Summary
The insect antennal lobe (AL) contains the first synapses of the olfactory system, where olfactory sensory neurons (OSNs) contact second order projection neurons (PNs). In Drosophila melanogaster, OSNs expressing specific receptor genes send stereotyped projections to one or two of about 50 morphologically defined glomeruli[1–3]. The mechanisms for this precise matching between OSNs and PNs have been studied extensively in D. melanogaster, where development is deterministic and independent of neural activity[4–6]. However, a number of insect lineages, most notably the ants, have receptor gene repertoires many times larger than D. melanogaster and exhibit more structurally complex antennal lobes[7–12]. Moreover, perturbation of OSN function via knockout of the odorant receptor (OR) co-receptor, Orco, results in drastic AL reductions in ants[13,14], but not in Drosophila[15]. Here, we characterize AL development in the clonal raider ant, Ooceraea biroi. We find that, unlike in Drosophila, ORs and Orco are expressed before the onset of glomerulus formation, and Orco protein is trafficked to developing axon terminals, raising the possibility that ORs play a role during ant AL development. Additionally, ablating ant antennae at the onset of pupation results in AL defects that recapitulate the Orco mutant phenotype. Thus, early loss of functional OSN innervation reveals latent structure in the AL that develops independently of peripheral input, suggesting that the AL is initially pre-patterned, and later refined in an OSN-dependent manner. This two-step process might increase developmental flexibility and thereby facilitate the rapid evolution and expansion of the ant chemosensory system.
eTOC blurb
Ryba et al. describe differences in antennal lobe development between ants and flies, suggesting that ants, with their complex olfactory systems, employ a different developmental logic than flies, where the system is simpler. Ants can therefore be used to explore how developmental strategies relate to sensory system complexity and evolution.
Results and Discussion
The glomerular map is specified over the first 10 days of pupal development
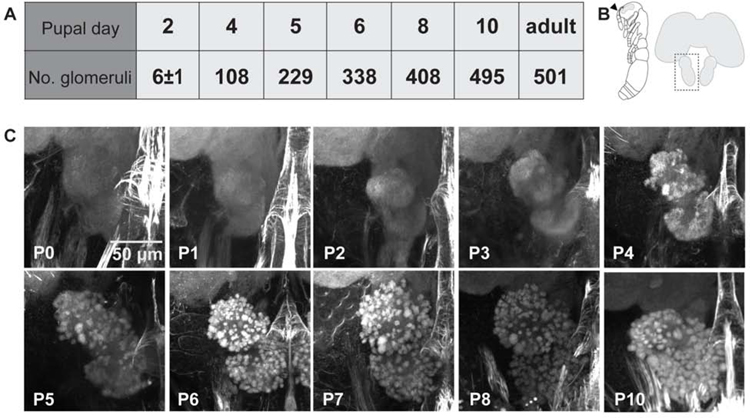
We characterized the timing of major events in O. biroi AL development by examining pupal brains using confocal microscopy. O. biroi larvae undergo a morphological transition upon pupation (P0), then remain as pupae for 14 days (P1–P14). We observed O. biroi AL morphology on each day between P0 and P8, as well as on P10 (Figure 1). Developing pupal antennal lobes increased in volume, and starting at P4, recognizable structure emerged as dense bundles of neuropil became visible. These early features strongly resemble olfactory glomeruli in the adult, but is unclear whether they are mature at the level of fine structure and represent functional olfactory synapses. Manual counts from reconstructed lobes showed that glomeruli formed rapidly, until by P10 the pupal antennal lobe contained as many glomeruli as a mature lobe (Figure 1A). Structure developed consistently from individual to individual, at least at the gross level, suggesting a stereotyped developmental program (Figure S1A). Position of a given glomerulus within the lobe did not seem to be associated with whether it appeared early or late in development. Instead, glomeruli formed in a diffuse manner across the entire lobe as development progressed (Figure 1C).
Figure 1: Antennal lobe development over the first 10 days of pupation.
(A) Manual counts of glomeruli from confocal stacks of pupal brains stained with phalloidin. Glomeruli were counted in the right antennal lobe from each brain. P2 n=6 brains with average 6±1 SD glomeruli; n=1 for all other pupal time-points; adult n=2 brains with 493 and 509 glomeruli (average 501; adult counts were previously published[13]). (B) Illustration of an ant pupa with the antenna marked by an arrowhead (left), and schematic of an ant brain with the left antennal lobe boxed to show the orientation of images in (C). (C) Representative projections of confocal z-stacks through the right antennal lobe of phalloidin stained pupal brains (see also Figure S1A).
Odorant receptor genes are expressed early in development
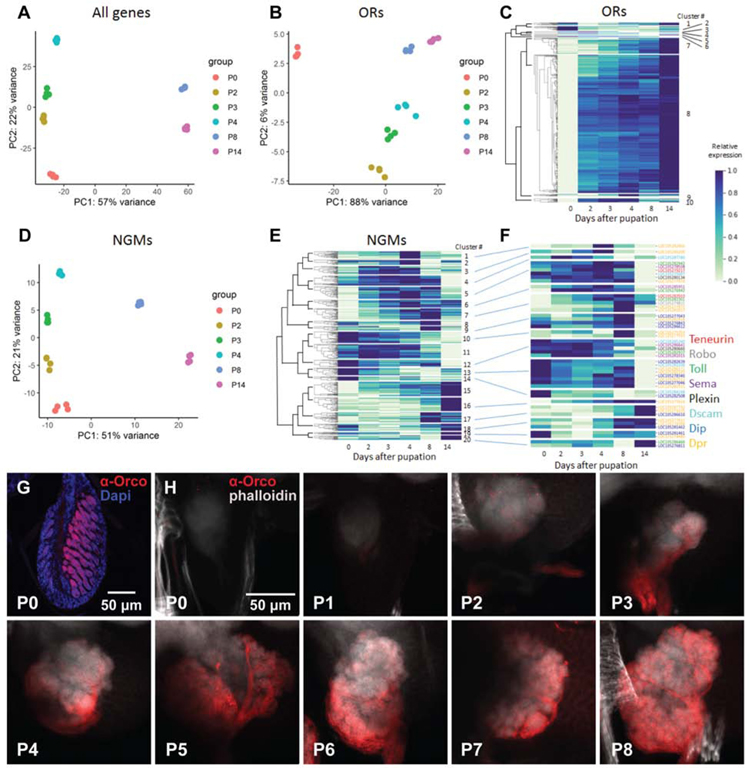
To examine genetic correlates of glomerulus formation, we performed RNA sequencing (RNA-seq) of pupal antennae. We collected antennae at P0, P2, P3, P4, P8, and P14, a range of ages that spans pupal development and captures early changes in particular. We found 11,425 genes expressed in the antennae (defined as at least 10 fragments detected in at least one library), of which 10,850 showed differential expression between time-points (LRT, adjusted P < 0.05). Principal component analysis (PCA) showed clear separation between all time-points along PC1 and PC2, which together explained 79% of the variance (Figure 2A). There was a striking separation along PC1 between P0–P4 and P8/P14. The top ten genes contributing to PC1 included several structural genes potentially involved in cuticle development, along with two putative neuropeptides (Table S1). The top ten genes contributing to PC2 also included several genes likely involved in cuticle development (Table S1). The extensive changes in structural and cuticle proteins are consistent with the large cuticular surface area of the ant antenna.
