Abstract
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), the causative agent of a potentially fatal disease named coronavirus disease 2019 (COVID-19), has raised significant public health concerns globally. To date, the COVID-19 pandemic has caused millions of people to be infected with SARS-CoV-2 worldwide. It has been known since the 2003 SARS epidemic that coronaviruses (CoVs) have large RNA genomes, the replication of which requires an RNA-dependent RNA replication/transcription complex. CoV nonstructural proteins (Nsps) play pivotal roles in the assembly of this complex and associated enzymatic functions in virus genomic replication. Several smaller nonenzymatic Nsps assist with RNA-dependent RNA polymerase function. In this study, we determined the structure of SARS-CoV-2 nonstructural protein 9 (nsp9), an RNA-binding protein that is essential for CoV replication. Its homotetrameric structure with two stable dimeric interfaces provids a structural basis for understanding the mechanisms of RNA-binding protein self-assembly, which may be essential for the regulation of viral RNA replication and transcription.
Keywords: SARS-CoV-2, COVID-19, nsp9, Tetramer
Introduction
Coronavirus disease 2019 (COVID-19), an acute respiratory distress syndrome caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), is currently a global pandemic that has been spreading across all continents since late 2019 [1–3]. According to the World Health Organization COVID-19 situation reports, the number of individuals with a confirmed SARS-CoV-2 infection stood at approximately 16 million worldwide as of July 27, 2020, with related fatalities standing at over 0.64 million [4]. SARS-CoV-2 is a coronavirus (CoV) strain that belongs to the family Coronaviridae [5]. Genomic analysis suggests that SARS-CoV-2 is a new member of the genus Betacoronavirus, which is most closely related to the SARS-like virus previously identified in wild bats [6]. Human betacoronaviruses, including SARS-CoV, Middle East respiratory syndrome coronavirus (MERS-CoV), and SARS-CoV-2, share more sequence similarities with one another than with other CoVs [7]. However, SARS-CoV-2 has very high transmissibility, exhibiting rapid spreading through human-to-human contact, causing the WHO to declare it as a worldwide health emergency [8].
The genome of SARS-CoVs consists of an approximately 30-kb non-segmented positive-sense RNA sequence with a 5′ untranslated region (UTR), followed by a single open reading frame and a short flanking 3′ UTR [9]. The life cycle of SARS-CoVs starts with its binding to the cell surface receptor angiotensin-converting enzyme 2 (ACE2) through its spike glycoproteins, resulting in receptor-mediated endocytosis [10]. Upon entry into the host cell, the virus expresses its replicase gene to initially generate two precursor polyproteins, pp1a and pp1ab. These then undergo cleavage to yield 16 mature nonstructural proteins (nsp1–16) as well as several intermediate precursors by two distinct viral proteinases, the papain-like proteinase within nsp3 and a 3C-like proteinase nsp5 [11]. These nsps includes replicative enzymes (e.g., nsp12/RdRp, and nsp13/helicase), a variety of subunits performing accessory functions for viral RNA synthesis (e.g., nsp8/primase, nsp14/exoribonuclesae, and nsp15/endoribonuclease NendoU) and several smaller nonenzymatic nsps (nsp7, nsp9, and nsp10) assist with RNA-dependent RNA polymerase function [12–17].
As a member of the oligosaccharide/oligonucleotide-binding (OB-fold) superfamily, the RNA-binding protein nsp9 is critical to the RNA-dependent RNA replicase machinery of CoVs [18] nsp9 is essential for efficient viral growth as it plays a pivotal role in the formation of the replication and transcription complex machinery [19]. Mutation of the SARS-CoV nsp9 gene through a reverse genetics approach was found to prevent the propagation of the virus [20]. The sequences of nsp9 homologs are highly conserved among betacoronaviruses especially between SARS-CoV and SARS-CoV-2 (~ 97% sequence homology), and several nsp9 homologous structures have been determined in different viral species, including SARS-CoV [19, 21–24].
To elucidate the molecular mechanisms of SARS-CoV-2 replication, which would aid in the diagnosis, treatment, and prevention of the COVID-19 pandemic, researchers have made significant efforts to characterize the Nsp structures in a short amount of time since the COVID-2019 outbreak [18]. In this paper, we report the crystal structure of SARS-CoV-2 nsp9 at 2.95 Å. This structure was revealed to be a horseshoe-like tetramer, which may play an essential role in nsp9 oligomerization and in the regulation of viral nucleic acid binding during the replication of the virus. Two significant contact surfaces stabilize the SARS-CoV-2 nsp9 tetramer. One interface is composed of a parallel association of the C-terminal alpha (α)-helices and the N-terminal loop, which are highly conserved across CoV species. Antiparallel beta (β)-strands form the other interface from both protomers of nsp9, zippering the two β-barrels together. The structure of SARS-CoV-2 nsp9 provides insights into its multimerization and the regulation of viral nucleic acid binding during CoV replication.
