Abstract
The importance of coronaviruses as human pathogen has been highlighted by the recent outbreak of SARS-CoV-2 leading to the search of suitable drugs to overcome respiratory infections caused by the virus. Due to the lack of specific drugs against coronavirus, the existing antiviral and antimalarial drugs are currently being administered to the patients infected with SARS-CoV-2. The scientists are also considering repurposing of some of the existing drugs as a suitable option in search of effective drugs against coronavirus till the establishment of a potent drug and/or vaccine. Computer-aided drug discovery provides a promising attempt to enable scientists to develop new and target specific drugs to combat any disease. The discovery of novel targets for COVID-19 using computer-aided drug discovery tools requires knowledge of the structure of coronavirus and various target proteins present in the virus. Targeting viral proteins will make the drug specific against the virus, thereby, increasing the chances of viral mortality. Hence, this review provides the structure of SARS-CoV-2 virus along with the important viral components involved in causing infection. It also focuses on the role of various target proteins in disease, the mechanism by which currently administered drugs act against the virus and the repurposing of few drugs. The gap arising from the absence of specific drugs is addressed by proposing potential antiviral drug targets which might provide insights into structure-based drug development against SARS-CoV-2.
Keywords: SARS-CoV-2, Coronavirus, Target proteins, Drug repurposing, Computational drug discovery, COVID-19, Clinical trials
Graphical abstract
1. Introduction
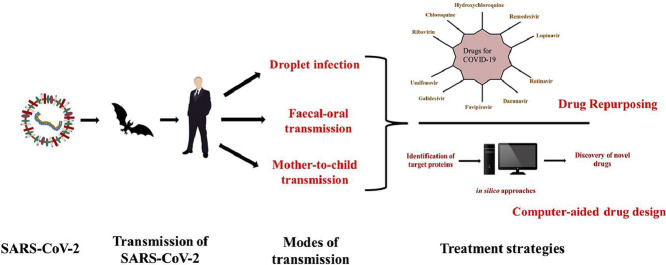
The pandemic of COVID-19 has led the world for an emergency search of potential drugs to combat such deadly viral diseases. As per the reports, drugs developed through repurposing of existing drugs are focused in an order to defeat pathogenicity of virus. Various target proteins of virus have been identified with specific mechanism of action. This review article will be focusing on the roles of target protein in disease and how the available antiviral drugs act upon such targets to combat the virus. Apart from that, new insights into the various strategies used for drug and vaccine development would be discussed.
Importance of computer-aided drug discovery has emerged as a potential tool to be used worldwide to work in the direction of structural biology utilizing structure of target proteins. In an attempt of drug repurposing, computational methods are participating in understanding the mechanism of virus at molecular level and giving a focused direction to the research with cost-effectiveness and economic viability to benefit the society. Hence, in this review, the approach of using computational tools to discover suitable drugs for COVID-19 is also discussed.
2. Origin of disease
The sudden emergence of viral pneumonia cases in Wuhan city of China in late December 2019 created havoc throughout the world. It is believed to have originated from Huanan Seafood Wholesale Market in Wuhan, China in December 2019 and since then it has spread worldwide. The causal microorganism, later identified as a novel coronavirus, was named as 2019-nCoV or SARS-CoV-2 by World Health Organization (WHO) and the novel coronavirus disease was termed as COVID-19 (Perrella et al., 2020; Zhou et al., 2020). SARS-CoV-2 is a zoonotic virus and it shares similarity with SARS-CoV which was responsible for severe acute respiratory syndrome (SARS) pandemic in early 2000s. The COVID-19 has infected millions of people across the globe causing high fatality especially among kids and elderly people. As per the reports of WHO, a worldwide total of 19,936,210 COVID-19 cases were reported till August 11, 2020 with 732,499 deaths. America reported the highest number of total positive cases (10,697,832) highest deaths (390,850) till that time followed by Europe with 3,606,373 total cases and 217,278 deaths. The mortality rate of this disease worldwide is approximately 3.7% till date which shows its high transmission rate (World Health Organization, 2020).
The origin of virus is animals from where it jumped to humans. It is thought the virus was transmitted to humans by bats but the host of the virus is still unclear. Other than bats, pangolins are also believed as hosts transmitting coronavirus to humans as the whole genome of pangolin-CoV shares 91.02% identity to SARS-CoV-2 genome (Zhang et al., 2020a,b). It is also suspected that the bats infected other animal, termed as intermediate hosts which transmitted virus to humans. This can be assumed based on similar cases of coronaviruses, SARS and MERS, wherein the viruses were transmitted from bats to an intermediate hosts before infecting humans (Wu et al., 2020a,b; Zhang et al., 2020a,b; Zheng, 2020).
3. Transmission of SARS-CoV-2
The first cases of COVID-19 disease indicated animal-to-human transmission of virus as the main mechanism of transmission of this disease. When the number of cases rose without getting exposed to any seafood market or direct contact with animal this theory was eliminated and it was concluded that the transmission of virus is human-to-human. It cannot be neglected that the virus is transmitted before any symptoms develop in the infected person as it has been reported that the asymptomatic individuals are also capable of transmitting the virus (Cascella et al., 2020). Hence, isolation or quarantine of the patient is the only way to reduce transmission of the disease.
The SARS-CoV-2 is transmitted through respiratory secretions in the form of droplets and by close contact with the infected person. It is also reported that the faeces of the infected patient also contains coronavirus. Hence, the virus can also be replicated in digestive tract of patients and exist there. This suggests the possibility of faecal-oral transmission of the virus (Holshue et al., 2020; Wu et al., 2020a,b). In some cases, the transmission of virus from mother to child post childbirth has also been reported. The new-born of the infected mother was found to be positive for viral nucleic acid but this study lacks scientific evidence (Zhu et al., 2020). The transmission and infection of CoV by consuming virus-infected food has not been reported till date (Wu et al., 2020a,b).
4. Structure of SARS-CoV-2
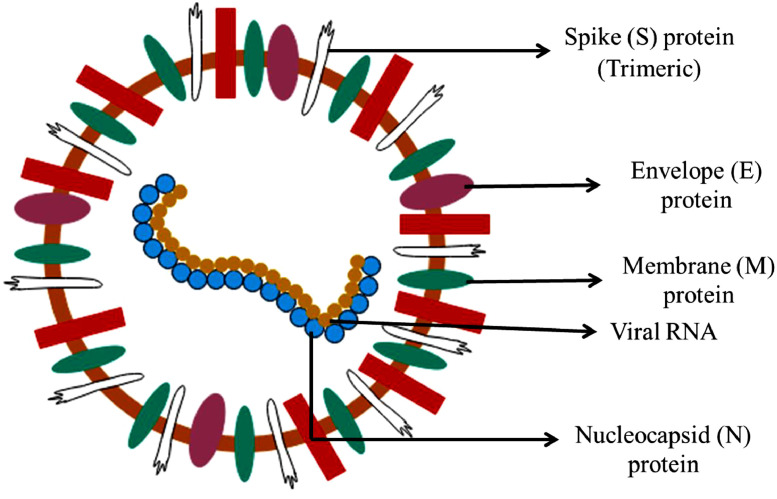
The structure of SARS-CoV-2 is similar to other coronaviruses. The coronaviruses consist of positive single-stranded RNA strands with nucleocapsid protein and are enveloped. The viral genome is approximately of 30 kb with a 3’-poly-A tail and a 5’-cap (Kim et al., 2020a,b). The SARS-CoV-2 consists of four structural proteins, namely, spike (S), envelope (E), membrane (M) and nucleocapsid (N) proteins (Figure 1 ). Spike protein is a transmembrane glycoprotein which protrudes from the surface of virus in a homotrimeric form. This acts as a key therapeutic target. The S-protein is divided into two subunits: S1 and S2. The S1 subunit consists of receptor binding domain (RBD) which binds to ACE2 receptor present in host cell (Wang et al., 2020). The S1 subunit shares 40% of amino acid identity with other SARS-CoVs. The S2 subunit consists of a fusion peptide which helps in the fusion of virus with cellular membrane, a transmembrane domain and a cytoplasmic domain. This subunit is highly conserved and hence it could be a potential target for antiviral drugs. The boundary of two subunits consists of a cleavage site which is cleaved by host proteases upstream of the fusion peptide (Andersen et al., 2020a,b). This cleavage helps in activation of proteins required to fuse with membrane by permanent conformational changes. The S-protein contains N-linked glycans which makes it highly glycosylated. The glycans are required for proper folding of S-protein. The fusion and entry of coronavirus into target cells is facilitated by receptor-binding and processing of S-protein by protease. The interaction of S1/S2 with ACE2 (Angiotensin Converting Enzyme 2) of host cells helps in the entry of virus into host cells (Cascella et al., 2020; Shang et al., 2020a,b).
Fig. 1.
The structure of SARS-CoV-2, causal microorganism of COVID-19
Among the other proteins, the envelope protein has a role in viral pathogenicity and helps in assembly and release of virus particles. The non-structural proteins (NSPs) help in blocking the innate immune response of the host cells (Lei et al., 2018; Cascella et al., 2020).
5. Mechanism of replication and infection of SARS-CoV-2
As the structure of SARS-CoV-2 is similar to SARS-CoV, its mechanism of replication and infection in a host cell is also postulated to be same. After entering into human body, the coronavirus binds to enterocytes and pneumocytes where it replicates and forms new virus particles. The virus also targets tubular epithelial cells of kidney, cerebral neuronal cells and immune cells. The spike protein of the virus helps in its attachment with the host-cell protein of target cells. Similar to SARS-CoV, SARS-CoV-2 interacts with angiotensin converting enzyme-2 (ACE-2) in the host cell (Li et al., 2003; Shang et al., 2020a,b). Once into the cell, the virus releases its genome along with nucleocapsid inside the host cells. The two polyproteins, pp1a and pp1b, are produced by ORF1a and ORF1b genes which enable the translation process of the virus using the host cells’ ribosomes (Stobart et al., 2013). Both the polyproteins help in the formation of replication-transcription complex (Te Velthuis et al., 2012). The polyproteins are processed by virally encoded protease, namely, chymotrypsin-like protease (3CLpro) or main protease (Mpro) and two-papain-like protease, and produces 16 non-structural proteins or NSPs. All these NSPs have specific function in replication and transcription of viral genome. For example, NSP1 and NSP2 suppress the expression of host gene whereas NSP3 forms a multidomain complex. NSP4 and NSP6 are transmembrane protein and NSP5 is a protease which has a role in replication (Stobart et al., 2013). NS7 and NSP8 act as primase enzyme (Te Velthuis et al., 2012). NSP9 is a RNA binding protein which helps in the dimerization of RNA. The dimeric form is important for viral infection. Disruption of dimerization inhibits the infection by the virus (Egloff et al., 2004; Hu et al., 2017). NSP10 is a cofactor for activation of replicative enzyme (Bouvet et al., 2014). NSP12 to NSP16 are enzymes involved in various processes of viral replication and transmission. NSP12 is RNA dependent RNA polymerase (RdRp) which plays an important role in viral replication and transcription and also is a primary drug for some antiviral drugs (Gao et al., 2020). NSP13 has helicase activity, NSP14 has exoribonuclease activity and NSP15 has endonuclease activity. NSP16 has methyltransferase activity (Alsaadi and Jones, 2019).