Figure 2: RNA-seq and Orco staining during pupal development.
(A) PCA of antennal gene expression for all genes at each developmental time-point. (B) PCA of odorant receptor (OR) gene expression in the antennae over the course of development (see also Figure S1B–D and Table S1). (C) Gene expression heatmap and clustering of OR genes throughout development. (D) PCA of neuronal guidance molecule (NGM) gene expression in antennae over the course of development. (E) Gene expression heatmap and clustering of NGM genes throughout development. (F) Gene expression heatmap for NGMs in gene families known to function in AL patterning in Drosophila melanogaster. Relationships to clusters from e) shown by connecting lines. In (C), (E), and (F), gene expression is represented relative to the minimum and maximum expression of each gene across all time-points. Four biological replicates were sequenced for each time-point. (See also Table S2 for raw read counts.) (G) Representative confocal image of P0 antennae sectioned at 12 µm and stained with Orco antibody and DAPI, showing that Orco is strongly labeled in OSN membranes. (H) Projections of confocal z-stacks through pupal brains harvested between P0 and P8 and stained with Orco antibody and phalloidin. Orco is localized to axon terminals, and staining marks OSN axons as they sprout to the antennal lobe. Staining is visible at the edge of the AL by P2, and OSN axons have spread across the surface by P4.
As it has been proposed that ORs themselves may be involved in AL patterning[14], we next examined OR expression over time. PCA of OR expression showed clear separation of time-points, with monotonic progression along PC1 (which explained 88% of the variance; Figure 2B). We found that nearly all ant ORs were expressed by P2 (Figure 2C, Figure S1B, Table S2). The time-course gene expression clustering algorithm DP_GP_cluster[16] partitioned OR genes into 10 clusters (Table S2). The vast majority of OR genes were assigned to a single cluster (cluster 8; 434 of 537 genes and putative pseudogenes), with expression starting at P2 and increasing steadily thereafter, peaking at P14, the final time-point we collected (Figure 2C). An additional 50 ORs were assigned to a cluster with a slight delay in expression onset (P3 instead of P2; cluster 7), while 23 ORs, including Orco, were assigned to clusters that peaked in expression around mid-pupal development (clusters 9 and 10). In the case of Orco, expression was already high at P0, and increased only slightly at P2–4 (Table S2). Thirty ORs were assigned to clusters with divergent expression patterns, many of which had high expression at P0. Therefore, nearly all ORs were expressed early in development, before the onset of glomerulus formation. Early OR expression in the ant stands in striking contrast with D. melanogaster, where OR gene expression begins only after AL patterning and glomerulus formation are well underway[15]. This opens the possibility that, unlike in Drosophila, OR expression or OSN activity are indeed required for ant AL patterning.
To investigate the possible contribution of other patterning factors, we identified 462 candidate neural guidance molecules (NGMs; see methods for identification criteria) and examined their expression during development (Figure 2D, E, Figure S1C). PCA of NGMs showed clustering of time-points resembling that observed with all genes (Figure 2D). DP_GP_cluster recovered 20 separate clusters with divergent expression patterns (Table S2). Clusters 1–6 contained 144 genes and showed peak expression levels at P3 and P4. Clusters 7–9 and 17 contained 80 genes and showed peak expression levels at P8. The remaining clusters showed peak expression at P0/P2 (clusters 10–14; 126 genes) or at P14 (clusters 15, 16, and 18–20; 112 genes). In Drosophila, genes in the Teneurin, Robo, Toll, Semaphorin, Plexin, Dip, Dpr, and Dscam gene families are involved in AL patterning[17–25]. Many genes in each of these families were expressed during glomerulus formation, at P3, P4, and P8, and subsequently downregulated at P14 (Figure 2F, Figure S1C). However, there was no enrichment for any gene family in any gene expression cluster (chi-square test, P = 0.14). These results thus reveal no clear association between any particular NGM gene family and glomerulus development.
Orco is expressed in OSNs and shuttled to axon terminals before the onset of glomerulus formation
Given the early onset of Orco gene expression and the previously reported developmental phenotypes in loss-of-function mutants, we explored the localization of Orco protein in pupal antennae and brains. We collected material from pupae on each day from P0 through P8, immunostained all samples with Orco antibody, and counterstained with DAPI in antennae and phalloidin in brains. Staining labeled Orco in cell bodies and dendrites of neurons in the antennae from the beginning of pupal development (Figure 2G). Intriguingly, whereas in D. melanogaster Orco is localized exclusively to dendrites and OSN cell bodies[15], we noted strong staining of OSN axons projecting from cell bodies in the antennae, as well as in axon terminals in the brain (Figure 2H). Orco is known to chaperone tuning ORs to the cell membrane in flies[15], but is also capable of forming homomeric membrane-localized channels[26]. The presence of Orco in axon terminals thus raises the intriguing possibility that this protein, possibly in combination with tuning ORs, plays a role in ant AL development.
The unique localization of Orco in ant sensory neurons also allowed us to track axons into developing antennal lobes. We found that OSN axons first arrive at the outer edge of the antennal lobe at P2, before spreading over its entire surface (Figure 2H). Coincident with the formation of obvious glomeruli at P4, OSN axons grow into the tissue and surround recognizable glomerular structures. These findings are consistent with a role of OSN axons in the formation of mature glomeruli.
