c) to the profile observed in POPC SLBs at 24 μg/mL (Fig. 3e), but the changes in harmonics became homogeneous at 24 μg/mL (Fig. 3d), indicative of complete transmembrane orientation. Taken together, the QCM results demonstrate that the explicit composition of StAMPs not only affects membrane selectivity but the mode of membrane engagement, with bacterial-selective StAMPs preferentially manifesting transmembrane insertion, consistent with a barrel stave or toroidal pore mechanism40, rather than a surface-bound orientation characteristic of a carpet mechanism37.
We next applied QCM to determine if the observed variation among StAMPs in inducing hemolysis was also influenced by the rigidity of mammalian membranes. We tested a panel of StAMPs with progressively less membrane selectivity in SLBs composed of POPC and increasing amounts of added cholesterol to induce a gradient of membrane rigidities (among other effects of cholesterol)41. In the context of pure POPC SLBs, the greatest changes for nonselective StAMPs were observed in the third harmonic, again indicating a primarily surface-bound orientation. As the amount of cholesterol was increased, StAMPs displaying a transmembrane-like harmonics profile showed reduced interaction with POPC SLBs, whereas the most hemolytic, nonselective StAMPs, namely Mag(i+4)14 and 18, maintained membrane engagement (Fig. 3g–j). These data indicate that a robust hydrophobic network on the hydrophobic surface of StAMPs results in a carpet-like mechanism of interaction that is agnostic to the rigidity of mammalian membranes. In contrast, StAMPs with a reduced hydrophobic network that rely on a transmembrane-insertion mode of interaction demonstrate little to no engagement of zwitterionic membranes, particularly those with rigid phospholipid bilayers.
A Stable, Potent and Selective Double-Stapled StAMP
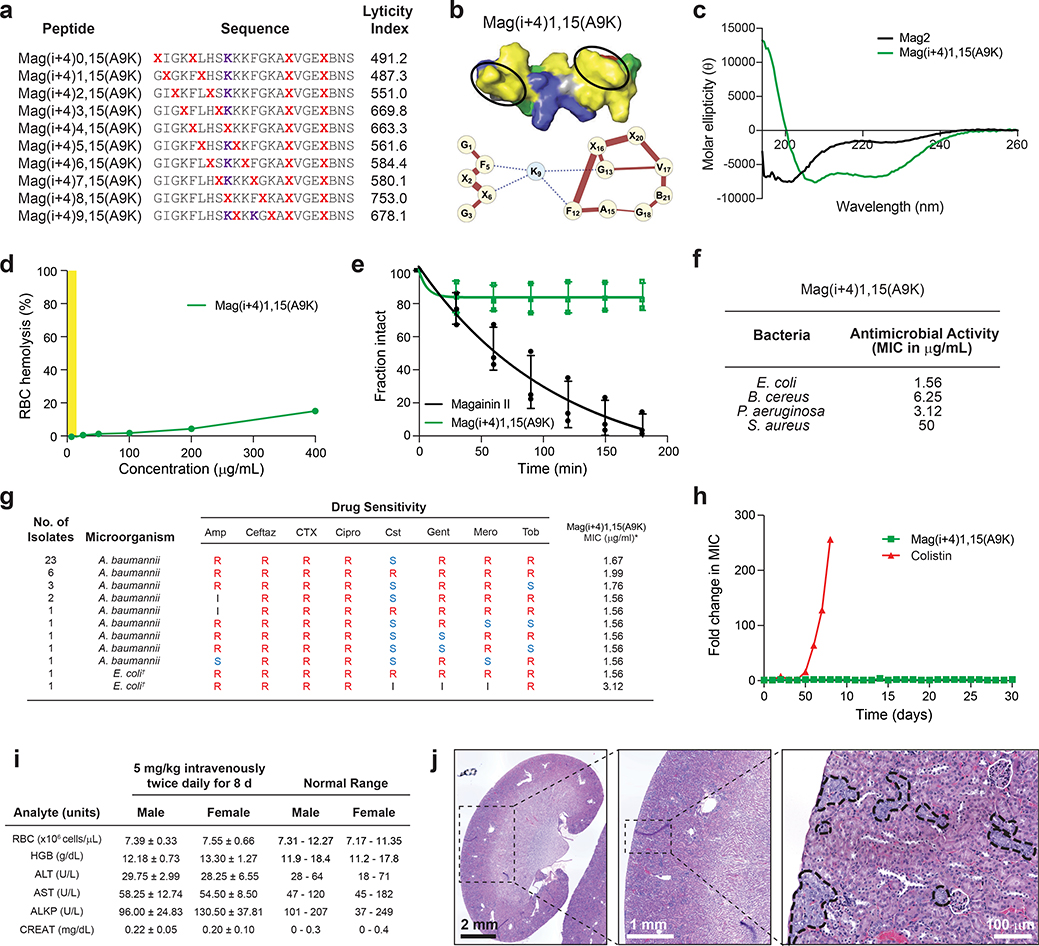
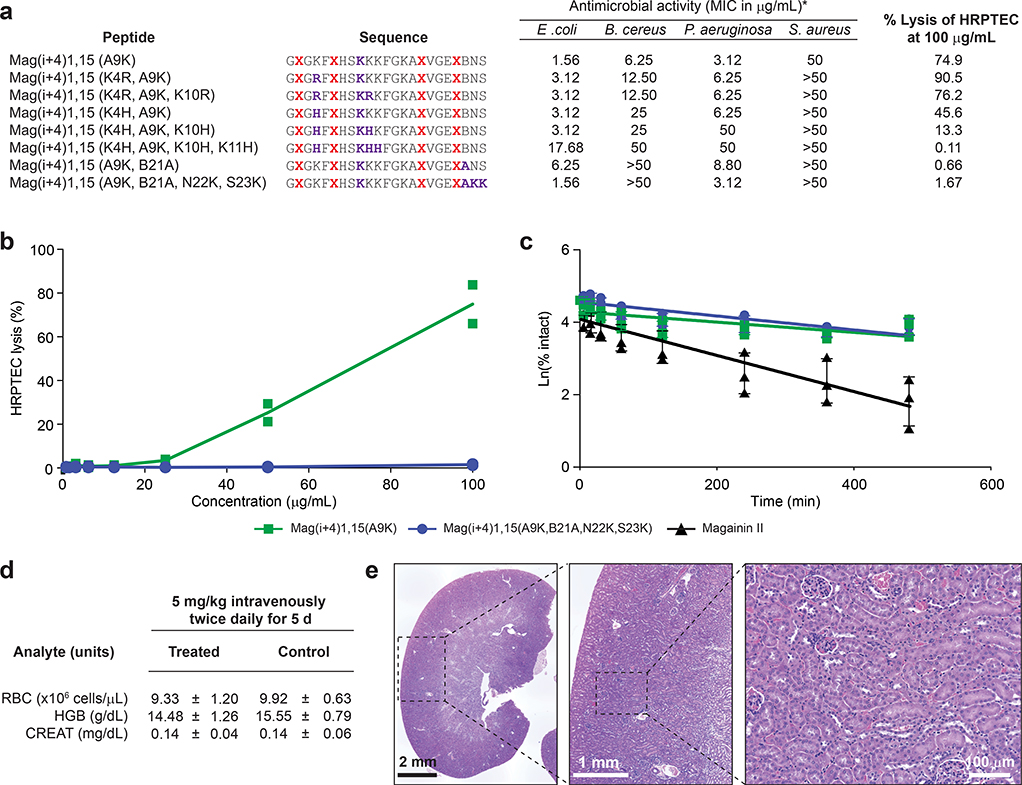
We applied our design algorithm and mechanistic findings to generate an optimized StAMP with features of α-helical structural stability, protease resistance, potent Gram-negative antimicrobial activity, and little to no treatment-induced hemolysis. We previously observed that insertion of two hydrocarbon staples into α-helical peptide sequences can maximize structural and proteolytic stability16, 24. Thus, we generated an in silico staple scanning library of Mag(i+4)15(A9K), an exemplary potent, Gram-negative selective StAMP, and chose the double-stapled peptide with the lowest LI, Mag(i+4)1,15(A9K), for synthesis and characterization (Fig. 4a–b). In contrast to the linear Mag2 peptide, the double-stapled Mag(i+4)1,15(A9K) peptide demonstrated a predominantly α-helical structure in solution as measured by circular dichroism spectroscopy (Fig. 4c) and exhibited >10 mg/mL solubility in saline solutions. In addition, insertion of the two i, i+4 staples conferred near complete protection of Mag(i+4)1,15(A9K) from broad-spectrum proteinase K digestion, whereas Mag2 was degraded over time such that no intact peptide was detectable after 3 hours of protease exposure (Fig. 4d).