After synthesis of proteins, M (Membrane), E (Envelope) and S (Spike) proteins enter into the endoplasmic reticulum- Golgi intermediate complex (ERGIC) where they make viral envelope or outer cover. The replicated genome binds N (Nucleocapsid) protein and forms ribonucleoprotein (RNP) complex (Narayanan et al., 2000). The viral particles are released from ERGIC by budding. The mature virions form vesicles which fuse with plasma membrane of host cells and release virus particles into the extracellular space (Nieto-Torres et al., 2011; de Wit et al., 2016). Upon infection, SARS-CoV produces excessive immune response in the host cells, also called as “cytokine storm”. This releases interleukin 6 which is produced by activated leukocytes and targets various cells and tissues. Interleukin 6 is a pro-inflammatory cytokine and can cause cytokine release syndrome (CRS). It is an acute systemic inflammatory syndrome which results in fever and multiple organ failure. These also damage lung tissue and, deteriorate function of lung which might result in lung failure in some cases (Ding et al., 2004; Du et al., 2009; Cascella et al., 2020; Prajapat et al., 2020).
6. Currently administered drugs and their mechanism of action against SARS-CoV-2
Due to the lack of specific drugs against SARS-CoV-2, commonly called as COVID-19, the currently available antiviral drugs are being administered to coronavirus-infected patients. These drugs target different pathways required for viral growth and infection by inhibiting certain proteins or important process of the pathways. Table 1 summarizes the site of inhibition of currently tested drugs for COVID-19.
Table 1.
Information of inhibitory properties of antiviral drugs.
 |
 |
Two of the common drugs administered worldwide belong to the family of antimalarial drugs, chloroquine and hydroxychloroquine. An aminoquinolone derivative, chloroquine (C18H26ClN3), was initially developed for the treatment of malaria and extraintestinal amebiasis. Later, it was repurposed to treat HIV, rheumatoid arthritis and systemic lupus erythematosus (Plantone and Koudriavtseva, 2018). It was also used to treat Zika virus (Li et al., 2017). Currently, this drug is undergoing clinical trials against COVID-19 (Colson et al., 2020). In the host cell, the drug diffuses through the cell membrane by passive diffusion and enters endosomes, lysosomes and Golgi vesicles. The chloroquine gets trapped into these organelles, gets converted into protonated form and raises the pH of the medium. As release of virus requires low pH and the pH of endosomes is raised, the virus particles cannot fuse and enter into cells. Chloroquine inhibits ACE2 glycosylation at its terminals. As ACE2 is the receptor by which SARS-CoV and SARS-CoV-2 enters cells, the unglycosylated form of ACE2 is unable to interact with SARS-CoV-2 spike protein and hence the entry of virus into the cells is prevented. (Ducharme and Farinotti, 1996; Vincent et al., 2005; Zhu et al., 2020) Chloroquine inhibits the actions of tumor necrosis factor (TSF), toll-like receptor 9 and glutathione S-transferase whereas it modulates the activity of Angiotensin-converting enzyme (ACE) 2.
Hydroxychloroquine (C18H26ClN3O) is also an aminoquinoline and consists of R and S enantiomers. Similar to the action of chloroquine, hydroxychloroquine also inhibits the formation and entry of virus particles by elevating the pH of endosomes. This drug is also been used for its efficacy against SARS-CoV-2 (Vincent et al., 2005; Zhu et al., 2020). Hydroxychloroquine cross links with human DNA, is antagonist of toll-like receptor 7 and toll-like receptor 9 and modulates the activity of angiotensin-converting enzyme 2 in humans.
Remdesivir, with a chemical formula of C27H35N6O8P, is also an antiviral drug which is currently being used as a treatment against SARS-CoV-2 (de Wit et al., 2020). Remdesivir is an analog of adenosine triphosphate which has a potential to treat RNA viruses by inhibiting the action of RNA polymerase. The nucleoside analog gets incorporated into RNA during elongation. This prevents further addition of nucleotides leading to termination of RNA transcription (Agostini et al., 2018). The antiviral activity of remdesivir has been demonstrated against Ebola (Warren et al., 2016) and viruses belonging to coronavirus family, SARS-CoV and MERS-CoV (Sheahan et al., 2017). The drug inhibits the action of replicase polyprotein 1 ab in SARS-CoV and RNA-directed RNA polymerase L in Zaire Ebolavirus (strain Mayinga-76).
Lopinavir (C37H48N4O5), originally tested for treatment of HIV-1 patients, is currently being investigated as a treatment regime against SARS-CoV-2 causing COVID-19 in combination with ritonavir (Harrison, 2020a,b). Lopinavir is an antiretroviral protease inhibitor and is used for the treatment of HIV-1 infection in adults. It is generally used in combination with other antiretroviral drugs due to the poor bioavailability and biotransformation property of the compound. Ritonavir inhibits the enzymes responsible for metabolism of lopinavir thereby increasing the exposure of lopinavir and enhancing its antiviral activity. The different stages of lifecycle of HIV (assembly, budding and maturation) are controlled by Gag polyprotein and its proteolytic products which is also the major structural protein of the virus. The HIV-1 protease enzyme which is a dimeric aspartic protease cleaves the Gag polyprotein and hence is important in viral lifecycle (Sundquist and Kräusslich, 2012). Lopinavir inhibits the action of HIV-1 protease enzyme as the hydroxyethylene scaffold present in the compound mimics the peptide linkage cleaved by HIV protease. As the hydroxyethylene scaffold cannot be cleaved by HIV-1 protease, the proteolysis of Gag polyprotein does not take place resulting in formation of immature and non-infectious viral particles (De Clercq, 2009). The drug, lopinavir, inhibits human immunodeficiency virus type 1 protease in HIV-1.
Another antiviral drug used for the treatment of HIV-1 infections is darunavir. Darunavir (C27H37N3O7S) is also a protease inhibitor which is used as a second generation drug in combination with other antiretroviral drugs to combat resistance to standard chemotherapy (De Meyer et al., 2005; Tremblay, 2008). The compound is currently under clinical trials for the treatment of COVID-19 (Harrison, 2020a,b). The mechanism of darunavir is similar to lopinavir in inhibiting the proteolysis of Gag polyprotein. Darunavir makes close contact with the active amino acids (Asp-29 and Asp-30) present in primary chain in protease. Hence this helps in treating resistant strains of HIV-1 (De Meyer et al., 2005). The drug binds at the active site of protease enzyme and on the surface of flaps in protease dimer. The molecules of the drug are flexible and hence it can also adapt to the changes in shape of the enzyme. This compound also inhibits human immunodeficiency virus type 1 protease in HIV-1.
Ritonavir (C37H48N6O5S2) is also HIV protease inhibitor but is generally used to increase the efficacy of other protease inhibitors. It is used, in combination to other therapies, to treat infections against hepatitis C virus (HCV) as it is a potent inhibitor of P450 CYP3A4 present in intestinal tract and liver (Hull and Montaner, 2011). Ritonavir limits the efflux and cellular transport of other protease inhibitors via MRP efflux channels and P-glycoprotein. The HIV-1 protease activity in HIV-1 is inhibited by this compound.
Favipiravir or favilavir (C5H4FN3O2) is an analog of pyrazine which was used for the treatment of influenza by targeting RNA-dependent RNA polymerase (Furuta et al., 2017). These enzymes are important for replication and transcription of viral genomes and hence favipiravir inhibits replication of influenza A and B (Shu and Gong, 2016; de Farias et al., 2017; Furuta et al., 2017). The compound has been investigated for treatment of Ebola virus and Lassa virus and is under investigation for COVID-19 (Madelain et al., 2016; Furuta et al., 2017; Rosenke et al., 2018). Favipiravir prevents the entry and exit of virus from cells. The active form of favipiravir, favipiravir-RTP (ribofuranosyl-5′-triphosphate), has shown to inhibit RNA polymerase thereby preventing viral genome replication. Favipiravir being a pyrazine analog gets incorporated into RNA strand and prevents its elongation and viral proliferation. Similarly, favirapir-RTP also contains purine analogs which inhibit RNA elongation by competing with purine nucleosides for polymerase binding (Furuta et al., 2017). As the catalytic domain of RNA-dependent RNA polymerase of RNA viruses is similar, this catalytic domain can be used by favipiravir to target broad-spectrum viruses. Favipiravir targets catalytic subunit of RNA-directed RNA polymerase of influenza A virus.
Galidesivir with a chemical structure of C11H15N5O3 is an adenosine analog used against Zaire Ebolavirus. The animal studies have reported its efficacy against a broad range of viruses, namely, Marburg, Yellow Fever, Ebola and Zika viruses (Tchesnokov et al., 2019). The in vitro studies have shown its activity against negative- and positive-sense RNA viruses including arenaviruses, filoviruses and coronaviruses. Due to its broad-spectrum activity, it is under clinical trials for viral infections as well as for COVID-19. Galidesivir binds viral RNA polymerase where the nucleotides bind for elongation. Due to alteration in electrostatic interaction from binding of galidesivir, the structural orientation of the viral enzyme changes, inhibiting the activity of RNA polymerase and terminating elongation of RNA strand (Westover et al., 2018; Eyer et al., 2019). Galidesivir inhibits the action of RNA-directed RNA polymerase L in Zaire Ebolavirus (strain Mayinga-76).
Umifenovir (C22H25BrN2O3S) is currently used for the treatment of influenza and respiratory viral infections (Blaising et al., 2014). It has been used for the treatment of infections caused by many enveloped and non-enveloped DNA and RNA viruses, such as, Zika virus, Lassa virus, Flavivirus, Ebola virus, Herpes simplex virus and foot-and-mouth disease (Fink et al., 2018; Haviernik et al., 2018; Li et al., 2018; Herod et al., 2019; Hulseberg et al. 2019) whereas in vitro studies have reported its efficacy against Hantaan virus, chikungunya virus, hepatitis B and C viruses, reovirus and coxsackie virus B5 (Blaising et al. 2014; Pecheur et al. 2016). It is being investigated for the treatment of infections caused by SARS-CoV-2 along with other HIV therapies (Lu, 2020; Wang et al., 2020). Umifenovir is an indole-based hydrophobic drug which directly acts as antiviral/host-targeting agents. The drug has direct virucidal activity and host-targeting activity by affecting the stages of viral life cycle. The dual activity of umifenovir might be the reason for its broad-spectrum antiviral activity (Blaising et al., 2014). Due to the hydrophobicity of umifenovir, it forms aromatic stacking interactions with tyrosine and tryptophan acting against viruses. The interaction of drug with plasma membrane might interfere with intracellular trafficking and clathrin-mediated exocytosis (Blaising et al., 2013) or with the lipid envelope of viruses. This may prevent entry of virus in cells (Teissier et al., 2011; Blaising et al., 2014). Its interaction with the viral lipids and proteins also interferes with the viral life cycle.