Ablating sensory neurons early in development phenocopies Orco loss-of-function mutants
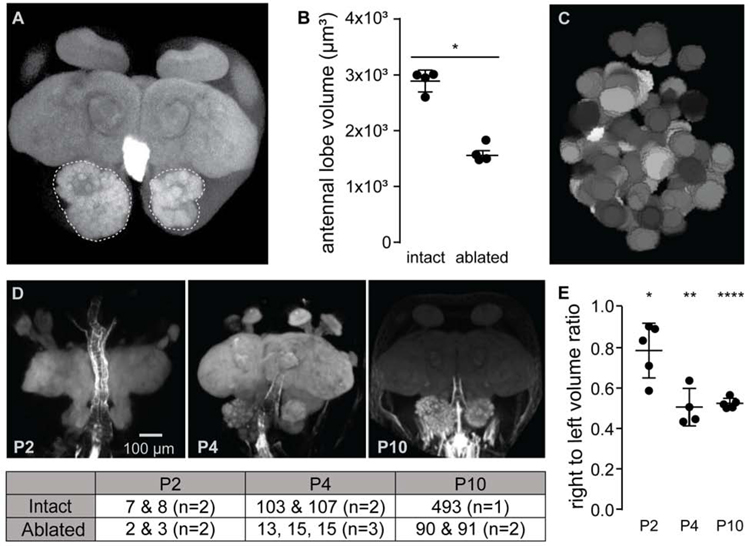
The appearance of observable AL structure coincides at P4 with the growth of OSN axons into the lobe, suggesting that glomerular division is contingent on OSNs. Indeed, previous findings suggest that this is the case in other insects[27–29]. We investigated the consequences of ablating OSNs at P0, before sensory neurons sprout to the antennal lobe. Most projections to the AL come from OSNs housed under sensory sensilla in the antennal club. Furthermore, projections from ant antennae are entirely ipsilateral[11]. Thus, we surgically removed the right antennal club at P0, unilaterally ablating sensory neurons. Aging manipulated animals to eclosion before collecting and imaging their brains allowed us to compare morphology between normally developing and deafferented ALs within single animals. Lobes that received no antennal input were markedly reduced in volume as compared to intact lobes, and had many fewer glomeruli (Figure 3A, B). In fact, reconstructions of aeafferented lobes from two animals showed 90 and 91 large glomeruli, respectively, which exactly replicates the reduced antennal lobe structure in Orco loss-of-function mutants[13] (Figure 3C).
Figure 3: Early antenna ablation rapidly retards size and complexity of the developing antennal lobe.
(A) Maximum projection of a phalloidin-stained adult brain from an ant with the right antenna surgically removed at P0. Antennal lobes are outlined for context. (See also Figure S4.) (B) Antennal lobe volumes in adults were significantly lower on the side where antennae had been ablated at P0 compared to the AL with intact innervation (t-test for difference between average volumes). (C) Representative manual reconstruction of the right antennal lobe in (A) showed 91 distinct glomeruli. Reconstruction of a second brain (not shown) revealed 90 glomeruli (see also Figures S3–S6). (D) Representative images of developing pupa brains with the right antenna surgically removed at P0, along with glomerular counts at each time-point for each antennal lobe listed in table below. (E) Ratio of antennal lobe volumes measured from confocal z-stacks at each time-point showed volume of intact antennal lobes rapidly outpacing that of denervated lobes (t-test for difference from a volume ratio of one). *: p <0.05, **: p <0.01, ****: p <0.00001.
In flies, OSNs from the palps innervate a sizeable fraction of AL glomeruli, as do OSNs from the antennae that express ionotropic receptors (IRs) and gustatory receptors (GRs), two other types of chemosensory receptors that do not rely on Orco[1–3]. One possible explanation of our results is therefore that the reduced AL structures we see in Orco mutants and surgically denervated animals arise because different populations of OSNs survive each manipulation. That is, the structures look similar largely by coincidence. For this to be true, some combination of ca. 90 populations of non-OR OSNs would have to survive Orco loss-of-function, whereas ca. 90 populations of AL-innervating OSNs in the palps would escape antenna ablation. However, analysis of previously published gene expression data[30] showed that only 4 IRs and GRs are expressed in ant heads (including palps, but excluding antennae), 13 in antennae, and 7 in both. Furthermore, backfills from antennae of wild-type adults showed that every glomerulus in the AL receives input from antennal neurons (Figure S2A), while backfills from Orco loss-of-function mutants showed that only six glomeruli in the T7 cluster continue to receive input from the antennae (Figure S2B). Finally, α-Orco staining of wild-type adult ALs revealed that all glomeruli except the T7 glomeruli are innervated by Orco expressing OSNs (Figure S3A). Taken together, these results suggest that only the six glomeruli in the T7 cluster are innervated by OSNs expressing non-OR chemosensory receptors. Our genomic analysis also showed that only 12 ORs are expressed in ant heads (including palps, but excluding antennae). Finally, α-Orco staining of brains from adults that received unilateral ablation of the right antenna at P0 showed no glomeruli innervated by Orco expressing OSNs (Figure S3B). These data, in combination with backfills from antennae in wild-type adults, suggest that few if any glomeruli are uniquely innervated by OSNs from the palps. Therefore, survival of non-OR and/or non-antennal OSNs can explain at most a small subset of the 90 glomerular structures in either Orco loss-of-function mutants or animals with antenna ablations. Instead, we conclude that the reduced AL morphologies that result from either manipulation are phenocopies of one another, probably resulting from an early loss of antennal OSNs in Orco mutants. Importantly, Orco loss-of-function also results in a significant loss of OR-expressing OSNs in the antennae of ants but not flies[13,14]. This suggests that the differences in AL phenotype between Orco mutants in the two species can at least partly be explained by the difference in OSN development or survival during the pupal stage.
The 90 remaining structures in deafferented antennal lobes could represent structures that develop before normal AL development arrests, or could result from aberrant, retarded development unique to the deafferented lobe. To compare early changes in normally developing and deafferented antennal lobes, we removed the right antennal club at P0, and then collected and imaged brains at P2, P4, and P10. In the absence of input from its ipsilateral antenna, the size and complexity of a developing antennal lobe was retarded early in development. Deafferented lobes showed reduced volume as compared to intact lobes as early as P2, a disparity that increased as pupae aged (Figure 3D). Additionally, by P4, the number of observable glomeruli in deafferented versus intact lobes was markedly reduced (Figure 3E). These results show that, in ants, proper AL development is contingent on OSN innervation. However, even in the absence of sensory neuron input, a reduced structure forms that ultimately contains ca. 90 glomeruli, perhaps a template from which OSN-dependent elaboration of antennal lobe structure would normally progress.