Figure 4. Biophysical, biological and safety profile of a double-stapled StAMP.
a, HNM analysis of a second staple-scanning library of Mag(i+4)15(A9K) revealed a spectrum of lyticity values and predicted optimal properties for the double-stapled Mag(i+4)1,15(A9K) construct, which was then synthesized and characterized in a battery of biophysical and biological analyses. b, The HNM of Mag(i+4)1,15(A9K) demonstrates stronger hydrophobic interactions within the established N-terminal hydrophobic patch, as compared to Mag(i+4)15(A9K), but preserves the key disconnection between N- and C-terminal highly hydrophobic surfaces. c, Mag(i+4)1,15(A9K) (green) demonstrates stabilized α-helical structure in solution, whereas Mag2 (black) is a random coil, as assessed by circular dichroism spectroscopy performed two times independently with similar results. d, Data showing the proteolytic degradation of Mag2 (black) versus the proteolytic resistance of Mag(i+4)1,15(A9K) (green) upon exposure to proteinase K. Data shown are the mean ± s.d. of three independent experiments (shown as dots). e, Mag(i+4)1,15(A9K) demonstrates potent antimicrobial activity against Gram-negative pathogens, such as E. coli and P. aeruginosa, with MIC values of <5 μg/mL. The MIC is a geometric mean of four independent experiments. f, Mag(i+4)1,15(A9K) shows no RBC hemolytic activity across a broad dose-effective range, and even when dosed as high as 400 μg/mL exhibited <20% hemolysis. RBC hemolysis testing was performed two times independently with similar results. The Gram-negative bactericidal dosing range is highlighted in yellow. g, Data showing antimicrobial potency of Mag(i+4)1,15(A9K) against multidrug-resistant Gram-negative clinical isolates from the United States Centers for Disease Control and Prevention, including colistin-resistant A. baumannii and MCR-1 plasmid-positive E. coli. S, susceptible; I, intermediate; R, resistant; Amp, ampicillin; Ceftaz, ceftazidime; CTX, ceftriaxone; Cipro, ciprofloxacin; Cst, colistin; Gent, gentamicin; Mero, meropenem; Tob, tobramycin. †MCR-1 plasmid carrier. h, Exposure of a colistin- and Mag(i+4)15(A9K)-sensitive strain of A. baumannii to sub-MIC dosing of the corresponding antibiotics for 30 d led to colisitin resistance within 7 d of exposure but persistent susceptibility to Mag(i+4)15(A9K). The experiment was performed two times independently with similar results. i, Data show the results of RBC parameters, and liver and kidney function testing, of mice treated with Mag(i+4)15(A9K) at an intravenous dose of 5 mg/kg administered twice daily for 8 d. Data are mean ± s.d. for testing performed on n=8 mice (4 male, 4 female). j, Data showing H&E stained kidney sections from a representative mouse of n=8 treated with Mag(i+4)15(A9K) at a dose of 5 mg/kg intravenously twice daily for 8 d. The tissue region demarcated by the dotted square is successively enlarged in the panel to the right. Focal areas of tubular degeneration are outlined in black in the rightmost panel.
In terms of antimicrobial potency, the double-stapled Mag(i+4)1,15(A9K) StAMP had a twofold and fourfold improved potency compared to the single-stapled Mag(i+4)15(A9K) against E. coli (1.56 μg/mL) and P. aeruginosa (3.12 μg/mL), respectively (Fig. 2d, 4e). Despite the enhanced lytic activity toward Gram-negative bacteria, no RBC lysis was observed in the dose-effective range (Fig. 4f). Even at a dose of 400 μg/mL (~160 μM), which is 250 and 130 times greater than the MICs for E. coli and P. aeruginosa respectively, there was less than 20% hemolysis, reflecting a potentially unprecedented membrane-specificity window for cationic α-helical AMPs. Correspondingly, HX-MS analysis showed strong interaction selectivity for anionic liposomes (Supplementary Fig. 6a) and the QCM results demonstrated homogenous changes across all harmonics in anionic POPC:POPG SLBs upon treatment with Mag(i+4)1,15(A9K) at low and high concentrations, indicative of transmembrane insertion (Supplementary Fig. 6b–c). In contrast, essentially no interaction was observed between zwitterionic POPC SLBs and Mag(i+4)1,15(A9K), even upon high dose treatment (Supplementary Fig. 6d–e) and across the spectrum of membrane rigidities (Supplementary Fig. 6f). These data provide mechanistic corroboration for the observed functional specificity of Mag(i+4)1,15(A9K) in lysing bacterial membranes and again support a transmembrane insertion model for the membrane-disruptive activity of bacterial-selective StAMPs.
Safety profile of a lead StAMP that kills MDR bacteria
The optimized profile of Mag(i+4)1,15(A9K) motivated us to advance this lead compound to broader testing in multidrug-resistant (MDR) Gram-negative clinical isolates. Mag(i+4)1,15(A9K) consistently exhibited potent activity against a variety of Gram-negative pathogens regardless of their drug resistance patterns (Supplementary Table 2). In addition, Mag(i+4)1,15(A9K) was effective against 39 clinical isolates of A. baumannii, including strains with prominent colistin resistance42, 43, and two E. coli strains that harbor the mobilized colistin resistance (MCR-1) plasmid42 (Fig. 4g). One of the potential benefits of the membrane-lytic mechanism of StAMPs is avoidance of acquired resistance, as the target is the bacterial membrane itself rather than a particular macromolecule. We evaluated acquired resistance of A. baumannii to colistin or Mag(i+4)1,15(A9K) during serial passaging at sub-MIC dosing over a 30-day period. While colistin resistance developed within one week, StAMP sensitivity was unaltered for the duration of the experiment (Fig. 4h). In addition, the induced colistin-resistant A. baumannii strain showed no cross-resistance to Mag(i+4)1,15(A9K) (MIC unchanged at 1.56). These results indicate that the unique membrane-lytic mechanism of StAMPs may indeed offer an advantage of avoiding drug resistance.