Results
Overall structure of the SARS-CoV-2 nsp9 protomer
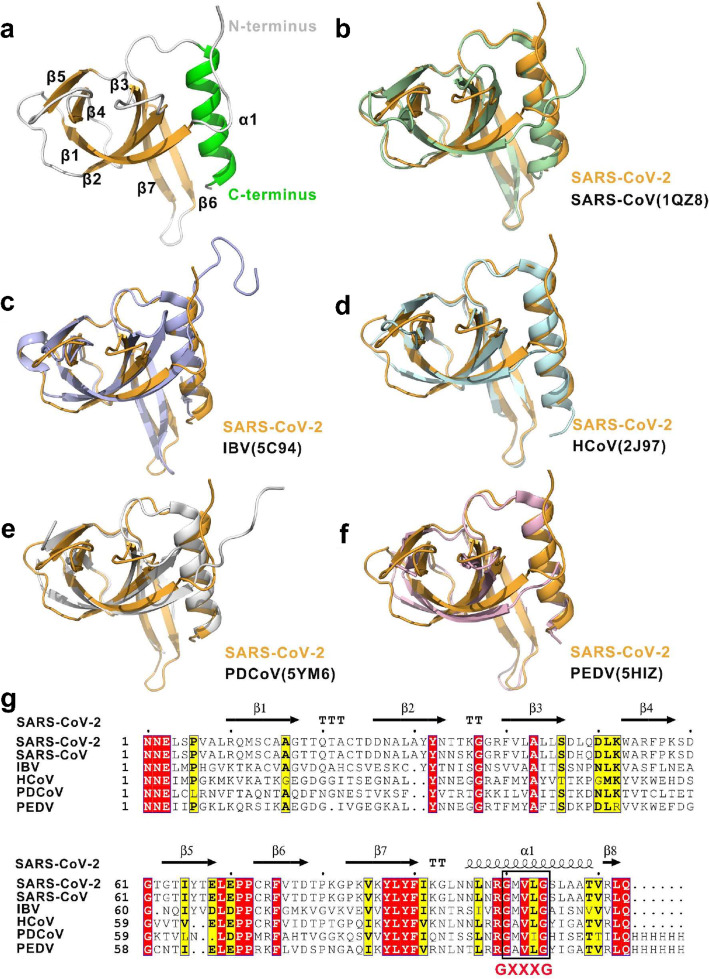
The SARS-CoV-2 nsp9 protomer is made up of seven β-strands (β1–β7) flanked by an N-terminal extension and a C-terminal α-helix (α1) (Fig. 1a). The core of the protein comprises a β-stranded barrel made up of two antiparallel β-sheets that are packed orthogonally. One of the sheets is formed by β1–β3 with a half strand of β7, and the other sheet is composed of the β4 and β5 strands. A tight β-hairpin involving β6 and β7 lies adjacent to the α-helix, extending out of the β-barrel. The elongated loops link the individual β-sheets of the barrel, along with the N-terminal β-strand and a C-terminal α-helix. The fold of this nsp9 protomer meets the structural features of OB-fold modules, one of which is a β-barrel consisting of six highly coiled antiparallel β-sheets [25]. Therefore, the structural assignment of the nsp9 monomer is the OB-fold, a compact structural motif in the OB-fold superfamily that is frequently used for nucleic acid recognition [26].
Fig. 1.
Overall structure of the SARS-CoV-2 nsp9 protomer. a Different views of the protomeric architecture of SARS-CoV-2 nsp9 with the secondary structure labeled. The α-helix, β-sheets, and loops are colored in green, orange, and white, respectively. b–f Superimposition of SARS-CoV-2 nsp9 with other nsp9 structures in the coronavirus family. The nsp9s from SARS-CoV-2, SARS-CoV, avian infectious bronchitis virus (IBV), human coronavirus 229E (hCoV-229E), porcine delta coronavirus (PDCoV), and porcine epidemic diarrhea virus (PDEV) are colored in orange, light blue, marine, white, and salmon, respectively. The PDB codes are indicated in the lower right corner. gSequence alignment of CoV nsp9 homologs. Comparison of the SARS-CoV, IBV, HCoV, PDCoV, and PDEV sequences with the SARS-CoV-2 sequence. Identical residues are highlighted in red, and conserved residues are shown in yellow. The table was produced with ESPript 3.0, using secondary structure elements for SARS-CoV-2 nsp9 assigned using DSSP. Residues boxed in red are completely conserved
The SARS-CoV-2 nsp9 protomeric structure is highly conserved among CoVs. To date, five nsp9 structures from various CoV species have been reported; namely, SARS-CoV nsp9 (PDB: 1QZ8), avian infectious bronchitis virus (IBV) nsp9 (PDB: 5C94), human coronavirus (hCoV) 229E nsp9 (PDB: 2 J97), porcine delta coronavirus (PDCoV) nsp9 (PDB: 5YM6), and porcine epidemic diarrhea virus (PEDV) nsp9 (PDB: 5HIZ) [21–24]. A superimposition of all protomeric nsp9 backbone coordinates showed that the root-mean-square deviation values between SARS-CoV-2 and SARS-CoV, IBV, HCoV, PDCoV, and PDEV are 0.562 Å for 88 Cα atoms, 0.70 Å for 73 Cα atoms, 1.311 Å for 50 Cα atoms, 3.940 Å for 74 Cα atoms, and 0.479 Å for 52 Cα atoms, respectively (Fig. 1b–f). These data suggest that the nsp9 structure of SARS-Cov-2 is highly similar to that of all CoVs, especially SARS-CoV nsp9 (Fig. 1g).