Ablating sensory neurons later in development arrests the formation of new glomeruli
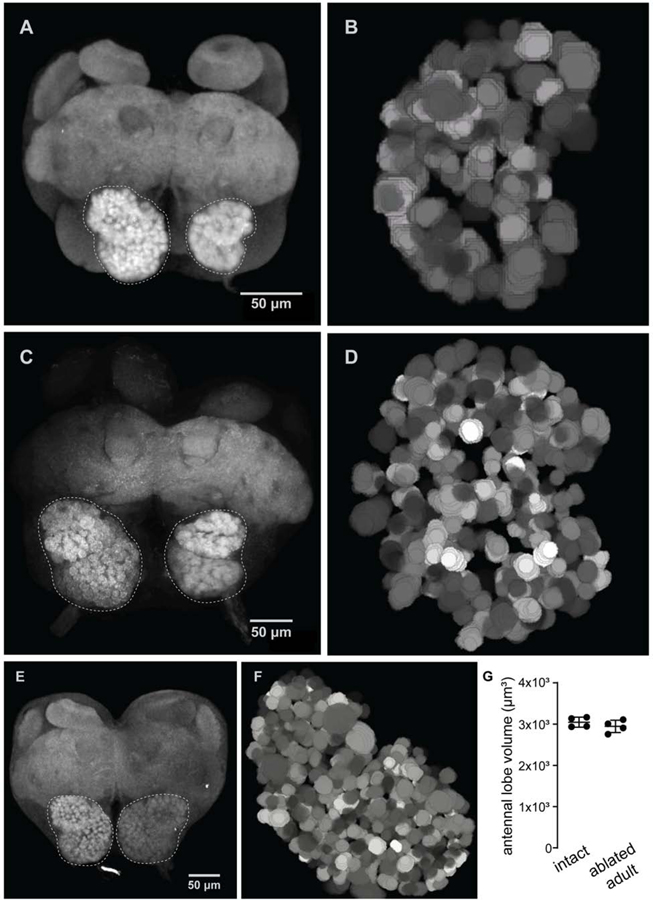
Based on our findings, OSN innervation may only be necessary for the initial formation of mature glomeruli, or it may be required continuously to maintain the glomerular map. To distinguish between these possibilities, we examined the effects of removing sensory afferents at later stages of development by surgically ablating antennae at P4 and P6, aging pupae to eclosion, and collecting brains from newly eclosed adults. Phalloidin stain in confocal images showed that, while reduced, the glomerular map is more complex in these animals than in animals that were denervated at P0 before any sensory neuron innervation of the AL (Figure 4A–D). In fact, a lobe deafferented at P4 contained 118 glomeruli (Figure 4B), while a lobe deafferented at P6 contained 290 glomeruli (Figure 4D), numbers that roughly corresponded to the number of glomeruli present at the corresponding time-point in developing pupae (Figure 1A). Therefore, upon loss of OSN innervation, AL development rapidly arrests, while already formed glomeruli are maintained. Interestingly, lobes deafferented at a given time-point displayed consistent structure from ant to ant, again suggesting that development of the glomerular map is stereotyped.
Figure 4: Antenna ablation arrests formation of the glomerular map.
(A, B) Representative image (from n=5 brains total) of a lobe denervated at P4 and visualized with phalloidin staining in a brain collected from an adult. Reconstruction from a different brain from the sample, 118 glomeruli were reconstructed. (C, D) The same pair of images from a brain ablated at P6 and collected from an adult (representative from n=5 brains total; 290 glomeruli reconstructed). (E, F) Brain image and AL reconstruction from an adult ant ablated one day after eclosion and aged for two weeks showed that the glomerular map is stable once formed, and its maintenance does not require sensory input (representative from n=2 brains total; 493 glomeruli reconstructed). (G) Antennal lobe volumes in adults with one antenna removed were not significantly different between sides (t-test for difference between average volumes).
Finally, the developmental arrest produced by this manipulation serves as a further point of comparison to Orco mutants and lobes for which sensory neuron input was removed at P0. Normally developing P4 pupal brains showed similar numbers of glomeruli as lobes deafferened at P0, raising the possibility that development simply arrested in manipulated animals and in Orco mutants at or around P4. However, when we removed antennae at P4, we observed a qualitatively different result than when we removed antennae at P0. Lobes deafferented at P4 had appreciably more glomeruli (Figure 4B). Additionally, lobes that lost antennal input later in development had well-defined, dense, and circular glomeruli (Figure 4A, C), whereas we observed large, sometimes amorphous glomeruli both when lobes were deafferented at P0 (Figure 3A) and in Orco mutants[13]. Taken in conjunction with our day-by-day examination of how deafferented lobes develop, these data suggest that if brains never receive sensory input, the antennal lobes develop on a unique trajectory.
We wondered whether continual sensory input was necessary for the maintenance of a stable glomerular map in ants. Therefore, we performed the same surgical ablation in newly eclosed callows that had been allowed to develop normally through pupation. We aged these callows for two weeks, before collecting and imaging brains. Deafferented antennal lobes showed no reduction in volume (Figure 4G). Reconstruction of a deafferented lobe from one animal showed 493 glomeruli (Figure 4E, F), a number consistent with previously published reconstructions of unmanipulated adults[13,14]. Like late-stage pupa ablations, surgical ablations in adults suggest that glomeruli are stable once specified, even if afferent OSN input is subsequently removed.
Ablating sensory neurons during early fly development prevents glomerulus formation
Our observations that OSNs are necessary to allow glomerulus formation are consistent with studies of deafferented ALs in bees and moths, and with characterizations of ALs in ato mutant flies that lack a large subset of OSNs[28,29,31]. To investigate whether our observation of OSN-independent AL structure in ants may apply to other insects, we examined the effects of ablating OSNs at the beginning of pupal development in D. melanogaster. We genetically ablated OSNs from developing fly pupae using +peb-GAL4, ey-FLP, UAS-FRT-w+-FRT-RTA, a line in which Ricin toxin A (RTA) is restricted to sensory neurons from the eye-antennal disc (including all OSNs)[22,32–35], and examined phalloidin (actin) and nc82 (synapses) stained confocal image stacks of the antennal lobe in newly eclosed adult flies. OSN-ablated flies lacked morphologically distinguishable glomeruli, although minimal substructure in the form of around five poorly differentiated blobs could be observed in some individuals (Figure S4). These structures qualitatively contrasted with the well-differentiated structures observed in surgically ablated and Orco knockout ants. The variability in the amount of sub-sectioning that we observed from brain to brain in the fly could be due to slight variations in timing of the onset of toxin expression or to the efficacy of FLP-mediated recombination of the RTA allele. However, we did observe a number of brains in which there was no obvious sub-structure to the lobe whatsoever (Figure S4A, B). These findings imply that the two-stage specification of AL glomeruli observed in ants is not a general phenomenon in insects. Work in D. melanogaster suggests that PN dendrites pre-pattern the antennal lobe prior to the arrival of OSN axons, and thus serve as a template to guide development of the glomerular map in flies[6,22]. Presumably, lack of OSN innervation in flies prevents the maturation of visible glomeruli from this pre-patterning stage. An intriguing possibility is that an analogous PN-driven structure develops in ants and is revealed by the death of OSNs early in development.