Because major roadblocks in transforming AMPs into antibiotics for internal use have included RBC hemolysis and kidney damage4, limiting their testing in clinical trials to topical administration only44, we performed both dose-finding and toxicity testing of Mag(i+4)1,15(A9K) in mice. Using a regimen of 5 mg/kg administered intravenously twice daily for 8 d, we observed no change in body weight among male or female mice (Supplementary Fig. 7) and the animals maintained RBC, liver, and kidney parameters within the normal range (Fig. 4i). No gross tissue abnormalities were observed upon necropsy other than variable inflammation at the sites of repeated tail vein injections. The only notable finding on histologic analysis was evidence of mild-to-moderate tubular degeneration of the kidneys in particular mice, which was seen on repeat evaluation of mice treated with the same regimen for 5 d (Fig. 4j, Supplementary Fig. 8). Although only a subclinical finding with no observed effect on serum creatinine, we sought to optimize Mag(i+4)1,15(A9K) further with the goal of eliminating the potential for kidney injury.
A further optimized StAMP for preclinical testing
Given the longstanding association of cationic antibiotics with kidney toxicity45, we generated a series of point-mutant analogs of Mag(i+4)1,15(A9K) to variably modulate hydrophobicity and/or the nature of cationic charge, as these factors have been shown to contribute to the renal toxicity of cationic antibiotics like colistin46, 47 (Fig. 5a). We then screened this library of StAMPs for lysis of human renal proximal tubular epithelial cells by lactate dehydrogenase (LDH) release assay. While Mag(i+4)1,15(A9K) demonstrated little to no hemolytic activity across a broad dosing range (Fig. 4f), this StAMP did induce lysis of the more sensitive HRPTECs at doses above 20 μg/mL (Fig. 5b). However, upon reducing the hydrophobicity by mutating norleucine (B; natively methionine, M) 21 to alanine, and then installing compensatory C-terminal positive charges (N22K and S23K) to restore antimicrobial potency48, HRPTEC toxicity was now effectively eliminated in addition to the absence of RBC hemolysis, yielding Mag(i+4)1,15(A9K,B21A,N22K,S23K) (Fig. 5a, b, Supplementary Fig. 9). We also found that both double-stapled StAMPs were markedly more stable in mouse plasma compared to the linear Mag2 template (Fig. 5c), consistent with the striking protease resistance observed in vitro upon introducing double staples (Fig. 4d). The proteolytic sensitivity (Fig. 4d) and plasma instability (Fig. 5c) of Mag2 necessitated intraperitoneal dosing to most fairly compare the in vivo activity of our lead StAMP to Mag2. First, we established the StAMP dose required to cure at least 75% of immunocompromised mice infected with A. baumannii (ATCC 19606) in a peritonitis-sepsis model (n=8 per arm)49. Mag(i+4)1,15(A9K,B21A,N22K,S23K) was capable of curing 88% of mice after intraperitoneal administration of only two 5 mg/kg doses (Supplementary Fig. 10a). We then applied this dosing regimen in the corresponding model using a colistin-resistant MDR strain of A. baumannii (AR-0303; Supplementary Table 2) and observed 75% survival with StAMP treatment and a 75% death rate for Mag2 by 24 h, with all vehicle-treated mice having succumbed to infection by 12 h (Supplementary Fig. 10b). These data highlight the potential therapeutic advantage of stapled vs. linear AMPs.
Figure 5. Optimized properties of a lead StAMP for preclinical development.
a, The antimicrobial activity (MIC in μg/mL) and percentage of LDH release from HRPTEC cells at 100 μg/mL were determined for a mutant panel of Mag(i+4)15(A9K)-based StAMPs. The MIC is a geometric mean of two independent experiments. Percent LDH release (lysis) data are the mean of two independent experiments. b, Comparative LDH release assay demonstrating elimination of HRPTEC lysis upon B21A/N22K/S23K mutagenesis of Mag(i+4)15(A9K). Data are the mean of two independent experiments (shown as dots). c, Plasma stability testing revealed that double-stapling increased the plasma half-life of the unstapled template peptide, Mag2, from 2 h to greater than 6 and 8 h for Mag(i+4)15(A9K) and Mag(i+4)15(A9K,B21A,N22K,S23K), respectively. Data are mean ± s.d. of three independent experiments (shown as dots). d, RBC parameters and serum creatinine levels for CD-1 mice treated with Mag(i+4)15(A9K,B21A,N22K,S23K) at an intravenous dose of 5 mg/kg administered twice daily for 5 d. Data are mean ± s.d. for testing performed on n=8 female mice. e, H&E stained kidney sections from a representative mouse of n=8 treated with Mag(i+4)15(A9K,B21A,N22K,S23K) at an intravenous dose of 5 mg/kg administered twice daily for 5 d revealed normal tissue and cellular architecture. The tissue region demarcated by the dotted square is successively enlarged in the panel to the right.
We next investigated whether Mag(i+4)1,15(A9K,B21A,N22K,S23K) retained the benign toxicity profile of Mag(i+4)1,15(A9K) and remedied the observed histopathologic changes of the kidney, as predicted by the HRPTEC assay (Fig. 5a, b). Upon repeating the in vivo toxicity analyses using the further optimized double-stapled StAMP, we again observed no change in weight (Supplementary Fig. 11), RBC parameters, or renal function tests (Fig. 5d), and observed no evidence of renal tubule degeneration (Fig. 5e, Supplementary Fig. 12). Taken together, these data indicate that peptide stapling can be effectively applied to generate structurally-reinforced, protease-resistant, bacterial membrane-selective, and plasma-stable Mag2-based StAMPs that can kill even the most resistant Gram-negative bacteria and be administered internally without the toxic side effects that have long thwarted the clinical translation of AMPs.