Ribavirin (C8H12N4O5) is a broad-spectrum antiviral activity against DNA and RNA viruses. It is a synthetic prodrug which is metabolized into guanosine nucleoside and interferes with the synthesis of viral mRNA. It is used for treating viral hemorrhagic fevers including Venezuelan hemorrhagic fever, Crimean-Congo hemorrhagic fever, Lassa fever, and Hantavirus infection and hepatitis C. The ribavirin prodrug is activated to ribavirin mon-, di-, and triphosphate metabolites by adenosine kinase. Ribavirin triphosphate binds to the active site of viral mRNA polymerase and inhibits its activity. Due to this, the replication of virus is reduced or defective virions are produced. Ribavirin also inhibits inosine monophosphate dehydrogenase in host cells resulting in decreased pool of GTP. This leads to reduced viral protein synthesis and limited replication of viral genomes (Te et al., 2007). Another mechanism of antiviral activity of ribavirin is that it causes mutation in virus that result in early termination of RNA in hepatitis C virus. Ribavirin also stimulates humoral immune response in host cells and down-regulates the genes involved in apoptosis and interferon inhibition (Martin and Jensen, 2008). Ribavirin inhibits catalytic subunit of RNA-directed RNA polymerase in influenza virus and targets RNA-directed RNA polymerase L in HPIV-2 and genome polyprotein in DENV-2.
7. Potential target proteins
Information about the structure and metabolic pathways of coronavirus and pathophysiology of diseases associated with SARS-CoV-2 help in the identification of potential target proteins to explore drugs (J Alsaadi et al., 2019). As mentioned earlier, the essential structural proteins of coronavirus are spike protein (S) which is a trimeric protein, membrane protein (M), envelop protein (E) and nucleocapsid protein (N). Glycoprotein, hemagglutinin esterase (HE) is also found in some viruses such as beta-CoVs (Hilgenfeld, 2014). Coronavirus RNA genome is conserved with seven genes in order of 5’ to 3’ direction: ORF1a, ORF1b, S, OEF3, E, M, N. ORF1a and ORF1b cover the two-third part of viral RNA genome and produces polyproteins PP1a and PP1ab which are two replicase proteins of virus. Processing of these polypeptides (PPs) further generate sixteen non-structural mature proteins (NSPs). These NSPs are responsible for participating in various viral functions which include replicase-transcriptase complex formation. The rest of the viral genome encodes mRNA which takes part in production of structural proteins such as spike protein, envelop protein, membrane protein, nucleocapsid and accessory proteins (McBride et al., 2014). HE protein is the envelope-associated protein and is another important protein that is expressed by some of the CoV strains only (Li, 2016). CoV RNA genome is packed inside the nucleocapsid and is further covered by envelope protein (Guo et al., 2008).
Originally some coronaviruses are found to cause enzootic diseases which are naturally limited to their animal hosts but it has crossed the species barrier of animal-human and proceeded towards the zoonotic infections in human (Rest and Mindell, 2003; Lau et al., 2005; Hon et al., 2008; Chan et al., 2013; Lu et al., 2015). The jumps of cross-species barriers accordingly allowed coronaviruses such as SARS-CoV and MERS-CoV to form virulent human viruses.
Evidently, the resoluteness of such infections and lack of approved and efficient prognosis highlight the urgency for detailed and thorough investigation of molecular biology of coronavirus majorly focusing on structural and accessory proteins (Masters, 2006; Kilianski et al., 2013; Kilianski and Baker, 2014; Liu et al., 2014; Lou et al., 2014). Fusion inhibitors, live and attenuated vaccines have proven as promising approaches in treatment but these also require in-depth information of molecular biology of CoV (Masters, 2006; DeDiego et al., 2007; Netland et al., 2010; Enjuanes et al., 2011; Graham et al., 2012; Heald-Sargent and Gallagher, 2012; Regla-Nava et al., 2015).
The encoding of majorly four structural proteins through coronaviral genome, namely, spike protein (S), nucleocapsid, membrane protein (M) and envelop protein (E), are required for producing a complete structure of viral particle (Mortola and Roy, 2004; Masters, 2006; Wang et al., 2017). The probable target proteins of SARS-CoV-2 (Bhatia et al., 2020) as reported in protein data bank has been enlisted with PDB IDs in Table 2 . Due to the similarity of SARS-CoV-2 genome with other coronaviruses, the knowledge of target proteins in them can also be used to develop drugs against SARS-CoV-2. Therefore, the target proteins of other coronaviruses, along with their PDB IDs, are mentioned in Table 3 .
Table 2.
Target Proteins with PDB ID for COVID-19.
| S. No. | PDB ID | Target Protein Name |
|---|---|---|
| 1. | 6W4B | The crystal structure of Nsp9 replicase protein of COVID-19 |
| 2. | 6LU7 | The crystal structure of COVID-19 main protease in complex with an inhibitor N3 |
| 3. | 6M03 | The crystal structure of COVID-19 main protease in apo form |
| 4. | 6Y84 | COVID-19 main protease with unliganded active site (2019-nCoV, coronavirus disease 2019, SARS-CoV-2) |
| 5. | 5R7Y | PanDDA analysis group deposition – Crystal Structure of COVID-19 main protease in complex with Z45617795 |
| 6. | 5R7Z | PanDDA analysis group deposition – Crystal Structure of COVID-19 main protease in complex with Z1220452176 |
| 7. | 5R80 | PanDDA analysis group deposition – Crystal Structure of COVID-19 main protease in complex with Z18197050 |
| 8. | 5R81 | PanDDA analysis group deposition – Crystal Structure of COVID-19 main protease in complex with Z1367324110 |
| 9. | 5R82 | PanDDA analysis group deposition – Crystal Structure of COVID-19 main protease in complex with Z219104216 |
| 10. | 5R83 | PanDDA analysis group deposition – Crystal Structure of COVID-19 main protease in complex with Z44592329 |
| 11. | 5R84 | PanDDA analysis group deposition – Crystal Structure of COVID-19 main protease in complex with Z31792168 |
| 12. | 6LXT | Structure of post fusion core of 2019-nCoV S2 subunit |
| 13. | 7BQY | The crystal structure of covid-19 main protease in complex with an inhibitor n3 at 1.7 angstrom |
| 14 | 6LU7 | The crystal structure of COVID-19 main protease in complex with an inhibitor N3 |
| 15 | 5R84 | PanDDA analysis group deposition – Crystal Structure of COVID-19 main protease in complex with Z31792168 |
| 16 | 5R7Y | PanDDA analysis group deposition – Crystal Structure of COVID-19 main protease in complex with Z45617795 |
| 17 | 5R82 | PanDDA analysis group deposition – Crystal Structure of COVID-19 main protease in complex with Z219104216 |
| 18 | 5R81 | PanDDA analysis group deposition – Crystal Structure of COVID-19 main protease in complex with Z1367324110 |
| 19 | 6W63 | Structure of COVID-19 main protease bound to potent broad-spectrum non-covalent inhibitor X77 |
| 20 | 6M03 | The crystal structure of COVID-19 main protease in apo form |
| 21 | 6W4B | The crystal structure of Nsp9 RNA binding protein of SARS CoV-2 |
| 22 | 7BUY | The crystal structure of COVID-19 main protease in complex with carmofur |
| 23 | 7BV2 | The nsp12-nsp7-nsp8 complex bound to the template-primer RNA and triphosphate form of Remdesivir (RTP) |
| 24 | 7BV1 | Cryo-EM structure of the apo nsp12-nsp7-nsp8 complex |
| 25 | 6YB7 | SARS-CoV-2 main protease with unliganded active site (2019-nCoV, coronavirus disease 2019, COVID-19) |
| 26 | 7BTF | SARS-CoV-2 RNA-dependent RNA polymerase in complex with cofactors in reduced condition |
| 27 | 6M71 | SARS-Cov-2 RNA-dependent RNA polymerase in complex with cofactors |
| 28 | 6LVN | Structure of the 2019-nCoV HR2 Domain |
| 29 | 6W9C | The crystal structure of papain-like protease of SARS CoV-2 |
| 30 | 5R8T | PanDDA analysis group deposition of ground-state model of SARS-CoV-2 main protease screened against DSI poised (Enamine), Fraglites and Peplites (Newcastle university), Mini Frags (Astex), York 3D (York university), electrophile cysteine covalent (Weizman institute) fragment libraries |
| 31 | 6WEN | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS-CoV-2 in the apo form |
| 32 | 6Y13 | The N-terminal RNA-binding domain of the SARS-CoV-2 nucleocapsid phosphoprotein |
Table 3.
Proteins with PDB ID targeting other Human coronavirus for COVID-19.
