Single-cell RNA-seq reveals increased numbers of ensheathing glia in Harpegnathos workers reprogrammed to a queen-like phenotype.
Abstract
In social insects, workers and queens arise from the same genome but display profound differences in behavior and longevity. In Harpegnathos saltator ants, adult workers can transition to a queen-like state called gamergate, which results in reprogramming of social behavior and life-span extension. Using single-cell RNA sequencing, we compared the distribution of neuronal and glial populations before and after the social transition. We found that the conversion of workers into gamergates resulted in the expansion of neuroprotective ensheathing glia. Brain injury assays revealed that activation of the damage response gene Mmp1 was weaker in old workers, where the relative frequency of ensheathing glia also declined. On the other hand, long-lived gamergates retained a larger fraction of ensheathing glia and the ability to mount a strong Mmp1 response to brain injury into old age. We also observed molecular and cellular changes suggestive of age-associated decline in ensheathing glia in Drosophila.
INTRODUCTION
Age-associated cognitive decline has an immense impact on human societies and is caused by genetic, epigenetic, and environmental factors, whose interplay is poorly understood. Social insects, including ants, provide a fascinating model to study the epigenetic regulation of brain function and longevity (1, 2). The division of labor typical of social insect colonies is based on the distinct physical and behavioral phenotypes of highly related individuals organized in separate social castes. These individuals carry out distinct specialized tasks such as reproduction, foraging, defense, and nest maintenance. A notable difference between social castes is their life expectancy: In most species reproductive queens live much longer—often up to an order of magnitude longer—than sterile workers from the same colony (3).
Brains of queens and various types of workers differ at a molecular level in the genes that they express (4–6) and often also at a structural level in the overall volume and relative size of anatomical substructures, such as the mushroom body (7–9). However, whether brains from different social castes have substantially different cellular compositions that might contribute to caste phenotype has not been investigated, partly because we still lack a comprehensive molecular description of the variety of cell types that constitute a social insect brain.
The ant Harpegnathos saltator offers a unique opportunity to study the epigenetic regulation of phenotypic plasticity (2). While in most ant species social castes are permanently established during the larval stage, adult Harpegnathos workers can acquire a queen-like phenotype and become reproductive individuals called “gamergates” (10). Although they are not morphological queens, workers that become gamergates display a remarkable switch in social behavior and an extensive molecular reprogramming of the brain with hundreds of differentially expressed genes (4, 11). Workers that become gamergates also attain queen-like longevity, with a fivefold increase in average life span from 7 months to 3 years (12).
Previous studies reported macroscopic changes in the brain of gamergates compared to workers, including a volumetric shrinkage of the optic lobe (13), suggesting that brain remodeling might accompany caste reprogramming. However, the questions of which cell types are most affected by the transition and most likely to contribute to the caste-specific regulation of behavior and longevity have not been explored.
We performed high-throughput single-cell RNA sequencing (RNA-seq) in brains from workers and gamergates at different ages and with or without brain injury. We compared these profiles with previously published single-cell data from the nonsocial insect Drosophila melanogaster, where castes do not exist (14, 15), as well as bulk RNA-seq from other social insects (6, 16). Our findings reveal caste-, aging-, and injury-associated cellular dynamics in the Harpegnathos brain and suggest that regulation of ensheathing glia numbers contributes to the longer life span of the reproductive caste.
RESULTS
Transcriptional types of neurons and glia in a social insect brain
We performed single-cell RNA-seq using 10× Genomics on brains harvested from workers (n = 6) and gamergates (n = 5) 30 days after initiating the caste transition (Fig. 1A). We previously identified differences in gene expression at a later time point (day 120) and noted that after 30 days of transition, most of the dueling interactions have ceased and the newly converted gamergates have started to lay eggs (4); therefore, we reasoned that this earlier time point would be adequate to detect caste-associated molecular differences between workers and gamergates. Despite the difference in technology, our single-cell RNA-seq at day 30 captured the differential regulation of a large portion of the caste-specific genes previously identified in bulk at day 120 (rS = 0.42, P = 0.0001) (fig. S1A).
Fig. 1. Single-cell transcriptomes from worker and gamergate brains.
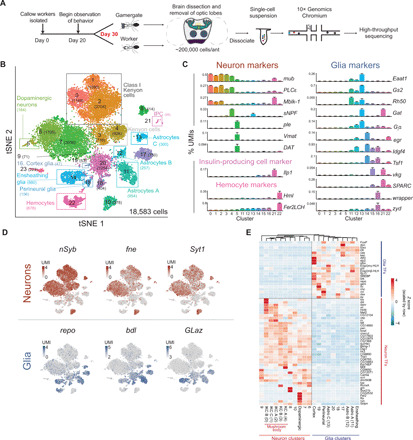
(A) Scheme of the experiment. Workers and gamergates were separated on the basis of behavior and ovary status. Brains were dissected and optic lobes removed. The central brain, including mushroom bodies (dark green), ellipsoid bodies (green), fan-shaped bodies (yellow), and antennal lobes (blue), plus the gnathal ganglion (purple) were dissociated into a single-cell suspension and processed for single-cell RNA-seq. (B) Annotated tSNE visualization of the clustering of 18,583 single-cell transcriptomes obtained by pooling all cells from six worker and five gamergate replicates. The number of cells in each cluster is indicated in parenthesis. IPC, insulin-producing cells. (C) Selected marker genes for the clusters annotated in (B). The y axis shows the collapsed pseudobulk expression in each cluster (as % of total cluster UMIs) for the indicated gene. Bars represent the means of 11 biological replicates + SEM. (D) Heatmap plotted over global tSNE showing normalized UMIs per cell for known neuronal markers (red) and glia markers (blue). (E) Heatmap for normalized expression levels (z score) for the indicated transcription factors (TFs) in collapsed single-cell clusters. Only transcription factors with a |log2(neurons/glia)| > 1 are shown, but the columns were clustered on all transcription factors. Astro A–C, astrocytes A–C.
To obtain a comprehensive description of cell types in the Harpegnathos brain, we first considered all samples, regardless of caste. We retained only cells with a minimum of 500 unique transcripts [as defined by unique molecular identifiers (UMIs)] over at least 200 different genes and obtained 18,583 cells, which gave us a >99% probability of detecting at least 50 cells from a population as rare as 0.5% (17). Globally, the filtered cells had a median of 987 UMIs per cell mapping to a total 11,926 genes and a UMI median of at least 815 in each biological replicate (n = 11; fig. S1B), which is in line with previous single-cell RNA-seq datasets from the Drosophila brain (table S1) (14, 15, 18).
Using the Seurat pipeline (19), we obtained 24 clusters (Fig. 1B and tables S2 to S4), which we annotated using known markers from Drosophila (Fig. 1C). Our clusters effectively separated neurons (0 to 10) and glial cells (11 to 20), confirming that we were able to capture characteristic transcriptomes of single cells (Fig. 1D). Neuron clusters were identified by the expression of previously defined markers nSyb, fne, and Syt1, whereas expression of known glia markers repo, bdl, and GLaz identified glia clusters (14, 15). Using additional established markers (see table S2 for references), we mapped cells from most of the clusters to corresponding types in the Drosophila brain (Fig. 1C and tables S2 and S3), including the following: Kenyon cells (KCs; mub and PLCε; clusters 0 to 4), dopaminergic neurons (ple and Vmat; cluster 5), three distinct clusters of astrocytes (Eaat1, Gs2, Rh50, Gat, and/or Giα; clusters 11 to 13), ensheathing glia (egr, Tsf1, and Idgf4; cluster 14), perineurial glia (vkg and SPARC; cluster 15), cortex glia (wrapper and zyd; cluster 16), insulin-producing cells (Ilp1; cluster 21), and hemocytes (Hml and Fer2LCH; cluster 22). Some clusters (clusters 6 to 10 and 17 to 20) were identified as neurons or glia on the basis of the expression of common markers (e.g., nSyb versus repo; Fig. 1D), but the lack of subtype-specific marker gene expression, possibly due to sequencing depth, precluded further classification.
Transcription factors are key specifiers of identity and function for all cells, including those in the brain. Cell type–specific transcriptional networks have been observed in other single-cell experiments (14, 18) and likely play a role in shaping the diversity and plasticity of cell types in the ant brain. We curated a panel of 423 transcription factors in the Harpegnathos genome based on their sequence conservation with Drosophila homologs and determined their expression pattern in single cells. Hierarchical clustering based on transcription factor expression alone separated neuron and glia clusters (Fig. 1E).
On the basis of our clustering, 27% of the single cells recovered from Harpegnathos brains are glia (fig. S1, C and D). Comparable single-cell RNA-seq studies estimated a 2 to 10% frequency of glia cells in Drosophila brains (fig. S1, C and D) (14, 15) or optic lobes (18). Although uneven capture rates among cell types make single-cell RNA-seq poorly suited to determine absolute numbers, these numbers are in the same range as the ~10% estimate for the frequency of glia in Drosophila obtained by two independent studies using immunofluorescence and genetic reporters (20, 21). Thus, with the caveat that comparisons across species and datasets from different laboratories must be interpreted with caution, our analyses suggest that Harpegnathos brains contain more glia than Drosophila brains.
Large mushroom bodies in the Harpegnathos brain
To better characterize neuronal populations in Harpegnathos, we reclustered in isolation all the transcriptomes from neuronal clusters 0 to 10 (Fig. 2A and tables S2 and S4) and found that most of neurons (54%, corresponding to 36% of all brain cells) expressed genes that mark KCs in Drosophila (mub and Pka-C1) and Apis mellifera (PLCε and E75) (Fig. 2B and fig. S2, A and B). KCs are the principal component of mushroom bodies, the center of learning and memory in insects (22), which are known to be markedly large in social Hymenoptera (7), including ants (23). Our single-cell RNA-seq confirms this conclusion, as KCs comprise a much larger fraction (54%) of neurons in Harpegnathos than in Drosophila (5 to 10%) (Fig. 2, C and D, and fig. S2C), even after accounting for differences in the datasets by equalizing read numbers and UMI distributions (fig. S2, D, F, H, and I). Immunofluorescence stainings for the KC marker Pka-C1 in Harpegnathos labeled structures with the anatomical features of mushroom bodies including a thick pedunculus (Fig. 2E, gray arrowhead) and prominent double cup-shaped calyces (Fig. 2E, white arrowhead), characteristic of Hymenoptera (7, 24). Consistent with the increased frequency of KCs in our single-cell data, Harpegnathos mushroom bodies appeared to occupy a larger relative volume in the ant brain as compared to the corresponding structures in Drosophila (Fig. 2E). Additional low signal was observed in other brain regions and likely explained by antibody cross-reactivity or low-level, scattered expression of Pka-C1 in non-KC cells (see Fig. 2B). Western blots for Pka-C1 from total brain extracts revealed higher relative levels of this protein in ants as compared to flies (Fig. 2F).
Fig. 2. Distinctive features of Harpegnathos neurons and glia.
