Abstract
Introduction:
Advances in ablation for atrial fibrillation (AF) continue to be hindered by ambiguities in mapping, even between experts. We hypothesized that convolutional neural networks (CNN) may enable objective analysis of intracardiac activation in AF, which could be applied clinically if CNN classifications could also be explained.
Methods:
We performed panoramic recording of bi-atrial electrical signals in AF. We used the Hilbert-transform to produce 175,000 image grids in 35 patients, labeled for rotational activation by experts who showed consistency but with variability (kappa=0.79). In each patient, ablation terminated AF. A CNN was developed and trained on 100,000 AF image grids, validated on 25,000 grids, then tested on a separate 50,000 grids.
Results:
In the separate test cohort (50,000 grids), CNN reproducibly classified AF image grids into those with/without rotational sites with 95.0% accuracy (CI 94.8–95.2%). This accuracy exceeded that of support vector machines, traditional linear discriminant and k-nearest neighbor statistical analyses. To probe the CNN, we applied Gradient-weighted Class Activation Mapping which revealed that the decision logic closely mimicked rules used by experts (C-statistic 0.96).
Conclusions:
Convolutional neural networks improved the classification of intracardiac AF maps compared to other analyses, and agreed with expert evaluation. Novel explainability analyses revealed that the CNN operated using a decision logic similar to rules used by experts, even though these rules were not provided in training. We thus describe a scaleable platform for robust comparisons of complex AF data from multiple systems, which may provide immediate clinical utility to guide ablation.
Keywords: Arrhythmia, Atrial Fibrillation, Deep Learning, Machine Learning
Introduction
The success of ablation therapy for atrial fibrillation (AF) remains at 40–70% despite advances in mechanistic understanding and technology1, 2. It has been difficult to improve upon pulmonary vein isolation (PVI) using anatomical lines or even posterior wall isolation1, 2. Intuitively, AF ablation could be personalized to intracardiac patterns, yet there is considerable ambiguity in interpreting AF electrograms (complex fractionated electrograms3, high dominant frequency4) or spatial maps of AF5–7. Machine learning (ML) offers the potential to reduce ambiguity in analyzing complex intracardiac data in AF, interpret mechanisms and guide therapy.
Numerous approaches are emerging to use intracardiac AF electrograms to identify ablation targets outside the pulmonary veins (PV) by dipole density mapping8, 9, electrographic flow mapping10, 11, CartoFinder12–14, Stochastic Trajectory Analysis of Ranked Signals15 and other approaches16–19. However, it is unclear how to reconcile differences in AF maps by these approaches. While some AF mapping systems have been compared10, 19, 20, and one validated directly against optical maps of human AF21, most have not been objectively compared. A major limitation of current AF mapping is that it requires human interpretation which introduces variability and is difficult to scale. A truly automated method that is as accurate as a group of human experts might improve the results of map guided AF ablation.
We hypothesized that machine learning (ML) provides a computational platform to classify AF maps and automatically identify potential ablation targets. ML is a rapidly developing branch of computer science which can reveal hidden data structures in complex data22, 23. ML has been applied to the ECG to diagnose arrhythmias24, 25,26, yet has rarely been applied to intracardiac AF data. Moreover, ML is often considered a ‘black box’ with unclear rationale behind its decisions22, 23, which may limit its clinical use. We further hypothesized that explainability analyses may reveal how ML makes its classification decisions.
We tested our hypotheses by developing supervised ML to classify panoramic intracardiac AF data, in a well characterized cohort of persistent AF patients in whom ablation terminated AF. To broaden applicability, we trained ML on AF maps created by a freely available mapping approach. We also compared results of a CNN to traditional statistics and ML methods. Finally, we applied novel methods from computer science to explain how trained machines arrive at their classification in an attempt to demystify the ‘black box’ and increase clinical confidence.
Methods
Patient Inclusions
We included 35 patients with persistent AF from the COMPARE-AF registry (COMParison of Algorithms for Rotational Evaluation in Atrial Fibrillation, NCT02997254) undergoing panoramic mapping of AF with ablation that terminated persistent AF to sinus rhythm or atrial tachycardia. Persistent AF was defined by guidelines18, and AF was refractory to ≥ 1 anti-arrhythmic medication.
Cases were performed after written informed consent of each subject under protocols approved by the Human Research Protection Program at each center. All methods were performed in accordance with relevant guidelines and regulations. Sample online data, videos, and non-proprietary AF mapping software code used in this project are available to researchers for the purposes of verifying the results or procedures on http://narayanlab.stanford.edu.
Electrophysiological Study and Ablation
Patients were studied in the post-absorptive state. Class I and III anti-arrhythmic medications were discontinued for > 5 half-lives (>30 days for amiodarone). Catheters were advanced to the right atrium (RA), coronary sinus and trans-septally to left atrium (LA). Basket catheters (64 poles, Abbott, St. Paul, MN) were positioned in right then left atria for AF mapping. These newer baskets capture >80% of atrial area in recent studies27.
Radiofrequency energy was delivered via an irrigated catheter (SmartTouch, Biosense-Webster; or Tacticath, Abbott, St. Paul, MN) at 25–35 watts. All patients had prospective ablation at regions identified by a clinical mapping system (Rhythmview™, Abbott, St. Paul, MN) that may reveal sites of sustained rotational activity in AF as validated in simultaneous optical mapping of human hearts21.
Ablation lesions were applied for 15–30 seconds at each site, successively, to cover areas of 2–3 cm2 as described by Miller et al.28 The precise electrode where prospective ablation acutely terminated persistent AF was labeled. PVI was performed comprising circumferential ablation of left and right PV pairs with verification of PV isolation using dedicated circular mapping catheters.
Data Flow in the Study
Figure 1 shows data flow in this study. Intracardiac voltage time-series data from the heart (fig. 1a) were recorded using basket catheters positioned in left then right atrium (fig. 1b). Unipolar electrograms were recorded at 0.05 to 500 Hz bandpass, at 1 kHz sampling with electroanatomic location turned off to reduce electromagnetic interference. Analysis used 1 minute of data focusing on the 4 seconds used clinically to guide ablation. Raw electrograms comprising 64 basket and other channels were exported from Bard (LabSystem Pro), Prucka (GE Cardiolab) or Siemens recorders. Raw basket recordings of AF are available at http://narayanlab.stanford.edu.
Figure 1: Data Acquisition and Preprocessing.

Study Flow. a. Left and Right Atria in the Heart and Torso. b. Basket Catheters used to record signals globally within both atria in a 67-year-old woman with persistent AF, showing atrial shell and therapy (ablation) site. c. AF termination on 64 electrodes inside the heart by treating site in b., terminating to normal (sinus) rhythm. d. Spatial maps of activation phase in AF in the entire left atrium, shown as 8×8 grids (interpolated for clarity) colorized from 0 to 2π. Sampled 4×4 tiles may show e. presence of rotational features, such as where therapy terminated AF, or their absence. f. Spatial maps of phase every 5 ms for 1000 ms (200 total per patient).
