Abstract
Most undergraduate neuroscience courses include a neurodevelopment component. Typically, the focus is on development of the mammalian central nervous system, including the concepts of neurulation, patterning of the neural tube, and differentiation of the various cells required to build a functional nervous system. However, it can be challenging to design an affordable undergraduate laboratory exercise to reinforce these concepts for students outside of lecture. Here we describe a laboratory exercise that takes advantage of the high level of conservation in neurodevelopmental pathways using Drosophila as a model organism to illuminate the connection between cell differentiation and nervous system function. Following a lesson discussing spinal cord development, students use Drosophila larvae to assess the effects of mutations in highly conserved motor neuron differentiation genes on motor behaviors such as crawling. As outcomes of this laboratory, students are able to master important neurodevelopmental concepts, connect neurodevelopment to nervous system function, and gain experience with experimental design and data analysis.
Keywords: Motor neuron differentiation, Neurodevelopment, Spinal cord development, Motor systems, Drosophila
Development of the mammalian nervous system is a standard topic of discussion in nearly all undergraduate neuroscience courses. Most courses focus on the embryological origins of the central nervous system (CNS), which arises from the ectoderm of the embryo during a process called neurulation. During neurulation, the neural ectoderm undergoes morphological changes that cause it to invaginate along the dorsal surface of the embryo, fold into a cylinder, and separate itself from the surrounding, non-neural ectoderm. Neurulation thus creates a cylinder of neural ectoderm that lies just below the dorsal body surface and extends from the anterior to the posterior of the embryonic body. This cylinder, the neural tube, is the precursor for the entire central nervous system. This initially simple structure undergoes profound changes to give rise to the complex cells and structures of the brain at its anterior end, and of the spinal cord throughout the trunk and abdomen of the body.
Development of the neural tube into the CNS is incredibly complex, requiring coordination of cell division, differentiation, and migration. The spinal cord is a good model for understanding these developmental processes. For example, extensive research has shown how diffusible signals produced by tissues proximal to the developing spinal cord, the notochord and the dorsal epidermal ectoderm, generate morphogen gradients that drive cell differentiation (Jessell, 2000; Lee and Pfaff, 2001; Lewis, 2006; Butler and Bronner, 2015; Gouti et al., 2015). A gradient of sonic hedgehog (Shh) arising from the notochord regulates differentiation of ventral cell types, while a gradient of bone morphogenetic protein (BMP) family ligands arising from the dorsal ectoderm regulates differentiation of dorsal cell types. Together, these opposing signaling gradients drive cell differentiation throughout the developing spinal cord, yielding a variety of neurons and glial cells organized into distinct functional compartments along the dorsal-ventral axis.
These early developmental steps are critical for building a functional nervous system, but may be difficult for students to grasp for a variety of reasons. First, like all aspects of mammalian development, they occur while the embryo is sheltered inside the mother’s uterus, and thus cannot be easily observed. Furthermore, events such as intercellular signaling and changes in gene expression are crucial developmental processes, but are essentially biochemical reactions that are invisible to the eye. Finally, the early development of the spinal cord, in which cells adopt distinct fates and developmental pathways, do not necessarily cause obvious external changes to the cell; for example, motor neurons are not immediately distinguishable from interneurons, and changes to cell morphology and tissue structure may not be apparent until much later in development. All of these challenges make it difficult for students to connect early developmental processes to the mature, functional nervous system with which they are more familiar.
We carried out a literature search to identify lab activities that would help students connect early development to adult nervous system function. Although it is easy to find innovative neuroscience labs aimed at undergraduate students (particularly those published in this journal), we discovered that few of these labs focus on neurodevelopment. Those we did find often utilized mammalian cell culture, a model that can be expensive and difficult to implement at institutions without proper equipment (such as cell culture hoods and incubators; e.g. Catlin et al., 2016 and Pemberton et al., 2018). We therefore designed lab that is inexpensive, easy to implement, and helps students connect developmental processes like cell differentiation to the structure and function of the mature nervous system.
We focused our lab exercise on the development of spinal motor neurons as they are responsible for governing movement, a trait that can be readily grasped by students and directly assessed in a lab. Furthermore, motor neuron differentiation pathways are highly conserved, making it possible for us to use an affordable model organism and simple tools and techniques, appropriate for labs with any number of students and being carried out at any institution, regardless of available resources.
As mentioned previously, Shh induces differentiation of ventral cell types within the developing spinal cord, including motor neurons. Intermediate levels of Shh activate expression of the transcription factors Pax6, Nkx6.1, Mnr2, and Olig2 within a ventral region of the spinal cord referred to as pMN, from which all spinal motor neurons will arise (Jessell, 2000; Price and Briscoe, 2004; Stifani, 2014; Zagorski et al., 2017). These proteins cooperatively activate a transcriptional program that drives motor neuron differentiation. An important step along that path is the activation of both Lhx3 and Islet1, which form a DNA-binding complex that directly activates genes specific to motor neuron function (Lee et al., 2012; Cho et al., 2014; Kania, 2014; Stifani, 2014). Once differentiated, motor neurons extend axons outward from the spinal cord toward the skeletal muscles of the body. Disruption of these developmental processes results in failed innervation of the skeletal muscles and thus impaired body movement.