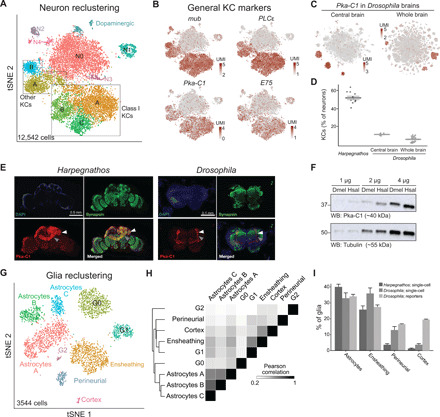
(A) Annotated tSNE visualization for the reclustering of neurons from six worker and five gamergate replicates at day 30. (B) Heatmap plotted over neuronal tSNE showing normalized UMIs for known mushroom body markers from Drosophila (left) and A. mellifera (right). (C) Heatmap plotted over neuronal tSNE for the KC marker Pka-C1 from two Drosophila single-cell RNA-seq datasets, one from the central brain after removing optic lobes [left; (15)] and one from the whole brain, inclusive of the optic lobes [right; (14)]. (D) Relative abundance of KCs as determined by percentage of neurons in clusters that express Pka-C1 in Harpegnathos brains and in the two Drosophila single-cell RNA-seq datasets. Horizontal bars indicate means ± SEM. (E) Immunofluorescence for the neuronal marker synapsin and the KC marker Pka-C1 in Harpegnathos (left) and Drosophila (right) with 4′,6-diamidino-2-phenylindole (DAPI) as nuclear counterstain. Gray arrowhead, pedunculus; white arrowhead, calyx. (F) Western blot (WB) for Pka-C1 in the indicated amount of total protein extract from Drosophila (Dmel) or Harpegnathos (Hsal) brains. Tubulin was used as loading control. (G) Annotated tSNE visualization for the reclustering of glia from six worker and five gamergate replicates at day 30. (H) Clustered heatmap showing the pairwise Pearson correlation score between collapsed transcriptomes (pseudobulk analysis) of glia clusters, considering only variable genes that were used to define the clusters by Seurat. (I) Relative abundance of key glia subsets in Harpegnathos single-cell RNA-seq, Drosophila single-cell RNA-seq from the whole brain (14), and as defined by glial subset-specific expression of GAL4 (20). Bars indicate means + SEM.
The larger mushroom body of Harpegnathos comprised a diverse repertoire of KC transcriptomes, as they separated into five clusters compared to the three clusters found in Drosophila (Fig. 2, A and C). KCs were first described in the honey bee A. mellifera, where they are divided into three classes (I to III) on the basis of their connectivity (7). This classification appears to hold and translate to distinct transcriptional types in the Harpegnathos brain, as cells expressing known markers for class I KCs (IKC) IP3R and Mblk-1 (table S2) clustered apart from other KCs (fig. S2E). Gene ontology (GO) terms classically associated with mushroom body function were selectively enriched in transcriptionally distinct subtypes of IKCs, including “olfactory learning” (all IKCs), “learning or memory” (IKC A), and “associative learning” (IKC B) (fig. S2G).
Thus, as typical of social insects, the Harpegnathos brain contains an expanded mushroom body. In addition, our data revealed a transcriptionally diverse repertoire of KCs.
Harpegnathos glia cells are diverse and include multiple subsets of astrocytes
Given the relative abundance of glia in Harpegnathos compared to Drosophila (fig. S1D), we decided to further analyze these cells. We reclustered and assigned identities to glia cells using genes homologous to known Drosophila markers (Fig. 2G and fig. S3, A to C; see table S2 for references). We identified cortex glia (wrapper and zyd), two clusters of astrocytes (astrocytes A and B in Fig. 2G; Eaat1 and Gat or Rh50), perineurial glia (vkg and Tret), and ensheathing glia (egr, Tsf1, and Idgf4).
Although egr, Tsf1, and Idgf4 were reported as markers for ensheathing glia by single-cell RNA-seq in Drosophila (14), we note that the use of the term “marker” in this case does not imply complete specificity, rather, enrichment. While egr is mostly confined to Harpegnathos ensheathing glia (with some expression in cluster G2 and hemocytes), Tsf1 and Idgf4 are expressed in other clusters too, albeit at lower levels (fig. S3C). Previous ensheathing glia studies in Drosophila have mostly relied on GAL4 driver lines. One of the most widely used is GMR56F03 (20, 25), which uses the cis-regulatory region of CG9657, a putative member of the SLC5 family of glucose transporters. The endogenous CG9657 along with its paralog CG6723 are among the most specific markers for ensheathing glia in Drosophila by single-cell RNA-seq (14). The closest Harpegnathos homolog for both of these genes is encoded by LOC105183203, which we have now renamed Slc5eg. This gene is the second most specific protein-coding marker for the ensheathing glia cluster after Tsf1 (fig. S3C and tables S2 and S5). Approximately 50% of cells in the ensheathing glia cluster express higher than average levels of egr, Tsf1, Idgf4, and Slc5eg, a combination that is barely detectable in any other cluster (fig. S3D).
One cluster displayed Eaat1 expression and a transcriptome closely related to those of the two other types of astrocytes (Fig. 2H), and we named it “astrocytes C.” In addition, three clusters (G0, G1, and G2) expressed multiple glia markers but could not be unequivocally assigned to known transcriptional types (Fig. 2G and table S5). The three distinct classes of Harpegnathos astrocytes comprised ~40% of total glia cells, which was comparable to the frequency of astrocytes in Drosophila (Fig. 2I), as measured by fluorescence microscopy using genetic reporters (20) and single-cell RNA-seq (14). Similarly, the proportion of ensheathing glia in Harpegnathos (24% of all glia) was comparable to their frequency in genetically labeled Drosophila brains (Fig. 2I), suggesting that single-cell RNA-seq protocols efficiently capture the transcriptomes of these two cell types. On the other hand, perineurial and cortex glia were present at different frequencies across datasets. Perineurial glia, which envelop the outer surface of the brain, were a threefold larger fraction of Drosophila (15%) than Harpegnathos (4%) glia (Fig. 2I). This difference might, in part, be due to the larger ant brain having a lower surface-to-volume ratio, but we cannot exclude technical issues specific to Harpegnathos in recovering these cells. Technical difficulties seemingly affected the recovery of cortex glia in previous Drosophila single-cell RNA-seq studies, as evidenced by the much smaller fraction recovered with this technology (3.7%) compared to imaging-based estimates (19%) (20). The former number is comparable to our quantifications by single-cell RNA-seq in Harpegnathos (Fig. 2I) and suggests that the honeycomb-like structure of this glia subtype (20) might hamper their recovery in single-cell suspensions in both species.
A separation of astrocytes into different types was not observed in previous studies of the Drosophila brain (14), which we reanalyzed to confirm that astrocyte transcriptomes formed a single cluster (fig. S3E). This distinction was independent of sequencing depth, as equalizing the number of glia cells and UMI distributions across datasets did not cause the three separate Harpegnathos clusters to merge (fig. S3G). Astrocyte diversity has not been explored in insects, but there are reports of regional and functional heterogeneity among mammalian astrocytes (26), which have been corroborated by transcriptomic analyses (27). The three Harpegnathos astrocyte clusters share expression of “common” astrocyte markers (Eaat1, Gs2, and Rh50; Fig. 1C) but differ in several other genes (tables S3 and S5) as well as in the associated enriched GO terms (fig. S3F). Our results show that Harpegnathos brains contain a large fraction of glia cells, many of which belong to glia types described in previously published studies of the Drosophila brain, while others display molecular fingerprints only thus far found in ants.
Caste-specific brain remodeling includes the expansion of ensheathing glia in gamergates
In social insects, worker and queen brains are functionally and structurally distinguishable, with visible anatomical differences between the castes, especially in the mushroom body (7–9). In Harpegnathos, bulk transcriptional differences accompany behavioral and reproductive changes in workers that transition to gamergate status (4). The occurrence of extensive brain remodeling during the caste transition is supported by morphometric analyses that show a loss in brain volume concentrated in the optic lobes (13), but whether changes in cell composition correspond to this molecular and anatomical differences in the brain is completely unexplored.
We separated and compared single-cell transcriptomes from brains of workers (n = 6) and gamergates (n = 5) after 30 days of transition. The clustering profiles were broadly similar, demonstrating the absence of major batch effects; however, KC type A, ensheathing glia, and perineurial glia displayed significant (adjusted P = 0.06, 0.03, and 0.03, respectively; Wald test) caste-specific changes in relative numbers (Fig. 3, A and B, and fig. S4A). KC A neurons were relatively more abundant in gamergates than in workers (Fig. 3, A and B, and fig. S4B) and were distinguishable from other KCs by their strong and specific expression of the neuropeptide sNPF (fig. S4C) (28, 29) and of Sema2a (fig. S4C), a gene involved in axon guidance and remodeling (30).
Fig. 3. Cellular remodeling in the Harpegnathos brain after the caste transition.
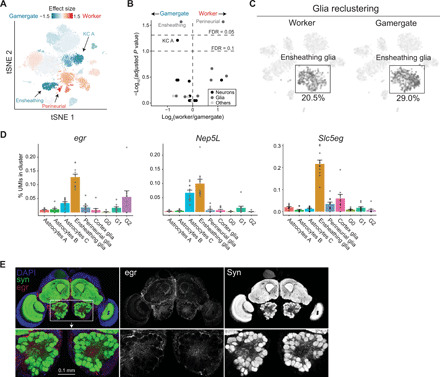
(A) Heatmap plotted over global tSNE showing the changes in cell type abundance in workers versus gamergate brains. The color scale indicates the effect size (mean/SD). The three clusters that are significantly (adjusted P < 0.1, Wald test) affected are indicated. (B) Volcano plot of log2(ratio) and −log10(adjusted P value from Wald test with Benjamini-Hochberg multiple test correction) for the relative frequency of each cluster in workers versus gamergates. Thresholds for false discovery rate (FDR) < 0.1 and FDR < 0.05 are shown. (C) Visualization and quantification of worker (left) and gamergate (right) contributions to the ensheathing glia cluster in the reclustered tSNE for glia only (see Fig. 2G). Worker and gamergate datasets were downsampled to include the same number of total cells for comparison. (D) Expression levels (% of cluster UMIs) across glia subsets for egr, Nep5L, and Slc5eg. Bars indicate means + SEM. (E) Immunofluorescence of a thick section of a Harpegnathos brain stained with antibodies against egr (red) and synapsin (green) and counterstained with DAPI (blue). The bottom panels show a magnification of the antennal lobes.
We also observed caste-specific changes in glia composition. The transition from worker to gamergate status resulted in a ~60% reduction in perineurial glia (fig. S4D) and a ~40% expansion of ensheathing glia (Fig. 3C). The latter observation was of particular interest to us because, in Drosophila, ensheathing glia cells are essential to brain health and homeostasis as they remove damaged neuronal structures generated by stress, injury, or programmed cell death (31, 32).