The design algorithm yields diverse StAMP prototypes
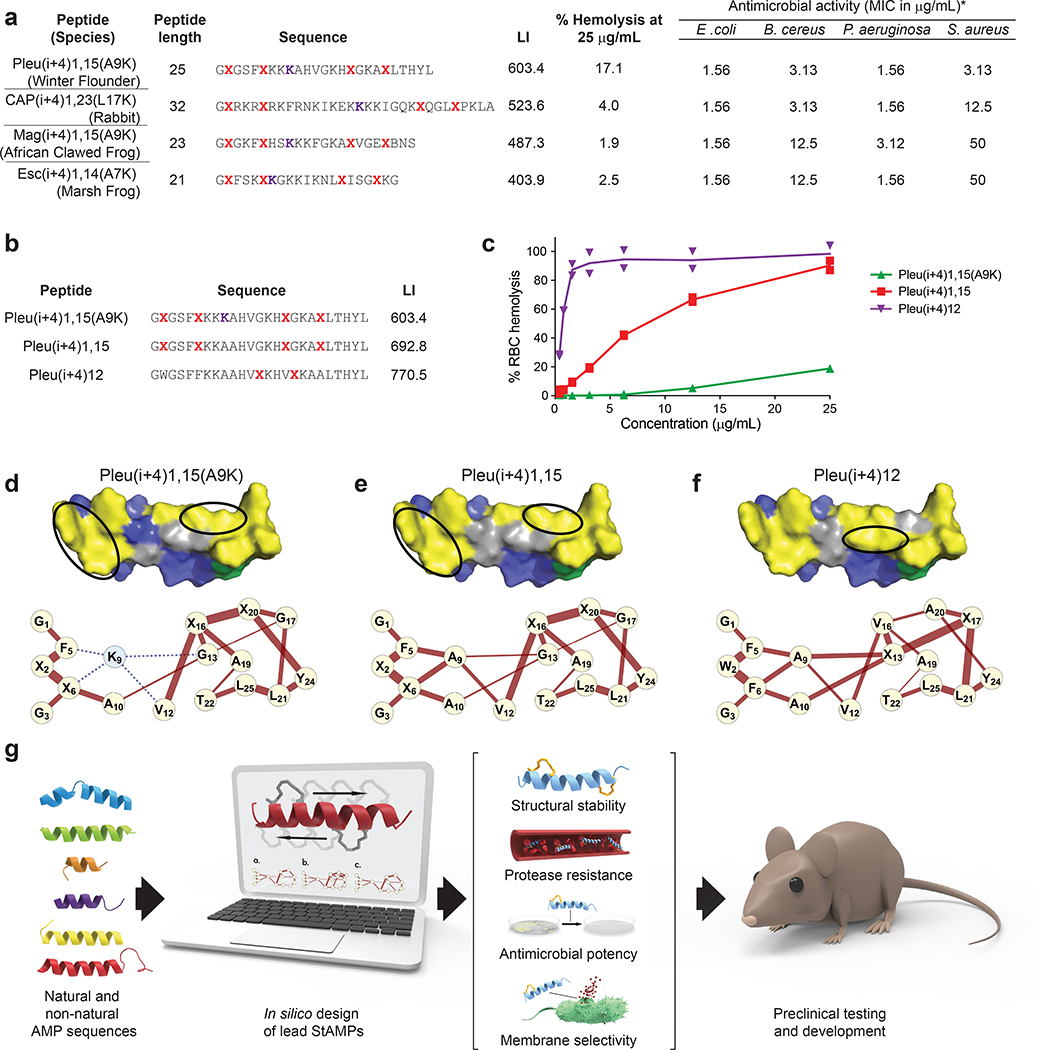
Having derived design principles and a predictive algorithm for generating bacterial membrane-selective StAMPs modeled after Mag2, we investigated whether our computational tool could be applied to rapidly provide lead StAMPs for preclinical development of diverse template sequences. We analyzed AMPs spanning different species (winter flounder, rabbit, marsh frog), peptide lengths (21–32 amino acids), and sequence compositions, and for each template synthesized StAMPs with the lowest predicted LI. In each case, the in silico-designed double-stapled StAMP exhibited RBC hemolytic activity below 20% at 25 μg/mL dosing and potent Gram-negative antimicrobial activity with an MIC range of 1.56–3.13 μg/mL (Fig. 6a).
Figure 6. Computational design of diverse membrane-selective StAMPs.
a, The antimicrobial activity (MIC in μg/mL) and percentage of RBC hemolysis at 25 μg/mL were determined for a series of pleurocidin (pleu), CAP18 (CAP), and esculentin (Esc)-based double-stapled StAMPs predicted to have low mammalian membrane lyticity based on the computational design algorithm. The MIC is a geometric mean of two independent experiments. The data for percentage hemolysis are the mean of three independent experiments. b, Sequence composition and LIs of single- and double-stapled pleurocidin-based StAMPs. c, Comparative dose-responsive hemolysis of pleurocidin StAMPs, highlighting the striking differences between compositions as predicted by the corresponding LIs in b. Data are the mean of two independent experiments (shown as dots). d-f, Surface views of the distinct hydrophobic faces of pleurocidin StAMPs and the corresponding hydrophobicity network maps (HNMs). Staple locations for each StAMP are indicated by a black oval. g, Workflow for the computational design of lead StAMPs for preclinical testing and clinical translation.
We then compared the hemolytic activity of pleuricidin-based StAMPs that were predicted to have a range of mammalian membrane lyticity and again observed excellent corroboration between the LI and the observed RBC hemolysis (Fig. 6b–c). As for Mag2 StAMPs, the distinct HNMs for each pleuricidin StAMP explain how the strength and extent of the hydrophobic network, and its interruption by a single polar residue, dictate the risk of mammalian membrane lysis (Fig. 6d–f), underscoring the fundamental design principle that the distribution rather than absolute level of hydrophobicity accounts for non-specific membrane toxicity. Indeed, a single staple that bridges and strengthens the hydrophobic network is more deleterious than installing two hydrophobic staples (Fig. 6e–f). Overall, the algorithm reproducibly produces bacterial-selective StAMP prototypes based on diverse AMP sequences, allowing for the rapid design and testing of a myriad of preclinical leads with potent antimicrobial activity (Fig. 6g).
DISCUSSION
There is an urgent need for new antibiotics that can eradicate MDR Gram-negative bacteria50, 51. Because resistance mechanisms rapidly emerge to specific drug targets and the outer membrane of Gram-negative bacteria can also block drug entry,52 the membrane-lytic mechanism of natural AMPs is a potentially promising therapeutic alternative53. However, the clinical translation of AMPs has been hampered by peptide instability and toxicity associated with mammalian membrane lysis,54 with indications even limited to topical use26, 44 having failed Phase II and III testing15, 55. Here, we applied all-hydrocarbon stapling to discern and encode design principles for the selective targeting of bacterial membranes, yielding computationally generated StAMP prototypes that are stable, protease-resistant, active against MDR bacteria, and capable of treating infection in mice without causing hemolytic or renal toxicity.
Cationic AMPs are typically unstructured in solution, and thus vulnerable to proteolysis, because alignment of positive charge upon α-helical folding is disfavored owing to charge repulsion until counter-balanced by the anionic complementarity of bacterial membranes1. Peptide stapling is an attractive platform for AMP-based antibiotic development because reinforcement of bioactive α-helical structure also renders the twisted amide bonds as poor substrates for proteases.16–18 Prior studies of stapled AMP peptides involved limited sampling of staple type and position within various template sequences, leading to mixed results and nonspecific membrane toxicity23–28. By applying a library approach, we determined that staples incorporated into established regions of hydrophobicity avoid conferring undesirable lytic properties. Further, because StAMPs are predominantly prefolded in solution, the nature of the hydrophobic interaction with mammalian membranes, as reflected by the three-dimensional spatial distribution and strength of the peptide’s hydrophobicity network, is predictable, enabling the development of HNMs and an index of lyticity that is generalizable and clinically relevant.
Applying the design algorithm, we rapidly identified Mag(i+4)1,15(A9K) as a selective StAMP candidate and demonstrated that it had very little hemolytic activity, lysed bacteria by transmembrane insertion, avoided proteolytic degradation, did not induce bacterial resistance, and was tolerated in mice using a twice daily intravenous dosing regimen. Fine tuning of this StAMP by point mutagenesis and testing in HRPTEC LDH release assays, yielded Mag(i+4)1,15(A9K,B21A,N22K,S23K), which did not elicit even subclinical, microscopic kidney tubule injury, providing an expanded therapeutic window for internal use. We applied our algorithm to three additional diverse AMP sequences, and generated StAMP prototypes with potent antimicrobial activity, little to no hemolytic toxicity, and without the need to synthesize or screen a library of analogs. We hope that our results will inspire the advancement of StAMPs as a novel class of antibiotics to combat MDR infections.