Structural basis for SARS-CoV-2 nsp9 multimerization
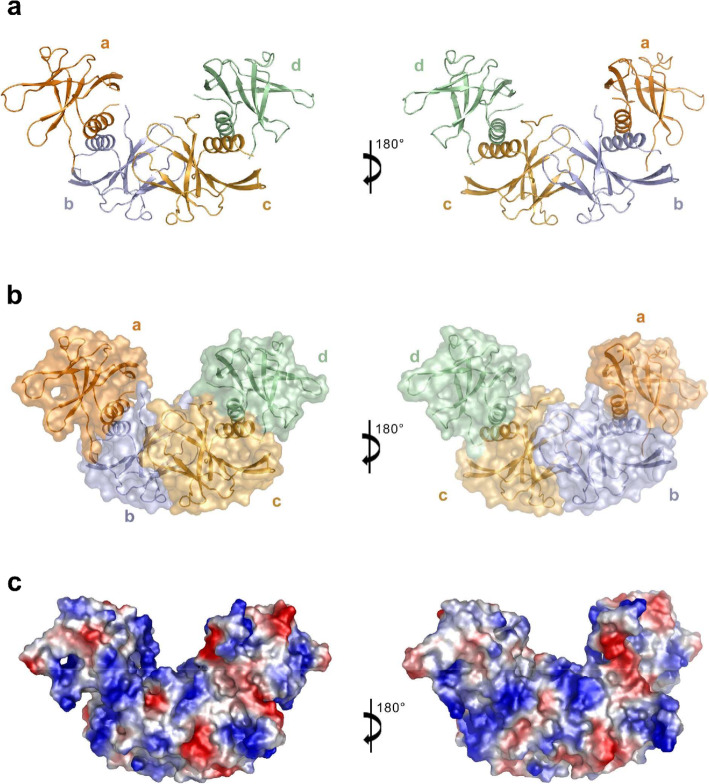
The initial structure solved by molecular replacement showed that six SARS-CoV-2 nsp9 protomers form an OB-fold cluster in an asymmetric unit (Supplementary Fig. 1a). The OB-fold cluster is stacked in three layers (I, II, and III). For the convenience of description, we have named the protomer in layer I as molecule a’, the three protomers in layer II as molecule b’/c’/d’, and the two protomers in layer III as molecule e’/f ’. To obtain more information about the protein interfaces and the likely biological assemblies of the OB-fold cluster, we calculated the structure of SARS-CoV-2 nsp9 using PDBePISA [27]. The buried area and binding energy results (Supplementary Table 1) indicated that there is one stable tetrameric form of SARS-CoV-2 nsp9 in the crystal lattice. Each protomer associated with the tetramer assembly comes from different asymmetric units in this structure (Supplementary Fig. 1b). In order to get the best view of the tetramer of nsp9 and to provid a general description of the structure, we refined the structure to get a new asymmetry unit that contains an intact tetrameric form of nsp9. In the new asymmetric unit, four of six protomers form a stable tetramer (molecule a/b/c/d) (Fig. 2a) while the other two protomers on the side of the tetramer, which forms tetramer with other two protomers in neighbor unit. Overall, four nsp9 protomers assemble into a horseshoe-like complex, with a buried area of close to 4638 Å2 (Fig. 2b and c).
Fig. 2.