The expression of the phagocytic receptor Draper (drpr), its associated adaptor protein dCed-6, and the tumor necrosis factor (Tnf) homolog eiger (egr) is required for the response to injury of ensheathing glia in Drosophila (32–34). The Harpegnathos glia population that expanded during the worker-gamergate transition expressed egr and Ced6, confirming their phagocytic nature (Fig. 3D and fig. S4E). Moreover, Harpegnathos ensheathing glia expressed the highest levels of a Hymenoptera-specific member of the neprilysin family, which we called neprilysin-5-like (Nep5L) (Fig. 3D). This cluster also showed strong preferential expression for Slc5eg (Fig. 3D), which is homologous to two ensheathing glia markers in Drosophila, one of which was used to generate an ensheathing glia driver line. Last, in Harpegnathos brains, cells expressing egr displayed an anatomical distribution remarkably similar to that observed in bona fide ensheathing glia in Drosophila (25, 32), especially around the glomeruli of the antennal lobes (Fig. 3E), confirming their identity. These data highly suggest that the adult caste transition in Harpegnathos results in plastic changes in cell composition, including an expansion of KC A neurons and of neuroprotective ensheathing glia.
Up-regulation of ensheathing glia markers in the reproductive caste across Hymenoptera
To determine whether the caste-specific expansion of ensheathing glia observed in Harpegnathos might be conserved in other social insects, we used publicly available data from an additional ant species with a worker-gamergate system, Dinoponera quadriceps, a carpenter ant with a more conventional social structure, Camponotus planatus, and the primitively eusocial wasp Polistes canadensis. Because no single-cell RNA-seq was available in these species, we reanalyzed the available bulk RNA-seq data (6, 16). We reasoned that changes in mRNA levels for genes expressed preferentially in ensheathing glia might be detectable even in bulk brain RNA.
First, we analyzed the expression of the three top and most abundant protein-coding ensheathing glia markers in Harpegnathos: Tsf1, Idgf4, and Prx2540-1 (fig. S5A, left). We found that in some cases these genes were up-regulated in the brains of reproductive individuals according to the bulk RNA-seq and in no case they were down-regulated (fig. S5A, right). We extended this analysis to the top 25 genes from our ensheathing glia marker list in Harpegnathos (table S3, cluster 14) and found that in both C. planatus and P. canadensis, more of these genes were up-regulated in the reproductive caste than expected by chance, although the same comparison in D. quadriceps was not significant (fig. S5B). When considering all 213 Harpegnathos ensheathing glia markers in aggregate, the trend toward up-regulation in reproductive individuals was statistically significant (Fisher’s exact test) in all three species (fig. S5C). Thus, although the bulk nature of the RNA-seq data prevents us from drawing a firm conclusion, we suggest that ensheathing glia might be expanded in the brains of reproductive individuals in other Hymenoptera species in addition to Harpegnathos.
Harpegnathos ensheathing glia respond to brain injury
Glia cells provide essential support to brain function and are required for damage control in response to injury in both mammals and insects (31, 35). In the mammalian brain, phagocytic microglia cells proliferate and migrate to sites of injury and are involved in clearing damaged tissue, modulating inflammation, and promoting remyelination in concert with oligodendrocyte precursors (36–38). In Drosophila, repair functions are carried out by ensheathing glia cells, which combine features of microglia and oligodendrocytes and activate conserved transcriptional networks and molecular pathways following injury (32, 34, 39).
Harpegnathos ensheathing glia expressed egr and ced-6 (Fig. 3D and fig. S4E), which are involved in the injury response in Drosophila; thus, we hypothesized that these cells would also respond to injury in Harpegnathos. We isolated young workers from stable, nontransitioning colonies and induced damage in their brains by puncturing them with a needle (Fig. 4A and fig. S6A), which, in Drosophila, results in glia activation and proliferation at the injury site (33). We performed Drop-seq (40) on dissected brains 1 and 3 days after injury (median UMI = 448; fig. S6B) and analyzed the glial compartment (Fig. 4B and tables S2 and S6). Most clusters recovered in the Drop-seq experiment mapped to the same cell types identified in the deeper 10× Genomics dataset obtained for the day 30 worker-gamergate comparison (fig. S6C).
Fig. 4. Brain injury causes activation of ensheathing glia.
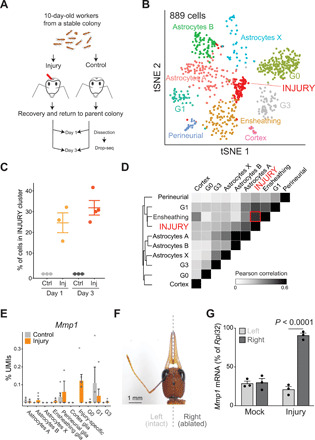
(A) Scheme of the experiment. Needle-stabbed brains and age-matched control were analyzed by Drop-seq at days 1 and 3 after injury. (B) Annotated tSNE visualization of glia-only reclustering from pooled control and injury samples at days 1 and 3 (n = 3 control and injury for day 1; n = 3 control and 4 injury for day 3). The injury-specific cluster is indicated. (C) Relative frequency of cells in the injury-specific cluster as % of glia. Horizontal lines indicate means ± SEM. (D) Clustered heatmap showing the pairwise Pearson correlation score between collapsed transcriptomes of glia clusters considering only variable genes that were used to define the clusters by Seurat. The high transcriptome-wide correlation between the injury-specific cluster and resting ensheathing glia is indicated by a red square. (E) Expression levels (as % of cluster UMIs) for the activation marker Mmp1. Bars indicate means + SEM. (F) Head of a 30-day-old Harpegnathos worker subjected to antenna amputation. Photo credit: Lihong Sheng, University of Pennsylvania. (G) RT-qPCR for Mmp1 mRNA in the left (light gray) or right (dark gray) brain hemisphere after antennal ablation. Bars represent means + SEM. P value is from one-way analysis of variance (ANOVA) and Holm-Sidak test.
One transcriptional type of glia recovered in this experiment was unique to the injured brains and accounted for one-third of all glia cells 3 days after injury (Fig. 4, B and C, and fig. S6D). These cells expressed the most specific markers for Harpegnathos ensheathing glia (egr, Slc5eg, and Nep5L) as well as the phagocytosis gene ced-6 (fig. S6E). When compared to all other glia clusters, their transcriptome was most similar to that of ensheathing glia from uninjured brains in both Drop-seq (Fig. 4D) and 10× (fig. S6C) datasets. Last, these injury-specific cells showed high expression of Mmp1 (Fig. 4E), which is known to be activated in Drosophila ensheathing glia upon brain injury (41). The injury-activated glia, despite comprising only 2.8% of total cells, was responsible for 44% of UMIs for Mmp1. Thus, we propose that the transcriptional type of glia found only in injured brains comprises activated ensheathing glia cells, although formal proof of this will require more sophisticated tracing experiments.
To confirm that the observed transcriptional response was occurring as a local response to brain injury, we used a different experimental paradigm, whereby surgical amputation of an antenna (Fig. 4F) results in axonal degeneration and localized damage in the antennal lobe, which is known to induce a local glial response in Drosophila (33). As a proxy of glia activation in response to injury, we used reverse transcription followed by quantitative polymerase chain reaction (RT-qPCR) to measure Mmp1 expression by RT-qPCR. Consistent with observations in Drosophila (41), antennal ablation caused the up-regulation of Mmp1 in the damaged but not in the contralateral hemisphere (Fig. 4G). No difference between right and left antennal lobes was observed in mock-treated animals. Together, these observations suggest that, similar to their function in Drosophila, cells that express markers of ensheathing glia respond to brain damage in Harpegnathos by up-regulating Mmp1.
Gamergates are resistant to aging-associated decline in ensheathing glia
A key difference between the castes of social insects is that queens live much longer (often 10-fold or more) than workers from the same species (3). Even more remarkably, the life span of a Harpegnathos individual is markedly affected by changes in its social status, as the worker-gamergate transition results in a fivefold increase in life span (12). Given the importance of glia in protecting brains from the insults of time and disease (42–44), we wanted to test whether an expanded population of ensheathing glia in gamergates might contribute to their longer life expectancy and whether the accelerated aging trajectory of workers might be accompanied by a change in the opposite direction.
Using Drop-seq, we profiled brains from 5-day-old pretransition individuals, as well as 30-, 90-, and 120-day-old workers (n ≥ 3 per time point), corresponding to 2.4, 14, 43, and 57% of their average 210-day life span (12). The number and transcriptional identity of the glia clusters in these experiments were similar to those observed in the 10× Genomics profiling and injury experiments above (fig. S7, A to D). The frequency of ensheathing glia cells among all glia experienced a marked decline during aging in Harpegnathos workers, especially between days 30 and 90, and they were at their lowest numbers in 120-day-old workers (Fig. 5, A and B, and fig. S7, E and F). However, the glia of 120-day-old gamergate brains contained significantly more ensheathing glial cells than in age-matched workers (2.6-fold increase, adjusted P = 5 × 10−5 Wald test; Fig. 5B). On the other hand, one cluster of astrocytes was enriched in workers compared to gamergates at day 120 (fig. S7E). Because its transcriptional profile did not match any of the clusters identified in the deeper 10× Genomics dataset (fig. S7C), we designated this cluster as A astrocytes X. Unlike our previous observations at day 30, we found no significant differences in the perineurial glia population (fig. S7E), but very few cells overall mapped to this cluster, likely because of the lower capture rate and coverage of Drop-seq compared to 10×.
Fig. 5. Differential decline of ensheathing glia during aging in Harpegnathos castes.
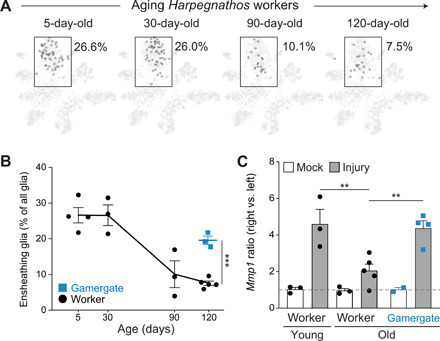
(A) Visualization and quantification of the contributions from workers of different ages to the ensheathing glia cluster in the Drop-seq tSNE. Cells from each time point were downsampled to the same total cell number for comparison. (B) Dynamics of the ensheathing glia cluster during aging. Each point represents a biological replicate. The line connects the means and error bars indicate ± SEM. The mean for the gamergate sample is shown as a thicker bar. ***P = 5 × 10−5 from a Wald test with multiple test correction. (C) Ratio of Mmp1 mRNA (RT-qPCR) in the right versus left brain hemisphere in ants subjected to ablation of the right antenna (gray) or a mock treatment as control (white). Activation of ensheathing glia as measured by up-regulation of Mmp1 was compared in young (30-day-old, left) and old (120- to 150-day-old, right) individuals. Bars represent means + SEM. The dashed line indicates a ratio of 1, i.e., no difference between right and left hemisphere. **P < 0.01 from one-way ANOVA and Holm-Sidak test.