In this lab, students used Drosophila melanogaster as a model to study the importance of motor neuron differentiation for movement and survival. We chose to use Drosophila as a model organism both because of its affordability and because it is easy to obtain mutants for most genes in its genome. Students worked with wild-type Drosophila, as well as Drosophila stocks carrying mutations in several genes that are critical for motor neuron differentiation in both mammals and Drosophila: Olig2, Lim3 (the Drosophila homolog of Lhx3), and Tailup (tup, the Drosophila homolog of Islet1) (Thor and Thomas, 1997; Thor et al., 1999; Certel and Thor, 2004; de Navascues and Modolell, 2010; Oyallon et al., 2012). These genes were selected because they act at multiple levels of the genetic hierarchy involved in motor system development, providing students with an opportunity to compare and contrast their phenotypes. In addition, loss of function mutations in any one of these genes in Drosophila results in overt motor neuron differentiation defects and failed innervation of target muscles, reflecting conservation of gene function between Drosophila and vertebrates (Thor and Thomas, 1997; Thor et al., 1999; Oyallon et al., 2012). Students both qualitatively and quantitatively assessed motility in larvae of these various genotypes. Students then completed lab reports in which they presented and analyzed their data, made conclusions about the requirement of motor neuron differentiation genes for larval mobility, and discussed the importance of developmental processes such as motor neuron differentiation for adult success and survival.
MATERIALS AND METHODS
Materials
Fly stocks (obtained from the Bloomington Drosophila Stock Center at Indiana University): Wild-type flies (Bloomington stock #5 or #3605; we used #3605), Olig2 mutant flies (Bloomington stock #59233), Tup (Islet homolog) mutant flies (Bloomington stock #2207), Lim3 mutant flies (Bloomington stock #3393)
Fly culture vials and plugs (available from Carolina Biological Supply, Catalog #173078).
Fly food (available from Carolina Biological Supply, Catalog #173210)
Incubator at 25 °C (optional)
Sharpies (one per group)
100 mm Petri dishes (empty; four per group)
20% sucrose solution (50 mLs per group)
Paintbrushes (one per group)
100 mm Petri dishes with 2% agarose (eight per group)
Graphing paper (0.2 cm2, four sheets per group)
Stereomicroscope (one per group)
Timer (one per group)
Plastic bags and freezer for disposal
Culture Preparation
Fly stocks were ordered in advance from the Bloomington Drosophila Stock Center at Indiana University (https://bdsc.indiana.edu/; allow one month for processing and delivery). Note that while the wild-type flies were homozygous, each of the mutant flies were delivered as heterozygous stocks due to homozygous lethality (Thor and Thomas, 1997; Thor et al., 1999; Oyallon et al., 2012). The adult flies for this lab had the following heterozygous genotypes: Olig2/CyO; Tup/CyO; and Lim3/SM6a. CyO and SM6a are balancer chromosomes, which are used in Drosophila fly husbandry to maintain heterozygous stocks. Balancer chromosomes help preserve gene mutations via two mechanisms: (1) balancer chromosomes contain inversions that impair genetic recombination, thereby preventing loss of the mutation due to genetic recombination; and (2) balancer chromosome themselves contain homozygous embryonic lethal mutations and thus can never become homozygous within the stock. As a result, a “balanced” stock will always include adult flies that are heterozygous for both the mutation of interest and the balancer chromosome. Readers can refer to the following website for further information if they are unfamiliar with fly genetics (https://bdsc.indiana.edu/stocks/balancers/balancer_intro.html).
Approximately three weeks before the experiments were to take place, fly stocks were expanded by serially transferring adults to a new vial every two days for a period of ten days to obtain healthy and abundant populations of each genotype (these vials can be stored at either room temperature or in a 25 °C incubator if available; see Table 1 for timeline). Five to six days before the experiments were to take place, adult flies (10–15 males and 10–15 females) were transferred to fresh vials of food. Each student group should have one vial of each genotype. Vials were stored at either 25 °C or at room temperature and adults were allowed to lay eggs in the vials for two-three days, until there are obvious signs of larvae within the vials. Adults were then removed, and vials were stored in the incubator until needed for lab to allow larvae to develop.
Table 1.