| S. No. | PDB ID | Target Protein Name |
|---|---|---|
| 1 | 4KXJ | Crystal structure of HCoV-OC43 N-NTD complexed with PJ34 |
| 2 | 3V3P | Crystal structure of N terminal domain of human coronavirus OC43 nucleocapsid protein |
| 3 | 4LM7 | Crystal structure of HCoV-OC43 N-NTD complexed with UMP |
| 4 | 4LI4 | Crystal structure of HCoV-OC43 N-NTD complexed with AMP |
| 5 | 4TWY | Structure of SARS-3CL protease complex with a phenylbenzoyl (S,R)-N-decalin type inhibitor |
| 6 | 4TWW | Structure of SARS-3CL protease complex with a Bromobenzoyl (S,R)-N-decalin type inhibitor |
| 7 | 4WY3 | Structure of SARS-3CL protease complex with a phenylbenzoyl (R,S)-N-decalin type inhibitor |
| 8 | 4OVZ | X-Ray Structural and Biological Evaluation of a Series of Potent and Highly Selective Inhibitors of Human Coronavirus Papain-Like Proteases |
| 9 | 3MJ5 | Severe Acute Respiratory Syndrome-Coronavirus Papain-Like Protease Inhibitors: Design, Synthesis, Protein-Ligand X-ray Structure and Biological Evaluation |
| 10 | 2FE8 | SARS coronavirus papain-like protease: structure of a viral deubiquitinating enzyme |
| 11 | 1UK4 | Crystal structure of SARS Coronavirus Main Proteinase (3CLpro) Complexed With An Inhibitor |
| 12 | 1UK2 | Crystal structure of SARS Coronavirus Main Proteinase (3CLpro) At pH8.0 |
| 13 | 1UK3 | Crystal structure of SARS Coronavirus Main Proteinase (3CLpro) At pH7.6 |
| 14 | 1UJ1 | Crystal structure of SARS Coronavirus Main Proteinase (3CLpro) |
| 15 | 3VB6 | Crystal structure of SARS-CoV 3C-like protease with C6Z |
| 16 | 3VB5 | Crystal structure of SARS-CoV 3C-like protease with C4Z |
| 17 | 3TLO | Crystal structure of HCoV-NL63 3C-like protease |
| 18 | 5ZUV | Crystal Structure of the Human Coronavirus 229E HR1 motif in complex with pan-CoVs inhibitor EK1 |
| 19 | 5ZVM | Crystal Structure of the Human Coronavirus SARS HR1 motif in complex with pan-CoVs inhibitor EK1 |
| 20 | 5X4S | Structure of the N-terminal domain (NTD)of SARS-CoV spike protein |
| 21 | 5WRG | SARS-CoV spike glycoprotein |
| 22 | 6Q05 | MERS-CoV S structure in complex with sialyl-lewisX |
| 23 | 6ACG | Trypsin-cleaved and low pH-treated SARS-CoV spike glycoprotein and ACE2 complex, ACE2-bound conformation 1 |
| 24 | 6ACK | Trypsin-cleaved and low pH-treated SARS-CoV spike glycoprotein and ACE2 complex, ACE2-bound conformation 3 |
| 25 | 3SCI | Crystal structure of spike protein receptor-binding domain from a predicted SARS coronavirus human strain complexed with human receptor ACE2 |
7.1. Spike protein
Spike protein is a type-I tetrameric (TM) protein with a clove shape (Pradesh et al., 2014). S protein facilitates the attachment of virus to surface receptors of host cell and further takes part in fusion between the virus and cell membrane of host and mediates the entry of virus into the host (Song et al., 2004; Siu et al., 2008; Kirchdoerfer et al., 2016). The expression of spike protein in some cell membranes facilitates cell-cell fusion among the infected and nearby uninfected cells. Such formation of large polynucleated cells is a proposed strategy for spreading of virus directly between cells, disrupting of antibodies responsible for virus-neutralisation (Glowacka et al., 2011; Qian et al., 2013; Fehr and Perlman, 2015).
The spike protein contains three segments which are ectodomain region (ED), trimeric region (TM) and an intracellular short tail domain (S1) (Pradesh et al., 2014). The S1 receptor-binding domain (3 S1 heads) and membrane fusion S2 subunit (TM stalk) together on C-terminal comprise the ectodomain. S1 domain acts as major antigen on the virus surface and consists of a receptor binding domain (RBD) (Ortego et al., 2007). S1 domain has two main components that are N and C terminal domains (NTD and CTD) (Ortego et al., 2007; Ou et al., 2020). RBD (consists 14 residues) of spike protein (SARS-CoV) interacts with the 18 residues of ACE2 (Fehr and Perlman, 2015). Amino acid residue R453 of RBD and K341 of ACE-2 play an important role for this contact. Point mutation of R441 or D454 has potential to disturb the binding capability with ACE-2. The S2 subunit consists of two heptad repeat regions known as HR 1 and HR 2 and a hydrophobic fusion peptide. The virus is called as CoV due to the gathering of spike proteins on the outer surface of virion in the trimeric form which gives it the resemblance of a crown. The spike protein supports the entry of virus into the host and is responsible for activating immune response of host cell against CoV (Gerna et al., 2007). The primary interaction of S1 domain and host receptor (for SARS-CoV: ACE2 receptor and for MERS-CoV: PP4) and further S2 segment mediates the fusion of viral membrane and host and subsequently allows the entry of RNA genome of coronavirus inside the host cells. Hence, these proteins are proposed as targets of importance for drug discovery (Gerna et al., 2007; Ortego et al., 2007; Woo et al., 2009; Fehr and Perlman, 2015).
7.2. Envelop protein (E)
The envelop protein is the most versatile and smallest (8.4–12 kDa size) of structurally major TM proteins of CoV family. E protein is expressed in abundance during the replication cycle in infected cell and very small portion of this protein gets incorporated into virion envelope (Narayanan et al., 2000; Escors et al., 2001; Venkatagopalan et al., 2015). It is comprised of two distinct domains which are a charged cytoplasmic tail and a hydrophobic domain (Narayanan et al., 2000). It is majorly localised at the intracellular trafficking site such as Golgi, ER and ERGIC (Corse and Machamer, 2000). Here, it takes part in assembly and budding of CoV (Nieto-Torres et al., 2011). Recombinant CoVs with the lack of E protein are found with reduced viral titres, incompetent progeny, significantly reduced yield propagation and crippled viral maturation which demonstrate the importance of envelope protein for production and maturation of virus (Curtis et al., 2002; Ortego et al., 2002, et al.,2007; Kuo and Masters, 2003; DeDiego et al., 2007). Hence, E protein has a significant role in morphogenesis of virus. According to studies, envelop protein has role in coordinating with other proteins and modulating protein activities (Narayanan et al., 2000). It acts as a virulent factor as well (Gil et al., 2020). E protein forms ion channel through oligomerisation and can be targeted by hexamethylene amiloride to block the associated E protein ion channel activities in mammalian cells which expressed SARS-CoV E protein (Bos et al., 1996; Corse and Machamer, 2003).
7.3. Membrane protein
The membrane protein, a structural protein of high abundance, has role in defining the shape of viral envelop (Neuman et al., 2011). It performs the job of shape maintenance through interacting with other major structural CoV proteins (Masters, 2006), incorporating Golgi complex into new virions and stabilizing N proteins (Nucleocapsid) (Corse and Machamer, 2000). M protein is referred as a central organiser for the assembly of coronavirus. Three TM domains characterizes the M protein with C terminal (long) inside and N-terminal (short) located at outside (Vennema et al., 1996; Baudoux et al., 1998). The driving force behind the formation of virion envelop is the homeotypic interaction between M proteins majorly and it is inadequate for virion formation alone (De Haan et al., 2000; Lim and Liu, 2001; Neuman et al., 2011). M protein plays an important role in intracellular homeostasis for viral assembly through various protein-protein interactions, such as, interaction of M-S, M-M and M-N proteins especially in viral assembly (Corse and Machamer, 2000). The interaction of M-S is necessary for retention of spike (S) protein in ERGIC complex (ER-Golgi intermediate compartment) which is also known as Golgi complex and later it gets incorporated into new progenies of virus (Opstelten et al., 1995; Mortola and Roy, 2004; Fehr and Perlman, 2015). The interaction of M-N is crucial for stabilizing RNP complex (Nucleocapsid-RNA complex), forms the internal core of virus and ultimately supports the completion of assembly process (Narayanan et al., 2000; Escors et al., 2001; Fehr and Perlman, 2015). The M and E are the major proteins in defining shape, which together forms the viral envelope and their interaction is enough to produce and release virus like particles (VLPs) (Bos et al., 1996; Vennema et al., 1996; Baudoux et al., 1998; Corse and Machamer, 2000; Corse and Machamer, 2003; Mortola and Roy, 2004).
Membrane protein also participates in host sensitization by the virus. M protein, via Toll-like receptor-dependent mechanism, is responsible for activating the IFN-β pathway and nuclear factor kappa pathway (NFK). A mutated membrane protein (V-68) failed in eliciting IFN-β response (Nieto-Torres et al., 2011). Vaccination of mice with SARS-M DNA induced and proliferated T-cell immune response and similar cytotoxic response of T-cells was also found when alveolar epithelial cells were infected with SARS-DNA (Opstelten et al., 1995).
7.4. Nucleocapsid protein (N)
The structure of N-protein is conserved amongst various members of the CoV family. N is the protein which primarily takes part in binding with CoV-RNA genome to make the nucleocapsid unlike other structurally major proteins (de Haan and Rottier, 2005). The three intrinsically disordered characteristic regions (IDRs) of nucleocapsid protein include N-arm (NTD), a central linker (CL) and a C-tail (CTD) (Bande et al., 2015). NTD and CTD form the major functional and structural domain of N-protein. N protein is majorly involved in functions related to the viral genome and also participates in other aspects of viral replication cycle and host cellular response on viral infection (McBride et al., 2014). RNA binding is the most important function of NTD and dimerization is the primary function of CTD (Vabret et al., 2008; Bande et al., 2015). As Central linker (CL) is serine and arginine rich region, hence it contains a high number of phosphorylation sites (Lou et al., 2014). Intrinsically disordered regions (IDRs) of C terminal play an important role in oligomerization of nucleocapsid protein and interactions of N-M proteins (Curtis et al., 2002).
Remarkably, localization of N protein into ER-Golgi complex region proposes its function in assembling and budding as well (Tooze et al., 1984; Klumperman et al., 1994). Transient expression of nucleocapsid shows increased production of VLPs in some coronaviruses, stating its requirement for the formation of envelope (Boscarino et al., 2008; Siu et al., 2008; Ruch and Machamer, 2011; Ruch and Machamer, 2012). RNP complex formation and maintenance are the major functions of N protein. Intrinsically disordered regions (IDRs) of C terminal plays an important role in the oligomerization of nucleocapsid protein and interactions of N-M proteins (Curtis et al., 2002). Apart from regulating transcription and replication of viral RNA, it inhibits the translation of protein via EF1 alpha-mediated action, alteration in host cell-cycle, host cell metabolism and apoptosis (Nucleocapsids are reported for CDK4 inhibition) in the host cell (Vabret et al., 2008; Lee, 2015). N protein takes part in inhibition of cytokinesis mediated cell proliferation in human peripheral blood (Ortego et al., 2002).
7.5. 3C-like protease
3C-like protease protein, also known as 3CLpro is found in homodimeric form. The active site of 3CLpro consists of a cystine-histidine (Cys-His) dyad which is responsible for its protease activity (Kilianski and Baker, 2014). Mutation on Ser139 and Phe140 eradicates the dimerization of this protease (3CLpro reported with PDB ID: 3F9G) (Verdiá-Báguena et al., 2012). The protease has the capability to cleave PP1a and PP1ab through 11 sites at P1 position to produce a mature protein which subsequently leads replication-transcription complex and release of mature NSPs (Verdiá-Báguena et al., 2013; Nieto-Torres et al., 2014; Lee, 2015).
7.6. Papain-like protease
Papain-like proteases (PLpro) is responsible for N-terminal cleavage of PP and generates three non-structural proteins (NSP 1, 2 and 3) (Surya et al., 2013; Lee, 2015). PLpro consists of a 316 amino acid long catalytic core domain responsible for cleaving substrates of replicase. In conjunction, LXGG sequence supports cleavage (Verdiá-Báguena et al., 2013). It is found that zinc and its conjugates, in higher doses, are capable for inhibiting SARS proteases of both the types, CLpro and PLpro (Hung and Sheng, 2002).