Consistent with the relative decline in ensheathing glia in their brains, old workers exhibited a weaker transcriptional response to neuronal damage compared to young workers, as shown by a decreased up-regulation of the glia activation marker Mmp1 (Fig. 5C). Upon antenna ablation, old gamergates showed a much stronger Mmp1 up-regulation than age-matched old workers (Fig. 5C), indicating that the increased frequency of ensheathing glia in gamergates retains the ability to respond to brain damage into old age.
Evidence of aging-associated ensheathing glia decline in Drosophila
To determine whether the decline in ensheathing glia is a conserved feature of aging brains, we analyzed the expression of ensheathing glia markers in brains of young (day 5) and old (day 70) Drosophila females. RT-qPCR for CG9657 and CG6723, two genes highly specific for ensheathing glia in Drosophila [Fig. 6A and fig. S8A, visualized using the SCope website; see (14)] and homologous to the Slc5eg marker gene in Harpegnathos, showed a threefold down-regulation in the whole brain (Fig. 6B and fig. S8B). Genome-wide transcriptome analyses of brains from young and old flies confirmed and extended these observations as most ensheathing glia markers were down-regulated in aged individuals (Fig. 6C and fig. S8C). The decline in expression was specific for markers of ensheathing glia, as no similar trends were observed when considering all genes, all genes identified as markers for any cell type by single-cell RNA-seq (14), or astrocyte marker genes (Fig. 6D).
Fig. 6. Age-associated decline of ensheathing glia in Drosophila.
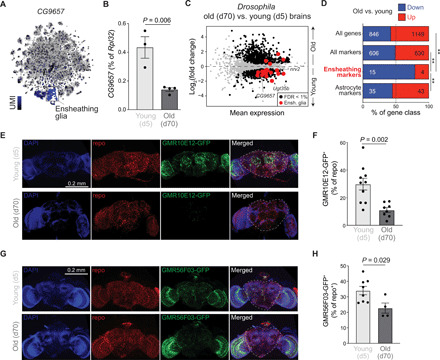
(A) Heatmap of UMI levels per cell plotted over tSNE for CG9657 in Drosophila brains obtained at the SCope website (http://scope.aertslab.org) with single-cell RNA-seq data from (14). (B) Abundance of CG9657 mRNA as determined by RT-qPCR (normalized to Rpl32) from dissected brains of day 5 (d5; young) and day 70 (old) Drosophila females. Bars represent means ± SEM. P value is from a Student’s t test. (C) MA plot of RNA-seq data from brains of young (day 5, n = 3) and old (day 70, n = 4) Drosophila females. All 4436 genes identified as cell type markers in (14) are shown. Differentially expressed (FDR < 1%) genes are shown in black. Differentially expressed (FDR < 1%) ensheathing glia marker genes are in red. (D) Proportion of indicated gene classes that were significantly up-regulated (red) or down-regulated (blue) in old versus young brains according to RNA-seq. **P <0.01 from one-sided Fisher’s test. (E) Visualization of ensheathing glia in young (top) and old (bottom) Drosophila brains using the GMR10E12-GAL4 (25) driver line crossed to a UAS-GFP reporter. Brains were costained with a repo antibody (red) to visualize all glia cells. The central brain region used for quantifications is outlined in the merged images with dashed lines. (F) Quantification of ensheathing glia cells (GFP+) in the central brain plotted as percentage of total glia cells (repo+). Each point is a biological replicate. Bars represent the means ± SEM. P value is from a Student’s t test. (G and H) Same as (E) and (F) but for line GMR56F03 (20).
To confirm these conclusions using a sequencing-independent method, we expressed green fluorescent protein (GFP) under the control of two GAL4 markers specific for ensheathing glia, GMR10E12 and GMR56F03 (20, 25). The two reporter lines differed in the expression patterns of GFP in the optic lobe, but in both cases, central brains from old flies contained a smaller proportions of glia cells (repo+) that expressed the ensheathing glia reporters (GFP+) than central brains from young (day 5) flies (Fig. 6, E to H). Although we cannot exclude that changes in GFP expression reflect changes in regulation of the promoter over time rather than a decrease in cellularity, together with our sequencing data, these results strongly suggest that the decline of ensheathing glia cells that we observed in Harpegnathos also occurs in aging Drosophila brains.
DISCUSSION
Social insects provide an exceptional opportunity to study how epigenetic pathways regulate brain function and longevity. While many studies, including ours, have reported differences in gene expression between brains from different castes in bulk RNA-seq, a single-cell RNA-seq approach was needed to analyze cellular plasticity and identify cell types most likely to contribute to caste-specific traits. We leveraged the unique phenotypic plasticity of Harpegnathos combined with the high-quality genomic resources in this ant species to obtain a comprehensive single-cell representation of the brain of a social insect. We found larger populations of mushroom body neurons and glia cells than in Drosophila and, within them, two subpopulations of cells that appear to expand as workers switch caste to become reproductive gamergates, short neuropeptide F (sNPF)–expressing KCs, and ensheathing glia. The latter showed an age-dependent decline in workers that was greatly attenuated in gamergates, suggesting that an enlarged ensheathing glia compartment provides heightened neuroprotection in the aging brain of gamergates, perhaps contributing to their fivefold longer life span.
Neuronal and glia cell types in the ant brain
Despite the evolutionary distance between ants and flies (>350 million years), we were able to assign identities to most single-cell clusters from Harpegnathos brains using known markers and transcription factors expressed in corresponding cell types in Drosophila (Fig. 1). The major distinction among neurons was between two types of KCs, IKCs (three clusters), as previously defined in honeybees, and other KCs (two clusters), which together account for a remarkable 54% of all Harpegnathos neurons outside of the optic lobe (Fig. 2). The observation that social Hymenoptera have larger mushroom body is far from novel (7), but our single-cell profiling also revealed unexpected complexities in this large population of KCs, which mapped to five different clusters. The single-cell transcriptomes for these different subsets of KCs, including the one uniquely affected during the caste transition (see below), provide molecular entry points to investigate the behavioral functions of different KC subtypes in social insects.
In addition to an enlarged mushroom body, our single-cell RNA-seq analyses revealed a prominent glia compartment in Harpegnathos (27%) compared to the 2 to 10% reported for Drosophila by single-cell RNA-seq (14, 15, 18) and imaging (20, 21) studies. We detected glia of known insect subtypes, including surface, cortex, and neuropile glia, the latter further divided into ensheathing glia and astrocyte-like cells (20). Three relatively small clusters (named G0, G1, and G2) did not express clearly identifiable markers and therefore constitute either ant-specific glial cells that do not exist in Drosophila or cells that exist in Drosophila but that have not been captured by existing studies. Additional evidence for the complexity of ant glia was found in the presence of three astrocyte subsets (Fig. 2), suggesting a subspecialization of this cell type in ants.
Caste-specific differences
Among the most remarkable properties of social insects is the marked difference in the behavioral and physiological phenotypes that originate from the same genome and give rise to the different social castes. Differences in the relative size of the brain and its anatomical compartments between different castes and even between individuals from the same caste allocated to different tasks (e.g., nursing versus foraging) have been reported (7–9, 45–48), but a comprehensive molecular comparison of cell types across castes was lacking. Here, we performed this comparison by taking advantage of the unique ability of Harpegnathos workers to convert to queen-like gamergates. We found evidence of caste-specific cellular plasticity in cell type composition of both neurons and glia, including an increase in the relative abundance of sNPF-expressing mushroom body KCs and ensheathing glia in gamergates accompanied by a relative reduction in perineurial glia (Fig. 3). While the caste- and age-associated dynamics of ensheathing glia were confirmed in an independent dataset (Fig. 5) and in a different species (Fig. 6), the relatively small changes in perineurial glia and KC A neurons were only detected in one experiment (single-cell RNA-seq at day 30), and therefore, future work will be required to confirm these findings and investigate their biological implications.
The changes in ensheathing glia caught our attention because they were substantial (40% increase at day 30 and 250% at day 120) and consistent across biological replicates, time points, and sequencing technology (10× Genomics and Drop-seq). Single-cell RNA-seq can only provide relative numbers based on relative capture efficiency, and therefore, we cannot formally exclude that the apparent increase of ensheathing glia in gamergates is the result of a simultaneous loss in absolute numbers of other cell types or that differences in recovery of this particular cell type from worker brains compared to gamergate brains might have affected our observations; however, the most parsimonious explanation is that ensheathing glia respond to caste reprogramming by increasing in numbers, most likely via proliferation. It is known that insect glia proliferate in response to injury (33, 34, 39), but to our knowledge, this is the first observation of a programmed, regulated expansion of a glia subset in otherwise healthy brains.
By reanalyzing published data, we detected caste-specific differences in the bulk expression of some ensheathing markers in three other social insects (fig. S5)—D. quadriceps and P. canadensis, both with a flexible worker caste, similar to Harpegnathos, and also in the ant C. planatus, which has a fixed social structure with developmentally defined workers and queens. It is important, however, to note that these results in other Hymenoptera must be interpreted with much caution as they are based on bulk RNA-seq for unconfirmed markers and not all comparisons were statistically significant. Together, these observations suggest that glia plasticity, and, more specifically, an increased frequency of ensheathing glia, accompany the worker-gamergate transition in Harpegnathos and might be a conserved feature in the reproductive castes of other social insects.
Ensheathing glia in injury and aging
In Drosophila, ensheathing glia play a critical role in the response to brain injury. They phagocytose degenerated axons after nerve damage via a well-studied molecular pathway that involves the phagocytic receptor Drpr, its adaptor protein dCed-6, and a signaling pathway that includes TRAF4, JNK, and STAT92E (49). Drpr knockout leads to neurodegeneration (50) as well as reduced life span in a Drosophila model of Alzheimer’s disease (51), supporting the notion that ensheathing glia have a neuroprotective function.
Three lines of evidence suggest that the analogous cell type in Harpegnathos retains this function. First, the cells that we identified as ensheathing glia expressed ced-6 as well as egr, which are required for the response to brain injury in Drosophila (33). Second, after acute brain damage in Harpegnathos workers, a subset of glial cells expressing a combination of markers specific for ensheathing glia (egr, Slc5eg, Nep5L) up-regulated the activation marker Mmp1 (Fig. 4), which is downstream of Drpr (51) and is expressed upon injury in Drosophila ensheathing glia (41). Third, the decline in the numbers of ensheathing glia during aging was accompanied by a reduced Mmp1 up-regulation in response to brain injury (Fig. 5). The functional homology of Harpegnathos ensheathing glia with neuroprotective glia in other animals might extend beyond insects, as microglia, the major glia cell type that responds to injury in mammalian brains (38, 52, 53), express Tnf, the mammalian homolog of the ensheathing glia marker egr. Furthermore, the ensheathing glia-specific Nep5L marker encodes a member of a metalloendopeptidase family that has been linked to the clearing of amyloid plaques in mouse brains (54).