Timeline of drosophila husbandry.
| Timeline | Task |
|---|---|
| At least six weeks before Lab 1 | Order fly stocks from the Bloomington Drosophila Stock Center |
| Three weeks before Lab 1 | Transfer adult flies into fresh vials to expand stocks |
| 19–13 days before Lab 1 | Transfer adult flies into fresh vials every two days to expand stocks (keep previous vials) |
| Lab 1 | Presentation and Pre-Lab assignment |
| Six days before Lab 2 | Transfer adult flies into fresh vials to prepare larval cultures |
| Three days before Lab 2 | Remove adult flies from vials; keep vials |
| Lab 2 | Carry out motor behavior experiment |
Week 1: Introduction And Pre-Lab Assignment
The basic concepts addressed in this lab, including neurulation, early CNS development, and cell type differentiation in the developing spinal cord, were covered as part of lecture. However, the first week of this lab exercise included an additional presentation providing more detailed background (see Appendices). The presentation reminded students specifically of the function of motor neurons within the nervous system and the genetic pathways that regulate their differentiation in the embryonic spinal cord, with a particular focus on the genes that are mutated in the fly stocks utilized for this lab: Olig2, Tup, and Lim3. This presentation also included an introduction to Drosophila as a model organism as well as videos depicting the movement of wild-type larvae to illustrate normal motor function (Oyallon et al., 2012; Nichols et al., 2012). Supplemental readings were posted to assist students with the completion of these pre-lab reports, which were due at the beginning of the following week’s lab (i.e., Price and Briscoe, 2004; Stifani, 2014; Gouti et al., 2015).
Students were then asked to complete a pre-lab assignment (see Appendices). The assignment asked students to summarize basic aspect of central nervous system development, such as neurulation and the roles of Shh and BMP in spinal cord patterning. These questions assessed student understanding of basic neurodevelopment. Additional questions asked students to explain the relationships amongst Shh, Olig2, Tup, and Lim3 to assess student understanding of the genetic networks involved in motor neuron differentiation. A final set of questions required students to apply that knowledge to larval behavior by predicting the effects of Olig2, Tup, or Lim3 gene mutations in larvae. As part of this assignment, students participated in experimental design by choosing qualitative parameters to assess during their larval observations the following week.
Week 2: Evaluation Of Larval Mobility
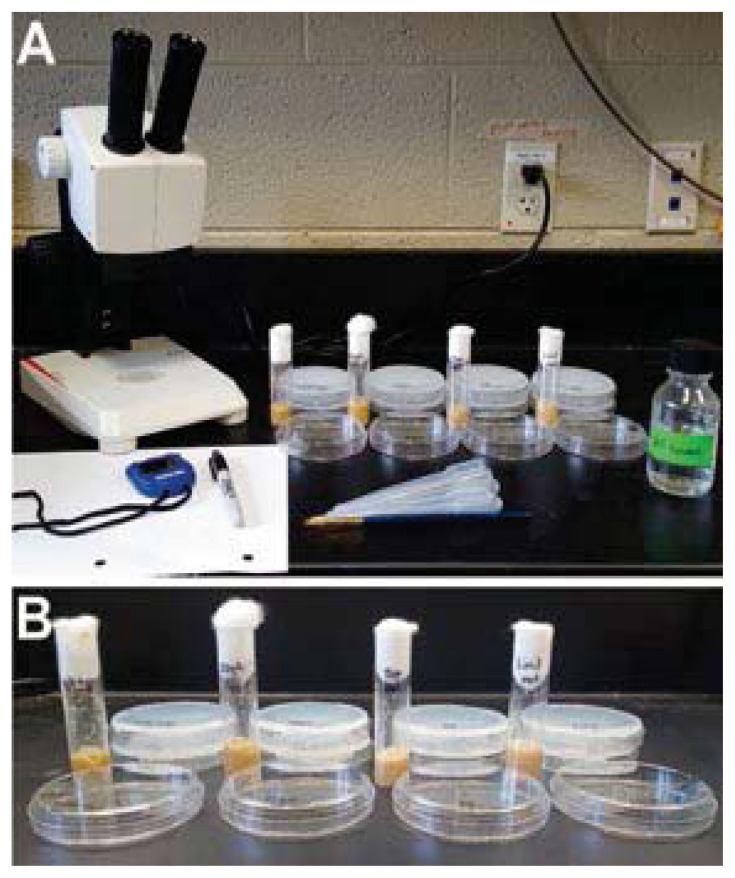
Experiments were carried out during the second week of lab. For that lab meeting, vials of larvae and additional materials including sucrose, transfer pipets, Petri dishes, paintbrushes, agar plates, graph paper, stereomicroscopes, and timers were assembled (Figure 1). Each group received one vial of each genotype (four total), four empty Petri dishes, and eight agar plates. Students were instructed to label Petri dishes and agar plates, with one Petri dish and two agar plates for each genotype being analyzed.
Figure 1.
(A) A typical laboratory setup including all necessary reagents. (B) Vials containing larvae of various genotypes (wild-type, Olig2, Tup, and Lim3), along with correspondingly labeled petri dishes (one empty, two with 2% agarose).