7.7. Hemagglutinin esterase
The hemagglutinin esterase enzyme is localized into the coronavirus envelope, specifically amid beta-coronaviridae. The HE enzyme is a marker of evolution of influenza and coronavirus. HE acts as a mediator for reversible attachment with O-acetylated-sialic-acids through the functioning as lectins and receptor-disrupting enzyme. Crystal structure of interaction of HE complexed with sialic acid is available to visualize in protein data bank with PDB ID: 3CL5 (Gerek et al., 2009).
7.8. NTPase/helicase
Helicase/NTpase has a prominent role in viral central dogma (Gerek et al., 2009). It is a member of SF1. The preferred substrates of HE are ATP, dCTP and dATP. Apart from that all NTPs are also hydrolysed by this (Jimenez-Guardeno et al., 2015). Toxicity and non-specificity are the major issues in developing helicase inhibitors as non-specificity leads to severe toxicity (Yang et al., 2005a,b). Although, instead of having theoretical limitations, HE is getting recognized increasingly as a druggable target for the various conditions of disease (Hogue and Machamer, 2008).
7.9. NSP15
Nsp15 is recently mapped and a conserved protein among the families of coronavirus. It is one of the essential proteins for viral lifecycle and virulence. As per earlier studies, it was supposed to be responsible for viral replication directly but recent studies proposed Nsp15 to help in viral replication through interfering the immune response of host (Kim et al., 2020a,b).
The non-structural protein15 from SARS-CoV-2 has 89 percent similarity with the protein of previous outbreak of SARS-CoV. The published studies of 2010 on SARS-CoV-1 reveal that the viral replication can be slowed down by inhibiting Nsp15. The studies available suggest the drug development targeting Nsp15 could be potential against COVID-19. Nsp15 is reported with PDB ID: 6VWW and revealed to scientific community at protein data bank (RCSB) on 3rd March, 2020.
8. Drug repurposing opportunities and challenges
Drug repurposing also known as drug repositioning and reprofiling is an efficient strategy of drug discovery from existing drugs. The approach significantly reduces time and cost as compared to discovery of novel drug / de novo drug designing and random clinical trials (Cheng et al., 2016a; Cheng et al., 2017, et al.,2019). Computer-aided drug discovery facilitates novel hypotheses to test efficient drug repurposing (Cheng et al., 2016a,b, et al.,2018). Conventional structure-based approaches have limitations where three-dimensional structures of target protein are not available. This is a major issue while targeting human and viral proteins. Because of the rapid evolution of viral genome, targeting single viral protein is not feasible (Zhou et al., 2020). Even though cellular factor of host is required for replication of human coronavirus during infection, efficient identification of systematic protein-protein interaction of host and virus offers a systematic way for elucidation of viral infection mechanism. Consequently, targeting viral proteins and antiviral targets of cell, that is, host-virus interactome can be a potentially novel strategy for exploring the treatment of viral infection (Dyall et al., 2014). Drug repurposing promises to identify a viable strategy, in a time-critical manner, for the screening of antiviral agents against SARS-CoV-2. It offers a combinational approach acting with ‘double-hit effect’ to combat coronavirus infection with more successful chances and clinical translation (Senanayake, 2020). The earlier screening of broad-spectrum antiviral agents (BSAA) have been introduced and tested safe in clinical trials and now been explored as promising repurposing candidates (Andersen et al., 2020a,b). Drug repurposing has been resulted in reaching for phase III clinical trials for drugs, such as, remdesivir, lopinavir, favipiravir and hydroxyquinone, etc. Conceptually, promiscuity of replication mechanism of virus and host-target interaction is taken as advantage to explore BSAAs (Bösl et al., 2019).
Another limiting dimension of phenotypic screening is the hit compounds with low potency as single agent, hence their dose with maximum tolerability is found subtherapeutic (Sun et al., 2016). To resolve such issues, two or more drugs are evaluated which are acting on various signalling pathways involved in viral replication with least redundancy. High-throughput screening to sought out synergistic combination from compound libraries could be an another strategy targeting host-virus interaction for emerging infectious diseases, which narrow down the research spectrum of individual agents (Bösl et al., 2019; Cheng et al., 2019). Such strategies are promising to address weakly-active BSAAs by potentially reducing dosage, improving efficiency, reducing time and drug development cost, lowering toxicity and minimizing occurrence of secondary resistance (Zheng et al., 2018). Notably, the lack of specificity of broad-spectrum antiviral agents leads to implications with highly virulent strains and emergence of drug resistance. An example of a non-structural coronavirus protein NSP14-exon with cofactor NSP10 acts on nucleotide-mismatch repair caused by the exposure of ribavirin (nucleoside analog) and hence has potential to negate antiviral efficiency of BSAA (Ferron et al., 2018). Simultaneously, the notion of determining drug repositioning strategy has competition with structure-based drug designing of small molecule therapeutics and preventative vaccines and off-targeted specificity. The interaction of coronavirus spike glycoprotein and human enzyme ACE2 as a port for cellular invasion has driven a way for rapid development of antibodies and vaccines (Salvatori et al., 2020). Such strategies possibly hindered through the genetic drift of viruses due to the potential of blunt antigenicity of epitope (Letko et al., 2020; Wan et al., 2020; Wrapp et al., 2020).
Another important factor in the quest of drug repurposing comprises issues with patent protection under international and national policies. COVID-19 global health emergency with the magnitude of such level seeks courageous international response from the political and government level. Also, rapid actions of regulatory authority are required for minimized financial hurdles and updated guidelines on drug licensing via reprofiling wherever necessary. Simultaneously, such efforts are vital acts behind the scene while seeking indications on existing compounds.
9. Computational drug discovery approach
The spike protein of coronavirus binds to the cell membrane of host via a receptor-mediated interaction which facilitates its entry in the host cell. The computational studies have indicated similarity in mechanism of SARS-CoV-2 with SARS virus. It has highest affinity with receptor ACE2 (Xu et al., 2020). Spike protein of SARS-CoV-2 has structural similarity with SARS virus spike protein where 73% region is conserved and variability is majorly in the region of protein for host cell interaction.
The other computation docking model is focusing on 3C like protease (3CLpro) which controls the various major viral functions. It consists of highly conserved SARS-CoV catalytic domain (Anand et al., 2003). Replication process is one of a major function of 3CLpro due to which it is prospected as ideal target for drug designing (Bacha et al., 2004). It is reported in protein data bank with PDB ID: 6LU7 (Liu et al., 2020). Remdesivir, zanamvir, saquinavir and indinavir are the potential hits inhibiting 3CLpro protease. Some off-label medications are also indicated through studies such as adeflavin, FAD (flavin adenine dinucleotide), coenzyme A. These can also be beneficial for the treatment of COVID-19.
Spike and protease, both the proteins are essential for the viral transmission and to cause virulence. Inhibition of both or any of these proteins can reduce the severity of infection. The efforts have been focusing on the competitive inhibition of binding to their natural substrates (Hall and Ji, 2020). As per the docking studies, RNA polymerase inhibitors and HIV protease inhibitors are also found to be promising to bind with SARS-CoV-2 target proteins. Subsequently, a protein synthesis inhibitor methisazone, CGP42112A as antigiotensin receptor agonist (ACE inhibitor) and compound ABT450 as NSP3-4A inhibitor have come up as suitable treatment options (Shah et al., 2020). According to another in silico study conducted, some of the commercial drugs such as valrubicin, colistin, icatibant, epirubicin, bepotastine, epoprostenol, aprepitant, vapreotide, perphanazine and caspofungin are also found to bind with same binding site of lopinavir/ritonavir (Cai et al., 2020).
Compound library of known bioactive agents from databases like ZINC15 (includes 3447 entries of FDA & Drug Bank approved compounds) facilitates to explore the potential inhibitors against the catalytic domains of these targets (Sterling and Irwin. 2015; Hall and Ji, 2020). Utilization of approved compounds from database leads to quicker trials and reduced efforts in getting approval from food and drug regulatory agencies. The drug reprofiling with the aid of computational drug discovery approaches helps in exploring therapeutics which can be promising for the treatment of coronavirus disease (Hall and Ji, 2020).
9.1. In silico studies on genomic sequence pattern analysis: Network-based studies on drug reprofiling for SARS-CoV-2
In a study, an integrated drug reprofiling methods with a network platform based upon pharmacology, drug targets in human protein-protein (P-P) interaction network and quantification of interplay within human coronavirus and host interactome (HCoV-Host) have been implemented using network strategies (Gordon et al., 2020a,b). It was observed through studies on 15 whole genome sequences of HCoV using phylogenetic analysis that the similarity between SARS-CoV-2 and SARS-CoV is 79.7%. The studies specified that the nucleocapsid and envelope proteins of novel coronavirus (SARS-CoV-2) are two evolutionary conserved regions with sequence similarities of 89.6% and 96% respectively with SARS-CoV. A network proximity study on hCoV-host interactions and drug targets using potential antiviral repurposable drugs were validated using enrichment analyses of human coronavirus induced transcriptomics data and drug-gene signature in human cell lines. The studies summarised rapid identification of potential drug combinations and candidate repositionable drugs against SARS-CoV-2 (Zhou et al., 2020).
9.2. Identification of sequence conservation pattern computationally
Studies of multiple sequence alignment of COVID-19 protease with orf1ab polyprotein and PDB template identification as 4MM3_B have revealed various residues which are highly conserved. This includes the ligand binding sites of COVID-19 protease (Thr 75, Arg 141, Gln175 and His 176).
Viral enzyme 3-chymotrypsin-like cysteine protease is essential for life cycle of coronavirus which controls the process of replication is a proven target for SARS-CoV and MERS-CoV. Computational studies revealed the similarity of genomic sequence of SARS-nCoV-2 with SARS-CoV (ul Qamar et al., 2020). Notably the re-emerging and new disease outbreaks like current pandemic of COVID-19 can potentially shake the global health system.
It is evident from the output of multiple sequence alignment that the ligand binding sites (Thr 75, Arg141, Gln 175 and His 176) were found to be conserved amongst the sequences of COVID-19 proteases. The current computational studies provide structural and molecular insight about COVID-19 proteases which might aid in deciphering drug-target interaction with some known potential protease inhibitors (Eleftheriou et al., 2020; Mothay and Ramesh, 2020).