In each patient, AF terminated acutely by ablation at one rotational activation site (fig. 1b,c). Multi-electrode arrays produced electrograms at 8 electrodes on each of 8 splines, shown in fig. 1c at the site of AF termination.
Data Processing and Featurization
We pre-processed AF signals at 64 sites (8X8 grids) in each atrium. Briefly, we subtracted a mean QRS complex (ventricular activation) from each atrial electrogram. Signals were filtered at 1.5–25 Hz to compute period (cycle length) in AF, which was used to recompose sinusoids, and the Hilbert transform was then used to compute phase maps.
In figure 1d, AF maps represent phase values (0 to 2π) for the entire atrium, created using a published non-proprietary algorithm which identifies areas of interest20, 29 with few false-positives30 (https://narayanlab.stanford.edu). Using this approach, each patient demonstrated 2.0±0.4 sites of rotational activity, defined as complete rotations (> 2 π radians) for a duration of >20% of the mapped segment20 (fig. 1e). Figure 1f indicates time sequences of these tiles. Notably, the variable expert interpretation of phase maps between clinicians may contribute to documented variability in ablation results between centers5–7.
We trained ML on spatial subregions designed to encompass the spatial scale of drivers shown in optical mapping studies21 of human AF. From each original 8X8 grid (X3 channels for RGB colors), we generated 25 overlapping 4×4 tiles (fig 1e) using a sliding window with stride 4 (spatial shift of 1 electrode). We repeated this process every 5 ms for 1000 ms to generate 200 maps for each tile, representing 5–7 cycles or beats of AF (fig. 1e), or 5000 tiles/patient (175,000 tiles for 35 patients). Each tile was assigned a binary label (1/0) based on whether rotational activity was present or not, based on the expert reviewer’s determination blind to termination status. We did not analyze sites of focal activation which may represent intramural breakthroughs from optical mapping of human AF21, and are a minority of potential drivers16, 17. To study reliability, a randomly selected subset of 1050 AF tiles from all patients were labeled by 3 blinded readers, showing 90% agreement yet some variability (Kappa = 0.79).
We analyzed all driver sites to avoid bias, as studies of different methods show AF termination at sites of stable and varying as well as transient repeating rotational activation16, 17, and because ablation at a non-terminating site could potentially facilitate AF termination at a subsequent ablation site29.
Convolutional Neural Networks
We first developed a CNN in Python consisting of 5 convolutional layers and 3 fully connected layers, an input layer, an output layer, as well as multiple activation, max-pooling, and batch normalization layers. We used the TensorFlow deep learning library adapted from a validated design31. Details of the architecture are provided in Supplemental methods, and Figure 2 shows each layer and its characteristics.
Figure 2: Detailed convolutional neural network (CNN) architecture.

showing all the layers in the network and their respective dimensions.
We trained the CNN on interpolated subsamples (13×13×3) of inputs, to provide a higher-resolution input to the CNN for better feature extraction. These inputs were fed through the 5 convolutional and 3 fully connected CNN layers to compute the forward pass output, which was backpropagated to compute gradients and update weights of the CNN.
For development and testing, labeled data were randomly partitioned into independent training (57%, 100,000 tiles, 20 patients), validation (14%, 25,000 tiles, 5 patients), and testing (29%, 50,000 tiles, 10 patients) sets, each from distinct patients. The ratio of rotational to non-rotational tiles in the training and test cohorts were balanced (training 1.16:1; testing 1:1.04). The validation set was used to tune hyperparameters to reduce over-fitting.
Traditional Statistical and Machine Learning Algorithms
We compared the accuracy of the CNN in classifying intracardiac AF data to traditional statistical analyses. We first used techniques to reduce data size, i.e. dimensionality. Increased dimensionality causes exponential increases in parameter space, which makes data sparse and reduces the robustness of results. We thus took steps to avoid this ‘curse of dimensionality’ for all statistical analyses32.
We first applied principal component analysis (PCA) to the AF mapping data and saved the principal components (PCs, also known as factor scores) for each tile to use as the input dataset. We used the first 36 PCs as they accounted for >95% of variance in the dataset, which we used to reduce dimensionality of the tiles32. The reduced dimensionality tiles, standardized using their absolute values, were used as inputs for all of the following methods.
We initially performed unsupervised k-means cluster analysis on the entire dimensionality reduced tiles as input data (175,000 samples), to examine the relationships of clusters (with varying values of k = 2 to 20 clusters) to the expert labels.
We used 3 traditional supervised approaches: linear discriminant analysis (LDA),k-nearest-neighbor analysis (k-NN), and support vector machines (SVM). LDA is a parametric procedure that seeks a weighted combination of variables to optimize prediction, k-NN seeks to optimize subsets based on proximity, and SVM creates a decision boundary to separate classes using a limited number of input examples (the support vectors). LDA was applied to determine the optimal linear combination of dimension-reduced tiles that separates them into 2 classes (non-rotational and rotational) based on the expert labels. The k-NN method was applied to the same input data with k varied to identify the optimal solution. The SVM was trained on the same input data using a linear kernel and a regularization factor (C = 1) to separate the classes.
Probing How CNNs Classify AF Electrical Patterns
Explaining ML classifications is critical for clinical use22, 23 yet rarely achieved. We used Gradient-weighted Class Activation Mapping (Grad-CAM)33, which has recently been applied to probe CNN classifications of images in fields outside medicine, to identify which input data within the heart were most critical to the final classification of trained CNNs.
Figure 3 shows our approach. Input tiles are first presented with known output labels to train the CNN. Explainability can be applied once the CNN is trained. We applied explainability to the final convolutional layer 5, which better represent partitions of the input data (higher-level semantics)33 than earlier convolution layers 1–4. We identified weights (i.e. output features) for layer 5, generated by forward propagation (figure 3a), and computed backpropagated gradients of the output with respect to the final convolutional layer 5, . The backpropagated weight, , of size (m × n × k, equivalent to the convolutional layer size) was averaged along the 1st (m, rows) and 2nd (n, columns) dimensions, producing a k × 1 array of averaged backpropagated weights. An inner product between the array and the output of the convolutional layer along the 3rd (k) dimension was then performed, resulting in an m × n matrix in which higher values represent inputs with greater significance in determining classification. For visualization, the matrix was normalized from 0 to 1, then plotted as a heatmap of the importance of input regions to the CNN classification (fig. 3c).
Figure 3: Explainability Analysis to Probe How the CNN Interprets Intracardiac AF Patterns.