Prior to beginning the experiment, students were reminded of the genotypes expected in the larval populations. Because the parental stocks are heterozygous, their progeny are produced in a 1:2:1 ratio of balancer chromosome homozygotes to heterozygotes to mutant homozygotes. However, balancer chromosome homozygotes die as embryos and thus are absent from larval populations. Larval populations could therefore contain a mixture of heterozygous and homozygous mutant individuals depending on homozygous mutant phenotypes. In the case of Tup, homozygous mutants die as embryos, and therefore all larvae are expected to be heterozygous Tup/CyO (Thor and Thomas, 1997). Lim3 mutants die in late larval or pupal stages, and therefore larval populations within this stock are expected to contain a roughly 2:1 ratio of Lim3/SM6a heterozygotes to Lim3/Lim3 homozygous mutants (Thor et al., 1999). Olig2 mutants die as pupae or adults, and therefore larval populations within this stock are expected to contain a 2:1 ratio of Olig2/CyO heterozygotes to Olig2/Olig2 homozygous mutants (Oyallon et al., 2012). Note that while the larval phenotypes of heterozygous Tup, Lim3, and Olig2 mutants have not previously been reported, our unpublished observations suggest that heterozygotes of all three mutants may exhibit mild to moderate motor defects. However, we did not share this information with students, instead allowing them to reflect on the heterozygous phenotypes as part of their post-lab assignment.
To begin their experiments, students first retrieved larvae from their vials. Sucrose solution was added to each vial using transfer pipets, enough to cover food with about one inch of liquid. This step encourages larvae to crawl out of food and into the liquid sucrose solution. After five minutes, after multiple larvae were observed in the sucrose solution, transfer pipets were used to move larvae from the sucrose solution into the appropriately labeled empty petri dish. Students then carried out a qualitative analysis of larval movement. They first used a paintbrush to transfer one wild-type larva to an appropriately labeled agar plate and observed its movement (Note: students should be directed to use the largest larvae for these assays; larval size increases with age and students should attempt to use larvae that are similar in age for all experiments). Students were asked to describe the normal movement of the wild-type larva, paying particular attention to the qualitative parameters they had chosen to assess in their pre-lab assignments. Their observations were recorded on their lab assignments (see Appendices). This process was repeated until a minimum of ten wild-type larvae had been observed. Students then repeated this procedure with the mutant larvae, observing at least ten mutant larvae of each genotype to account for the presence of both heterozygous and homozygous mutants in some genotypes (see Oyallon et al., 2012; and Appendices for examples of mutant larval movement).
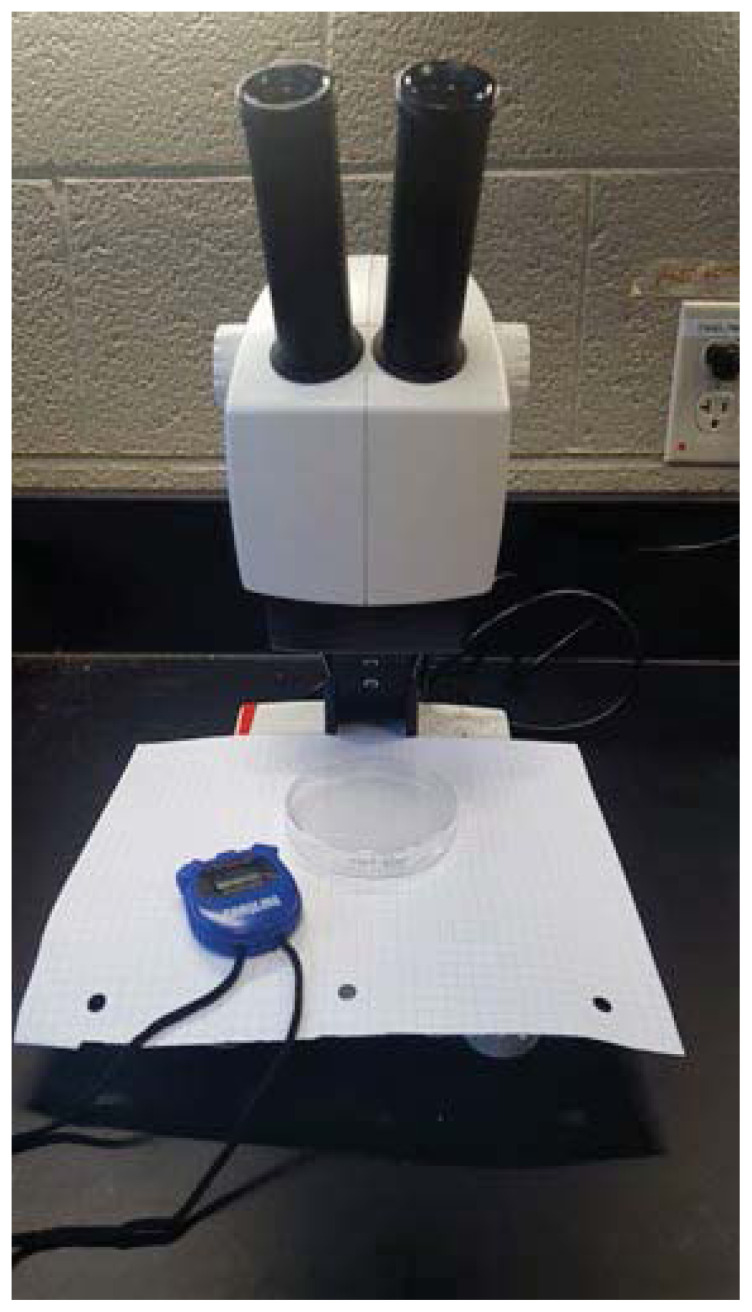
In the next part of the lab, students assessed larval movement in a more quantitative fashion. For this experiment, the remaining agar plates, one for each genotype, were placed on top of the graphing paper, and students prepared their timers in order to measure the rate of movement of each larval genotype (Figure 2). Students set the timer for one minute, transferred a wild-type larva to the appropriate agar plate, started the timer, and then counted how many grid lines the larva crossed before the timer indicated that a minute had passed. Larvae were counted as crossing grid lines when the anterior end of the larvae crossed into a new grid square. Students recorded their measurement on their lab assignments or notebooks, then repeated with nine additional larvae, for a total of ten measurements. Students then performed the same analysis with mutant larvae, again assessing ten larvae from each group.