Conclusion
Here we present a detailed molecular and anatomic description of antennal lobe development in ants. We found that in ants, as opposed to flies, ORs are expressed in OSNs prior to glomerulus formation. Furthermore, at least the conserved OR co-receptor, Orco, is present at the axon terminals as they invade the AL coincident with glomerulus formation. We also showed that OSN innervation is necessary for glomerulus maturation throughout early development, but not for continued maintenance of glomeruli. However, even without any OSN innervation from the antennae, ca. 90 glomerular structures form in the ant AL. This suggests a two-step process, in which the gross AL structure is established independently of OSNs, but the formation of mature glomeruli depends on OSN innervation. We observed no such phenomenon in flies, which have aglomerular ALs following genetic denervation. At this point, we can only speculate about the nature of the ca. 90 remaining structures in ants. However, because they develop in the absence of OSN input, they are unlikely to constitute a class of functional glomeruli that survive Orco loss-of-function, as has been suggested previously[14,36]. Instead, this reduced AL structure might represent pre-patterning that is revealed in the absence of OSN innervation. A two-step developmental logic of this sort could facilitate rapid expansion of the sensory periphery, while allowing downstream circuitry to remain developmentally compact. For example, like in Drosophila[37], PNs could fall into a moderate number of molecularly defined classes. Unlike in Drosophila, however, increased complexity in the mature glomerular map could then be driven by sensory neuron innervation. The mouse olfactory system shows remarkably similar glomerular organization in the olfactory bulb, the brain area analogous to the insect AL, but must contend with around a thousand afferent OSN classes. Mice ORs indeed instruct olfactory bulb development[38], and this sort of developmental plasticity likely affords mammalian olfactory systems developmental robustness and evolutionary flexibility, allowing for rapid flux in OR repertoires[39,40]. If AL patterning in the second stage of ant development were likewise directly mediated by ORs, this could facilitate similar flexibility by allowing single-step evolution of novel olfactory circuits. Although all of our findings are consistent with a role of OSNs in refining ant AL patterning, they also suggest that the developmental defects observed in Orco knockouts in fact result from OSN death early in development. It therefore remains unclear whether ORs themselves are involved in patterning the AL beyond promoting OSN survival. Our findings highlight the usefulness of a comparative approach to studying sensory development and establish ants as a system in which to explore further how downstream wiring strategies relate to evolution at the sensory periphery.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Daniel J.C. Kronauer (dkronauer@rockefeller.edu)
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
RNA-seq reads are deposited in the National Center for Biotechnology Information sequence read archive (Bioproject accession #PRJNA632608).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Ants
Large stock colonies of Ooceraea biroi were maintained at 25°C in Tupperware containers (32 × 14 cm) with a plaster of Paris floor ca. 2 cm thick. During the brood care phase, colonies were fed 3 times weekly with fire ant (Solenopsis invicta) brood, and cleaned and watered once per week as necessary.
Flies
Flies were maintained on cornmeal-molasses (“R”) food (Lab Express, Ann Arbor, MI) in a humidified incubator at 25°C on a 12:12 light:dark cycle.
METHOD DETAILS
Collection of material for brain imaging
Late-stage larvae were isolated from stock colonies and placed in circular Petri dishes (50 mm diameter, 9 mm height, ca. 5 mm thick plaster floor) with an equal number of workers. Sufficient numbers of larvae were isolated such that pupae could be collected and processed at each of the focal stages of development to assemble a complete time course from a single cohort of larvae. Petri dishes were cleaned and watered, and colonies were fed as needed until larvae pupated. Colonies then remained without further maintenance until pupae were collected. For the wild-type time course, pupae were collected in the morning of the appropriate day. Depending on the experiment, heads or antennae were immediately dissected in cold PBS and moved to 4% PFA in PBS-T (1X PBS with 0.5% Triton-X) to fix at room temperature for 2 hours. Tissues were washed 3X in cold PBS and then stored at 4°C in PBS until all material for a complete time course could be processed in parallel.
More larvae were staged for the ablation experiments, as only ca. 40–80% of pupae survived antenna ablation (survival was low for early-stage ablations and increased for late-stage ablations). On the morning of the appropriate day, pupae were collected, and the right antenna was cut at the scape using a set of sharpened forceps sterilized with Neosporin solution. Pupae were gently transferred to a clean Petri dish with a plaster floor, and an equal number of workers were added to care for the ablated individuals until they were collected for dissection. At the appropriate day post-ablation, heads were dissected and fixed as above. To collect brains from callow (newly eclosed) ants, individuals were identified by their much lighter color and were collected one day post eclosion. Brains were dissected out of the head case in cold PBS, fixed in 4% PFA in PBS (no Triton), washed, and held in PBS until processing.
Neuroanatomy
Phalloidin stain for anatomy time course
Fixed brains were dissected out of the cuticle, and heads from a single age-cohort were pooled in PBS-T with 1:50 Alexa Fluor 555 phalloidin. Heads were incubated overnight on a shaker at room temperature, and then washed 3X in PBS. The tissue was dehydrated on a fresh gradient of propanol: 30, 50, 70, 90, 100, 100%, cleared and mounted with methyl salicylate, and stored in the dark at 4°C until imaging. Brains were imaged on a Zeiss LSM 880 confocal microscope with a 40X water objective. Z-stacks were obtained of the entire brain, and were further processed in the Fiji[41] distribution of ImageJ unless otherwise noted. The final unmanipulated time course sampled n=3 brains at each time point except for P4, where n=2 (Figure S1A). For tracking pupal development post-ablation, we collected n=10 brains at P2 and P4, and n=3 brains for all other sampled time points (P5, P6, P8, and P10).
Immunohistochemistry in pupal brains
Fixed brains were dissected and pooled as above, and incubated in blocking solution (5% normal goat serum in PBS-T) for 1 hour on a shaker at room temperature. Brains were then transferred to a 1:500 dilution of mouse anti-Orco primary antibody in blocking solution, and incubated overnight on a shaker at room temperature, then washed 3X in PBS, and incubated in donkey anti-mouse Alexa Fluor 594 secondary antibody with 1:50 Alexa Fluor 647 phalloidin for two hours on a shaker at room temperature. Brains were washed 3X in PBS, mounted in DAKO, and stored at 4°C until imaging. Brains were imaged as above.
Immunohistochemistry in antennae
Antennae were processed as described previously[13]. Briefly, fixed antennae were incubated overnight in a solution of 25% sucrose in PBS-T at 4°C, then embedded in Tissue-Tek OCT and frozen. Antennae were sectioned at 12 µ m at −20°C, and after being mounted on a glass slide, the sections were dried at room temperature for 3 hours. Sections were then fixed in 4% paraformaldehyde in PBS-T at room temperature for 30 min, and then rinsed with PBS. Then they were incubated in blocking solution and in mouse anti-Orco as described above, followed by rinsing and incubation with donkey anti-mouse Alexa Fluor 594 with 1µg/ml DAPI. Finally, antennae were washed 3X in PBS, mounted in DAKO, and stored at 4°C until imaging.