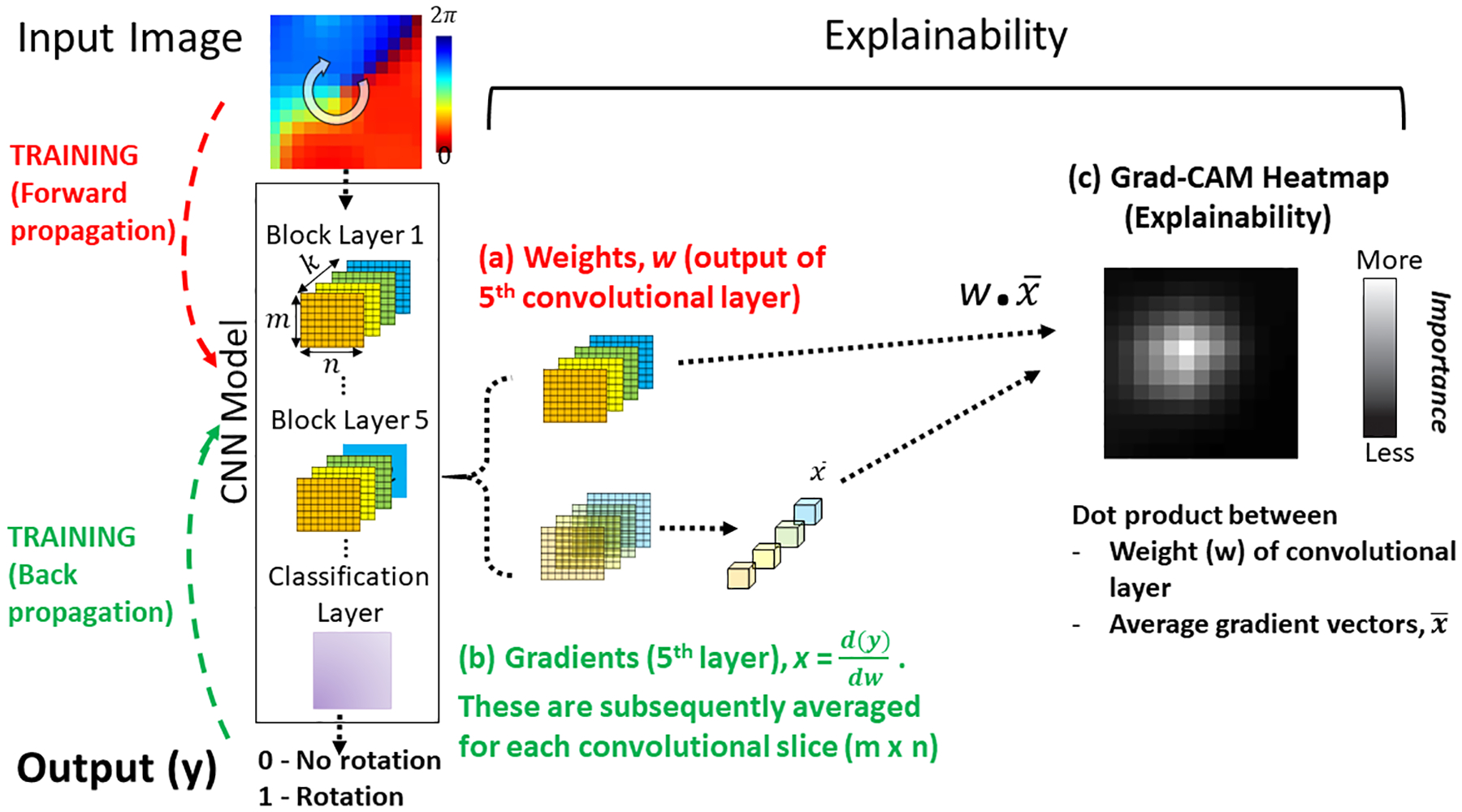
In training, the ML uses forward propagation of an input tile, creating weights w (red), then backward propagation to update internal weights using gradients x (green). This training process matches each input with its known output label (0,1). Explainability can be applied once the CNN is trained. (a) Weights w and (b) Gradients x of the output of the 5th convolutional layer are combined by the dot-product operation. (c) Grad-CAM Heatmap plots the importance of each input pixel to the CNN classification. Brighter (higher value) pixels have a greater influence on the CNN.
Heatmaps were compared to 1050 tiles graded by all expert reviewers, and all heatmap regions were compared to the majority diagnosis between experts.
Statistics
Continuous clinical data are represented as mean ± standard deviation (SD) or as median [quartiles]. Normality was evaluated using the Kolmogorov-Smirnov test. Nominal values were expressed as n (%). Comparisons between the expert rater and each classification scheme were made by quantifying decision statistics (sensitivity, specificity, positive and negative predictive value, and accuracy) with their exact 95% confidence intervals and quantified by kappa. Group comparisons were made using chi-square, ANOVA, or Kruskal-Wallis test as appropriate. Differences in mean accuracy per patient were tested using t-tests for independent samples. Correlations with mean accuracy were computed using Pearson’s or Spearman’s correlation as appropriate. A probability of < 0.05 was considered statistically significant. Agreement of the consensus of expert raters with Grad-CAM heatmap values was evaluated using a receiver operating characteristic analysis setting a cut-point at the maximum Youden index.
Results
Clinical details of the population are presented in Table 1. Of the 175,000 total tiles, 49.5% were expert-classified as rotational and 50.5% were classified as non-rotational. Of these tiles, 55% represented sites where AF termination was achieved by ablation.
Table 1.
Clinical Demographics
| Entire Cohort (n=35) | Training (n=20) | Validation (n=5) | Testing (n=10) | P-Value | |
|---|---|---|---|---|---|
| Age (years) | 65 ± 9.3 | 66 ± 9.3 | 64 ± 10.0 | 65 ± 10.0 | 0.941 |
| Male N (%) | 23 (66%) | 12 (60%) | 3 (60%) | 8 (80%) | 0.530 |
| Persistent AF N (%) | 35 (100%) | 20 (100%) | 5 (100%) | 10 (100%) | 1.000 |
| Prior AF Ablation N (%) | 16 (46%) | 10 (50%) | 3 (60%) | 3 (30%) | 0.460 |
| AF History (days) | 96 [31–133] | 100 [38–130] | 47 [28–734] | 120 [15–830] | 0.544 |
| Hypertension N (%) | 22 (63%) | 13 (65%) | 3 (60%) | 6 (60%) | 0.955 |
| LA diameter (mm) | 46 ± 8.1 | 45 ± 9.6 | 51 ± 5.3 | 47 ± 4.7 | 0.494 |
| LV ejection fraction (%) | 55 ± 10 | 56 ± 10 | 48 ± 8.8 | 57 ± 9.1 | 0.235 |
| CHADSVASc score | 2 ± 1.3 | 2 ± 1.4 | 2 ± 2.1 | 2 ± 1.2 | 0.999 |
| Coronary disease N (%) | 6 (17%) | 4 (20%) | 1 (20%) | 1 (10%) | 0.778 |
Values are mean ± SD, median [quartiles], or N (%)
Supervised CNN to Identify AF Regions of Interest
We first determined the data requirements for CNN to classify intracardiac AF data, which is previously unreported. Classification accuracy of CNN for the training set converged to 100% and loss converged to zero (fig 4a) for training sets of >30,000 image tiles. Notably, accuracy of the CNN in the test cohort varied with the size of the previous training cohort. In figure 4b, CNN had <80% accuracy in the test cohort when models had been trained on 10,000 tiles (10% of training cohort), but rapidly passed 90% accuracy when models had been trained on >20,000 tiles.