ONLINE METHODS
Solid phase peptide synthesis
Fmoc-based solid-phase peptide synthesis was used to synthesize StAMPs in accordance with our reported methods for generating all-hydrocarbon stapled peptides56. To achieve the various staple lengths, α-methyl, α-alkenyl amino acids were installed at i, i+4 positions using two S-pentenyl alanine residues (S5) and at i, i+7 positions by inserting R-octenyl alanine (R8) at the i position and S5 at the i+7 position. For the stapling reaction, Grubbs 1st generation ruthenium catalyst dissolved in dichloroethane was added to the resin-bound peptides. To ensure maximal conversion, 3–5 rounds of stapling were performed. The peptides were then cleaved off of the resin using trifluoroacetic acid, precipitated using a hexane:ether (1:1) mixture, air dried, and purified by LC-MS. All peptides were quantified by amino acid analysis. TFA-HCl exchange was performed on the peptides used in animal studies at the Dana-Farber Cancer Institute. Peptide compositions and their experimental applications are listed in Supplementary Table 3.
Bacterial specimens
StAMPs were screened against a panel of bacteria that included E. coli (ATCC 25922), B. cereus (ATCC 14579), P. aeruginosa (ATCC 27853), S. aureus (ATCC 25923), and A. baumannii (ATCC 19606). MDR Gram-negative bacteria (strains RB001, RB002, RB019, RB020, RB040, RB013, RB197, RB206) were provided by Drs. Deborah Hung and Roby Bhattacharya of the Broad Institute (Cambridge, MA). MDR A. baumannii and MCR-1 plasmid-carrying E. coli panels were obtained from the Antimicrobial Resistance Isolate Bank of the Centers for Disease Control and Prevention.
Microbroth dilution assay
Bacterial cells were grown overnight at 37°C in Mueller-Hinton broth (MHB), minimally refined by anion exchange (Q Sepharose Fast Flow) to both ensure peptide solubility and maintain comparable bacterial growth, and then diluted and allowed to grow for an additional 5 hours. Serial dilutions of peptide stocks in water (10 μl) were prepared using clear round-bottom polypropylene 96-well plates, and 90 μl of bacteria in refined MHB were added to yield a final inoculum of 5×105 CFU/ml. The plates were then covered with porous tape to reduce evaporation, and incubated for 20–24 hours at 37°C. The reported MIC is the minimum peptide concentration at which no visible growth was observed.
RBC hemolysis assay
Human whole blood specimens (Innovative Research) were centrifuged to isolate RBCs that were then washed and suspended in phosphate-buffered saline to yield a 1% (v/v) suspension. The suspension was added to serial dilutions of peptide stocks in water in clear round-bottom polypropylene 96-well plates and the plates incubated for 1 h at 37°C. The plates were then centrifuged and the supernatant isolated to determine the amount of hemoglobin released using a spectrophotometer (570 nm). The percentage of hemolysis was calculated as: ([Treated Absorbance – Untreated Control Absorbance] x 100)/(1% Triton X-100 Treated Absorbance – Untreated Control Absorbance).
Circular dichroism spectroscopy
Peptides were dissolved in either 5 mM potassium phosphate solution at pH 7 or distilled H2O to achieve concentrations of 25–50 μM. Circular dichroism (CD) spectra were obtained on a spectropolarimeter (Aviv) using standard measurement parameters of temperature, 37°C; wavelength, 190–260 nm; step resolution, 0.5 nm; speed, 20 nm/min; accumulations, 10; response, 1 sec; bandwidth, 1 nm; path length, 0.1 cm. The α-helical content of each peptide was calculated by dividing the mean residue ellipticity by the reported value for a model helical decapeptide57.
Lyticity index and hydrophobicity network map algorithm
The LI was calculated according to the formula below, in which the summation is computed over all hydrophobic amino acids:
Built-in assumptions include (1) the peptide has α-helical structure, (2) hydrophobic amino acids within this structure interact to form a network, (3) each vertex is a hydrophobic amino acid (Hi) and the edges are formed by the interactions with the structurally closest hydrophobic amino acids (i, i+3 and i, i+4 relationships), (4) each edge is weighted by summing the hydrophobicity of the amino acid vertices that form the edge. The hydrophobicity values (Hi) employed were based on Kovacs et al. (Table III, Column 4)58. The stapling amino acid S-pentenyl alanine was assigned an Hi of 29.5. In the resultant network plot for each peptide, named the HNM, vertices represent the hydrophobic amino acids and the thickness of the edges are proportional to the weight and contribution of each network edge to the lyticity index (see also Supplementary Fig. 3). A user interface for this algorithm is available as a web application at http://www.walenskylab.org/HNM.
Hydrogen-deuterium exchange mass spectrometry
Deuterium exchange occurred at the MIC for each StAMP peptide (Fig. 1a). Peptide stock solutions at 64x MIC were prepared in liposome buffer (10 mM sodium phosphate pH 7.4, 100 mM NaCl, H2O). A total of one μL of each peptide stock solution was added to 3 μL of liposome buffer containing the anionic, bacterial-mimetic liposomes (POPC:POPG; 4:1) or zwitterionic, mammalian-mimetic liposomes (POPC:Chol; 10:1), which were preequilibrated at 21°C for 5 min before the addition of peptide. The resultant mixture was incubated for an additional 5 min at 21°C to allow for peptide-liposome association. Deuterium labeling was initiated by adding 60 μL (15-fold) labeling buffer (10 mM sodium phosphate pD 7.4, 100 mM NaCl, D2O). After 10 sec, the reaction was quenched with ice-cold acidic buffer (100 mM potassium phosphate pH 2.5) and then 400 pmoles of peptide was immediately injected for mass analysis. Undeuterated controls for each StAMP were prepared as described above except for 15-fold dilution into liposome buffer instead of labeling buffer. Quenched samples were injected onto a 2 × 20 mm analytical guard column (Alltech) packed in-house with POROS 20-R2 reversed-phase media. Desalting was performed for 10 sec by manually flushing 500 μL of 0.05% TFA over the trapping column. The injection valve, column, and all associated tubing were submerged in ice to minimize back-exchange. The peptides were eluted from the column using a Shimadzu prominence HPLC (LC-20AD) with a 3 min 15–98% acetonitrile gradient at a flow rate of 50 μl/min. Eluent was directed into a Waters XevoG2 mass spectrometer equipped with a standard ESI source for mass analysis. Raw spectra from a 10 m/z window for the +3 charge state for each peptide were processed with HXExpress 3.0 software59 to determine deuterium incorporation.