Figure 2.
Experimental setup for quantitative assessment of larval movement. Agar plate should be placed over graphing paper and prepared for observation under stereomicroscope. Larva can then be transferred onto agar plate and its movement can be assessed for one minute (using timer to determine when a minute has elapsed).
After experiments had been completed, most experimental materials were discarded as non-hazardous waste in normal waste bins. Flies and larvae, and all materials that may have larvae on them, were placed in plastic bags, stored overnight in a −20 °C freezer to euthanize all flies, larvae, and eggs, and then discarded in a normal waste bin the following day.
Data Analysis And Evaluation
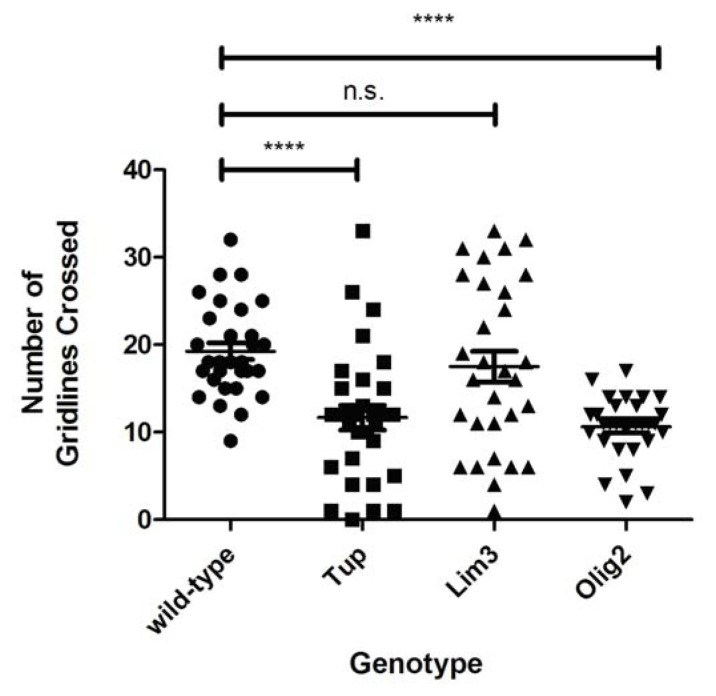
Following data collection, each student group calculated the average gridlines crossed per minute for each of the three mutant groups and the wild-type larval group. The calculated averages for the wild-type and mutant larvae from each student group (minimum of three student groups) were then compiled for the entire class, generating an overall average for the wild-type, Tup, Olig2, and Lim3 mutants (see Figure 3 for an example of student data). Students were also asked to complete a series of post-lab questions to summarize their data and draw conclusions for each mutant. A final question asked students to consider whether their data matched their predictions or not, and to propose explanations for any discrepancies between their hypothesized and actual results.
Figure 3.
Results of quantitative analysis of motor behavior in motor mutant larvae. The average number of gridlines crossed per minute are shown for wild-type (WT), Tup1, Olig2, and Lim3 motor mutant larvae. Averages were calculated by compiling data from three student lab groups; each group assessed ten larvae/genotype. Error bars represent standard error of the mean. Data were analyzed using t-tests; **** indicates p<0.0001; n.s. indicates p>0.05
RESULTS AND DISCUSSION
Although neurodevelopment is an important topic in undergraduate neuroscience courses, it can be a challenge to design and implement an easy, affordable lab exercise that allows students to explore embryonic development of the nervous system and directly connect the events of embryogenesis to the functional, adult nervous system. Most published neuroscience lab exercises focus on aspects of adult nervous system function rather than development, and those that do feature development may be expensive or difficult to implement at schools with limited resources (such as Catlin et al., 2016; Pemberton et al., 2018). The exercise we have described here is based on one of the most basic concepts of neurodevelopment, differentiation of functional cell types within the embryonic central nervous system, and helps students understand how this process is required for adult processes such as movement. In addition, our lab uses a simple model organism and readily available reagents, making it easy to implement at any undergraduate institution.