Figure 4. Neural Network Accuracy for Intracardiac AF Electrical patterns in training and test cohorts.
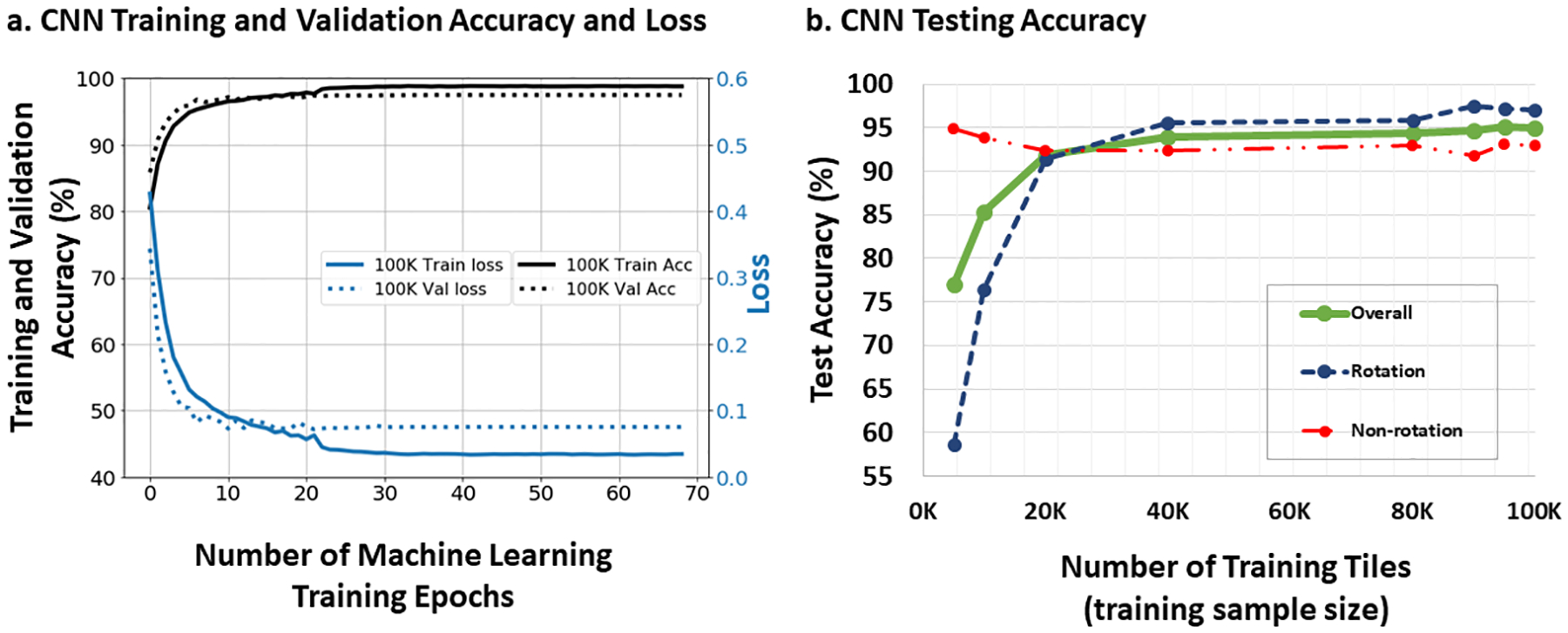
a. CNN training and validation set accuracy and loss as a function of training iterations (epochs). When trained with 100,000 input tiles, CNN accuracy and loss converged to 100% and 0, respectively. In the validation cohort, accuracy and loss converged to 98% and 0, respectively. b. Network Accuracy in the independent Test Cohort Varies with the Size of Prior Training Cohorts. Accuracy for the desired outputs (region of interest Y/N) increased dramatically with the number of tiles used in training, exceeding 90% at >20,000 training tiles and exceeding 95% at >90,000 training tiles.
The CNN model trained on the complete training set was used, as it provided highest accuracy (figure 4b). Accuracy of classification in the test cohort was 95.0% (CI 94.8–95.2%; Table 2). Misclassified tiles (5.0%; 2508/50,000) were present in all test patients (251±173 tiles per patient).
Table 2.
Accuracy of Classification Models for Regions of Interest in AF
| Classification | Kappa† | Sensitivity | Specificity | PPV | NPV | Accuracy |
|---|---|---|---|---|---|---|
| Method | % | % | % | % | % | |
| Supervised Convolutional Neural Network | ||||||
| CNN, | 0.982 | 99.6 | 98.5 | 98.7 | 99.5 | 99.1 |
| Train | (99.5–99.7) | (98.4–98.6) | (98.6–98.8) | (99.5–99.6) | (99.0–99.2) | |
| CNN, | 0.966 | 96.7 | 99.4 | 98.8 | 98.4 | 98.5 |
| Validation | (96.3–97.1) | (99.3–99.5) | (98.5–99.0) | (98.2–98.6) | (98.3–98.7) | |
| CNN, | 0.900 | 97.0 | 93.0 | 93.1 | 97 | 95 |
| Test | (96.8–97.1) | (92.7–93.3) | (92.7–93.4) | (96.8–97.2) | (94.8–95.2) | |
| Unsupervised Cluster Analysis | ||||||
| k-means, k=2 | 0.530 | 69.1 | 92.0 | 94.9 | 58.3 | 76.4 |
| All data | (68.8–69.3) | (91.8–92.3) | (94.7–95.0) | (58.0–58.6) | (76.2–76.6) | |
| k-means, k=12 | 0.589 | 77.0 | 82.3 | 83.5 | 75.5 | 79.4 |
| All data | (76.7–77.2) | (82.0–82.6) | (83.2–83.7) | (75.2–75.8) | (79.3–79.6) | |
| Supervised Classical Machine Learning Methods | ||||||
| LDA, | 0.617 | 85.8 | 76 | 77.9 | 84.4 | 80.9 |
| Train | (85.0–86.1) | (75.6–76.3) | (77.6–78.2) | (84.1–84.7) | (80.6–81.1) | |
| LDA, | 0.595 | 85.0 | 74.6 | 76.4 | 83.7 | 79.7 |
| Test | (84.6–85.5) | (74.1–75.1) | (75.9–76.9) | (83.3–84.2) | (79.4–80.1) | |
| k-NN, k=109 | 0.647 | 77.0 | 88.2 | 87.9 | 77.4 | 82.3 |
| Train | (76.7–77.4) | (87.9–88.4) | (87.6–88.2) | (77.0–77.7) | (82.0–82.5) | |
| k-NN, k=109 | 0.615 | 83.4 | 86 | 94.9 | 62.4 | 84 |
| Validation | (82.8–83.9) | (85.1–86.9) | (94.6–95.2) | (61.3–63.4) | (83.5–84.5) | |
| k-NN, k=109 | 0.576 | 75.3 | 84.0 | 87.1 | 70.3 | 78.9 |
| Test | (74.8–75.7) | (83.5–84.5) | (86.7–87.5) | (69.8–70.9) | (78.5–79.2) | |
| SVM, | 0.601 | 85.8 | 74.3 | 79.4 | 81.5 | 80.3 |
| Train | (85.1–85.8) | (73.9–74.7) | (79.1–79.7) | (81.1–81.9) | (80.0–80.5) | |
| SVM, | 0.638 | 74.7 | 88.7 | 76.8 | 87.5 | 84 |
| Validation | (73.8–75.7) | (88.2–89.1) | (75.9–77.7) | (86.9–87.9) | (83.5–84.4) | |
| SVM, | 0.595 | 82.9 | 76.7 | 77.4 | 82.3 | 79.7 |
| Test | (82.4–83.4) | (76.1–77.2) | (76.9–77.9) | (81.8–82.8) | (79.4–80.1) | |
Values are percent (95% confidence intervals)
p-value for all <0.001
Univariate analysis using t-tests and correlation revealed no significant relationship between CNN accuracy and any clinical and demographic variable in table 1 (all p>0.15). CNN accuracy was 97.3% for tiles in which the raters were in unanimous agreement and 85.1% in more difficulty cases where only a majority of raters agreed in the reliability study. Thus, the model was able to interpret even ambiguous intracardiac AF maps. CNN accuracy in the test set was similar for tiles containing termination sites (95.6%) compared to other sites (94.2%).