Glomerulus counts and antennal lobe volumes
Three-dimensional projections were created from confocal image stacks using the maximum Z-projection feature in Fiji. Reconstructions of glomeruli were produced by manually segmenting the confocal stacks using the Segmentation Editor plugin. Images of the reconstructions were generated from the plugin. To calculate antennal lobe volumes, confocal image stacks were loaded into Imaris, a three-dimensional region of interest was manually defined, and a volume of the region was calculated within the software. Volumes were not normalized to a marker structure, as the relevant data were ratios between the two lobes and were therefore internally normalized.
Backfills from antennae
Ants were immobilized in viscoelastic liquid silicone (Silly Putty™) at 4°C and the antennae were severed at the base of the flagellum using a razor blade. A droplet of microruby (rhodamine dextran with biotin, 3,000 MW, D7162; Thermo Fisher Scientific) dissolved in distilled water was immediately applied to the stump. Ants were incubated like this overnight in a humidified box in the dark at 4°C. Brains were subsequently dissected from the head in PBS and fixed in 4% PFA for four hours. Brains were then serially dehydrated in ethanol before mounting in methyl-salicylate and imaging as described above.
Gene expression
Gene expression time course during pupal development
P0 pupae from a single large stock colony were isolated as above and divided into four separate colonies. Ten pupae were collected from each colony on days zero, two, three, four, eight, and 14 (days which correspond to major anatomical changes in the AL) and frozen at −80°C. Antennae were then dissected from the pupae directly into Trizol™ reagent (Thermo Fisher). All antennae from each colony and time-point were pooled for extraction and sequencing, resulting in four biological replicates for each time-point. Samples were then homogenized using a Qiagen TissueLyser II, and the aqueous phase extracted using PhaseLock columns (5PRIME) and purified using the Qiagen RNeasy kit following the manufacturer’s protocol. Illumina libraries were then prepared and sequenced on an Illumina NextSeq 500 at the Rockefeller University Genomics Resource Center.
RNA-seq reads were mapped to clonal raider ant genome release v5.4 (NCBI accession GCF_003672135.1), including junctions from the latest RefSeq annotation (Ooceraea biroi annotation release 100; note that numbering was restarted following the transfer of the clonal raider ant from the genus Cerapachys to the genus Ooceraea, thus the O. biroi annotation release 100 is not the same as the C. biroi annotation release 100, despite both being RefSeq annotations of the same species), along with previously published manual annotations of chemosensory genes[42]. RefSeq gene annotations with coding sequence overlaps to manually annotated genes were excluded. Reads mapping to each gene were then quantified using HTSeq[43]. Differential expression analysis was performed using DESeq2[44]. To determine which genes were differentially expressed between different time-points, each time-point was coded as a factor in a DESeq2 model, and a likelihood ratio test was used to compare this model to a model with no differences (“reduced = ~1”). Functions in the DESeq2 package were also used to perform VST normalization of expression data for clustering analyses. Genes were assigned to clusters by their expression over the time course using a Dirichlet process Gaussian process model implemented in DP_GP_cluster[16]. The MPEAR criterion was used to assess optimal clustering. Genes within each cluster were further hierarchically clustered for display purposes using nearest point clustering by correlation implemented by the cluster.hierarchy.linkage function from scipy[45], and each cluster was then hierarchically clustered using the mean gene expression for each time-point for all genes within the cluster.
Quantification of gene expression in adult antennae and heads
Previously published RNA-seq data from McKenzie et al.[30] was downloaded from the NCBI SRA database (BioProject accession PRJNA230648) and processed as described above. Fragments per kilobase per million mapped reads (FPKM) was calculated using a custom python script, and genes with expression greater than 2 FPKM were considered as expressed in a given tissue.
Identification of candidate neural guidance molecule genes
All genes from the latest NCBI RefSeq annotation of the clonal raider ant (NCBI Ooceraea biroi annotation release 100) were functionally annotated with GO terms, IPR protein domains, transmembrane domains, and signal peptides by using the InterProScan[46] program to run Phobius[47] and query the SMART, ProSiteProfiles, Pfam, Superfamily, PANTHER, PRINTS, and CDD databases. A gene was considered as a candidate neural guidance molecule (NGM) coding gene if 1) it contained either a signal peptide or a transmembrane domain and 2) it contained either an EGF-like domain (IPR000742), an IGG-like domain (IPR007110), a cell-adhesion GO annotation (GO:0007155), or a protein binding GO annotation (GO:0005515). We also used blastp against functionally annotated Drosophila melanogaster genes and the NCBI conserved domains database to identify members of protein families known to function in OSN axon guidance in D. melanogaster (DIPs, DPRs, Dscams, Tenurins, Tolls, Semaphorins, Plexins, and Robo). All members of these protein families were annotated as candidate NGM genes in our InterProScan analysis, validating this approach for identification of NGMs.
Fly ablations
Experimental Genotypes
| Peb-Gal4, eyFLP; CyO/attp40; UAS-shi[ts]/+ |
| Peb-Gal4, eyFLP; CyO/UAS>w+>RTA; UAS-shi[ts]/+ |
The scheme for ablating olfactory neurons at the placode stage and independent of OR versus IR expression was based on[22], and the Peb-Gal4, eyFLP recombinant was a gift of Liqun Luo. All animals analyzed were female due to genetic constraints. Animals with eyFLP, Peb-Gal4, and UAS>w+>RTA largely lacked eyes and had visibly affected antennae. shi[ts] was included in the genotype due to exigencies of the crossing strategy. We did not rear flies at the elevated temperature where shi[ts] has an effect on synaptic vesicle release (usually 29°C or higher), and thus do not expect it to have affected the results, which were stark and consistent in additional experiments with ChAT staining where the shi[ts] allele was not included. Brains were dissected at 2–7 days post-eclosion and stained and mounted as described previously[31]. We analyzed 5–10 brains per genotype per batch, and derived two data-points from each brain corresponding to the two hemispheres. Antibodies were mouse anti-ChAT 9E10 (DSHB, 1:200), mouse nc82 anti-brp (DSHB, 1:40). Brains were counterstained with Phalloidin-568 (Invitrogen, 1:40 or 1:80) and DAPI (1µ g/mL). Images were collected on a Leica SP8 confocal microscope with 1 µm spacing in Z (along the anterior-posterior axis) and ~200 nm axial resolution.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analyses are described in the main manuscript. Analysis tools for gene expression are described and cited in the main text. T-test analyses in Figure 3B and 3E were done using GraphPad Prism version 8.0.0 for Windows, GraphPad Software, San Diego, California USA, www.graphpad.com.