9.3. In silico binding site analysis of COVID-19 protease inhibitors
Multiple sequence alignment is a potential computational techniques for the identification of conserved domains of COVID-19 proteases. It was carried out using orf1ab polyprotein from pneumonia virus of Wuhan seafood market (YP_009724389.1). The best template was identified with PDB using Clustal Omega from SWISS MODEL server (Sievers et al., 2011). Five protease inhibitors were subjected to dock with predicted model of protease. The studies on molecular interaction revealed that 3D protease model had Arg 141, Thr 75, His 176 and Gln 175 as potential binding sites for drugs. More than one binding sites were observed with remdesivir (Thr 75 and His 176) (Mothay and Ramesh, 2020).
9.4. Molecular docking studies for drug target interaction
Computational molecular docking studies are of great help in deciphering the interaction of repurposed drugs on novel coronavirus target proteins. In silico docking studies performed on spike glycoprotein complexed with ACE2 (PDB ID: 6CS2) revealed the participation of central pocket of protein for ligand interaction. The main protease of COVID-19 with PDB ID: 6LU7 (complexed with inhibitor N3) has two chains (chain A and chain B) which have found to interact with ligand. It was further shown that the oxygen from carbonyl group in ester side chain of oseltamivir, an antiviral drug, interacts with amino acids Lys 102 and Ser 158 of COVID-19 main protease (complexed with inhibitor N3). van der Waals interactions were found at Val 104, Gln 107, Gln 110, Asn 151, Asp 153, Thr 111, Ile 106, Thr 292 and Phe 294. Molecular docking of ritonavir with protease revealed the high affinity of drug with chain A of main protease of COVID-19 complexed with N3 inhibitor. The amide group oxygen of ritonavir showed hydrogen bond interaction with Thr 111. The benzene ring showed pi-pi and pi-anion interactions with Phe 294 and Asp153, respectively. Remdesivir took part in interaction with main protease via hydrogen bonding between cyano group nitrogen with Phe 294 and tetrahydrofuran ring oxygen with Gln 110. Aromatic ring has shown some pi sigma interactions with Val 104 and Ile 249. Docking studies conducted for chloroquine with main protease and spike glycoprotein revealed significantly comparable interactions. Binding interaction with main protease was observed in chain C binding pocket with pi-alkyl interactions between Ile 106 and Val 104 with chlorobenzene ring. The interaction with spike glycoprotein-ACE2 complex was attributed with pi alkyl and pi-pi interactions between chlorine (attached with benzene) and benzene with Phe 952 and Tyr 738, respectively. Hence it is concluded that chloroquine prefers interacting with spike glycoprotein-human ACE2 complex over main protease complexed with N3 inhibitor.
Computer-aided drug discovery techniques in deciphering drug repurposing reveal the high affinity of antiviral drugs such as ritonavir, oseltamivir, remdesivir and favipiravir with COVID-19 main protease in complex with N3 while antimalarial drugs including hydroxyquinone and chloroquine show high affinity interaction with spike glycoprotein-human ACE2 complex. Hence, possibly anti-COVID-19 activity of antiviral drugs is due to the interaction with main protease while antimalarial drugs attributed with high affinity with spike glycoprotein-human ACE2 complex (Narkhede et al., 2020).
10. Drug discovery approaches targeting major SARS-CoV-2 proteins
10.1. Strategy against Spike protein and its interactions
Receptor binding domain is majorly targeted for drug design studies for spike proteins. A study reported with a similar peptide sequence of RBD is capable of hampering ACE2 interaction of S1-RBD domain which has role in preventing viral entry in the host cell (studies conducted on Vero cells, IC50 - 40 μM) (Hu et al., 2005; Han et al., 2006; Du et al., 2009). Another peptide sequence (OC43-HR2P) derived from HR2 (heptad repeat) region of S2 domain (HCoV-OC43) optimized as EK1 has exhibited fusion inhibitory role among pan-CoV (Xia et al., 2019). Spike protein specific monoclonal antibodies CR301 and 80R can block the interaction of spike protein with ACE2, hence neutralize the SARS-CoV infection (Du et al., 2009). M396 monoclonal antibody is found with competitive role for receptor binding domain (RBD) (Prabakaran et al., 2006).
10.2. Drug discovery strategy targeting N protein
Formation of RNA-Nucleocapsid protein complex and its maintenance is the crucial function of N protein. It has role in regulation of transcription and replication of viral genome (RNA). It acts as an inhibitor of protein translation of host via EF1α-mediated action (McBride et al., 2014), alteration of metabolism of host cell, cell cycle (inhibits CDK4) and apoptosis. N protein also inhibits cytokinesis thus inhibits cellular proliferation in the peripheral blood system of human. N-terminal domain of N protein has sites for RNA binding. Inhibitors are designed for RNA binding sites of the NTD of N protein (Lin et al., 2014, Chang et al., 2016). Herbal products such as polyphenolic compounds like gallocatechin gallate and catechin are found to show inhibitory activity targeting NTD against HCoV (Roh, 2012). Similarly, C-terminal domain (CTD) of Nucleocapsid (N) protein helps in oligomerization on C-terminal. As per the studies, competition with the process of oligomerization via a C-terminal sequence of tail peptide (N377-389) has significant inhibitory role on virus (titer concentration of 300 μM) (Lo et al., 2013).
10.3. Strategies on proteases
The SARS coronavirus genome encodes numerous proteins. A major component of coronavirus genome is replicase gene, encodes 16 non-structural proteins (NSPs) in two large forms of PPs (PP1a and PP1ab). Cysteine proteases of two types act on PPs and release NSPs. The cleavage of C terminal of PPs is done by chymotrypsine-like cysteine protease (3Cpro or Mpro-main protease). The Mpro, also known as papain-like protease (PLpro), processes the N-terminal region (Lindner et al., 2005). PLpro cuts the first three cleavage site of PPs and CLpro cleaves the rest eleven sites. This ultimately release 16 NSPs (Jo et al., 2020). Some peptidomimetic compounds and metal conjugates showed inhibitory action against 3CLpro. Various small molecules also act as an inhibitor such as arylboronic acid, thiophenecarboxylate, derivatives of quinolinecarboxylate and analogs of phthalhyrazide-substituted ketoglutamate (Hsu et al., 2005). Inhibition of Mpro is reported with some flavonoids (Jo et al., 2020). Crystal structure of Mpro with N3 inhibitor complex is reported with PDB ID: 2AMQ (Yang et al., 2005a,b). HIV protease inhibitors ritonavir and lopinavir also has inhibitory activity on Mpro.
11. Status of drugs under clinical trials
Lopinavir/ritonavir drug combination therapy to target viral protease and a membrane fusion inhibitor umifenovir which targets viral entry, together is approved for the treatment of HIV and influenza. Currently, different combination of these drugs are considered for phase IV clinical trials for COVID-19 associated with pneumonia (ClinicalTrials.gov ID: NCT04255017) (Chang et al., 2016; Trials, 2020a,b,c). Remdesivir, an RNA-dependent RNA polymerase inhibitor was under investigation level of phase III for moderate and mild SARS-CoV-2 infection (ClinicalTrials.gov Identifier: NCT04252664) (Trials, 2020a,b,c). This study has been suspended in China on April 2020 due to the lack of eligible patients. Preclinical studies on remdesivir have shown activity against SARS-CoV and MERS-CoV (Gordon et al., 2020a,b; Harrison, 2020a,b). It has shown remarkable studies in a controlled and randomised trial on Ebola virus infection and demonstrated antiviral effects (Mulangu et al., 2019). Alongside other agents evaluated at Phase III level for combination therapy include hydroxychloroquine (antimalarial) according to promising data of in vitro studies which has been completed in April 2020 (ClinicalTrials.gov Identifier: NCT04261517) (Trials, 2020a,b,c). Apart from immunomodulating properties of choloroquine, it exhibits antiviral action at entry and after-entry stages of coronavirus infection. It has shown potential synergistic role with BSAAs and found to enhance antiviral effect of remdesivir (Wrapp et al., 2020). At the initial stages, favipiravir, an RNA polymerase inhibitor in combination therapy, is also on phase II of clinical trial for pneumonia associated with novel coronavirus disease (Chinese Clinical Trial Registry Identifier: ChiCTR2000029544) (Registry, 2020). Apart from that, preclinical studies on ribonucleic analog ribavirin have also shown activity against SARS-CoV-2 under in vitro investigations (Wrapp et al., 2020).
12. Conclusion
Solving and mapping three-dimensional structure of coronavirus helps scientists to conclude how a viral replication process in human cells can be interfered. It is now clear that complete assembly of structural proteins of coronavirus is not required to form an infectious and complete virion. This suggests the dispensability of some structural proteins or possibly, coronavirus might encode some proteins which overlap compensatory functions additionally. However, each protein plays a primary role as a structural component of virus particle, but these have also found to participate in some of the aspects of its replication cycle. Some viral proteins have been participating in a flexible multienzyme complex system which facilitates compensatory low fidelity of viral replication. There are viable strategies to enable proofreading of coronavirus RNA and maintain genetic integrity and, consequently, rendering some degree of evolution or freedom to mutate.
Here, definitely high-throughput screening and virtual screening come to rescue (Fischer et al., 2020). Hence, it is a need to screen drugs with their affinity to perturb the functionality of such systems in CoV. This includes the possibility of targeting catalytic subunits such as NSP14-exon and critical sites with allosteric effects where conformational changes are caused by conserved residues in the entire RNA repair complex of virus. Henceforth, the search should be focusing on identification of inhibitors for viral nuclease.
Computer-aided drug discovery techniques indicated high affinity of antiviral drugs such as ritonavir, oseltamivir, remdesivir and favipiravir with COVID-19 main protease in complex with N3 while antimalarial drugs (hydroxyquinone and chloroquine) show high affinity interaction with spike glycoprotein-human ACE2 complex, which suggest that abovementioned molecules can be used as suitable against target proteins of SARS-CoV-2. We can antedate the notion of drug reprofiling for emerging coronavirus disease through scrutinization based on these studies. Conclusively, this is not only a fight against COVID-19 but also for the various emerging viral diseases and evolutionarily arising microbial and viral infections. The sole concept of new antiviral and medicinal products, that is, one drug for one virus vs. one drug for multiple virus and multiple drugs for one virus are the challenges for their clinical indications.
Declaration of Competing Interest
Authors declare no conflict of interest.
Acknowledgement
We are thankful to National Institute of Technology Raipur India for providing the necessary facilities to prepare the manuscript and permission to publish it.