Traditional Statistics and Machine Learning to Classify AF Data
Table 2 summarizes unsupervised and supervised classification of AF data for 36 principal components.
In k-means clustering with k=2 and k=12, each cluster was classified as rotational or non-rotational, whichever represented a higher proportion of the two expert label classes. Accuracy (cluster purity) was 76.4% for 2 clusters and highest for 12 clusters (79.4%).
Applying linear discriminant analysis (LDA) to dimensionality-reduced tiles34 separated the two classes with an accuracy of 79.7% in the test set, slightly higher than the best k-means clustering result (k = 12).
k-nearest neighbors (k-NN) was also applied to dimensionality reduced tiles, analogous to k-means and LDA analyses. We tuned the setting of k based on the performance on the validation set; we found that k = 109 yielded the best validation accuracy (84.0%). In the test cohort, this model achieved an accuracy of 78.9% with a sensitivity and specificity of 75.3% and 84.0%, respectively (Table 2).
The SVM model achieved an accuracy of 80.3% and 84.0% in training and validation, respectively. In the test cohort, the SVM model achieved an accuracy of 79.7% with a sensitivity and specificity of 82.9 and 76.7%, respectively (Table 2).
Probing the “Black Box” of trained Machine Learning Models
Grad-CAM33 heatmaps were used to probe trained CNNs to identify which input regions were most critical to classification. Heatmaps in Figures 3 and 5 show the last convolutional layer. Fig. 5 shows rotational activation during AF for 3 patients and lack of rotation in a 4th. In each case, Grad-CAM identified functional units in the trained CNN (layer 5) that mapped to the precise location of rotational sites identified by experts in the input data. This was true whether tiles showed one (figure 5a) or multiple concurrent (figs 5b, 5c) sites.
Figure 5: Grad-CAM Heatmaps of trained CNN empirically detect AF features identified by experts with domain knowledge.

(a) Input vector showing site of interest in AF in a 49-year-old female. The heatmap site in Conv 5 coincides with the precise location in the heart coded by experts as a site of rotation. (b) AF in a 63-year-old female with AF, showing two concurrent regions of interest. (c) AF in a 64-year-old man, showing 3 regions of interest. (d) AF in 74 year-old-female showing no region of interest. In each case, Grad-CAM heatmaps empirically identified tile regions identified by experts with physiological knowledge, although CNN were not explicitly trained in expert rules.
Comparing Explainability Analyses of CNN to Experts
We explored Grad-CAM in a subset of 105 tiles (3 selected randomly per patient) in which 3 expert reviewers marked the precise location(s) of rotational activity. Reviewers identified 58 tiles as showing rotational elements, some showing 2 or more for a total of 77 sites. Figure 5 shows that Grad-CAM heatmaps correlated with input regions used by experts to code a driver (i.e. rotational core).
Gradings by Grad-CAM were compared to expert consensus based on input tile grid location. Grad-CAM matched expert-identified sites with an area under the receiver-operating curve (C-statistic) of 0.961 (95% CI 0.939–0.984). Setting a cut-point using the maximum Youden index yielded a sensitivity of 93.5% (85.4–97.5) and a specificity of 90.8% (89.8–91.7) for the expert identified features, respectively.
Discussion
Convolutional neural networks can classify complex intracardiac AF data to identify organized regions from disorganized activity, including sites where ablation terminates persistent AF. We show that this approach was superior to support vector machines, and to several traditional statistical approaches. We used novel explainability analyses from computer sciences to show that, during training, CNNs developed a logic similar to rules applied by experts. This is notable since these rules were neither codified nor used for ML training. These data provide a potentially scaleable foundation to analyze complex intracardiac AF data, by defining data sizes and structures for ML, by comparing ML architectures, and by showing interpretation of its ‘black box’. These results may be used directly to reduce ambiguity in interpreting current AF mapping approaches and ultimately to improve clinical care.
Convolutional Neural Networks and Explainability in Heart Rhythms
CNNs have reliably identified data structures in fields as complex as voice recognition, image classification, and robot-motion planning22, 23, yet have not previously been used to interpret electrical patterns from inside the heart. Recent studies show that CNNs can detect AF from the ECG24, 25,26, although these studies did not pinpoint regions of interest for therapy, nor of potential pathophysiological interest, and were degraded by noise25 which is common in intracardiac electrograms. This sensitivity to non-physiological factors, and the inherent ambiguity of AF signals, emphasizes the need to explain ML classification prior to clinical use.
Explainability analysis used Grad-CAM, which does not require retraining or post-hoc modifications to the CNN. Earlier layers of the CNN extract lower level features such as lines, edges or specific colors such as red, green, or blue. Later convolutional layers extract higher level features such as circular patterns or color progression. Grad-CAM uses these trained weights to identify the location where features most relevant for classification are present in the input. Other explainability approaches could also be used. One approach is to visualize the layer activations and/or weights for the different convolutional layers in the network35, 36. Sensitivity analyses, such as LIME37, have also been proposed, in which portions of the input are systematically omitted, and their importance inferred by examining the resulting changes in classification. Such methods may be limited as multiple concurrent regions are difficult to probe simultaneously, yet achieved by Grad-CAM in figure 5b and c, and because omitting portions of the input may create artifacts that impact classification. Moreover, altering the representation may fundamentally change confidence in network predictions38.