In mammals, glia composition and properties display developmental and age-related dynamics (44, 55, 56) including a shift to a more neuroprotective function as the brain ages (57), and restoration of the phagocytic function of microglia in aged mice improves their cognitive performance (58). In Drosophila, glia are thought to play a role in regulating health and life span (43), and ensheathing glia from aged individuals are less effective in clearing accumulated neuronal debris (41), suggesting that declining ensheathing glia might be a cause rather than a consequence of aging. Given the extreme difference in life span between workers and gamergates in Harpegnathos, we hypothesized that the glia composition from the different castes would display divergent kinetics during aging. Aging workers exhibited a progressive loss of ensheathing glia cells (Fig. 5, A and B), whereas in gamergates, they remained a larger proportion of total glia cells over the same time span (Fig. 5B). Consistent with this, old gamergates retained the ability to mount a response to neuronal damage, which was lost in old workers (Fig. 5C). Last, transcriptomic analyses and imaging of genetic reporters in Drosophila revealed that ensheathing glia marker genes are down-regulated in older brains (Fig. 6), suggesting that aging-associated decline of ensheathing glia is a conserved phenomenon.
Conclusions and outlook
Single-cell RNA-seq profiling of Harpegnathos brains as they undergo the adult caste transition from worker to gamergate revealed plastic events in neurons and glia. Although they were not explored here, the dynamics observed in KC and other cell types are noteworthy and deserve to be further analyzed in future studies. Neuroprotective ensheathing glia cells were substantially expanded in gamergates as early as day 30 of the transition, and their frequency declined in old workers but not age-matched gamergates. Although our results cannot prove a causal link, we propose that a homeostatic expansion of ensheathing glia in Harpegnathos gamergates promotes healthy brain aging by increasing its repair capacity, thus contributing to a prolonged life span. Along similar lines, we speculate that ensheathing glia play a broader role in regulating life-span differences between castes in other social insects and that elevated numbers and proper function of phagocytic glia are required for healthy brain aging across the animal kingdom.
METHODS
Worker-gamergate transitions
Harpegnathos colonies were housed in plastic boxes with a plaster nest chamber in a temperature- and humidity-controlled ant facility on a 12-hour light cycle and maintained on a live cricket diet as previously described (4). To induce worker-gamergate transitions, we transferred 20 callow females (3 to 4 days old; backgrounds DR-91, TL, DR-105, and DR-101) from mature colonies to a new nest with four males. Each ant was individually painted with a unique two-color combination. After 20 days, ants were monitored for signs of caste-specific behavior, including foraging activity to identify workers and egg laying to identify for gamergates. Ten days later (for a total of 30 days of transition), individuals were euthanized and brains were collected by dissection. The caste of each individual was confirmed by inspecting ovaries for the presence of mature oocytes. Only individuals who laid eggs and had mature oocytes in their ovaries by day 30 were classified as gamergates, whereas only individuals who foraged and had inactivated ovaries were considered workers.
Single-cell dissociation
For consistency with our previous study (4), we removed the optic lobes during all brain dissections. To dissociate cells, 1 ml of papain solution [5-ml Dulbecco’s modified Eagle’s medium (DMEM) per vial] with 45 μM actinomycin D to block new transcription during processing was added to each brain, which was rotated at room temperature for 12 min. Brains were washed twice with DMEM, once with phosphate-buffered saline (PBS)–B (PBS + 0.01% bovine serum albumin) and resuspended in 200-μl PBS-B, pipetting up and down with 200-μl tips about 20 times and gel loading tips about 30 times on ice. Samples were incubated for 5 min at room temperature, and pipetting was repeated. Samples were filtered on a 40-μm Flowmi strainer, and live cells were counted by trypan blue exclusion using both a Countess II and a hemocytometer.
10× Genomics
For each biological replicate, three to four brains (cell viability >80%) were pooled. Samples were mixed with Drosophila S2 cells at a 10:1 cell ratio as a quality control. Cells were added to the RT mix with the aim of capturing the transcriptomes of ~5000 to 10,000 cells. All downstream complementary DNA (cDNA) synthesis (12 PCR cycles), library preparation, and sequencing were carried out as instructed by the manufacturer (10× Genomics Chromium Single Cell 3’ Reagent Kit v2 User Guide RevD), with minor modifications. Custom barcodes were used as indexes. After PCR, libraries were sequenced on an Illumina NextSeq 500. The read configuration was 26 (read 1), 8 (index 1), and 58 base pairs (bp) (read 2). Libraries were loaded at 2.0 pM.
Immunofluorescence
Harpegnathos brains were dissected in DMEM and fixed in 4% paraformaldehyde overnight at 4°C. Fly heads were dissected in 1× PBS and fixed in 4% paraformaldehyde for 2 hours at room temperature. Washed in PBS twice and embedded with 4% agarose gel in PBS. Thick sections (100 μm) were obtained with a Vibratome (Leica VT1000S), permeabilized in PBST (1× PBS + 0.5% Triton X-100) two times for 10 min, and then blocked with 5% goat serum in PBST for at least 1 hour at room temperature. Sections were incubated with primary antibodies overnight at 4°C in 5% goat serum in PBST, washed in PBST three times for 20 min, incubated with appropriate fluorescently labeled secondary antibodies for 2 hours in PBST, washed six times with PBST, and stained with 4′,6-diamidino-2-phenylindole (DAPI) for 10 min. Last, samples were washed three times for 5 min and then mounted in antifade mounting medium (Vector Laboratories, H-1000). Sections were imaged with a Leica SPE laser scanning confocal microscope. Antibodies used were as follows: anti-Synapsin: DSHB 3C11 (1:20); anti–Pka-C1: Cusabio (1:1000); anti-egr (1:500, a gift from M. Miura) (59); anti-mouse immunoglobulin G (IgG) (H+L), Alexa Fluor 488: ThermoFisher Scientific (1:500), anti-rabbit IgG (H+L), Alex Fluor 568: ThermoFisher Scientific (1:500).
Western blots
Harpegnathos and Drosophila brains were dissected, separated from the optic lobes, homogenized in protein extraction reagent (ThermoFisher Scientific, no. 89822) with protease inhibitors, frozen in −80°C for at least 30 min, and then thawed for 15 min at room temperature with gentle rocking. Lysates were centrifuged at 16,000g for 20 min to remove insoluble material. Antibodies used were as follows: anti-tubulin: DSHB E7 (1:500), anti–Pka-C1: Cusabio (1:2000). Anti-mouse IgG (H+L): Jackson Immune Research (1:5000), Anti-rabbit IgG (H+L): Jackson Immune Research (1:5000).
Stabbing brain injury
Ten-day-old callow workers were separated from stable colonies and briefly anesthetized on dry ice. Each ant was punctured with a tin sterile insect pin (Fine Science Tools no. 26002-15) in the proximity of the ocelli. Controls were age-matched callow workers that were subjected to the same experimental procedure minus the injury. After 30 min of recovery, ants were returned to their colony. Brains were collected 1 or 3 days after injury.
Antenna ablation and RT-qPCR
Thirty- and 120-day-old workers were collected from stable colonies and briefly anesthetized on dry ice. The right antenna was surgically removed at the base of the head using scissors heat-sterilized with a Germinator 500 (Braintree Scientific). As a control, age-matched workers were subjected to the same experimental procedure, including anesthesia, but the antenna was not removed (mock treatment). Thirty minutes after amputation, the ants were returned to their colony. Brains were harvested 3 days after injury, and the two hemispheres were separated. RNA was purified with TRIzol from each hemisphere, and expression of Mmp1 was quantified by RT-qPCR with primers CGCTATCCAGGCCTTGTACG and CCTTGGAGTTGAACATCGCG using Power SYBR Green RNA-to-CT (ThermoFisher Scientific). Rpl32 was used as a reference gene (4); primer sequences, CGTAGGCGATTTAAGGGTCA and TTTCGGAAGCCAGTTGGTAG.
Drop-seq (injury and aging)
For each sample, two brains were pooled. Ant cells were mixed with Drosophila S2 cells at a 10:1 ratio as a quality control. The final cell suspension was diluted to 100 cells/μl in PBS, and 1.5 ml of the cell suspension was loaded for each Drop-seq run. Barcoded beads were resuspended in freshly made lysis buffer, composed of 200 mM tris-HCl (pH 8.0RT), 20 mM EDTA, 6% Ficoll PM-400 (GE Healthcare), 0.2% sarkosyl (Sigma-Aldrich), and 50 mM dithiothreitol, at a concentration of 120 beads/ml. The flow rate for cells and beads were set to 4000 ml/hour, while the droplet generation oil (Bio-Rad) was flown at 15,000 ml/hour. Droplets were generated and collected in a 50-ml Falcon tube for a run time of 15 min. Droplet breakage was performed by with perfluoro-1-octanol (Sigma-Aldrich), followed by reverse transcription, exonuclease I treatment, and amplification of cDNA according to the Drop-seq protocol from the McCarroll laboratory (http://mccarrolllab.org/dropseq/), with minor modifications. cDNA was amplified using the Terra PCR Direct Polymerase Mix Kit. After two rounds of purification with 0.6× SPRIselect beads, we tagmented 600 pg of DNA using the Nextera XT DNA Sample Preparation Kit (Illumina, catalog no. FC-131-1096). Libraries were further amplified with 12 PCR cycles using custom P5-TSO hybrid and custom Nextera-compatible primers with different indexes. Libraries were loaded at 2.0 pM. Custom read 1 and index 2 primers were used, and libraries were sequenced on an Illumina NextSeq 500. The read configuration was 20 (read 1), 8 (index 1), 8 (index 2), and 56 bp (read 2).
Aging workers and gamergates
Newly eclosed ants from four stable colonies (backgrounds DR-91, TL, DR-105, and DR-101) were painted individually with a unique two-color combination and returned to the colony of origin. Brains were collected after 5, 30, 90, and 120 days. Although spontaneous conversion of workers to gamergates in stable colonies is rare, the ovaries of the collected individuals were inspected for the absence of mature oocytes to confirm worker status. Gamergates (120-day-old) were collected from stable colonies, and their ovaries were inspected to confirm their gamergate status.
Gene expression in young and old Drosophila brains
For the aging experiment, y1w* flies were raised at 25°C and 50% humidity on a 12-hour light/dark cycle using standard Bloomington Drosophila Medium (Nutri-Fly). Newly eclosed flies, over a 24-hour period, were transferred into fresh food vials. After 48 hours, 10 mated females were collected under mild CO2 anesthesia and transferred into fresh food vials. Flies were maintained in the same conditions as described above and transferred to new food vials twice a week. After 5 (young) and 70 days (old), brains were dissected and RNA was purified with TRIzol. RNAs were quantified using the Power SYBR Green RNA-to-CT 1-step kit.