Quartz crystal microbalance
Lipids were purchased from Avanti Polar Lipids (Alabaster, AL), including POPC, POPG and cholesterol. The lipids were dissolved to achieve a concentration of 10 mg/mL in CHCl3:MeOH (v/v 3:1) and stored at 20°C until use for up to six months. QCM experiments were carried out at room temperature on an E4 QCM-D system using 5 MHz SiO2 QCM-D chips (Q-sense AB; Västra Frölunda, Sweden). Small unilamellar vesicles of various compositions were prepared from lipid films by suspension in 20 mM Tris containing 150 mM NaCl and 5 mM MgCl2 (pH 7.4) at a concentration of 0.2 mg/mL and then sonicated until a clear solution was formed. After flowing 2 mL Tris buffer over the QCM chips at a flow rate of 250 μL/min, and attaining a stable F and D reading across all harmonics, the small unilamellar vesicle suspension was flowed across the chips at a rate of 100 μL/min for 3 min or until a stable SLB had formed, as determined by a constant F and D value. Buffer was then flowed across the chips for 1 min at 100 μL/min to remove any unbound lipid. StAMPs were then flowed over the chips at the indicated concentrations at a flow rate of 100 μL/min. After 5 min, the flow was stopped for 10 min, and then 2% SDS, EtOH and H2O were sequentially flowed over the chip at a flow rate of 1 mL/min to remove peptide and bound lipid from the surface of the chip. Data were analyzed using the Qtools (Q-Sense) software package.
Protease resistance assay
Proteolytic digestion was measured by LC-MS (Agilent 1260) using the following parameters, as reported20: 20 μl injection, 0.4 ml flow rate, 15 min run time consisting of a gradient of water (0.1% trifluoroacetic acid) to 15%−75% acetonitrile (0.1% trifluoroacetic acid) over 10 min, 4 min wash to revert to starting gradient conditions, and 0.5 min post-time. The diode array detector signal was set to 210 nm with an 8 nm bandwidth and mass selective detector set to scan mode with one channel at (M+2H)/2, ± 1 mass units, and the other at (M+3H)/3, ± 1 mass units, where M is mass and H is the hydrogen ion. Reaction samples were composed of 50 μl peptide in 50 mM Tris HCl buffer (pH 7.4, 2 mg/mL stock) and 150 μl of 50 mM Tris HCl buffer (pH 7.4). Upon injection of the 0-hour time sample, 2.5 μl of 4 ng/μl proteinase K (Roche) was added, and the amount of intact peptide quantitated by serial injection over time. The mass selective detector area versus time plots gave an exponential decay curve that was derived by nonlinear regression analysis using Prism software (GraphPad).
In vitro resistance acquisition
Sequential culturing of A. baumannii (ATCC 19606) in the presence of sub-inhibitory levels of antimicrobial treatment was conducted over a period of 30 d. Bacteria were grown in 1 mL of refined MHB containing either Mag(i+4)1,15(A9K) or colistin at 0.25x, 0.5x, 1x, 2x and 4x MIC concentrations. Every 24 hours, cultures from the second highest concentration that allowed for growth were diluted 1:100 into fresh media containing the above concentrations. Dosing was incrementally increased if the elevation in MIC was confirmed by broth microdilution after passaging overnight in drug-free media.
Cell culture
HRPTECs (SA7K Clone, Sigma) were maintained in MEM (Invitrogen) supplemented with RPTEC Complete Supplement (Sigma). Cells were verified to be mycoplasma-free using the MycoAlert mycoplasma detection kit (Lonza Biologics Inc).
HRPTEC Lysis Assay
HRPTECs were plated in 96-well format (2 × 104 cells per well), and after overnight incubation, full media was replaced with MEM (Invitrogen) supplemented with RPTEC Tox Supplement (Sigma). Serial dilutions of StAMPs from a 4 mg/mL water stock, or vehicle, were added to the HRPTECs in a final volume of 100 μL. After incubating at 37 °C for 90 min, 50 μL of cell culture media was transferred to a clear plate (Corning), incubated with 50 μL of LDH reagent (Roche) for 10 min, and absorbance measured at 490 nm on a microplate reader (SpectraMax M5 Microplate Reader, Molecular Devices).
Plasma stability testing
Peptide stability was tested in freshly drawn mouse plasma collected in Lithium heparin tubes. Triplicate incubations were set up with 500 μl of plasma spiked with 10 μM of the individual peptides. Samples were gently shaken in an orbital shaker at 37° C and 25 μl aliquots were removed at 0, 5, 15, 30, 60, 240, 360, and 480 minutes and added to 100 μl of a mixture containing 10% methanol:10% water:80% acetonitrile to stop further degradation of the peptides. The samples were allowed to sit on ice for the duration of the assay and then transferred to a MultiScreen Solvinert 0.45 μm low binding hydrophilic PTFE plate (Millipore). The filtrate was directly analyzed by LC-MS/MS. The peptides were detected as double or triple charged ions using the mass transition of 840.4 to 834 for Mag(i+4)1,15(A9K), 507.1 to 84 for Mag(i+4)1,15(A9K,B21A,N22K,S23K), and 617.4 to143.1 for Mag2 using a Sciex 5500 mass spectrometer. The percentage of remaining peptide was determined by the decrease in chromatographic peak area and log transformed to calculate the half-life.
In vivo toxicity analyses
Eight CD-1 mice (4 male, 4 female) were dosed at 5 mg/kg twice daily for 8 d by intravenous bolus injection of Mag(i+4)1,15(A9K) dissolved in saline at a concentration of 0.5 mg/mL. The second daily dose was administered approximately 8 h after the first dose. On day 9, blood was withdrawn for evaluation of RBC parameters and serum chemistries, and the animals euthanized for necropsy and histologic analysis. Harvested tissues were embedded in paraffin, sectioned, mounted on glass slides and stained with hematoxylin and eosin. The study was conducted in compliance with all applicable ethical regulations by Charles River Laboratories under Protocol No. 20102661 that was approved by the Charles River Testing Facility’s Institutional Animal Care and Use Committee. Mag(i+4)1,15(A9K) and Mag(i+4)1,15(A9K, B21A,N22K,S23K) toxicity were also tested in CD-1 mice (female, n=8 per arm) by dosing at 5 mg/kg twice daily for 5 days by intravenous bolus injection of the peptide dissolved in saline at a concentration of 0.5 mg/mL. Saline injections were performed for the vehicle control arm. On day 6, blood was withdrawn for evaluation of RBC parameters and serum creatinine, and the animals euthanized for necropsy and histologic analysis. Harvested tissues were embedded in paraffin, sectioned, mounted on glass slides and stained with H&E. The study was conducted in compliance with all applicable ethical regulations at the Dana-Farber Cancer Institute under Protocol No. 06–004, which was approved by the Dana-Farber Institutional Animal Care and Use Committee.
Peritonitis-sepsis model in immunocompromised mice
Female C57BL/6 mice (n=8 per arm) were rendered neutropenic by intraperitoneal administration of cyclophosphamide at 150 mg/kg and 100 mg/kg doses on day 1 and day 4 of the study, respectively. On day 5, 107 colony-forming units of an A. baumannii strain (ATCC 19606 or AR-0303) were injected into the peritoneal cavity (left side) to initiate infection. Mice were treated with vehicle (saline), Mag(i+4)1,15(A9K, B21A,N22K,S23S), or Mag2 by IP injection (right side) at 0 and 2h post-infection. Mice were monitored for 24–48 h and euthanized promptly if they became moribund. This study was conducted in compliance with all applicable ethical regulations at the Dana-Farber Cancer Institute under Protocol No. 06–004, which was approved by the Dana-Farber Institutional Animal Care and Use Committee.