Future studies may delineate additional cardiovascular applications in which machine learning classifies physiological data in a manner analogous to experts, increasing confidence for clinical use. Conversely, such studies may also identify disease states or specific questions for which trained ML diverges from expert logic. While less immediately applicable clinically, this may provide a starting point for novel mechanistic studies.
Importance of Mapped Features for Atrial Fibrillation
We selected to apply ML to identify organized AF regions in this study because their subjective interpretation may contribute to varied results of mapping and ablation between centers5–7. It would ultimately be useful to apply ML directly to AF electrograms. Currently, however, there is little consensus on interpreting raw AF electrograms. Electrogram dispersion introduced by Seitz et al.39 is promising, but requires additional validation such as for unipolar and bipolar signals or other technical factors. Alternatively, one may examine organized electrograms near drivers, yet such sites also may represent passive activation40. Similarly, disorganized electrograms may lie near the core of drivers41 yet could also represent wavefront collision or noise42.
AF drivers have been shown directly by optical imaging of human AF21, yet meta-analyses of ablation vary from excellent to dismal5–7. Intention-to-treat analysis of the recent REAFFIRM trial showed no benefit of ablating drivers, yet many off-protocol strategies were used which may have caused both intention-to-treat groups to overlap. Indeed, on-treatment analysis of REAFFIRM revealed a trend that PVI+driver ablation was superior to PVI alone (77.7% vs 65.5% success; p=0.09)43.
Nevertheless, subjectivity in interpreting AF maps is a real limitation of existing technologies3, 5, 6. These factors motivated our hypothesis to use ML to analyze AF maps by open source methods. The numerical computation of phase renders such sites ideal for probing how they are learned by CNN. While meta-analyses suggest that phase mapping may be superior to activation mapping of AF7, this also remains controversial.
Our results provide a scaleable foundation to analyze complex AF maps by multiple approaches, and could be applied to several systems. We therefore focused on results from a non-proprietary freely available system in this study. Future studies could also tailor results to clinical data from individual patients, such as demographic, metabolic and structural elements.
Limitations
Further studies should extend to patients in whom therapy was not acutely successful. Such studies may analyze if differences can be detected in intracardiac AF mapping features and/or patient characteristics. AF termination does not always predict long-term success, yet it is one of the few acute markers of ablation success for persistent AF. Future studies should apply ML to predict long term outcome, which is difficult to assess in this observational registry in which subjects were treated by different protocols. For this study, we elected to analyze and train to AF maps, where there is some consensus on what constitutes an organized rotation, or focal activation, or no organization. This provides a practical tool to reconcile AF maps for emerging AF mapping systems. Future studies should examine raw intracardiac electrograms, acknowledging that this introduces separate limitations44. The multi-electrode recording basket has spatial resolution limits, although it may be sufficient to map organized AF drivers27. Future studies should extend to higher resolution catheters. Further research should be done to explore other ML architectures such as deeper CNN networks with residual connections, other optimization methods, such as Adam, RMSProp, or other stochastic optimization techniques.
Supplementary Material
Sources of Funding:
AJR acknowledges research funding from NIH (F32HL144101). JABZ acknowledges funding from a Fulbright-British Heart Foundation fellowship. TB acknowledges funding from a Josephson-Wellens Heart Rhythm Society Fellowship grant and NIH (K23 HL145017). MZ and PB acknowledge affiliate members and other supporters of the Stanford DAWN project---Ant Financial, Cisco, Facebook, Google, Infosys, Intel, Microsoft, NEC, SAP, Teradata, and VMware---as well as Toyota Research Institute, Keysight Technologies, Amazon Web Services, and the NSF under CAREER grant CNS-1651570. SMN and WJR report research grants from NIH (HL103800, HL83359, HL122384, HL149134).
Non-Standard Abbreviations and Acronyms
- AF
Atrial Fibrillation
- CNN
Convolutional Neural Networks
- Grad-CAM
Gradient-weighted Class Activation Mapping
- k-NN
K-Nearest-Neighbor analysis
- LA
Left Atrium
- LDA
Linear Discriminant Analysis
- ML
Machine learning
- PCA
Principal Component Analysis
- PCs
Principal Components
- PV
Pulmonary Vein
- PVI
Pulmonary Vein Isolation
- RA
Right Atrium
- SVM
Support Vector Machines
Footnotes
Clinical Trial Registration:
Competing Interests:
MIA reports intellectual property rights from Stanford University. SMN reports consulting from Beyond.ai Inc, TDK Inc., Up to Date, Abbott Laboratories, and the American College of Cardiology Foundation (all modest); Intellectual Property Rights from University of California Regents and Stanford University. WJR reports Intellectual Property Rights from the University of California Regents.
FA, PC, JABZ, TB, and AJR report no disclosures.