Structure of the SARS-CoV-2 nsp9 tetramer. a Architecture and assembly of the nsp9 tetramer. Molecules a, b, c, and d in the stable tetramer are colored in orange, light blue, bright orange and pale green, respectively, and depicted in cartoon representations. b The stable tetramer is colored, labeled as Fig. 2a and depicted in cartoon and surface representations. c The electrostatic potential surface of the stable tetramer. Blue, positive charge (+ 70 kBT); Red, negative charge (− 70 kBT)
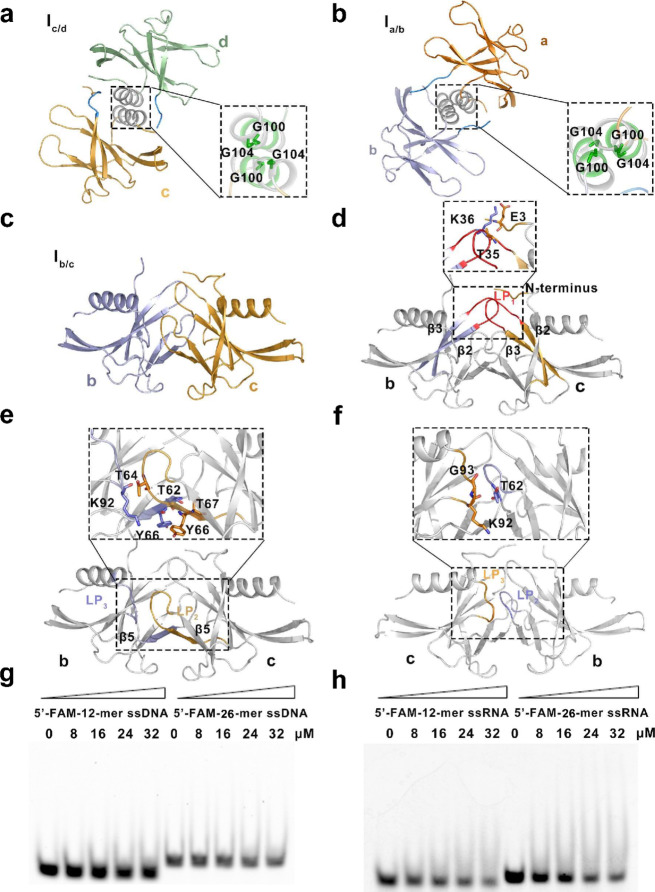
Three interfaces (Ia/b, Ib/c, and Ic/d) are engaged in the tetrameric structure in two combination modes (helix interface and sheet interface). Interfaces Ia/b (between molecules a and b) and Ic/d (between molecules c and d) share the same combination mode (i.e., the helix interface), having a buried area of 626 Å2 and 652 Å2, respectively. Each helix interface involves the conserved C-terminal α-helices (residues 96–109) that are associated in parallel and the N-terminus of nsp9 (residues 1–9) (Fig. 3a and b). The parallel α-helices allow two molecules to be packed together by a conserved protein–protein interaction motif (GXXXG). Residues G100 and G104, which are located on the helices from each subunit, play a pivotal role in tetramer stabilization. Further stabilization of this tetrameric interface is given by residues 6–10 in the N-terminus, clipping to the edge of β6 from its neighboring partner.
Fig. 3.
Details of structure of the SARS-CoV-2 nsp9 tetramer and function in nucleic acid binding. a, b Details of Ic/d and Ia/b in the helix interface. The molecules in these two interfaces are shown as cartoons and colored and labeled as in Fig. 2a. The expanded boxed area to the right shows a detailed view of the critical residues in the Ic/d and Ia/b interfaces, which are shown as sticks. c Molecules involved in Ib/c. The molecules in this interface are shown as cartoons and colored and labeled as in Fig. 2a. d–f Detailed views of the first, second, and third contact regions in interface Ib/c, respectively. The secondary structures of each contact region are colored and labeled according to Fig. 2a and the critical residues in each region are shown as sticks. g Single-stranded DNA (ssDNA)-binding abilities of SARS-CoV-2 nsp9, as determined by electrophoretic mobility shift assay. h Single-stranded RNA (ssRNA)-binding abilities of SARS-CoV-2 nsp9. The length of ssDNA, the state of SARS-CoV-2 nsp9, and the concentration of proteins used in the reaction system are indicated above the gel
Compared with previously reported CoV nsp9 structures, interface Ib/c (between molecules b and c) is a unique binding surface located in the center of the SARS-CoV-2 nsp9 tetrameric structure (Fig. 3c). This interactive region adopts a sheet-interface style formed by β5 (residues 64–68) and three connection loops (LP1, LP2, and LP3) from both molecules b and c. The buried area of the Ib/c interface is 1040 Å2, which is the largest buried area compared with those of interfaces Ic/d and Ia/b in the SARS-CoV-2 nsp9 tetramer. There are three major contact regions in the Ib/c interface. The first contact region associates with two LP1 loops (residues 33–42), connecting β2 and β3 from each of the subunits of c and b. Part of the N-terminal tail (residues 2–5) from molecule c is also involved in this contact surface. Two hydrogen bonds and one salt bond are involved in the interface between residues E3 and T35 in molecule c and residue K36 in molecular b (Fig. 3d). Additionally, residues G38, F40, V41, and L44 make a significant contribution to this hydrophobic contact surface. The second contact region consists of connecting LP2 (residues 58–63) and LP3 loops (residues 91–96) and β5 (residues 64–68) from each of the subunits. The LP2 loop on molecule c comes in direct contact with LP3 on molecule b. An antiparallel β-sheet located at the center of the sheet interface is composed of two β5 strands. In this hydrophobic contact surface, three residues (T62, T64, and Y66) on molecule c form four hydrogen bonds with residues K92, T67, and Y66 on molecule b, respectively (Fig. 3e). The third contact region involved in interface Ib/c is composed of the LP3 loop on molecule c and the LP2 loop on molecule b. Residues K92 and G93 on molecule c form two hydrogen bonds with residue T62 on molecule b (Fig. 3f). These three contact surfaces in interface Ib/c contribute a hydrophobic base with eight hydrogen bonds and one salt bridge, making the SARS-CoV-2 nsp9 tetramer extremely stable in the crystal structure.