References
- Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., Smith E., Case J., Feng J., Jordan R. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. MBio. 2018;9 doi: 10.1128/mBio.00221-18. e00221-00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen P.I., Ianevski A., Lysvand H., Vitkauskiene A., Oksenych V., Bjørås M., Telling K., Lutsar I., Dampis U., Irie Y. Discovery and development of safe-in-man broad-spectrum antiviral agents. Int. J. Infect. Dis. 2020 doi: 10.1016/j.ijid.2020.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacha U., Barrila J., Velazquez-Campoy A., Leavitt S.A., Freire E. Identification of novel inhibitors of the SARS coronavirus main protease 3CLpro. Biochemistry. 2004;43:4906–4912. doi: 10.1021/bi0361766. [DOI] [PubMed] [Google Scholar]
- Bande F., Arshad S.S., Hair Bejo M., Moeini H., Omar A.R. Progress and challenges toward the development of vaccines against avian infectious bronchitis. J. Immunol. Res. 2015;2015 doi: 10.1155/2015/424860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudoux P., Carrat C., Besnardeau L., Charley B., Laude H. Coronavirus pseudoparticles formed with recombinant M and E proteins induce alpha interferon synthesis by leukocytes. J. Virol. 1998;72:8636–8643. doi: 10.1128/jvi.72.11.8636-8643.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia R., Narang R.K., Rawal R.K. A summary of viral targets and recently released PDB IDs of SARS-CoV-2. Open Virol. J. 2020;14:7–8. [Google Scholar]
- Blaising J., Levy P., Polyak S., Stanifer M., Boulant S., EI P. Arbidol inhibits viral entry by interfering with clathrin-dependent trafficking. Antiviral Res. 2013;100:215–219. doi: 10.1016/j.antiviral.2013.08.008. [DOI] [PubMed] [Google Scholar]
- Blaising J., Polyak S., Pecheur E. Arbidol as a broad-spectrum antiviral: an update. Antiviral Res. 2014;107:84–94. doi: 10.1016/j.antiviral.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos E.C., Luytjes W., Van der Meulen H., Koerten H.K., Spaan W.J. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology. 1996;218:52–60. doi: 10.1006/viro.1996.0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscarino J.A., Logan H.L., Lacny J.J., Gallagher T.M. Envelope protein palmitoylations are crucial for murine coronavirus assembly. J. Virol. 2008;82:2989–2999. doi: 10.1128/JVI.01906-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bösl K., Ianevski A., Than T.T., Andersen P.I., Kuivanen S., Teppor M., Zusinaite E., Dumpis U., Vitkauskiene A., Cox R.J. Common nodes of virus–host interaction revealed through an integrated network analysis. Front. Immunol. 2019;10:2186. doi: 10.3389/fimmu.2019.02186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet M., Lugari A., Posthuma C.C., Zevenhoven J.C., Bernard S., Betzi S., Imbert I., Canard B., Guillemot J.-C., Lécine P. Coronavirus Nsp10, a critical co-factor for activation of multiple replicative enzymes. J. Biol. Chem. 2014;289:25783–25796. doi: 10.1074/jbc.M114.577353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q., Yang M., Liu D., Chen J., Shu D., Xia J., Liao X., Gu Y., Cai Q., Yang Y. Experimental treatment with favipiravir for COVID-19: an open-label control study. Engineering. 2020 doi: 10.1016/j.eng.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascella M., Rajnik M., Cuomo A., Dulebohn S.C., Di Napoli R. StatPearls Publishing; 2020. Features, Evaluation and Treatment Coronavirus (COVID-19) StatPearls [Internet]. [PubMed] [Google Scholar]
- Chan J.F.-W., To K.K.-W., Tse H., Jin D.-Y., Yuen K.-Y. Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol. 2013;21:544–555. doi: 10.1016/j.tim.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.-k., Jeyachandran S., Hu N.-J., Liu C.-L., Lin S.-Y., Wang Y.-S., Chang Y.-M., Hou M.-H. Structure-based virtual screening and experimental validation of the discovery of inhibitors targeted towards the human coronavirus nucleocapsid protein. Mol. Biosyst. 2016;12:59–66. doi: 10.1039/c5mb00582e. [DOI] [PubMed] [Google Scholar]
- Cheng F., Desai R.J., Handy D.E., Wang R., Schneeweiss S., Barabási A.-L., Loscalzo J. Network-based approach to prediction and population-based validation of in silico drug repurposing. Nat. Commun. 2018;9:1–12. doi: 10.1038/s41467-018-05116-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F., Hong H., Yang S., Wei Y. Individualized network-based drug repositioning infrastructure for precision oncology in the panomics era. Brief. Bioinform. 2017;18:682–697. doi: 10.1093/bib/bbw051. [DOI] [PubMed] [Google Scholar]
- Cheng F., Murray J.L., Rubin D.H. Drug repurposing: new treatments for zika virus infection? Trends Mol. Med. 2016;22:919–921. doi: 10.1016/j.molmed.2016.09.006. [DOI] [PubMed] [Google Scholar]
- Cheng F., Murray J.L., Zhao J., Sheng J., Zhao Z., Rubin D.H. Systems biology-based investigation of cellular antiviral drug targets identified by gene-trap insertional mutagenesis. PLoS Comput. Biol. 2016;12 doi: 10.1371/journal.pcbi.1005074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y.-S., Williamson P.R., Zheng W. Improving therapy of severe infections through drug repurposing of synergistic combinations. Curr. Opin. Pharmacol. 2019;48:92–98. doi: 10.1016/j.coph.2019.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson P., Rolain J.-M., Raoult D. Chloroquine for the 2019 novel coronavirus. Int. J. Antimicrob. Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corse E., Machamer C.E. Infectious bronchitis virus E protein is targeted to the Golgi complex and directs release of virus-like particles. J. Virol. 2000;74:4319–4326. doi: 10.1128/jvi.74.9.4319-4326.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corse E., Machamer C.E. The cytoplasmic tails of infectious bronchitis virus E and M proteins mediate their interaction. Virology. 2003;312:25–34. doi: 10.1016/S0042-6822(03)00175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis K.M., Yount B., Baric R.S. Heterologous gene expression from transmissible gastroenteritis virus replicon particles. J. Virol. 2002;76:1422–1434. doi: 10.1128/JVI.76.3.1422-1434.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E. Anti-HIV drugs: 25 compounds approved within 25 years after the discovery of HIV. Int. J. Antimicrob. Agents. 2009;33:307–320. doi: 10.1016/j.ijantimicag.2008.10.010. [DOI] [PubMed] [Google Scholar]
- de Farias S.T., dos Santos Junior A.P., Rêgo T.G., José M.V. Origin and evolution of RNA-dependent RNA polymerase. Front. Genet. 2017;8:125. doi: 10.3389/fgene.2017.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., Rottier P.J. Molecular interactions in the assembly of coronaviruses. Adv. Virus Res. 2005;64:165–230. doi: 10.1016/S0065-3527(05)64006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Haan C.A., Vennema H., Rottier P.J. Assembly of the coronavirus envelope: homotypic interactions between the M proteins. J. Virol. 2000;74:4967–4978. doi: 10.1128/jvi.74.11.4967-4978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Meyer S., Azijn H., Surleraux D., Jochmans D., Tahri A., Pauwels R., Wigerinck P., de Béthune M.-P. TMC114, a novel human immunodeficiency virus type 1 protease inhibitor active against protease inhibitor-resistant viruses, including a broad range of clinical isolates. Antimicrob. Agents Chemother. 2005;49:2314–2321. doi: 10.1128/AAC.49.6.2314-2321.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., Feldmann F., Cronin J., Jordan R., Okumura A., Thomas T., Scott D., Cihlar T., Feldmann H. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc. Natl. Acad. Sci. 2020;117:6771–6776. doi: 10.1073/pnas.1922083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeDiego M.L., Álvarez E., Almazán F., Rejas M.T., Lamirande E., Roberts A., Shieh W.-J., Zaki S.R., Subbarao K., Enjuanes L. A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J. Virol. 2007;81:1701–1713. doi: 10.1128/JVI.01467-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., He L., Zhang Q., Huang Z., Che X., Hou J., Wang H., Shen H., Qiu L., Li Z. Organ distribution of severe acute respiratory syndrome (SARS) associated coronavirus (SARS‐CoV) in SARS patients: implications for pathogenesis and virus transmission pathways. J. Pathol. 2004;203:622–630. doi: 10.1002/path.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., He Y., Zhou Y., Liu S., Zheng B.-J., Jiang S. The spike protein of SARS-CoV—a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducharme J., Farinotti R. Clinical pharmacokinetics and metabolism of chloroquine. Clin. Pharmacokinet. 1996;31:257–274. doi: 10.2165/00003088-199631040-00003. [DOI] [PubMed] [Google Scholar]
- Dyall J., Coleman C.M., Hart B.J., Venkataraman T., Holbrook M.R., Kindrachuk J., Johnson R.F., Olinger G.G., Jahrling P.B., Laidlaw M. Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrob. Agents Chemother. 2014;58:4885–4893. doi: 10.1128/AAC.03036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff M.-P., Ferron F., Campanacci V., Longhi S., Rancurel C., Dutartre H., Snijder E.J., Gorbalenya A.E., Cambillau C., Canard B. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc. Natl. Acad. Sci. 2004;101:3792–3796. doi: 10.1073/pnas.0307877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eleftheriou P., Amanatidou D., Petrou A., Geronikaki A. In silico evaluation of the effectivity of approved protease inhibitors against the main protease of the novel SARS-CoV-2 virus. Molecules. 2020;25:2529. doi: 10.3390/molecules25112529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjuanes L., Nieto-Torres J., Jimenez-Guardeno J., DeDiego M. 2011. Replicating Vaccines: Birkhauser Advances in Infectious Diseases. [Google Scholar]
- Escors D., Ortego J., Laude H., Enjuanes L. The membrane M protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability. J. Virol. 2001;75:1312–1324. doi: 10.1128/JVI.75.3.1312-1324.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyer L., Nougairède A., Uhlířová M., Driouich J.-S., Zouharová D., Valdés J.J., Haviernik J., Gould E.A., De Clercq E., de Lamballerie X. An E460D substitution in the NS5 protein of tick-borne encephalitis virus confers resistance to the inhibitor Galidesivir (BCX4430) and also attenuates the virus for mice. J. Virol. 2019;93 doi: 10.1128/JVI.00367-19. e00367-00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Perlman S. Springer; 2015. Coronaviruses: An Overview of their Replication and Pathogenesis, Coronaviruses; pp. 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferron F., Subissi L., De Morais A.T.S., Le N.T.T., Sevajol M., Gluais L., Decroly E., Vonrhein C., Bricogne G., Canard B. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl. Acad. Sci. 2018;115:E162–E171. doi: 10.1073/pnas.1718806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink S., Vojtech L., Wagoner J., Slivinski N., Jackson K., Wang R., Khadka S., Luthra P., Basler C., Polyak S. The Antiviral Drug Arbidol Inhibits Zika Virus. Sci. Rep. 2018;8:8989. doi: 10.1038/s41598-018-27224-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A., Sellner M., Neranjan S., Smieško M., Lill M.A. Potential inhibitors for novel coronavirus protease identified by virtual screening of 606 million compounds. Int. J. Mol. Sci. 2020;21:3626. doi: 10.3390/ijms21103626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta Y., Komeno T., Nakamura T. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc. Jpn. Acad. Series B. 2017;93:449–463. doi: 10.2183/pjab.93.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerek Z.N., Keskin O., Ozkan S.B. Identification of specificity and promiscuity of PDZ domain interactions through their dynamic behavior. Proteins Struct. Funct. Bioinf. 2009;77:796–811. doi: 10.1002/prot.22492. [DOI] [PubMed] [Google Scholar]