References
- 1.Clarnette JA, Brooks AG, Mahajan R, Elliott AD, Twomey DJ, Pathak RK, Kumar S, Munawar DA, Young GD, Kalman JM, Lau DH and Sanders P. Outcomes of persistent and long-standing persistent atrial fibrillation ablation: a systematic review and meta-analysis. Europace. 2018;20:f366–f376. [DOI] [PubMed] [Google Scholar]
- 2.Lee JM, Shim J, Park J, Yu HT, Kim T-H, Park J-K, Uhm J-S, Kim J-B, Joung B, Lee M-H, Kim Y-H, Pak H-N and Investigators P-A. 2019. JACC: Clinical Electrophysiology. The Electrical Isolation of the Left Atrial Posterior Wall in Catheter Ablation of Persistent Atrial Fibrillation;in press. [DOI] [PubMed] [Google Scholar]
- 3.Providencia R, Lambiase PD, Srinivasan N, Ganesh Babu G, Bronis K, Ahsan S, Khan FZ, Chow AW, Rowland E, Lowe M and Segal OR. Is There Still a Role for Complex Fractionated Atrial Electrogram Ablation in Addition to Pulmonary Vein Isolation in Patients With Paroxysmal and Persistent Atrial Fibrillation? Meta-Analysis of 1415 Patients. Circ Arrhythm Electrophysiol. 2015;8:1017–29. [DOI] [PubMed] [Google Scholar]
- 4.Atienza F, Almendral J, Ormaetxe JM, Moya A, Martinez-Alday JD, Hernandez-Madrid A, Castellanos E, Arribas F, Arias MA, Tercedor L, Peinado R, Arcocha MF, Ortiz M, Martinez-Alzamora N, Arenal A, Fernandez-Aviles F, Jalife J and Investigators R-A. Comparison of Radiofrequency Catheter Ablation of Drivers and Circumferential Pulmonary Vein Isolation in Atrial Fibrillation: A Noninferiority Randomized Multicenter RADAR-AF Trial. J Am Coll Cardiol. 2014;64:2455–67. [DOI] [PubMed] [Google Scholar]
- 5.Ramirez FD, Birnie DH, Nair GM, Szczotka A, Redpath CJ, Sadek MM and Nery PB. Efficacy and safety of driver-guided catheter ablation for atrial fibrillation: A systematic review and meta-analysis. J Cardiovasc Electrophysiol. 2017;28:1371–1378. [DOI] [PubMed] [Google Scholar]
- 6.Baykaner T, Rogers AJ, Meckler GL, Zaman J, Navara R, Rodrigo M, Alhusseini M, Kowalewski CAB, Viswanathan MN, Clopton P, Narayan SM, Heidenreich PA and Wang PJ. Clinical Implications of Ablation of Drivers for Atrial Fibrillation: A Systematic Review and Meta-Analysis. Circ Arrhythm Electrophysiol. 2018;11:e006119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin CY, Lin YJ, Narayan SM, Baykaner T, Lo MT, Chung FP, Chen YY, Chang SL, Lo LW, Hu YF, Liao JN, Tuan TC, Chao TF, Te ALD, Kuo L, Vicera JJB, Chang TY, Salim S, Chien KL and Chen SA. Comparison of phase mapping and electrogram-based driver mapping for catheter ablation in atrial fibrillation. Pacing Clin Electrophysiol. 2019;42:216–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grace A, Willems S, Meyer C, Verma A, Heck P, Zhu M, Shi X, Chou D, Dang L, Scharf C, Scharf G and Beatty G. High-resolution noncontact charge-density mapping of endocardial activation. JCI Insight. 2019;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willems S, Verma A, Betts TR, Murray S, Neuzil P, Ince H, Steven D, Sultan A, Heck PM, Hall MC, Tondo C, Pison L, Wong T, Boersma LV, Meyer C and Grace A. Targeting Nonpulmonary Vein Sources in Persistent Atrial Fibrillation Identified by Noncontact Charge Density Mapping. Circ Arrhythm Electrophysiol. 2019;12:e007233. [DOI] [PubMed] [Google Scholar]
- 10.Swerdlow M, Tamboli M, Alhusseini MI, Moosvi N, Rogers AJ, Leef G, Wang PJ, Rillig A, Brachmann J, Sauer WH, Ruppersberg P, Narayan SM and Baykaner T. Comparing phase and electrographic flow mapping for persistent atrial fibrillation. Pacing Clin Electrophysiol. 2019;42:499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bellmann B, Zettwitz M, Lin T, Ruppersberg P, Guttmann S, Tscholl V, Nagel P, Roser M, Landmesser U and Rillig A. Velocity characteristics of atrial fibrillation sources determined by electrographic flow mapping before and after catheter ablation. Int J Cardiol. 2019. [DOI] [PubMed] [Google Scholar]
- 12.Verma A, Sarkozy A, Skanes A, Duytschaever M, Bulava A, Urman R, Amos YA and Potter T. Characterization and significance of localized sources identified by a novel automated algorithm during mapping of human persistent atrial fibrillation. J Cardiovasc Electrophysiol. 2018;29:1480–1488. [DOI] [PubMed] [Google Scholar]
- 13.Daoud EG, Zeidan Z, Hummel JD, Weiss R, Houmsse M, Augostini R and Kalbfleisch SJ. Identification of Repetitive Activation Patterns Using Novel Computational Analysis of Multielectrode Recordings During Atrial Fibrillation and Flutter in Humans. J Am Coll Cardiol: Clinical Electrophysiology. 2017;3. [DOI] [PubMed] [Google Scholar]
- 14.Honarbakhsh S, Schilling RJ, Dhillon G, Ullah W, Keating E, Providencia R, Chow A, Earley MJ and Hunter RJ. A Novel Mapping System for Panoramic Mapping of the Left Atrium: Application to Detect and Characterize Localized Sources Maintaining Atrial Fibrillation. JACC Clin Electrophysiol. 2018;4:124–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Honarbakhsh S, Hunter RJ, Ullah W, Keating E, Finlay M and Schilling RJ. Ablation in Persistent Atrial Fibrillation Using Stochastic Trajectory Analysis of Ranked Signals (STAR) Mapping Method. JACC Clin Electrophysiol. 2019;5:817–829. [DOI] [PubMed] [Google Scholar]
- 16.Narayan SM, Krummen DE, Shivkumar K, Clopton P, Rappel W-J and Miller J. Treatment of Atrial Fibrillation by the Ablation of Localized Sources: The Conventional Ablation for Atrial Fibrillation With or Without Focal Impulse and Rotor Modulation: CONFIRM Trial. J Am Coll Cardiol. 2012;60:628–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haissaguerre M, Hocini M, Denis A, Shah AJ, Komatsu Y, Yamashita S, Daly M, Amraoui S, Zellerhoff S, Picat MQ, Quotb A, Jesel L, Lim H, Ploux S, Bordachar P, Attuel G, Meillet V, Ritter P, Derval N, Sacher F, Bernus O, Cochet H, Jais P and Dubois R. Driver Domains in Persistent Atrial Fibrillation. Circulation. 2014;130:530–8. [DOI] [PubMed] [Google Scholar]