SARS-CoV-2 nsp9 with nucleic acid-binding ability
Our previous research on nsp9 from IBV showed that the multimerization of this protein was necessary for protein–nucleic acid interaction [23]. In this present study, we observed the nucleic acid-binding ability of SARS-CoV-2 nsp9, using the electrophoretic mobility shift assay. Under favorable conditions, the nsp9 wild type was incubated with single-stranded nucleic acids. As shown in Fig. 3g and h, the free single-stranded DNA and RNA bands in the lanes associated with nsp9 were slightly reduced with the increasing concentration of nsp9. Additionally, we observed the ability of nsp9 to interact with a range of sizes and types of single-stranded nucleic acids. Based on these results, we can conclude that nsp9 prefers to interact with the single-stranded nucleic acid.
Discussion
In this study, we first revealed the horseshoe-like tetrameric structure of nsp9 encoded by the genomic RNA of SARS-CoV-2. This homotetrameric structure comprised two significant styles of dimeric conformation that were found in nsp9 structures published in the last decade [19, 21]. These structural results from independent research groups revealed that the dimerization of nsp9 might occur biologically and could play a pivotal role in the nucleic acid-binding function of the protein. However, because the dimerized nsp9 structure involves different dimeric forms, an interesting question is which of these is the major unit that interacts with nucleic acids. In the horseshoe-like tetrameric structure of SARS-CoV-2 nsp9, we found that two dimeric forms exist. One of them, named the helix interface and first reported by Egloff et al. (2004) [21], is organized by the parallel association of the C-terminal α-helix and the N-terminal loop. The second dimeric form, named the sheet interface and first mentioned by Sutton et al. (2004) [19], is formed by the zippering of each β5 strand from both subunits. The sheet interface is much more stable in the tetrameric structure of SARS-CoV-2 nsp9 than in SARS-CoV nsp9. Aside from the β5 strand, there are three loops (LP1, LP2, and LP3) involved in the nsp9 tetrameric interface. Therefore, we propose that these two dimeric forms are the fundamental units for building the high-order oligomers of nsp9. According to two different research groups that studied nsp9 mutants, the mutation of conserved residues located on the C-terminal α-helix (e.g., SARS-CoV nsp9 G104E (PDB: 3EE7)) or the unexpected appearance of a disulfide on β5 (hCoV-229E nsp9 wild type (PDB: 2 J97)) could significantly change the dimer interface of nsp9 and its ability to bind with DNA or RNA [20, 22]. In summary, the two dimeric interfaces that occurred in all crystal structures of nsp9 are reasonable and likely contribute to the formation of the higher oligomeric states of the protein.
Materials and methods
Plasmid construction, protein expression, and purification
The SARS-CoV-2 nsp9 gene (genome nucleotides 12,686–13,024; Gene ID: 43740578) was synthesized and cloned into the pET-28a (+)-SUMO vector using BamHI and XhoI sites (Qingke Biotech, China). The recombinant plasmids, which were confirmed by DNA sequencing (Sangon Biotech, China), were then used to transform Escherichia coli BL21 (DE3) (Transgene, China). Cells were cultured in Luria broth medium at 37 °C until the optical density at 600 nm reached 0.6–0.8. Protein expression was then induced by incubating the cells with 0.5 mM isopropyl β-d-1-thiogalactopyranoside at 16 °C for 18 h. The nsp9 protein was labeled with a His-SUMO tag with a Ulp1 cleavage site, purified by nickel-nitrilotriacetic acid affinity chromatography (Qiagen, Germany), and then cleaved with Ulp1 protease. The cleaved and tag-removed protein was loaded onto a Resource S chromatography column (GE Healthcare, USA) and eluted with a linear gradient of 10 mM to 1 M NaCl. The peak fraction containing the target protein was pooled, then concentrated to 1 mL using Amicon Ultra concentrators (cutoff size of 10 kDa; Millipore, USA), and finally loaded onto a Superdex 200 column (GE Healthcare, USA) for further purification with a buffer composed of 20 mM Tris (pH 8.0), 150 mM NaCl, 1 mM dithiothreitol, and 10% glycerol (v/v). The purity of the proteins was greater than 95%, as confirmed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (Supplementary Fig. 2a and 2b).