Statistics
Linear regression analyses for StAMP biophysical parameters vs. hemolytic activity, nonlinear regression analyses for the protease resistance assay, and calculations of mean ± s.d. for hemolysis and mouse blood testing parameters were performed using Prism software (Graphpad, v. 7.04). MICs were calculated as geometric means of independent experiments using Excel software (Microsoft, v. 2016). The nonlinear regression and R2 calculations for the LI vs. hemolytic activity were performed using Mathematica (WolframAlpha). For the murine studies, statistical significance was calculated by unpaired two-tailed Student’s t-test (animal weights and blood parameters) and the log rank test (Kaplan-Meier survival curve analysis) to assess the entire survival distribution. Exact binomial power calculations for the comparisons of overall survival indicate that with 8 mice per treatment group, there was 80% power to distinguish 85% overall survival (OS) from 15% OS, testing at the 0.05 one-sided significance level. In addition, 8 mice per group provide 86% power to identify a 1.25 SD difference between groups in quantitative parameters such as body weights and blood parameters.
DATA AVAILABILITY STATEMENT
The datasets generated for the current study are included in the published article and its supplementary information. Requests for any further information can be made to the corresponding author.
CODE AVAILABILITY STATEMENT
The LI and HNM user interface, and the associated source code, can be accessed at www.walenskylab.org/HNM/ and is subject to a user agreement (Supplementary Note). The content and design of the LI and HNM user interface are protected by US and international copyright laws. The LI and HNM user interface or any portion thereof may not be reproduced, distributed, altered or used in any manner, beyond the user’s personal, academic and noncommercial use under the user agreement, without prior written consent from the Dana-Farber Cancer Institute. ‘Dana-Farber Cancer Institute’ is a service mark of Dana-Farber Cancer Institute, Inc., 450 Brookline Avenue, Boston, MA 02215. Inquiries regarding further information on a permitted use or a license to use any content should be addressed to the Office of General Counsel, BP 376, Dana-Farber Cancer Institute, 450 Brookline Avenue, Boston, MA 02215, USA.
Supplementary Material
ACKNOWLEDGMENTS
We thank D. Hung and R. Bhattacharyya for providing clinical strains of multidrug-resistant bacteria, K. Kean and P. Barendse for coding assistance, K. Hanford and M. Godes for technical support, A. Watts for helpful discussions, M. Cameron for plasma stability analyses, S. Breegi, C. Sypher and the Dana-Farber Cancer Institute Animal Resource Facility for technical assistance with in vivo studies, D. Neuberg for biostatistical support, and R. Bronson for histopathology services and expert slide review. We are also grateful to the Centers for Disease Control and Prevention (CDC) Antimicrobial Resistance Isolate Bank, which provided access to panels of multidrug-resistant bacteria. This research was supported by NIH grant R35CA197583 and a Leukemia and Lymphoma Society (LLS) Scholar Award to L.D.W. Additional support to L.D.W. was generously provided by the LaTorre Family, the Wolpoff Family Foundation, and the Todd J. Schwartz Memorial Fund. The Charles River toxicity study was supported in part by an option agreement between the Dana-Farber Cancer Institute and Aileron Therapeutics. This research was also supported by NIH grant R01GM101135 to J.R.E. and a research collaboration between J.R.E. and the Waters Corporation. R.M.’s doctoral research was supported in part by a Doctoral Foreign Study Award (no. DFS-134963) from the Canadian Institutes of Health Research and an IDEA-squared award from the Harvard-MIT Division of Health Sciences and Technology.
Footnotes
COMPETING INTERESTS STATEMENT
L.D.W. is a scientific advisory board member and consultant for Aileron Therapeutics.
REFERENCES
- 1.Brogden KA Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol 3, 238–250 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Brown KL & Hancock RE Cationic host defense (antimicrobial) peptides. Curr Opin Immunol 18, 24–30 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Hancock RE & Sahl HG Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol 24, 1551–1557 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Fjell CD, Hiss JA, Hancock RE & Schneider G Designing antimicrobial peptides: form follows function. Nat Rev Drug Discov 11, 37–51 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Theuretzbacher U Global antimicrobial resistance in Gram-negative pathogens and clinical need. Curr Opin Microbiol 39, 106–112 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Woodford N & Wareham DW Tackling antibiotic resistance: a dose of common antisense? J Antimicrob Chemother 63, 225–229 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Blair JM, Webber MA, Baylay AJ, Ogbolu DO & Piddock LJ Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol 13, 42–51 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Steiner H, Hultmark D, Engstrom A, Bennich H & Boman HG Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature 292, 246–248 (1981). [PubMed] [Google Scholar]
- 9.Zasloff M Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc Natl Acad Sci U S A 84, 5449–5453 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wade D et al. All-D amino acid-containing channel-forming antibiotic peptides. Proc Natl Acad Sci U S A 87, 4761–4765 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braunstein A, Papo N & Shai Y In vitro activity and potency of an intravenously injected antimicrobial peptide and its DL amino acid analog in mice infected with bacteria. Antimicrob Agents Chemother 48, 3127–3129 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y et al. Role of peptide hydrophobicity in the mechanism of action of alpha-helical antimicrobial peptides. Antimicrob Agents Chemother 51, 1398–1406 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang Z, Vasil AI, Gera L, Vasil ML & Hodges RS Rational design of alpha-helical antimicrobial peptides to target Gram-negative pathogens, Acinetobacter baumannii and Pseudomonas aeruginosa: utilization of charge, ‘specificity determinants,’ total hydrophobicity, hydrophobe type and location as design parameters to improve the therapeutic ratio. Chem Biol Drug Des 77, 225–240 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mant CT et al. De Novo Designed Amphipathic alpha-Helical Antimicrobial Peptides Incorporating Dab and Dap Residues on the Polar Face To Treat the Gram-Negative Pathogen, Acinetobacter baumannii. J Med Chem (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordon YJ, Romanowski EG & McDermott AM A review of antimicrobial peptides and their therapeutic potential as anti-infective drugs. Curr Eye Res 30, 505–515 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bird GH et al. Hydrocarbon double-stapling remedies the proteolytic instability of a lengthy peptide therapeutic. Proc Natl Acad Sci U S A 107, 14093–14098 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schafmeister CE, Po J & Verdine GL An All-Hydrocarbon Cross-Linking System for Enhancing the Helicity and Metabolic Stability of Peptides. JACS 122 5891–5892 (2000). [Google Scholar]
- 18.Walensky LD et al. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science 305, 1466–1470 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bird GH et al. Biophysical determinants for cellular uptake of hydrocarbon-stapled peptide helices. Nat Chem Biol 12, 845-+ (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walensky LD & Bird GH Hydrocarbon-Stapled Peptides: Principles, Practice, and Progress. J Med Chem 57, 6275–6288 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bird GH et al. Mucosal delivery of a double-stapled RSV peptide prevents nasopulmonary infection. J Clin Invest 124, 2113–2124 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meric-Bernstam F et al. Phase I trial of a novel stapled peptide ALRN-6924 disrupting MDMX- and MDM2-mediated inhibition of WT p53 in patients with solid tumors and lymphomas. J Clin Oncol 35, Suppl; abstr 2505 (2017). [Google Scholar]