- 18.Rodrigo M, Guillem MS, Climent AM, Pedron-Torrecilla J, Liberos A, Millet J, Fernandez-Aviles F, Atienza F and Berenfeld O. Body Surface Localization of Left and Right Atrial High Frequency Rotors in Atrial Fibrillation Patients: A Clinical-Computational Study. Heart Rhythm. 2014;11:1584–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Metzner A, Wissner E, Tsyganov A, Kalinin V, Schluter M, Lemes C, Mathew S, Maurer T, Heeger CH, Reissmann B, Ouyang F, Revishvili A and Kuck KH. Noninvasive phase mapping of persistent atrial fibrillation in humans: Comparison with invasive catheter mapping. Ann Noninvasive Electrocardiol. 2018;23:e12527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alhusseini M, Vidmar D, Meckler GL, Kowalewski C, Shenasa F, Wang PJ, Narayan SM and Rappel W-J. Two Independent Mapping Techniques Identify Rotational Activity Patterns at Sites of Local Termination during Persistent Atrial Fibrillation. J Cardiovasc Electrophys. 2017;28:615–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen BJ, Zhao J, Li N, Zolotarev A, Zakharkin S, Wang Y, Atwal J, Kalyanasundaram A, Abudulwahed SH, Helfrich KM, Bratasz A, Powell KA, Whitson B, Mohler PJ, Janssen PML, Simonetti OP, Hummel JD and Fedorov VV. Human Atrial Fibrillation Drivers Resolved With Integrated Functional and Structural Imaging to Benefit Clinical Mapping. JACC Clin Electrophysiol. 2018;4:1501–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krittanawong C, Johnson KW, Rosenson RS, Wang Z, Aydar M, Baber U, Min JK, Tang WHW, Halperin JL and Narayan SM. Deep learning for cardiovascular medicine: a practical primer. Eur Heart J. 2019;40:2058–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med. 2019;25:44–56. [DOI] [PubMed] [Google Scholar]
- 24.Bumgarner JM, Lambert CT, Hussein AA, Cantillon DJ, Baranowski B, Wolski K, Lindsay BD, Wazni OM and Tarakji KG. Smartwatch Algorithm for Automated Detection of Atrial Fibrillation. J Am Coll Cardiol. 2018;71:2381–2388. [DOI] [PubMed] [Google Scholar]
- 25.Tison GH, Sanchez JM, Ballinger B, Singh A, Olgin JE, Pletcher MJ, Vittinghoff E, Lee ES, Fan SM, Gladstone RA, Mikell C, Sohoni N, Hsieh J and Marcus GM. Passive Detection of Atrial Fibrillation Using a Commercially Available Smartwatch. JAMA cardiology. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajpurkar P, Hannun A, Haghpanahi M, Bourn C and Ng A. Cardiologist level arrhythmia detection with convolutional neural networks. 2017;arXiv:1707.01836 [cs.CV] [Google Scholar]
- 27.Honarbakhsh S, Schilling RJ, Providencia R, Dhillon G, Sawhney V, Martin CA, Keating E, Finlay M, Ahsan S, Chow A, Earley MJ and Hunter RJ. Panoramic atrial mapping with basket catheters: A quantitative analysis to optimize practice, patient selection, and catheter choice. J Cardiovasc Electrophysiol. 2017;28:1423–1432. [DOI] [PubMed] [Google Scholar]
- 28.Miller JM, Das MK, Jain R, Garlie J, Brewster J and Dandamudi G. Clinical Benefit of Ablating Localized Sources for Human Atrial Fibrillation: The Indiana University FIRM Registry. J Am Coll Cardiol. 2017;69:1247–56. [DOI] [PubMed] [Google Scholar]
- 29.Kowalewski CAB, Shenasa F, Rodrigo M, Clopton P, Meckler G, Alhusseini MI, Swerdlow MA, Joshi V, Hossainy S, Zaman JAB, Baykaner T, Rogers AJ, Brachmann J, Miller JM, Krummen DE, Sauer WH, Peters NS, Wang PJ and Narayan SM. Interaction of Localized Drivers and Disorganized Activation in Persistent Atrial Fibrillation: Reconciling Putative Mechanisms Using Multiple Mapping Techniques. Circ Arrhythm Electrophysiol. 2018;11:e005846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuklik P, Zeemering S, Maesen B, Maessen J, Crijns HJ, Verheule S, Ganesan AN and Schotten U. Reconstruction of instantaneous phase of unipolar atrial contact electrogram using a concept of sinusoidal recomposition and Hilbert transform. IEEE Trans Biomed Eng. 2015;62:296–302. [DOI] [PubMed] [Google Scholar]
- 31.Krizhevsky A, Sutskever I and Hinton GE. Imagenet classification with deep convolutional neural networks. Advances in neural information processing systems. 2012. [Google Scholar]
- 32.Altman N and Krzywinski M. The curse(s) of dimensionality. Nature methods. 2018;15:399–400. [DOI] [PubMed] [Google Scholar]
- 33.Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D and Batra D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-based Localization. Arxiv. 2017. [Google Scholar]
- 34.Jombart T, Devillard S and Balloux F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC genetics. 2010;11:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yosinski J, Clune J, Nguyen A, Fuchs T and Lipson H. Understanding Neural Networks Through Deep Visualization 31st International Conference on Machine Learning; 2015(Deep Learning Workshop,). [Google Scholar]
- 36.Zeiler MD and Fergus R. Visualizing and Understanding Convolutional Networks. Computer Vision – ECCV 2014. 2014:818–833. [Google Scholar]
- 37.Ribeiro MT, Singh S and Guestrin C. “Why Should I trust you?” Explaining the Predictions of Any Classifier. KDD 2016; San Francisco, CA, USA: 2016. [Google Scholar]
- 38.Alvarez-Melis D and Jaakkola TS. On the Robustness of Interpretability Methods. arXiv preprint arXiv:180608049. 2018. [Google Scholar]
- 39.Seitz J, Bars C, Theodore G, Beurtheret S, Lellouche N, Bremondy M, Ferracci A, Faure J, Penaranda G, Yamazaki M, Avula UM, Curel L, Siame S, Berenfeld O, Pisapia A and Kalifa J. AF Ablation Guided by Spatiotemporal Electrogram Dispersion Without Pulmonary Vein Isolation: A Wholly Patient-Tailored Approach. J Am Coll Cardiol. 2017;69:303–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalifa J, Tanaka K, Zaitsev AV, Warren M, Vaidyanathan R, Auerbach D, Pandit S, Vikstrom KL, Ploutz-Snyder R, Talkachou A, Atienza F, Guiraudon G, Jalife J and Berenfeld O. Mechanisms of Wave Fractionation at Boundaries of High-Frequency Excitation in the Posterior Left Atrium of the Isolated Sheep Heart During Atrial Fibrillation. Circulation. 2006;113:626–633. [DOI] [PubMed] [Google Scholar]
- 41.Zlochiver S, Yamazaki M, Kalifa J and Berenfeld O. Rotor meandering contributes to irregularity in electrograms during atrial fibrillation. Heart Rhythm. 2008;5:846–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Narayan SM, Wright M, Derval N, Jadidi A, Forclaz A, Nault I, Miyazaki S, Sacher F, Bordachar P, Clementy J, Jais P, Haissaguerre M and Hocini M. Classifying Fractionated Electrograms in Human Atrial Fibrillation Using Monophasic Action Potentials and Activation Mapping: Evidence for Localized Drivers, Rate Acceleration and Non-Local Signal Etiologies. Heart Rhythm. 2011a;8:244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brachmann J, Hummel J, Wilber D, Sarver A, Rapkin J, Shpun S and Szili-Törok T. Prospective randomized trial of conventional with and without driver ablation for persistent atrial fibrillation: the REAFFIRM study (Late Breaking Clinical Trial abstract). Heart Rhythm. 2019;in press. [Google Scholar]
- 44.Zaman JAB, Sauer WH, Al-Husseini MI, Baykaner T, Borne RT, Kowalewski CA, Busch S, Zei PC, Park S, Viswanathan MN, Wang PJ, Brachman J, Krummen DE, Miller JM, Rappel W-J, Narayan SM and Peters NS. Identification and Characterization of Sites Where Persistent Atrial Fibrillation is Terminated by Localized Ablation. Circulation Arrhyth/Electrophys. 2018;11:e005258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