Protein crystallization and optimization
Using the hanging-drop vapor diffusion method, the nsp9 protein was quantified using absorbance readings at A280 nm (Thermo NanoDrop 2000, USA) and concentrated to 17 mg/mL for the crystallization trials, by mixing 1 μL of protein with 1 μL of reservoir solution at 20 °C. Commercial crystallization kits (Hampton Research, UK) were used to obtain the initial crystallization conditions. The protein crystals were obtained in a solution containing 100 mM citric acid (pH 3.0), 200 mM potassium sodium tartrate tetrahydrate, and 2.0 M (NH4)2SO4 after growth for 2 days at 20 °C (Supplementary Fig. 2c and 2d). The crystals were then transferred to a solution containing 100 mM citric acid (pH 3.0), 200 mM sodium metformin, and 2.5 M (NH4)2SO4, and the mixture was then frozen and stored in liquid nitrogen for future data collection.
Data collection, processing, and structure determination
The nsp9 crystal diffraction data were collected at − 196 °C, using the SSRF Beamline BL18U1 apparatus (Shanghai, China) at a wavelength of 0.97930 Å. Data were processed and scaled using the HKL2000 package (Supplementary Fig. 2e) [28]. The nsp9 structure was solved by Phaser in the CCP4 program suite using molecular replacement with the structure of SARS-CoV nsp9 RNA-replicase (PDB: 1QZ8) as an initial search model [29]. Cycles of refinement and model building were carried out using the REFMAC5, Phenix, and COOT software programs [30, 31]. Model geometry was verified using MolProbity. The single crystals of SARS-CoV-2 nsp9 are in space group C2221, with cell dimensions of a = 88.7 Å, b = 134.5 Å, c = 167.0 Å, and ɑ = β = γ = 90°. Both the Matthews coefficient estimation and the self-rotation function suggested the presence of six molecules per asymmetric unit. The final structure was refined to 2.95 Å resolution. The final Rwork and Rfree for the refined structure were 21.3% and 29.9%, respectively. In total, 96.1% of the amino acid residues fell in the most-favored region and the additionally allowed region of the Ramachandran plot. The structural figures were drawn using PyMOL [32]. The data collection and refinement statistics are shown in Table 1.
Table 1.
SARS-CoV-2 nsp9 data collection and refinement statistics
| SARS-CoV-2 nsp9 | |
|---|---|
| Data collection | |
| Resolution (Å) | 2.95 |
| Space group | C2221 |
| Unit-cell parameters (Å, °) | a = 88.7, b = 134.5 c = 167.0, α = β = γ = 90 |
| Resolution (Å) | 50.00–2.95 (3.04–2.95) |
| Rmergea (%) | 22.1 (59.3) |
| Rpimb (%) | 6.6 (17.0) |
| Average I/σ(I) | 11.6 (4.5) |
| No. of observed reflections | 40,346 (3400) |
| No. of unique reflections | 21,270 (1762) |
| Completeness (%) | 99.55 (97.3) |
| Multiplicity | 12.4 (2.3) |
| Matthews coefficient (Å3Da−1) | 3.26 |
| Solvent content (%) | 62.3 |
| Molecules per asymmetric unit | 6 |
| Refinement | |
| Resolution (Å) | 44.49–2.95 |
| Rwork/Rfree | 0.21/0.30 |
| Ramachandran favored (%) | 88.31 |
| Ramachandran outliers (%) | 3.32 |
| No. of atoms | |
| Protein | 4963 |
| Water | 66 |
| Wilson B value | 87.32 |
| Root-mean-square deviations | |
| Bond length (Å) | 0.010 |
| Bond angle (°) | 1.293 |
aRmerge = ∑hkl∑i ∣ Ii(hkl) – 〈I(hkl)〉 ∣ /∑hkl∑iIi(hkl), where Ii(hkl) is an individual intensity measurement and 〈I(hkl)〉 is the average intensity for all i reflections
bRpim is approximately estimated by multiplying the Rmerge value by the factor [1/(N − 1)]1/2, where N is the overall redundancy of the data set
Electrophoretic mobility shift assay
The 5′-FAM-labeled 12-mer single-stranded DNA oligonucleotide 5′-GCTTTGATTTCG-3′, 5′-FAM-labeled 26-mer single-stranded DNA oligonucleotide 5′-GCTTTGATTTCGTGCATCTATGGAGC-3′, 5′-FAM-labeled 12-mer single-stranded RNA oligonucleotide 5′-GCUUUGAUUUCG-3′ and 5′-FAM-labeled 26-mer single-stranded RNA oligonucleotide 5′-GCUUUGAUUUCGUGCAUCUAUGGAGC-3′ were used for the electrophoretic mobility shift assay. Initially, 2 nmol DNA was incubated with different concentrations of nsp9 protein (in the native), in a solution containing 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (pH 8.0), 50 mM KCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 0.05% Triton-X-100, and 5% glycerol, for 30 min at 4 °C. Thereafter, a 10× loading buffer (250 mM Tris-HCl (pH 7.9) and 40% glycerol) was added to the mixture. Then, samples were electrophoresed on 6.5% non-denaturing Tris–borate–EDTA (TBE) polyacrylamide gels for 40 min at a voltage of 100 V, and the results were determined with a ChemiDoc Touch Imaging system (Bio-Rad, USA).