The primary goal of this lab exercise was for students to meet important learning objectives for the course. For example, students should be able to describe basic neurodevelopmental processes such as neurulation and explain how various cell types form during development of the nervous system. This lab exercise also required students to apply those basic concepts by predicting how disruption of embryonic development affects the function of the adult nervous system. Finally, students completing this lab gained experience in experimental design, data collection and analysis, and critical evaluation of results. Student mastery of these learning objectives and skills was assessed in pre- and post-lab assignments.
Student Outcomes
The pre-lab assignment assessed student mastery of several important learning objectives: (1) Describe early development of the spinal cord; (2) Describe the genetic networks that control motor neuron differentiation; (3) Predict the effects of gene mutations on motor neuron development and larval nervous system function; (4) Design a qualitative experiment to assess larval nervous system function. Evaluation of the pre-lab assignments suggests that students mastered these learning objectives by the end of this lab exercise. Lab Assignment 1, Questions #1–2 assessed the first learning objective; 100% of students answered these questions correctly by describing the process of neurulation accurately and clearly explaining how Shh and BMP act as morphogens to pattern the dorsal-ventral axis of the spinal cord. Lab Assignment 1, Questions #3–4 assessed the second learning objective; 82% of students answered these questions correctly by describing the correct roles of Shh, Olig2, Tup, and Lim3 in motor system development. Lab Assignment 1, Question #5 assessed the third learning objective; 100% of students answered this question correctly by predicting that loss of function mutations would result in failed motor neuron differentiation and defective larval movement. Importantly, 70% of students predicted that loss of Olig2 would have more severe effects than loss of either Tup or Lim3 because of its higher placement in the genetic hierarchy, further suggesting that students had mastered this learning objective. Lab Assignment 1, Question #6 assessed the fourth learning objective; 90% of students answered this question correctly by suggesting qualitative measures such as slow, uncoordinated movement and incomplete peristalsis. These responses suggest that students successfully mastered key neurodevelopmental concepts and were able to connect embryonic development to larval behavior.
Students also gained valuable experience with skills such as data analysis and critical evaluation of results. Lab Assignment 2 assessed our final learning objective: (5)
Collect and evaluate data in order to either accept or reject your predictions about how gene mutations would affect motor neuron development and larval motor function. All students were able to successfully collect quantitative and qualitative data in Lab Assignment 2. Students were then asked to evaluation their results in Lab Assignment 2, Question #4. All students responded that they had observed altered motor behavior within all mutant larval populations, as expected. Examples of observed altered behavior included fewer gridlines crossed, frequent directional changes, difficulty initiating movement, and dragging of the abdomen. Furthermore, most students (73%) noted that the data were variable, the phenotypic effects were not as severe as they had predicted, and suggested that these findings may have been due to the presence of heterozygous larvae in all samples. Student performance on these post-lab questions suggested that they were able to successfully participate in data collection and critically evaluate the data they collected.
Suggestions for Implementation
The lab exercise as described here was successful in achieving our desired learning outcomes. We have several recommendations for implementation to ensure success. While instructors themselves can take advantage of videos of mutant larvae to help recognize abnormal larval movement, they should refrain from showing those videos to students to avoid influencing student observations. In addition, because of the mixed genotypes present in some of the larval populations, it is imperative that students assess at least ten larvae per genotype per group, and we recommend pooling class data to obtain a larger dataset. However, the presence of mixed genotypes with different phenotypes presents a valuable opportunity to have students connect genotype to phenotype; instructors that wish to emphasize this relationship can ask students to graph their data in a dot-plot format (as shown in Figure 3) to better enable the identification of distinct phenotypic subgroups within their dataset, possibly reflecting the distinct genotypes present.
Beyond the applications described here, this basic lab structure could easily be modified to address additional learning objectives if desired. For example, instructors who would like to incorporate more quantitative data analysis could have students assess more than ten larvae to increase the number of data points and carry out statistical analyses of their results. Instructors could also expand the lab by including additional Drosophila motor mutants and/or including additional motor assays (additional motor assay protocols described in Nichols et al., 2012). An instructor could choose to spend more time exploring evolution of the central nervous system as part of this lab exercise if desired. Although we did not focus on the differences between the Drosophila and human nervous systems in our course, one could readily do so. The instructor could assign additional readings describing the structure of the Drosophila central nervous system, along with the reviews we assigned describing the mammalian central nervous system, and ask students to compare and contrast the roles of Olig2, Tup, and Lim3 in the development of these two species.
Finally, the approaches and methods used in this lab activity could be applied to other neuroscience topics such as diseases of the nervous system. Multiple neurological disorders affect movement, and some of those diseases have well-established Drosophila models, including spinobulbar muscular atrophy (SBMA), spinal muscular atrophy (SMA), and amyotrophic lateral sclerosis (ALS; Watson et al., 2008; Lloyd and Taylor., 2010; Grice et al., 2011; Casci and Pandey, 2015; Xu et al., 2015). The same experimental techniques could be used to help students investigate the phenotypes associated with these diseases. Therefore, this lab exercise not only effectively reinforces concepts in neurodevelopment but also provides a flexible template to be used for other undergraduate neuroscience labs.