For library preparation, polyA+ RNA was purified using Dynabeads Oligo(dT)25 (ThermoFisher Scientific) beads and constructed into strand-specific libraries using the deoxyuridine triphosphate method. Uridine triphosphate–marked cDNA was end-repaired using end-repair mix (Enzymatics, MA), tailed with deoxyadenine using Klenow exo- (Enzymatics), and ligated to custom dual-indexed adapters with T4 DNA ligase (Enzymatics). Libraries were size-selected with SPRIselect beads (Beckman Coulter, CA) and quantified by qPCR before and after amplification. Sequencing was performed in a paired-end mode on a NextSeq 500 (Illumina, CA).
Quantification of ensheathing glia in Drosophila using genetic reporters
GMR10E12-Gal4 (46517, Bloomington Drosophila Stock Center) and GMR56F03-Gal4 (39157, Bloomington Drosophila Stock Center) were used to mark ensheathing glia by crossing with the reporter line UAS-GFP (4775, Bloomington Drosophila Stock Center). Flies were raised at 25°C and 50% humidity on a 12-hour light/dark cycle using standard Bloomington Drosophila Medium (Nutri-Fly). Newly eclosed flies, over a 24-hour period, were transferred into fresh food vials. After 48 hours, 10 mated females were collected under mild CO2 anesthesia and transferred into fresh food vials. Flies were maintained under the same conditions as described above and transferred to new food vials twice a week. After 5 (young) and 70 days (old), brains were dissected in 1× PBS and fixed in 4% paraformaldehyde for 45 min at room temperature. Washed in PBS twice and permeabilized in PBST (1× PBS + 0.5% Triton X-100) three times for 15 min and then blocked with 5% goat serum in PBST for at least 1.5 hours at room temperature. Brains were incubated with primary antibodies overnight at 4°C in 5% goat serum in PBST, washed in PBST three times for 20 min, incubated with appropriate fluorescently labeled secondary antibodies for 2 hours in PBST, washed four times with PBST, and stained with DAPI for 10 min. Last, samples were washed three times for 5 min, then mounted in antifade mounting medium (Vector Laboratories, H-1000), and imaged with a Leica SPE laser scanning confocal microscope. Antibodies used were anti-repo, DSHB 8D12 (1:5), anti-GFP (1:500; ab13970), anti-chicken IgG (H+L), Alexa Fluor 488 (1:500; ThermoFisher Scientific), and anti-mouse IgG (H+L), Alexa Fluor 568 (1:500; ThermoFisher Scientific). FIJI and ilastik software were used for imaging processing and quantification. The central brain [also referred to as nonvisual brain (4) or midbrain (15) in other studies] were quantified in GMR10E12-GFP and GMR56F03-GFP flies as shown by the dashed lines in Fig. 6 (E and G). Total glial cells were measured by quantifying anti-repo fluorescence signal, and ensheathing glia were measured by quantifying anti-GFP fluorescence signal. In all cases, quantification was done in all Z planes of ≥ 4 brains for each condition.
Computational analyses
Single-cell data processing
Sequencing files from both 10× Genomics and Drop-seq data were processed using Drop-seq tools v2.0.0, developed by the McCarroll Lab (https://github.com/broadinstitute/Drop-seq). Sequencing data were aligned to a combined reference of the current H. saltator assembly (11) (Hsal_v8.5) and the D. melanogaster genome assembly (assembly Release 6 plus ISO1 MIT). Reads mapping to features from the Harpegnathos annotation (Hsal_v8.5) were counted, and a UMI matrix was produced for all cells with a minimum number of 150 genes.
Comparison of single-cell to bulk sequencing
Data from bulk sequencing of Harpegnathos nonvisual brains from 11 workers and 12 gamergates at day 120 (4) were downloaded from GSE83807 and aligned to the Hsal_v8.5 reference. Differentially expressed genes were called using DESeq2 (60). For the single-cell sequencing data, digital gene expression (DGE) matrices were made using the Drop-seq tools v2.0.0, developed by the McCarroll Lab (https://github.com/broadinstitute/Drop-seq), but using a cutoff of one gene per cell. This resulted in DGE matrices containing all information from the single-cell data (every read mapped to the exon of a gene, including from cells not used in clustering). The fold change between worker and gamergate of the top 100 differentially expressed genes in bulk sequencing was compared between bulk and single-cell data.
10× Genomics data analysis
Analysis of the UMI matrix produced by Drop-seq tools was performed with Seurat v2.3.4 (19). Cells with more than 200 genes and 500 UMIs were retained, with each gene being detected in at least three cells. Cells from each sample were log-normalized with a scale factor of 10,000. Data were then scaled so the mean of each gene across cells was 0 and the variance across cells was 1. As the data were generated in three separate experiments (experiment 1, 2 workers and 1 gamergate; experiment 2, 2 workers and 2 gamergates; experiment 3, 2 workers and 2 gamergates), experiment was regressed out along with nUMI using a linear model during the scaling step. After normalizing and scaling, variable genes were found using the FindVariableGenes function in Seurat with default parameters except x.low.cutoff = 0.0125, x.high.cutoff = 3, y.cutoff = 0.5. A total of 1447 variable genes were detected using this method.
Clustering and visualization of the data were accomplished using Seurat’s linear dimension reduction (principal components analysis) followed by t-distributed stochastic neighbor embedding (tSNE). Principal components selected for use in the tSNE were chosen by running a JackStraw resampling test and choosing all components until a component had a P > 0.05. Cells were clustered using a resolution of 1.
For reclustering of neurons and glia, cells were selected from full clustering by average cluster expression of neuron markers and glia markers. Reclustering was performed as above, but with a resolution of 0.3 for glia and 0.6 for neurons. Reclustering the glia resulted in one cluster with a far lower median UMI compared to other glia cells—these cells (marked as “***” in fig. S3, A and B) were discarded as potential low-quality cells. Marker genes for each cluster were determined by the Seurat FindMarkers function, with the parameters only.pos = T and min.pct = 0.25.
Pseudobulk expression measurements by sample and cluster
For cluster-level pseudobulk expression analyses (e.g., Fig. 1C), the number of UMI in for each gene from all cells in the cluster from each sample were added together and normalized by the total number of UMI detected in cells from that sample and cluster.
Transcription factors
Homologs for all Drosophila genes in the “transcription factors” gene group (FBgg0000745) from FlyBase were considered for analysis. The pseudobulk expression from each cluster (see above) for each transcription factor detected in single-sequencing was compared between neurons and glia to determine transcription factors specific for one of the cell types. All transcription factors were used to hierarchically cluster neuron and glia clusters, but only those with a | log fold change | > 1 were included in the heatmap generated by pheatmap (scaled by row) in Fig. 1E.
Reanalysis of Drosophila single-cell data
DGE matrices for Drosophila midbrain (15) and Drosophila brain (14) were downloaded from eLife (figure 1, source data 1) and GSE107451, respectively. Analysis of these DGE matrices was performed as above with modifications.
For the midbrain dataset, the minimum UMI threshold was set at 800, as in (15), and replicate was regressed out along with UMI during the scaling step. The resulting 10,305 cells were clustered using a resolution of 2.5. Neurons and glia were separated using neuron and glia markers, including nSyb and bdl (fig. S1C). A resolution of 2.5 was also used for reclustering the 9916 neurons. Very few cells were in clusters marked by glia-specific genes (in total, 373 glia); as in (15), we were unable to recover known glia clusters other than astrocytes upon reclustering glia.
For the full brain dataset, the minimum UMI threshold was 500 UMIs, yielding 56,875 cells, similar to the 56,902 cells analyzed in (14). Cells were clustered with a resolution of 2.0, and neurons and glia were separated as above (fig. S1C), resulting in 46,944 neurons and 3614 glia [comparable to the 3600 glia reported in (14)]. Neurons were reclustered with a resolution of 0.6, while glia were reclustered at a resolution of 0.2.
Subsampling of single-cell data
To compare Harpegnathos and Drosophila on equal footing from a technology perspective, we equalized the number of cells in the datasets while matching the UMI distribution. To accomplish this, we first reduced each dataset to the same number of cells. We then ordered the cells by number of UMI, and for cells at each index sampled the UMI to be equal to the same number as the cell at that index with the lowest number of UMI. We then renormalized and scaled the data as described above.
GO analysis
An in-house script (available upon request) was used for GO enrichment analysis. GO terms were assigned to Harpegnathos genes based off of GO terms assigned to their Drosophila and/or human homologs. For each ontology, enrichment for a gene set was calculated using the number of genes annotated with a particular GO term compared to genes not in the set. The universe of genes considered included only genes detected in the relevant cell subset (e.g., only neurons or only glia). Gene sets for neuron clusters or astrocyte clusters consisted of marker genes determined per cluster by Seurat (see above). GO analyses from the “biological process” category were visualized in heatmaps of –log10(P values) for each term in each cluster using pheatmap, scaled by row, to determine whether GO terms were broadly enriched or specific for certain clusters. Only terms with a Benjamini-Hochberg–adjusted P value of <0.1 were visualized on heatmaps.
Statistics for cell type frequency shifts between castes
P values were calculated for cell type frequency shifts between workers and gamergates using a similar procedure to the test for changes in cell type proportions recently implemented by Regev and colleagues (61). Cell type counts were modeled using a negative binomial distribution, with the total number of cells in each sample as an offset variable and the caste identity of the individual (worker or gamergate) as a covariate. This procedure was implemented in R using the glm.nb function from the MASS package, and P values were calculated using a Wald test on the regression coefficient. P values were adjusted for multiple testing using the Benjamini-Hochberg procedure.
Drop-seq data analysis
Drop-seq data were processed as above, but with cutoffs of 200 UMIs and a resolution of 0.6 (for full clustering and glia reclustering). For the injury experiment (Fig. 4), Drop-seq data for aging experiment (day 5, day 30, day 90, and day 120 workers and day 120 gamergates) and injury experiment (injury and age-matched controls) were processed together, but only control/injury cells were shown in tSNE visualization and used for the subsequent analyses.
Analysis of published D. quadriceps, C. planatus, and P. canadensis RNA-seq
For RNA-seq from other Hymenoptera, homologs were identified using BLAST. Protein sequences were directly obtained from National Center for Biotechnology Information (NCBI) annotations for D. quadriceps (ASM131382v1) and P. canadensis (ASM13138v1). Because of the lack of genome and annotation for C. planatus, we took advantage of its close evolutionary relationship with C. floridanus (16) and mapped against NCBI annotation for that species [Cflo v7.5 (11)]. These protein sequences were queried against a BLASTp database created from Harpegnathos protein sequences [Hsal_v8.5 (11)]. Homologs were defined as the gene with the best match, with an e-value threshold of 10−5. RNA-seq FASTQ files were downloaded from Gene Expression Omnibus (GEO) (D. quadriceps and P. canadensis, GSE59525; C. planatus, PRJNA472392) and were mapped against the assemblies above using STAR v2.4.1d. Gene-level counts and reads per kilobase per million (RPKMs) were calculated using GenomicRanges.