Supplementary Material
Table S2. Expression (raw mapped counts) for each gene at all time-points throughout pupal development, as well as clustering information for ORs and NGMs. Related to Figure 2A–F.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-Orco | Gift from Vanessa Ruta | [48] |
| Invitrogen Donkey anti-mouse AlexaFluor 647 | Thermo Fisher Scientific | Cat#A-31571 |
| Invitrogen Donkey anti-mouse AlexaFluor 594 | Thermo Fisher Scientific | Cat#A-21203 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| AlexaFluor Plus 555 phalloidin | Thermo Fisher Scientific | Cat#A30106 |
| AlexaFluor 647 phalloidin | Thermo Fisher Scientific | Cat#A22287 |
| DAPI | Thermo Fisher Scientific | Cat# D1306 |
| Rhodamine dextran with biotin 3000 MW | Thermo Fisher Scientific | Cat# D7162 |
| Critical Commercial Assays | ||
| RNeasy kit | Qiagen | Cat# 71404 |
| NextSeq 500 | Illumina | SY-451-1002 |
| Deposited Data | ||
| RNA-seq data | [11] | PRJNA230648 |
| RNA-seq data | This paper | PRNJA632608 |
| Experimental Models: Organisms/Strains | ||
| D. melanogaster: Peb-Gal4 (X) | Liqun Luo/Bloomington Drosophila Stock Center | BDSC 80570, [22] |
| D. melanogaster: Ey-flp (X) | Liqun Luo/ Bloomington Drosophila Stock Center | BDSC 5580, [35] |
| D. melanogaster: UAS>w+>RTA (II) | Liqun Luo/ Bloomington Drosophila Stock Center | BDSC 29000, [22] |
| D. melanogaster: empty attp40 (II) | Bloomington Drosophila Stock Center/ TRIP collection | BDSC 36304, [35] |
| D. melanogaster: UAS-shi[ts] | Liqun Luo | [22] |
| Software and Algorithms | ||
| HTseq | [43] | https://htseq.readthedocs.io/en/master/ |
| DEseq2 | [44] | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| ImageJ (Fiji) | [41] | https://fiji.sc/ |
| scipy | [45] | https://www.scipy.org/ |
| DP_GP_cluster | [16] | https://github.com/PrincetonUniversity/DP_GP_cluster |
| InterProScan | [46] | https://ebi.ac.uk |
| Phobius | [47] | https://phobius.sbc.su.se |
| GraphPad Prism, version 8.0.0 for Windows | GraphPad Prism | https://www.graphpad.com |
Highlights.
Ant odorant receptors show unique early expression and localization in pupae
Sensory neuron innervation is necessary for proper antennal lobe development
Sensory neuron ablation reveals latent antennal lobe structure in ants but not flies
Insects with complex olfactory systems may employ a different developmental logic
Acknowledgements
Research reported in this publication was supported by grants from the Faculty Scholars Program of the Howard Hughes Medical Institute and the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R35GM127007 (both to DJCK). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Howard Hughes Medical Institute. SKM was supported by NIH National Research Service Award Training Grant GM066699 and a Marie Skłodowska-Curie Individual Fellowship (ID 797969) from the European Commission. EJC was supported by start-up funds from the University of Michigan. Liqun Luo provided fly strains. This is Clonal Raider Ant Project paper #12.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- 1.Laissue PP, Reiter C, Hiesinger PR, Halter S, Fischbach KF, and Stocker RF (1999). Three-dimensional reconstruction of the antennal lobe in Drosophila melanogaster. Journal of Comparative Neurology 405, 543–552. [PubMed] [Google Scholar]
- 2.Couto A, Alenius M, and Dickson BJ (2005). Molecular, Anatomical, and Functional Organization of the Drosophila Olfactory System. Current Biology 15, 1535–1547. [DOI] [PubMed] [Google Scholar]
- 3.Vosshall LB (2000). Olfaction in Drosophila. Current Opinion in Neurobiology 10, 498–503. [DOI] [PubMed] [Google Scholar]
- 4.Sardana J, Organisti C, and Kadow ICG (2018). Eph Receptor Effector Ephexin Mediates Olfactory Dendrite Targeting in Drosophila. Developmental Neurobiology 78, 873–888. [DOI] [PubMed] [Google Scholar]
- 5.Technau GM ed. (2008). Brain development in Drosophila melanogaster (New York : Austin, Tex: Springer Science+Business Media ; Landes Bioscience; ). [Google Scholar]
- 6.Jefferis GSXE, Vyas RM, Berdnik D, Ramaekers A, Stocker RF, Tanaka NK, Ito K, and Luo L (2004). Developmental origin of wiring specificity in the olfactory system of Drosophila. Development 131, 117–130. [DOI] [PubMed] [Google Scholar]
- 7.Robertson HM, and Wanner KW (2006). The chemoreceptor superfamily in the honey bee, Apis mellifera: Expansion of the odorant, but not gustatory, receptor family. Genome Res 16, 1395–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salecker I, and Boeckh J (1995). Embryonic development of the antennal lobes of a hemimetabolous insect, the cockroach periplaneta americana: Light and electron microscopic observations. Journal of Comparative Neurology 352, 33–54. [DOI] [PubMed] [Google Scholar]
- 9.Gascuel J, and Masson C (1991). A quantitative ultrastructural study of the honeybee antennal lobe. Tissue and Cell 23, 341–355. [DOI] [PubMed] [Google Scholar]
- 10.d’Ettorre P (2016). Genomic and brain expansion provide ants with refined sense of smell. PNAS 113, 13947–13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKenzie SK, Fetter-Pruneda I, Ruta V, and Kronauer DJC (2016). Transcriptomics and neuroanatomy of the clonal raider ant implicate an expanded clade of odorant receptors in chemical communication. Proceedings of the National Academy of Sciences 113, 14091–14096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zube C, Kleineidam CJ, Kirschner S, Neef J, and Rössler W (2008). Organization of the olfactory pathway and odor processing in the antennal lobe of the ant Camponotus floridanus. Journal of Comparative Neurology 506, 425–441. [DOI] [PubMed] [Google Scholar]
- 13.Trible W, Olivos-Cisneros L, McKenzie SK, Saragosti J, Chang N-C, Matthews BJ, Oxley PR, and Kronauer DJC (2017). orco Mutagenesis Causes Loss of Antennal Lobe Glomeruli and Impaired Social Behavior in Ants. Cell 170, 727–735.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan H, Opachaloemphan C, Mancini G, Yang H, Gallitto M, Mlejnek J, Leibholz A, Haight K, Ghaninia M, Huo L, et al. (2017). An Engineered orco Mutation Produces Aberrant Social Behavior and Defective Neural Development in Ants. Cell 170, 736–747.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, and Vosshall LB (2004). Or83b Encodes a Broadly Expressed Odorant Receptor Essential for Drosophila Olfaction. Neuron 43, 703–714. [DOI] [PubMed] [Google Scholar]