APPENDIX 1. MOTOR DEVELOPMENT LAB
Overview
The purpose of this lab is to expand your understanding of nervous system development, organization, and function. We will be using an assay to assess motor behavior in the larval form of the model organism, Drosophila. The contribution of specific spinal motor neuron differentiation genes to development of motor behaviors will be tested in wildtype (WT), heterozygous (HET), and homozygous (HOM) larvae for the following developmental genes: Olig2, Tup (homolog to vertebrate Islet2), and Lim3 (homolog to vertebrate Lim3/Lhx3).
Week 1: An introduction to the lab will be provided and you will be given a series of questions that should be completed prior to next week’s lab.
Week 2: Bring the completed Week 1 lab assignment with you. During this lab session, each group will assess crawling behavior in WT and mutant larvae, recording both quantitative and qualitative measures of crawling behavior.
Week 1 Questions
The mutant flies we are studying during this lab have loss-of-function mutations in genes involved in spinal motor neuron differentiation and development. An understanding of key steps in early development of spinal motor neurons is essential to understanding the model organism and behavioral assays we are using. Please answer the following questions related to the stages of neural development:
Briefly explain the process of neurulation and formation of the neural tube.
Explain the role of the morphogens Sonic Hedgehog (Shh) and Bone morphogenic protein (BMP) in differentiation of spinal cord cells.
Explain the relationship between Shh and the transcription factor Olig2.
Explain the relationship between Olig2 and the genes Tup and Lim3.
Predict how loss-of-function mutations in Olig2, Tup, and Lim3 would affect larval behavior.
Review the videos from the Lab Introduction lecture and observe the WT larvae. Decide what qualitative descriptors you might use to assess the phenotypes of HET/HOM mutant larvae.
APPENDIX 2. MOTOR DEVELOPMENT LAB PROCEDURES
Supplies And Reagents At Each Station
Stereo Microscope
2 pieces of 0.2cm2 graph paper per group
Vials of wild-type and mutant larvae
Sucrose solution
Transfer pipette (for getting larvae out of sucrose)
Empty petri dish
Paint brush for transferring larvae
60mm petri dishes with 2% agarose
Timer
Procedures
Begin with WT larvae:
Add enough sucrose solution to vial to cover in ~1 inch of liquid; allow to sit until larvae appear in liquid solution.
Transfer ~10–15 larvae to empty petri dish using transfer pipets.
Use brush to transfer individual larva to agar plate.
First observe larva under stereomicroscope to assess general crawling behavior; describe qualitatively using parameters described in your pre-lab assignment and record your observations in table below.
Place plate over graph paper under stereomicroscope.
Next quantify crawling behavior by counting number of gridlines crossed in 1 minute and record result in table below. Repeat 9 times.
| Group | # of Gridlines crossed in 1 minute | Description of crawling behavior |
|---|---|---|
| WT-1 | ||
| WT-2 | ||
| WT-3 | ||
| WT-4 | ||
| WT-5 | ||
| WT-6 | ||
| WT-7 | ||
| WT-8 | ||
| WT-9 | ||
| WT-10 |
Repeat steps 1–6 using mutant larvae and record your observations in tables below. Make sure at least 10 larvae are assessed per genotype.
Discard larva in plastic bags provided, making sure to seal the bag.
| Group | # of Gridlines crossed in 1 minute | Description of crawling behavior |
|---|---|---|
| Group | # of Gridlines crossed in 1 minute | Description of crawling behavior |
|---|---|---|
| Group | # of Gridlines crossed in 1 minute | Description of crawling behavior |
|---|---|---|
Post Lab Questions
Summarize your qualitative descriptions of WT and mutant larval crawling behavior.
Use your group’s data to calculate the average number of gridlines crossed in 1 minute for each genotype.
Now use class data to calculate the average number of gridlines crossed in 1 minute for each genotype.
Based on your qualitative and quantitative data, did the mutants exhibit the types of motor defects you predicted in your pre-lab assignment? Propose explanations for your data and conclusions.