Bulk RNA-seq analyses in Drosophila brains
RNA-seq reads were mapped to the D. melanogaster assembly BDGP6 preindexed with transcript models from Ensembl 87 using STAR 2.5.0b with default parameters except --alignIntronMax set to 10,000. Aligned reads were assigned to gene models using the summarizeOverlaps function of the GenomicRanges R package. RPKMs were calculated with a slight modification, whereby only reads assigned to annotated protein-coding genes were used in the denominator, to minimize batch variability due to different amounts of contaminant ribosomal RNA. Differential expression was determined using the DESeq2 package and visualized by conventional MA plots using the log2(fold change) calculated from RPKMs. Because Davie et al. (14) identified two slightly different clusters of ensheathing glia cells (referred to as ens-A and ens-B), we only considered markers those genes expressed in both.
Other datasets used
Bulk sequencing data from Harpegnathos workers and gamergates at day 120 (4) were downloaded from GSE83798. Single-cell RNA-seq from Drosophila brains was downloaded from supplementary material from (15) and GSE107451 (14). Bulk RNA-seq data for D. quadriceps and P. canadensis were downloaded from GSE59525 (6). Bulk RNA-seq data for C. planatus were downloaded from PRJNA472392 (16).
Supplementary Material
Acknowledgments
We thank M. Miura for the gift of the egr antibody; T. Christopher and C. Brady for technical support; E. Torre for help in setting up Drop-seq; and J. Bozler, B. Kacsoh, L. Yu, D. Reinberg, and C. Desplan for comments on the manuscript. Funding: R.B. was supported, in part, by the Searle Scholars Program (15-SSP-102) and an NIH New Innovator Award DP2MH107055. This project was supported by a New Initiative Research Grant from the Charles E. Kaufman Foundation to R.B. and A.R. (KA2016-85223). A.R. was also supported by a NIH Center for Photogenomics grant (RM1 HG007743) and an NIH Transformative Research Award (R01GM137425). Funding for this work in S.L.B.’s laboratory was provided by the NIH (R01 AG055570). Author contributions: R.B., L.S., and E.J.S. designed the study. L.S. performed most experiments with help from J.G. and generated all data. E.J.S. performed all bioinformatic analyses. P.R., S.L., and A.R. helped with microscopy. K.M.G. provided aged flies. E.J.S., L.S., and R.B. wrote the manuscript with help from J.G., K.M.G., and S.L.B. Competing interests: A.R. receives royalties from LGC, Biosearch Technologies related to Stellaris RNA fluorescence in situ hybridization probes. The other authors declare that they have no competing interests. Data and materials availability: RNA-seq data generated for this study have been deposited in the NCBI GEO with accession no. GSE149668. An interactive web application to explore our single-cell RNA-seq data is available online at http://antSCout.bonasiolab.org. All other data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/34/eaba9869/DC1
REFERENCES AND NOTES
- 1.Robinson G. E., Fernald R. D., Clayton D. F., Genes and social behavior. Science 322, 896–900 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonasio R., Emerging topics in epigenetics: Ants, brains, and noncoding RNAs. Ann. N. Y. Acad. Sci. 1260, 14–23 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Keller L., Genoud M., Extraordinary lifespans in ants: A test of evolutionary theories of ageing. Nature 389, 958–960 (1997). [Google Scholar]
- 4.Gospocic J., Shields E. J., Glastad K. M., Lin Y., Penick C. A., Yan H., Mikheyev A. S., Linksvayer T. A., Garcia B. A., Berger S. L., Liebig J., Reinberg D., Bonasio R., The neuropeptide corazonin controls social behavior and caste identity in ants. Cell 170, 748–759.e12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Libbrecht R., Oxley P. R., Keller L., Kronauer D. J., Robust DNA methylation in the clonal raider ant brain. Curr. Biol. 26, 391–395 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patalano S., Vlasova A., Wyatt C., Ewels P., Camara F., Ferreira P. G., Asher C. L., Jurkowski T. P., Segonds-Pichon A., Bachman M., González-Navarrete I., Minoche A. E., Krueger F., Lowy E., Marcet-Houben M., Rodriguez-Ales J. L., Nascimento F. S., Balasubramanian S., Gabaldon T., Tarver J. E., Andrews S., Himmelbauer H., Hughes W. O. H., Guigó R., Reik W., Sumner S., Molecular signatures of plastic phenotypes in two eusocial insect species with simple societies. Proc. Natl. Acad. Sci. U.S.A. 112, 13970–13975 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fahrbach S. E., Structure of the mushroom bodies of the insect brain. Annu. Rev. Entomol. 51, 209–232 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Withers G. S., Fahrbach S. E., Robinson G. E., Selective neuroanatomical plasticity and division of labour in the honeybee. Nature 364, 238–240 (1993). [DOI] [PubMed] [Google Scholar]
- 9.Ehmer B., Gronenberg W., Mushroom body volumes and visual interneurons in ants: Comparison between sexes and castes. J. Comp. Neurol. 469, 198–213 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Peeters C., Liebig J., Hölldobler B., Sexual reproduction by both queens and workers in the ponerine ant Harpegnathos saltator. Insectes Soc. 47, 325–332 (2000). [Google Scholar]
- 11.Shields E. J., Sheng L., Weiner A. K., Garcia B. A., Bonasio R., High-quality genome assemblies reveal long non-coding RNAs expressed in ant brains. Cell Rep. 23, 3078–3090 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghaninia M., Haight K., Berger S. L., Reinberg D., Zwiebel L. J., Ray A., Liebig J., Chemosensory sensitivity reflects reproductive status in the ant Harpegnathos saltator. Sci. Rep. 7, 3732 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gronenberg W., Liebig J., Smaller brains and optic lobes in reproductive workers of the ant Harpegnathos. Naturwissenschaften 86, 343–345 (1999). [Google Scholar]
- 14.Davie K., Janssens J., Koldere D., de Waegeneer M., Pech U., Kreft Ł., Aibar S., Makhzami S., Christiaens V., Bravo González-Blas C., Poovathingal S., Hulselmans G., Spanier K. I., Moerman T., Vanspauwen B., Geurs S., Voet T., Lammertyn J., Thienpont B., Liu S., Konstantinides N., Fiers M., Verstreken P., Aerts S., A single-cell transcriptome atlas of the aging drosophila brain. Cell 174, 982–998.e20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Croset V., Treiber C. D., Waddell S., Cellular diversity in the Drosophila midbrain revealed by single-cell transcriptomics. eLife 7, e34550 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chandra V., Fetter-Pruneda I., Oxley P. R., Ritger A. L., McKenzie S. K., Libbrecht R., Kronauer D. J. C., Social regulation of insulin signaling and the evolution of eusociality in ants. Science 361, 398–402 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.C. Hafemeister, R. Satija (2019); https://satijalab.org/howmanycells.
- 18.Konstantinides N., Kapuralin K., Fadil C., Barboza L., Satija R., Desplan C., Phenotypic convergence: Distinct transcription factors regulate common terminal features. Cell 174, 622–635.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R., Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kremer M. C., Jung C., Batelli S., Rubin G. M., Gaul U., The glia of the adult Drosophila nervous system. Glia 65, 606–638 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jenett A., Rubin G. M., Ngo T. T. B., Shepherd D., Murphy C., Dionne H., Pfeiffer B. D., Cavallaro A., Hall D., Jeter J., Iyer N., Fetter D., Hausenfluck J. H., Peng H., Trautman E. T., Svirskas R. R., Myers E. W., Iwinski Z. R., Aso Y., DePasquale G. M., Enos A., Hulamm P., Lam S. C. B., Li H. H., Laverty T. R., Long F., Qu L., Murphy S. D., Rokicki K., Safford T., Shaw K., Simpson J. H., Sowell A., Tae S., Yu Y., Zugates C. T., A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2, 991–1001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heisenberg M., Mushroom body memoir: From maps to models. Nat. Rev. Neurosci. 4, 266–275 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Gronenberg W., Heeren S., Hölldobler B., Age-dependent and task-related morphological changes in the brain and the mushroom bodies of the ant Camponotus floridanus. J. Exp. Biol. 199, 2011–2019 (1996). [DOI] [PubMed] [Google Scholar]
- 24.Farris S. A., Evolution of insect mushroom bodies: Old clues, new insights. Arthropod Struct. Dev. 34, 211–234 (2005). [Google Scholar]
- 25.Wu B., Li J., Chou Y.-H., Luginbuhl D., Luo L., Fibroblast growth factor signaling instructs ensheathing glia wrapping of Drosophila olfactory glomeruli. Proc. Natl. Acad. Sci. U.S.A. 114, 7505–7512 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ben Haim L., Rowitch D. H., Functional diversity of astrocytes in neural circuit regulation. Nat. Rev. Neurosci. 18, 31–41 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Zeisel A., Hochgerner H., Lönnerberg P., Johnsson A., Memic F., van der Zwan J., Häring M., Braun E., Borm L. E., la Manno G., Codeluppi S., Furlan A., Lee K., Skene N., Harris K. D., Hjerling-Leffler J., Arenas E., Ernfors P., Marklund U., Linnarsson S., Molecular architecture of the mouse nervous system. Cell 174, 999–1014.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee K.-S., Kwon O.-Y., Lee J. H., Kwon K., Min K.-J., Jung S.-A., Kim A.-K., You K.-H., Tatar M., Yu K., Drosophila short neuropeptide F signalling regulates growth by ERK-mediated insulin signalling. Nat. Cell Biol. 10, 468–475 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Castillo P., Pietrantonio P. V., Differences in sNPF receptor-expressing neurons in brains of fire ant (Solenopsis invicta Buren) worker subcastes: Indicators for division of labor and nutritional status? PLOS ONE 8, e83966 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matthes D. J., Sink H., Kolodkin A. L., Goodman C. S., Semaphorin II can function as a selective inhibitor of specific synaptic arborizations. Cell 81, 631–639 (1995). [DOI] [PubMed] [Google Scholar]