- 23.Chapuis H et al. Effect of hydrocarbon stapling on the properties of alpha-helical antimicrobial peptides isolated from the venom of hymenoptera. Amino Acids 43, 2047–2058 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Dinh TT, Kim DH, Luong HX, Lee BJ & Kim YW Antimicrobial activity of doubly-stapled alanine/lysine-based peptides. Bioorg Med Chem Lett 25, 4016–4019 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Migon D, Neubauer D & Kamysz W Hydrocarbon Stapled Antimicrobial Peptides. Protein J 37, 2–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pham TK, Kim DH, Lee BJ & Kim YW Truncated and constrained helical analogs of antimicrobial esculentin-2EM. Bioorg Med Chem Lett 23, 6717–6720 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Stone TA, Cole GB, Nguyen HQ, Sharpe S, Deber CM Influence of hydrocarbon stapling on membrane interactions of synthetic antimicrobial peptides. Bioorg Med Chem (2017). [DOI] [PubMed] [Google Scholar]
- 28.Stone TA, Cole GB, Nguyen HQ, Sharpe S & Deber CM Influence of hydrocarbon-stapling on membrane interactions of synthetic antimicrobial peptides. Bioorg Med Chem (2017). [DOI] [PubMed] [Google Scholar]
- 29.Bechinger B, Zasloff M & Opella SJ Structure and interactions of magainin antibiotic peptides in lipid bilayers: a solid-state nuclear magnetic resonance investigation. Biophys J 62, 12–14 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zasloff M, Martin B & Chen HC Antimicrobial activity of synthetic magainin peptides and several analogues. Proc Natl Acad Sci U S A 85, 910–913 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gesell J, Zasloff M & Opella SJ Two-dimensional 1H NMR experiments show that the 23-residue magainin antibiotic peptide is an alpha-helix in dodecylphosphocholine micelles, sodium dodecylsulfate micelles, and trifluoroethanol/water solution. J Biomol NMR 9, 127–135 (1997). [DOI] [PubMed] [Google Scholar]
- 32.Reisser S, Strandberg E, Steinbrecher T & Ulrich AS 3D Hydrophobic Moment Vectors as a Tool to Characterize the Surface Polarity of Amphiphilic Peptides. Biophysical Journal 106, 2385–2394 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuzaki K et al. Mechanism of synergism between antimicrobial peptides magainin 2 and PGLa. Biochemistry 37, 15144–15153 (1998). [DOI] [PubMed] [Google Scholar]
- 34.Williams RW et al. Raman spectroscopy of synthetic antimicrobial frog peptides magainin 2a and PGLa. Biochemistry 29, 4490–4496 (1990). [DOI] [PubMed] [Google Scholar]
- 35.Engen JR Analysis of protein conformation and dynamics by hydrogen/deuterium exchange MS. Anal Chem 81, 7870–7875 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi XE et al. Hydrogen exchange-mass spectrometry measures stapled peptide conformational dynamics and predicts pharmacokinetic properties. Anal Chem 85, 11185–11188 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mechler A et al. Specific and selective peptide-membrane interactions revealed using quartz crystal microbalance. Biophys J 93, 3907–3916 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joshi T, Voo ZX, Graham B, Spiccia L & Martin LL Real-time examination of aminoglycoside activity towards bacterial mimetic membranes using Quartz Crystal Microbalance with Dissipation monitoring (QCM-D). Biochim Biophys Acta 1848, 385–391 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Wang KF, Nagarajan R & Camesano TA Differentiating antimicrobial peptides interacting with lipid bilayer: Molecular signatures derived from quartz crystal microbalance with dissipation monitoring. Biophys Chem 196, 53–67 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Murray B, Pearson CS, Aranjo A, Cherupalla D & Belfort G Mechanism of Four de Novo Designed Antimicrobial Peptides. J Biol Chem 291, 25706–25715 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Molugu TR & Brown MF Cholesterol Effects on the Physical Properties of Lipid Membranes Viewed by Solid-state NMR Spectroscopy. Adv Exp Med Biol 1115, 99–133 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Liu YY et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16, 161–168 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Moffatt JH et al. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob Agents Chemother 54, 4971–4977 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fox JL Antimicrobial peptides stage a comeback. Nat Biotechnol 31, 379–382 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Zavascki AP & Nation RL Nephrotoxicity of Polymyxins: Is There Any Difference between Colistimethate and Polymyxin B? Antimicrob Agents Chemother 61 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roberts KD et al. Antimicrobial Activity and Toxicity of the Major Lipopeptide Components of Polymyxin B and Colistin: Last-line Antibiotics against Multidrug-Resistant Gram-negative Bacteria. ACS Infect Dis 1, 568–575 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gallardo-Godoy A et al. Activity and Predicted Nephrotoxicity of Synthetic Antibiotics Based on Polymyxin B. J Med Chem 59, 1068–1077 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ge Y et al. In vitro antibacterial properties of pexiganan, an analog of magainin. Antimicrob Agents Chemother 43, 782–788 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zuluaga AF et al. Neutropenia induced in outbred mice by a simplified low-dose cyclophosphamide regimen: characterization and applicability to diverse experimental models of infectious diseases. BMC Infect Dis 6, 55 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.WHO (2017).
- 51.Lee JH, Jeong SH, Cha SS & Lee SH A lack of drugs for antibiotic-resistant gram-negative bacteria. Nat Rev Drug Discov 6, 938–939 (2007). [Google Scholar]
- 52.Zgurskaya HI, Lopez CA & Gnanakaran S Permeability Barrier of Gram-Negative Cell Envelopes and Approaches To Bypass It. ACS Infect Dis 1, 512–522 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boman HG Antibacterial peptides: basic facts and emerging concepts. J Intern Med 254, 197–215 (2003). [DOI] [PubMed] [Google Scholar]
- 54.Marr AK, Gooderham WJ & Hancock RE Antibacterial peptides for therapeutic use: obstacles and realistic outlook. Curr Opin Pharmacol 6, 468–472 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Gomes A, Teixeira C, Ferraz R, Prudencio C & Gomes P Wound-Healing Peptides for Treatment of Chronic Diabetic Foot Ulcers and Other Infected Skin Injuries. Molecules 22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bird GH, Crannell WC & Walensky LD Chemical synthesis of hydrocarbon-stapled peptides for protein interaction research and therapeutic targeting. Curr Protoc Chem Biol 3, 99–117 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Juretic D, Vukicevic D, Ilic N, Antcheva N & Tossi A Computational design of highly selective antimicrobial peptides. J Chem Inf Model 49, 2873–2882 (2009). [DOI] [PubMed] [Google Scholar]
- 58.Kovacs JM, Mant CT & Hodges RS Determination of intrinsic hydrophilicity/hydrophobicity of amino acid side chains in peptides in the absence of nearest-neighbor or conformational effects. Biopolymers 84, 283–297 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guttman M, Weis DD, Engen JR & Lee KK Analysis of overlapped and noisy hydrogen/deuterium exchange mass spectra. J Am Soc Mass Spectrom 24, 1906–1912 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for the current study are included in the published article and its supplementary information. Requests for any further information can be made to the corresponding author.