- 16.McDowell IC, Manandhar D, Vockley CM, Schmid AK, Reddy TE, and Engelhardt BE (2018). Clustering gene expression time series data using an infinite Gaussian process mixture model. PLOS Computational Biology 14, e1005896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong W, and Luo L (2014). Genetic control of wiring specificity in the fly olfactory system. Genetics 196, 17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong W, Mosca TJ, and Luo L (2012). Teneurins Instruct Synaptic Partner Matching in an Olfactory Map. Nature 484, 201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jhaveri D, Saharan S, Sen A, and Rodrigues V (2004). Positioning sensory terminals in the olfactory lobe of Drosophila by Robo signaling. Development 131, 1903–1912. [DOI] [PubMed] [Google Scholar]
- 20.Ward A, Hong W, Favaloro V, and Luo L (2015). Toll Receptors Instruct Axon and Dendrite Targeting and Participate in Synaptic Partner Matching in a Drosophila Olfactory Circuit. Neuron 85, 1013–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lattemann M, Zierau A, Schulte C, Seidl S, Kuhlmann B, and Hummel T (2007). Semaphorin-1a controls receptor neuron-specific axonal convergence in the primary olfactory center of Drosophila. Neuron 53, 169–184. [DOI] [PubMed] [Google Scholar]
- 22.Sweeney LB, Couto A, Chou Y-H, Berdnik D, Dickson BJ, Luo L, and Komiyama T (2007). Temporal target restriction of olfactory receptor neurons by Semaphorin-1a/PlexinA-mediated axon-axon interactions. Neuron 53, 185–200. [DOI] [PubMed] [Google Scholar]
- 23.Barish S, Nuss S, Strunilin I, Bao S, Mukherjee S, Jones CD, and Volkan PC (2018). Combinations of DIPs and Dprs control organization of olfactory receptor neuron terminals in Drosophila. PLOS Genetics 14, e1007560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hummel T, and Zipursky SL (2004). Afferent induction of olfactory glomeruli requires N-cadherin. Neuron 42, 77–88. [DOI] [PubMed] [Google Scholar]
- 25.Hummel T, Vasconcelos ML, Clemens JC, Fishilevich Y, Vosshall LB, and Zipursky SL (2003). Axonal targeting of olfactory receptor neurons in Drosophila is controlled by Dscam. Neuron 37, 221–231. [DOI] [PubMed] [Google Scholar]
- 26.Jones PL, Pask GM, Rinker DC, and Zwiebel LJ (2011). Functional agonism of insect odorant receptor ion channels. PNAS 108, 8821–8825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tolbert LP, and Sirianni PA (1990). Requirement for olfactory axons in the induction and stabilization of olfactory glomeruli in an insect. Journal of Comparative Neurology 298, 69–82. [DOI] [PubMed] [Google Scholar]
- 28.Gascuel J, and Masson C (1991). Developmental study of afferented and deafferented bee antennal lobes. Journal of Neurobiology 22, 795–810. [DOI] [PubMed] [Google Scholar]
- 29.Oland L, Orr G, and Tolbert L (1990). Construction of a protoglomerular template by olfactory axons initiates the formation of olfactory glomeruli in the insect brain. The Journal of Neuroscience 10, 2096–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKenzie SK, Oxley PR, and Kronauer DJ (2014). Comparative genomics and transcriptomics in ants provide new insights into the evolution and function of odorant binding and chemosensory proteins. BMC Genomics 15, 718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jhaveri D, and Rodrigues V (2002). Sensory neurons of the Atonal lineage pioneer the formation of glomeruli within the adult Drosophila olfactory lobe. Development 129, 1251–1260. [DOI] [PubMed] [Google Scholar]
- 32.Newsome TP, Asling B, and Dickson BJ (2000). Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development 127, 851–860. [DOI] [PubMed] [Google Scholar]
- 33.Allen MJ, O’Kane CJ, and Moffat KG (2002). Cell ablation using wild-type and cold-sensitive ricin-A chain in Drosophila embryonic mesoderm. Genesis 34, 132–134. [DOI] [PubMed] [Google Scholar]
- 34.Kitamoto T (2001). Conditional modification of behavior in Drosophila by targeted expression of a temperature-sensitive shibire allele in defined neurons. Journal of Neurobiology 47, 81–92. [DOI] [PubMed] [Google Scholar]
- 35.Gao XJ, Riabinina O, Li J, Potter CJ, Clandinin TR, and Luo L (2015). A Transcriptional Reporter of Intracellular Ca2+ in Drosophila. Nat Neurosci 18, 917–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan H, Jafari S, Pask G, Zhou X, Reinberg D, and Desplan C (2020). Evolution, developmental expression and function of odorant receptors in insects. J. Exp. Biol 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Horns F, Wu B, Xie Q, Li J, Li T, Luginbuhl DJ, Quake SR, and Luo L (2017). Classifying Drosophila Olfactory Projection Neuron Subtypes by Single-Cell RNA Sequencing. Cell 171, 1206–1220.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lodovichi C, and Belluscio L (2012). Odorant Receptors in the Formation of the Olfactory Bulb Circuitry. Physiology 27, 200–212. [DOI] [PubMed] [Google Scholar]
- 39.Jiang Y, and Matsunami H (2015). Mammalian odorant receptors: functional evolution and variation. Curr Opin Neurobiol 34, 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adipietro KA, Mainland JD, and Matsunami H (2012). Functional Evolution of Mammalian Odorant Receptors. PLOS Genetics 8, e1002821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nature Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKenzie SK, and Kronauer DJC (2018). The genomic architecture and molecular evolution of ant odorant receptors. Genome Res Available at: http://genome.cshlp.org/content/early/2018/10/19/gr.237123.118 [Accessed February 4, 2020]. [DOI] [PMC free article] [PubMed]
- 43.Anders S, Pyl PT, and Huber W (2015). HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, Burovski E, Peterson P, Weckesser W, Bright J, et al. (2020). SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nature Methods. [DOI] [PMC free article] [PubMed]
- 46.Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Käll L, Krogh A, and Sonnhammer ELL (2007). Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server. Nucleic Acids Res 35, W429–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Butterwick JA, Del Mármol J, Kim KH, Kahlson MA, Rogow JA, Walz T, and Ruta V (2018). Cryo-EM structure of the insect olfactory receptor Orco. Nature 560, 447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S2. Expression (raw mapped counts) for each gene at all time-points throughout pupal development, as well as clustering information for ORs and NGMs. Related to Figure 2A–F.
Data Availability Statement
RNA-seq reads are deposited in the National Center for Biotechnology Information sequence read archive (Bioproject accession #PRJNA632608).