- 31.Freeman M. R., Drosophila central nervous system glia. Cold Spring Harb. Perspect. Biol. 7, a020552 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doherty J., Logan M. A., Tasdemir O. E., Freeman M. R., Ensheathing glia function as phagocytes in the adult Drosophila brain. J. Neurosci. 29, 4768–4781 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kato K., Awasaki T., Ito K., Neuronal programmed cell death induces glial cell division in the adult Drosophila brain. Development 136, 51–59 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Kato K., Forero M. G., Fenton J. C., Hidalgo A., The glial regenerative response to central nervous system injury is enabled by pros-notch and pros-NFκB feedback. PLOS Biol. 9, e1001133 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuchero J. B., Barres B. A., Glia in mammalian development and disease. Development 142, 3805–3809 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin X., Yamashita T., Microglia in central nervous system repair after injury. J. Biochem. 159, 491–496 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Salter M. W., Stevens B., Microglia emerge as central players in brain disease. Nat. Med. 23, 1018–1027 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Prinz M., Priller J., Microglia and brain macrophages in the molecular age: From origin to neuropsychiatric disease. Nat. Rev. Neurosci. 15, 300–312 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Kato K., Losada-Perez M., Hidalgo A., Gene network underlying the glial regenerative response to central nervous system injury. Dev. Dyn. 247, 85–93 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Macosko E. Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A. R., Kamitaki N., Martersteck E. M., Trombetta J. J., Weitz D. A., Sanes J. R., Shalek A. K., Regev A., McCarroll S. A., Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Purice M. D., Ray A., Münzel E. J., Pope B. J., Park D. J., Speese S. D., Logan M. A., A novel Drosophila injury model reveals severed axons are cleared through a Draper/MMP-1 signaling cascade. eLife 6, e23611 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galloway D. A., Phillips A. E. M., Owen D. R. J., Moore C. S., Phagocytosis in the brain: Homeostasis and disease. Front. Immunol. 10, 790 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kounatidis I., Chtarbanova S., Role of glial immunity in lifespan determination: A Drosophila perspective. Front. Immunol. 9, 1362 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soreq L.; UK Brain Expression Consortium; North American Brain Expression Consortium, Rose J., Soreq E., Hardy J., Trabzuni D., Cookson M. R., Smith C., Ryten M., Patani R., Ule J., Major shifts in glial regional identity are a transcriptional hallmark of human brain aging. Cell Rep. 18, 557–570 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mysore K., Subramanian K. A., Sarasij R. C., Suresh A., Shyamala B. V., VijayRaghavan K., Rodrigues V., Caste and sex specific olfactory glomerular organization and brain architecture in two sympatric ant species Camponotus sericeus and Camponotus compressus (Fabricius, 1798). Arthropod Struct. Dev. 38, 485–497 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Kuebler L. S., Kelber C., Kleineidam C. J., Distinct antennal lobe phenotypes in the leaf-cutting ant (Atta vollenweideri). J. Comp. Neurol. 518, 352–365 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Muscedere M. L., Traniello J. F., Division of labor in the hyperdiverse ant genus Pheidole is associated with distinct subcaste- and age-related patterns of worker brain organization. PLOS ONE 7, e31618 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith A. R., Seid M. A., Jimenez L. C., Wcislo W. T., Socially induced brain development in a facultatively eusocial sweat bee Megalopta genalis (Halictidae). Proc. Biol. Sci. 277, 2157–2163 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu T. Y., MacDonald J. M., Neukomm L. J., Sheehan A. E., Bradshaw R., Logan M. A., Freeman M. R., Axon degeneration induces glial responses through Draper-TRAF4-JNK signalling. Nat. Commun. 8, 14355 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Draper I., Mahoney L. J., Mitsuhashi S., Pacak C. A., Salomon R. N., Kang P. B., Silencing of drpr leads to muscle and brain degeneration in adult Drosophila. Am. J. Pathol. 184, 2653–2661 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ray A., Speese S. D., Logan M. A., Glial draper rescues Aβ toxicity in a Drosophila model of Alzheimer’s disease. J. Neurosci. 37, 11881–11893 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Szalay G., Martinecz B., Lénárt N., Környei Z., Orsolits B., Judák L., Császár E., Fekete R., West B. L., Katona G., Rózsa B., Dénes Á., Microglia protect against brain injury and their selective elimination dysregulates neuronal network activity after stroke. Nat. Commun. 7, 11499 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Herzog C., Pons Garcia L., Keatinge M., Greenald D., Moritz C., Peri F., Herrgen L., Rapid clearance of cellular debris by microglia limits secondary neuronal cell death after brain injury in vivo. Development 146, dev174698 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marr R. A., Hafez D. M., Amyloid-beta and Alzheimer’s disease: The role of neprilysin-2 in amyloid-beta clearance. Front. Aging Neurosci. 6, 187 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Puñal V. M., Paisley C. E., Brecha F. S., Lee M. A., Perelli R. M., Wang J., O’Koren E. G., Ackley C. R., Saban D. R., Reese B. E., Kay J. N., Large-scale death of retinal astrocytes during normal development is non-apoptotic and implemented by microglia. PLOS Biol. 17, e3000492 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anderson S. R., Roberts J. M., Zhang J., Steele M. R., Romero C. O., Bosco A., Vetter M. L., Developmental apoptosis promotes a disease-related gene signature and independence from CSF1R signaling in retinal microglia. Cell Rep. 27, 2002–2013.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hickman S. E., Kingery N. D., Ohsumi T. K., Borowsky M. L., Wang L.-C., Means T. K., El Khoury J., The microglial sensome revealed by direct RNA sequencing. Nat. Neurosci. 16, 1896–1905 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pluvinage J. V., Haney M. S., Smith B. A. H., Sun J., Iram T., Bonanno L., Li L., Lee D. P., Morgens D. W., Yang A. C., Shuken S. R., Gate D., Scott M., Khatri P., Luo J., Bertozzi C. R., Bassik M. C., Wyss-Coray T., CD22 blockade restores homeostatic microglial phagocytosis in ageing brains. Nature 568, 187–192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.lgaki T., Pastor-Pareja J. C., Aonuma H., Miura M., Xu T., Intrinsic tumor suppression and epithelial maintenance by endocytic activation of Eiger/TNF Signaling in Drosophila. Developmental Cell. 16, 458–465 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haber A. L., Biton M., Rogel N., Herbst R. H., Shekhar K., Smillie C., Burgin G., Delorey T. M., Howitt M. R., Katz Y., Tirosh I., Beyaz S., Dionne D., Zhang M., Raychowdhury R., Garrett W. S., Rozenblatt-Rosen O., Shi H. N., Yilmaz O., Xavier R. J., Regev A., A single-cell survey of the small intestinal epithelium. Nature 551, 333–339 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grams R., Korge G., The mub gene encodes a protein containing three KH domains and is expressed in the mushroom bodies of Drosophila melanogaster. Gene 215, 191–201 (1998). [DOI] [PubMed] [Google Scholar]
- 63.Skoulakis E. M., Kalderon D., Davis R. L., Preferential expression in mushroom bodies of the catalytic subunit of protein kinase A and its role in learning and memory. Neuron 11, 197–208 (1993). [DOI] [PubMed] [Google Scholar]
- 64.Suenami S., Paul R. K., Takeuchi H., Okude G., Fujiyuki T., Shirai K., Kubo T., Analysis of the differentiation of kenyon cell subtypes using three mushroom body-preferential genes during metamorphosis in the honeybee (Apis mellifera L.). PLOS ONE 11, e0157841 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Paul R. K., Takeuchi H., Kubo T., Expression of two ecdysteroid-regulated genes, Broad-Complex and E75, in the brain and ovary of the honeybee (Apis mellifera L.). Zoolog. Sci. 23, 1085–1092 (2006). [DOI] [PubMed] [Google Scholar]
- 66.Kamikouchi A., Takeuchi H., Sawata M., Ohashi K., Natori S., Kubo T., Preferential expression of the gene for a putative inositol 1,4,5-trisphosphate receptor homologue in the mushroom bodies of the brain of the worker honeybeeApis melliferaL. Biochem. Biophys. Res. Commun. 242, 181–186 (1998). [DOI] [PubMed] [Google Scholar]
- 67.Takeuchi H., Kage E., Sawata M., Kamikouchi A., Ohashi K., Ohara M., Fujiyuki T., Kunieda T., Sekimizu K., Natori S., Kubo T., Identification of a novel gene, Mblk-1, that encodes a putative transcription factor expressed preferentially in the large-type Kenyon cells of the honeybee brain. Insect Mol. Biol. 10, 487–494 (2001). [DOI] [PubMed] [Google Scholar]
- 68.Friggi-Grelin F., Coulom H., Meller M., Gomez D., Hirsh J., Birman S., Targeted gene expression in Drosophila dopaminergic cells using regulatory sequences from tyrosine hydroxylase. J. Neurobiol. 54, 618–627 (2003). [DOI] [PubMed] [Google Scholar]
- 69.Greer C. L., Grygoruk A., Patton D. E., Ley B., Romero-Calderon R., Chang H. Y., Houshyar R., Bainton R. J., DiAntonio A., Krantz D. E., A splice variant of the Drosophila vesicular monoamine transporter contains a conserved trafficking domain and functions in the storage of dopamine, serotonin, and octopamine. J. Neurobiol. 64, 239–258 (2005). [DOI] [PubMed] [Google Scholar]
- 70.Bang S., Hyun S., Hong S.-T., Kang J., Jeong K., Park J.-J., Choe J., Chung J., Dopamine signalling in mushroom bodies regulates temperature-preference behaviour in Drosophila. PLOS Genet. 7, e1001346 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peco E., Davla S., Camp D., M. Stacey S., Landgraf M., van Meyel D. J., Drosophila astrocytes cover specific territories of the CNS neuropil and are instructed to differentiate by Prospero, a key effector of Notch. Development 143, 1170–1181 (2016). [DOI] [PubMed] [Google Scholar]
- 72.Richier B., Vijandi C. M., Mackensen S., Salecker I., Lapsyn controls branch extension and positioning of astrocyte-like glia in the Drosophila optic lobe. Nat. Commun. 8, 317 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Durkee C. A., Covelo A., Lines J., Kofuji P., Aguilar J., Araque A., Gi/o protein-coupled receptors inhibit neurons but activate astrocytes and stimulate gliotransmission. Glia 67, 1076–1093 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stork T., Sheehan A., Tasdemir-Yilmaz O. E., Freeman M. R., Neuron-glia interactions through the Heartless FGF receptor signaling pathway mediate morphogenesis of Drosophila astrocytes. Neuron 83, 388–403 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.DeSalvo M. K., Hindle S. J., Rusan Z. M., Orng S., Eddison M., Halliwill K., Bainton R. J., The Drosophila surface glia transcriptome: Evolutionary conserved blood-brain barrier processes. Front. Neurosci. 8, 346 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Volkenhoff A., Weiler A., Letzel M., Stehling M., Klämbt C., Schirmeier S., Glial glycolysis is essential for neuronal survival in Drosophila. Cell Metab. 22, 437–447 (2015). [DOI] [PubMed] [Google Scholar]
- 77.Coutinho-Budd J. C., Sheehan A. E., Freeman M. R., The secreted neurotrophin Spätzle 3 promotes glial morphogenesis and supports neuronal survival and function. Genes Dev. 31, 2023–2038 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Melom J. E., Littleton J. T., Mutation of a NCKX eliminates glial microdomain calcium oscillations and enhances seizure susceptibility. J. Neurosci. 33, 1169–1178 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Goto A., Kadowaki T., Kitagawa Y., Drosophila hemolectin gene is expressed in embryonic and larval hemocytes and its knock down causes bleeding defects. Dev. Biol. 264, 582–591 (2003). [DOI] [PubMed] [Google Scholar]
- 80.Gonzalez-Morales N., Mendoza-Ortiz M. A., Blowes L. M., Missirlis F., Riesgo-Escovar J. R., Ferritin is required in multiple tissues during Drosophila melanogaster development. PLOS ONE 10, e0133499 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/34/eaba9869/DC1