Protein data Bank accession codes
The structure factors and atomic coordinates have been deposited in the Protein Data Bank under the PDB ID codes 7BWQ.
Supplementary information
Additional file 1: Figure S1. The OB-fold cluster of SARS-CoV-2 nsp9. (a) The different layers are colored orange, light blue, and pale green, respectively, and protomers in every layer are depicted in cartoon representations and labeled with uppercase letters. (b) Molecules d’, c’, asym, and bsym in the stable tetramer are colored in bright orange, pale green, light blue, and orange, respectively, and depicted in cartoon and surface representations.
Additional file 2: Figure S2. Purification and X-ray data collection of SARS-CoV-2 nsp9. (a) The chromatography of the purification of SARS-CoV-1 nsp9 with a Superdex200™ 10/300 column. (b) SDS-PAGE analysis result of SARS-CoV-2 nsp9. M is the protein marker. (c) The primary crystal state of SARS-CoV-2 nsp9 in the commercial crystallization kit. (d) The crystal for X-ray data collection of SARS-CoV-2 nsp9 after crystallization condition optimized. (e) X-ray diffraction pattern for structure analysis (left) and X-ray diffraction point atlas of SARS-CoV-2 nsp9 (right).
Additional file 3: Table S1. PDBePISA calculation of interface parameters of the quaternary structure of SARS-CoV-2 nsp9.
Acknowledgments
We sincerely appreciate the staff of the State Key Laboratory of Biotherapy, Sichuan University, who assisted in our research work during the COVID-19 epidemic. The X-ray diffraction experiments were carried out at the Shanghai Synchrotron Radiation Facility (SSRF) BL18U1. We gratefully thank the beamline staff for their technical help and Dr. Qingxiang Sun for sharing with us the beamline schedule. We appreciate Dr. Guangwen Lu and Dr. Shengce Tao for providing us with the associated plasmids and Dr. Qiang Chen for gifting us valuable DNA. This work was supported by grants from the National Key Research and Development Program of China (2017YFA0505903, 2018YFC1312300), the research fund on COVID-19 of Sichuan Province (2020YFS0010), and the project on COVID-19 of West China Hospital, Sichuan University (HX-2019-nCoV-044).
Code availability
Not applicable.
Abbreviations
- CoV
Coronavirus
- COVID-19
Coronavirus disease 2019
- hCoV
Human coronavirus
- LP
Loop protein
- OB-Fold superfamily
oligosaccharide/oligonucleotide-binding superfamily
- nsp9
Nonstructural protein 9
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus-2
Authors’ contributions
DS and CZ designed the study. CZ purified and characterized the proteins, cultivated the protein crystals, collected X-ray diffraction data, and managed the biological experiments. DS solved the crystal structure. YY. assisted with the biological experiments and provided essential data for this study. DS and YC took the lead in writing the manuscript. LL designed and created all the Figs. YC, CC, and JH contributed to the data analysis and the final version of the manuscript. DS had final approval of the version to be published.
Funding
This work was supported by grants from the National Key Research and Development Program of China (2017YFA0505903, 2018YFC1312300), the research fund on COVID-19 of Sichuan Province (2020YFS0010), and the project on COVID-19 of West China Hospital, Sichuan University (HX-2019-nCoV-044).
Availability of data and materials
All data generated or analysed during this study are included in this published article [and its supplementary information files].
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
Changhui Zhang, Yiping Chen, Li Li, Yan Yang, Jun He, Cheng Chen, and Dan Su declare that they have no conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s43556-020-00005-0.
References
- 1.Coronaviridae Study Group of the International Committee on Taxonomy of V The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5(4):536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hui DS. E IA, Madani TA, Ntoumi F, Kock R, Dar O, et al. the continuing 2019-nCoV epidemic threat of novel coronaviruses to global health - the latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis. 2020;91:264–6. 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed]
- 4.WHO. WHO, Coronavirus disease (COVID-2019) situation reports. World Health Organization: Geneva, Switzerland. 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports of subordinate document. Accessed 27 July 2020.