REFERENCES
- Butler SJ, Bronner ME. From classical to current: analyzing peripheral nervous system and spinal cord lineage and fate. Dev Biol. 2015;398:135–146. doi: 10.1016/j.ydbio.2014.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casci I, Pandey UB. A fruitful endeavor: modeling ALS in the fruit fly. Brain Res. 2015;1607:47–74. doi: 10.1016/j.brainres.2014.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catlin R, Taylor A, Ditchek L, Burnett S, Khan S, Todd O, Adams M, Touhey E, Wynkoop A, Ryan J. Using cultured mammalian neurons to study cellular processes and neurodegeneration: a suite of undergraduate lab exercises. J Undergrad Neurosci Educ. 2016;14:A132–7. [PMC free article] [PubMed] [Google Scholar]
- Certel SJ, Thor S. Specification of Drosophila motoneuron identity by the combinatorial action of POU and LIM-HD factors. Development. 2004;131:5429–5439. doi: 10.1242/dev.01418. [DOI] [PubMed] [Google Scholar]
- Cho HH, Cargnin F, Kim Y, Lee B, Kwon RJ, Nam H, Shen R, Barnes AP, Lee JW, Lee S, Lee SK. Isl1 directly controls a cholinergic neuronal identity in the developing forebrain and spinal cord by forming cell type-specific complexes. PLoS Genet. 2014;10:e1004280. doi: 10.1371/journal.pgen.1004280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouti M, Metzis V, Briscoe J. The route to spinal cord cell types: a tale of signals and switches. Trends Genet. 2015;31:282–289. doi: 10.1016/j.tig.2015.03.001. [DOI] [PubMed] [Google Scholar]
- Grice SJ, Sleigh JN, Liu JL, Sattelle DB. Invertebrate models of spinal muscular atrophy: insights into mechanisms and potential therapeutics. Bioessays. 2011;33:956–965. doi: 10.1002/bies.201100082. [DOI] [PubMed] [Google Scholar]
- Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- Kania A. Concocting cholinergy. PLoS Genet. 2014;10:e1004313. doi: 10.1371/journal.pgen.1004313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Cuvillier JM, Lee B, Shen R, Lee JW, Lee SK. Fusion protein Isl1-Lhx3 specifies motor neuron fate by inducing motor neuron genes and concomitantly suppressing the interneuron programs. Proc Natl Acad Sci USA. 2012;109:3383–3388. doi: 10.1073/pnas.1114515109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Pfaff SL. Transcriptional networks regulating neuronal identity in the developing spinal cord. Nat Neurosci. 2001;(4 Suppl):1183–1191. doi: 10.1038/nn750. [DOI] [PubMed] [Google Scholar]
- Lewis KE. How do genes regulate simple behaviors? Understanding how different neurons in the vertebrate spinal cord are genetically specified. Philos Trans R Soc Lond B Biol Sci. 2006;361:45–66. doi: 10.1098/rstb.2005.1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd TE, Taylor JP. Flightless flies: Drosophila models of neuromuscular disease. Ann N Y Acad Sci. 2010;1184:e1–20. doi: 10.1111/j.1749-6632.2010.05432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Navascues J, Modolell J. The pronotum LIM-HD gene tailup is both a positive and negative regulator of the proneural genes achaete and scute of Drosophila. Mech Dev. 2010;127:393–406. doi: 10.1016/j.mod.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Nichols CD, Becnel J, Pandey UB. Methods to assay Drosophila behavior. J Vis Exp. 2012;61:3795. doi: 10.3791/3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyallon J, Apitz H, Miguel-Aliaga I, Timofeev K, Ferreira L, Salecker I. Regulation of locomotion and motoneuron trajectory selection and targeting by the Drosophila homolog of Olig family transcription factors. Dev Biol. 2012;369:261–276. doi: 10.1016/j.ydbio.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton K, Mersman B, Xu F. Using ImageJ to assess neurite outgrowth in mammalian cell cultures: research data quantification exercises in undergraduate neuroscience lab. J Undergrad Neurosci Educ. 2018;16:A186–194. [PMC free article] [PubMed] [Google Scholar]
- Price SR, Briscoe J. The generation and diversification of spinal motor neurons: signals and responses. Mech Dev. 2004;121:1103–1115. doi: 10.1016/j.mod.2004.04.019. [DOI] [PubMed] [Google Scholar]
- Stifani N. Motor neurons and the generation of spinal motor neuron diversity. Front Cell Neurosci. 2014;8:293. doi: 10.3389/fncel.2014.00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thor S, Thomas JB. The Drosophila islet gene governs axon pathfinding and neurotransmitter identity. Neuron. 1997;18:397–409. doi: 10.1016/s0896-6273(00)81241-6. [DOI] [PubMed] [Google Scholar]
- Thor S, Andersson SG, Tomlinson A, Thomas JB. A LIM-homeodomain combinatorial code for motor-neuron pathway selection. Nature. 1999;397:76–80. doi: 10.1038/16275. [DOI] [PubMed] [Google Scholar]
- Watson MR, Lagow RD, Xu K, Zhang B, Bonini NM. A Drosophila model for amyotrophic lateral sclerosis reveals motor neuron damage by human SOD1. J Biol Chem. 2008;283:24972–24981. doi: 10.1074/jbc.M804817200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Tito AJ, Rui YN, Zhang S. Studying polyglutamine diseases in Drosophila. Exp Neurol. 2015;274:25–41. doi: 10.1016/j.expneurol.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagorski M, Tabata Y, Brandenberg N, Lutolf MP, Tkacik G, Bollenbach T, Briscoe J, Kicheva A. Decoding of position in the developing neural tube from antiparallel morphogen gradients. Science. 2017;356:1379–1383. doi: 10.1126/science.aam5887. [DOI] [PMC free article] [PubMed] [Google Scholar]