- 5.Ji W, Wang W, Zhao X, Zai J, Li X. Cross-species transmission of the newly identified coronavirus 2019-nCoV. J Med Virol. 2020;92(4):433–440. doi: 10.1002/jmv.25682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang YZ, Holmes EC. A genomic perspective on the origin and emergence of SARS-CoV-2. Cell. 2020;181(2):223–227. doi: 10.1016/j.cell.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han Q, Lin Q, Jin S, You L. Coronavirus 2019-nCoV: a brief perspective from the front line. J Inf Secur. 2020;80(4):373–377. doi: 10.1016/j.jinf.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020. 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed]
- 11.van Hemert MJ, van den Worm SH, Knoops K, Mommaas AM, Gorbalenya AE, Snijder EJ. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog. 2008;4(5):e1000054. doi: 10.1371/journal.ppat.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.te Velthuis AJ, Arnold JJ, Cameron CE, van den Worm SH, Snijder EJ. The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic Acids Res. 2010;38(1):203–214. doi: 10.1093/nar/gkp904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee NR, Kwon HM, Park K, Oh S, Jeong YJ, Kim DE. Cooperative translocation enhances the unwinding of duplex DNA by SARS coronavirus helicase nsP13. Nucleic Acids Res. 2010;38(21):7626–7636. doi: 10.1093/nar/gkq647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li S, Zhao Q, Zhang Y, Zhang Y, Bartlam M, Li X, et al. New nsp8 isoform suggests mechanism for tuning viral RNA synthesis. Protein Cell. 2010;1(2):198–204. doi: 10.1007/s13238-010-0028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bouvet M, Imbert I, Subissi L, Gluais L, Canard B, Decroly E. RNA 3’-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc Natl Acad Sci U S A. 2012;109(24):9372–9377. doi: 10.1073/pnas.1201130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joseph JS, Saikatendu KS, Subramanian V, Neuman BW, Buchmeier MJ, Stevens RC, et al. Crystal structure of a monomeric form of severe acute respiratory syndrome coronavirus endonuclease nsp15 suggests a role for hexamerization as an allosteric switch. J Virol. 2007;81(12):6700–6708. doi: 10.1128/JVI.02817-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.te Velthuis AJ, van den Worm SH, Snijder EJ. The SARS-coronavirus nsp7+nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res. 2012;40(4):1737–1747. doi: 10.1093/nar/gkr893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, et al. Structure of M (pro) from COVID-19 virus and discovery of its inhibitors. Nature. 2020. 10.1038/s41586-020-2223-y.
- 19.Sutton G, Fry E, Carter L, Sainsbury S, Walter T, Nettleship J, et al. The nsp9 replicase protein of SARS-coronavirus, structure and functional insights. Structure. 2004;12(2):341–353. doi: 10.1016/j.str.2004.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miknis ZJ, Donaldson EF, Umland TC, Rimmer RA, Baric RS, Schultz LW. Severe acute respiratory syndrome coronavirus nsp9 dimerization is essential for efficient viral growth. J Virol. 2009;83(7):3007–3018. doi: 10.1128/JVI.01505-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Egloff MP, Ferron F, Campanacci V, Longhi S, Rancurel C, Dutartre H, et al. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc Natl Acad Sci U S A. 2004;101(11):3792–3796. doi: 10.1073/pnas.0307877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ponnusamy R, Moll R, Weimar T, Mesters JR, Hilgenfeld R. Variable oligomerization modes in coronavirus non-structural protein 9. J Mol Biol. 2008;383(5):1081–1096. doi: 10.1016/j.jmb.2008.07.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu T, Chen C, Li H, Dou Y, Zhou M, Lu D, et al. Structural basis for dimerization and RNA binding of avian infectious bronchitis virus nsp9. Protein Sci. 2017;26(5):1037–1048. doi: 10.1002/pro.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeng Z, Deng F, Shi K, Ye G, Wang G, Fang L, et al. Dimerization of coronavirus nsp9 with diverse modes enhances its nucleic acid binding affinity. J Virol. 2018;92(17). 10.1128/JVI.00692-18. [DOI] [PMC free article] [PubMed]
- 25.Murzin AG. OB (oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J. 1993;12(3):861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Theobald DL, Mitton-Fry RM, Wuttke DS. Nucleic acid recognition by OB-fold proteins. Annu Rev Biophys Biomol Struct. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 28.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 29.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, et al. PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rigsby RE, Parker AB. Using the PyMOL application to reinforce visual understanding of protein structure. Biochem Mol Biol Educ. 2016;44(5):433–437. doi: 10.1002/bmb.20966. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. The OB-fold cluster of SARS-CoV-2 nsp9. (a) The different layers are colored orange, light blue, and pale green, respectively, and protomers in every layer are depicted in cartoon representations and labeled with uppercase letters. (b) Molecules d’, c’, asym, and bsym in the stable tetramer are colored in bright orange, pale green, light blue, and orange, respectively, and depicted in cartoon and surface representations.
Additional file 2: Figure S2. Purification and X-ray data collection of SARS-CoV-2 nsp9. (a) The chromatography of the purification of SARS-CoV-1 nsp9 with a Superdex200™ 10/300 column. (b) SDS-PAGE analysis result of SARS-CoV-2 nsp9. M is the protein marker. (c) The primary crystal state of SARS-CoV-2 nsp9 in the commercial crystallization kit. (d) The crystal for X-ray data collection of SARS-CoV-2 nsp9 after crystallization condition optimized. (e) X-ray diffraction pattern for structure analysis (left) and X-ray diffraction point atlas of SARS-CoV-2 nsp9 (right).
Additional file 3: Table S1. PDBePISA calculation of interface parameters of the quaternary structure of SARS-CoV-2 nsp9.
Data Availability Statement
All data generated or analysed during this study are included in this published article [and its supplementary information files].