- Gerna G., Percivalle E., Sarasini A., Campanini G., Piralla A., Rovida F., Genini E., Marchi A., Baldanti F. Human respiratory coronavirus HKU1 versus other coronavirus infections in Italian hospitalised patients. J. Clin. Virol. 2007;38:244–250. doi: 10.1016/j.jcv.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil C., Ginex T., Maestro I., Nozal V., Barrado-Gil L., Cuesta-Geijo M.A., Urquiza J., Ramírez D., Alonso C., Campillo N.E. COVID-19: drug targets and potential treatments. J. Med. Chem. 2020 doi: 10.1021/acs.jmedchem.0c00606. [DOI] [PubMed] [Google Scholar]
- Glowacka I., Bertram S., Müller M.A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T.S., He Y., Gnirss K. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon C.J., Tchesnokov E.P., Feng J.Y., Porter D.P., Götte M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 2020;295:4773–4779. doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020:1–13. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R.L., Becker M.M., Eckerle L.D., Bolles M., Denison M.R., Baric R.S. A live, impaired-fidelity coronavirus vaccine protects in an aged, immunocompromised mouse model of lethal disease. Nat. Med. 2012;18:1820. doi: 10.1038/nm.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Korteweg C., McNutt M.A., Gu J. Pathogenetic mechanisms of severe acute respiratory syndrome. Virus Res. 2008;133:4–12. doi: 10.1016/j.virusres.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D.C., Jr., Ji H.-F. A search for medications to treat COVID-19 via in silico molecular docking models of the SARS-CoV-2 spike glycoprotein and 3CL protease. Travel Med. Infect. Dis. 2020 doi: 10.1016/j.tmaid.2020.101646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D.P., Penn-Nicholson A., Cho M.W. Identification of critical determinants on ACE2 for SARS-CoV entry and development of a potent entry inhibitor. Virology. 2006;350:15–25. doi: 10.1016/j.virol.2006.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison C. Coronavirus puts drug repurposing on the fast track. Nat. Biotechnol. 2020 doi: 10.1038/d41587-020-00003-1. [DOI] [PubMed] [Google Scholar]
- Harrison C. Drug researchers pursue new lines of attack against COVID-19. Nat. Biotechnol. 2020 doi: 10.1038/d41587-020-00013-z. [DOI] [PubMed] [Google Scholar]
- Haviernik J., Stefanik M., Fojtikova M., Kali S., Tordo N., Rudolf I., Hubalek Z., Eyer L., Ruzek D. Arbidol (Umifenovir): a broad-spectrum antiviral drug that inhibits medically important arthropod-borne flaviviruses. Viruses. 2018;10 doi: 10.3390/v10040184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald-Sargent T., Gallagher T. Ready, set, fuse! The coronavirus spike protein and acquisition of fusion competence. Viruses. 2012;4:557–580. doi: 10.3390/v4040557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herod M., Adeyemi O., Ward J., Bentley K., Harris M., Stonehouse N., Polyak S. The broad-spectrum antiviral drug arbidol inhibits foot-and-mouth disease virus genome replication. J. Gen. Virol. 2019;100:1293–1302. doi: 10.1099/jgv.0.001283. [DOI] [PubMed] [Google Scholar]
- Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogue B.G., Machamer C.E. Coronavirus structural proteins and virus assembly, Nidoviruses. Am. Soc. Microbiol. 2008:179–200. [Google Scholar]
- Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon C.-C., Lam T.-Y., Shi Z.-L., Drummond A.J., Yip C.-W., Zeng F., Lam P.-Y., Leung F.C.-C. Evidence of the recombinant origin of a bat severe acute respiratory syndrome (SARS)-like coronavirus and its implications on the direct ancestor of SARS coronavirus. J. Virol. 2008;82:1819–1826. doi: 10.1128/JVI.01926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu M.-F., Kuo C.-J., Chang K.-T., Chang H.-C., Chou C.-C., Ko T.-P., Shr H.-L., Chang G.-G., Wang A.H.-J., Liang P.-H. Mechanism of the maturation process of SARS-CoV 3CL protease. J. Biol. Chem. 2005;280:31257–31266. doi: 10.1074/jbc.M502577200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Li L., Kao R.Y., Kou B., Wang Z., Zhang L., Zhang H., Hao Z., Tsui W.H., Ni A. Screening and identification of linear B-cell epitopes and entry-blocking peptide of severe acute respiratory syndrome (SARS)-associated coronavirus using synthetic overlapping peptide library. J. Comb. Chem. 2005;7:648–656. doi: 10.1021/cc0500607. [DOI] [PubMed] [Google Scholar]
- Hu T., Chen C., Li H., Dou Y., Zhou M., Lu D., Zong Q., Li Y., Yang C., Zhong Z. Structural basis for dimerization and RNA binding of avian infectious bronchitis virus nsp9. Protein Sci. 2017;26:1037–1048. doi: 10.1002/pro.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull M.W., Montaner J.S. Ritonavir-boosted protease inhibitors in HIV therapy. Ann. Med. 2011;43:375–388. doi: 10.3109/07853890.2011.572905. [DOI] [PubMed] [Google Scholar]
- Hulseberg C., Fénéant L., Szymańska-de Wijs K., Kessler N., Nelson E., Shoemaker C., Schmaljohn C., Polyak S., White J. Arbidol and other low-molecular-weight drugs that inhibit Lassa and Ebola viruses. J. Virol. 2019;93 doi: 10.1128/JVI.02185-18. e02185-02118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung A.Y., Sheng M. PDZ domains: structural modules for protein complex assembly. J. Biol. Chem. 2002;277:5699–5702. doi: 10.1074/jbc.R100065200. [DOI] [PubMed] [Google Scholar]
- J Alsaadi E.A., Jones I.M. Membrane binding proteins of coronaviruses. Fut. Virol. 2019;14:275–286. doi: 10.2217/fvl-2018-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Guardeno J.M., Regla-Nava J.A., Nieto-Torres J.L., DeDiego M.L., Castano-Rodriguez C., Fernandez-Delgado R., Perlman S., Enjuanes L. Identification of the mechanisms causing reversion to virulence in an attenuated SARS-CoV for the design of a genetically stable vaccine. PLoS Pathog. 2015:11. doi: 10.1371/journal.ppat.1005215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S., Kim S., Shin D.H., Kim M.-S. Inhibition of SARS-CoV 3CL protease by flavonoids. J. Enzyme Inhib. Med. Chem. 2020;35:145–151. doi: 10.1080/14756366.2019.1690480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilianski A., Baker S.C. Cell-based antiviral screening against coronaviruses: developing virus-specific and broad-spectrum inhibitors. Antiviral Res. 2014;101:105–112. doi: 10.1016/j.antiviral.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilianski A., Mielech A., Deng X., Baker S.C. Assessing activity and inhibition of MERS-CoV papain-like and 3C-like proteases using luciferase-based biosensors. J. Virol. 2013:02105–02113. doi: 10.1128/JVI.02105-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Lee J.-Y., Yang J.-S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181 doi: 10.1016/j.cell.2020.04.011. 914-921.e910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Jedrzejczak R., Maltseva N.I., Wilamowski M., Endres M., Godzik A., Michalska K., Joachimiak A. Crystal structure of Nsp15 endoribonuclease NendoU from SARS‐CoV‐2. Protein Sci. 2020 doi: 10.1002/pro.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Cottrell C.A., Wang N., Pallesen J., Yassine H.M., Turner H.L., Corbett K.S., Graham B.S., McLellan J.S., Ward A.B. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumperman J., Locker J.K., Meijer A., Horzinek M.C., Geuze H.J., Rottier P. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 1994;68:6523–6534. doi: 10.1128/jvi.68.10.6523-6534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L., Masters P.S. The small envelope protein E is not essential for murine coronavirus replication. J. Virol. 2003;77:4597–4608. doi: 10.1128/JVI.77.8.4597-4608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.-W., Wong B.H., Wong S.S., Leung S.-Y., Chan K.-H., Yuen K.-Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Porcine epidemic diarrhea virus: an emerging and re-emerging epizootic swine virus. Virol. J. 2015;12:193. doi: 10.1186/s12985-015-0421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J., Kusov Y., Hilgenfeld R. Nsp3 of coronaviruses: Structures and functions of a large multi-domain protein. Antiviral Res. 2018;149:58–74. doi: 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Zhu X., Ji X., Quanquin N., Deng Y.-Q., Tian M., Aliyari R., Zuo X., Yuan L., Afridi S.K. Chloroquine, a FDA-approved drug, prevents Zika virus infection and its associated congenital microcephaly in mice. EBioMedicine. 2017;24:189–194. doi: 10.1016/j.ebiom.2017.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, function, and evolution of coronavirus spike proteins. Ann. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Liu Y., Wei F., Shen M., Zhong Y., Li S., Chen L., Ma N., Liu B., Mao Y., Li N., Hou W., Xiong H., Yang Z. Antiviral activity of arbidol hydrochloride against herpes simplex virus I in vitro and in vivo. Int. J. Antimicrob. Agents. 2018;51:98–106. doi: 10.1016/j.ijantimicag.2017.09.001. [DOI] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K., Liu D. The missing link in coronavirus assembly retention of the avian coronavirus infectious bronchitis virus envelope protein in the pre-Golgi compartments and physical interaction between the envelope and membrane proteins. J. Biol. Chem. 2001;276:17515–17523. doi: 10.1074/jbc.M009731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.-Y., Liu C.-L., Chang Y.-M., Zhao J., Perlman S., Hou M.-H. Structural basis for the identification of the N-terminal domain of coronavirus nucleocapsid protein as an antiviral target. J. Med. Chem. 2014;57:2247–2257. doi: 10.1021/jm500089r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner H.A., Fotouhi-Ardakani N., Lytvyn V., Lachance P., Sulea T., Ménard R. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 2005;79:15199–15208. doi: 10.1128/JVI.79.24.15199-15208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Fung T.S., Chong K.K.-L., Shukla A., Hilgenfeld R. Accessory proteins of SARS-CoV and other coronaviruses. Antiviral Res. 2014;109:97–109. doi: 10.1016/j.antiviral.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Zhang B., Jin Z., Yang H., Rao Z. The Crytal Structure of 2019-nCoV Main Protease in Complex with an Inhibitor N3. RCSB Protein Data Bank; 2020. [Google Scholar]
- Lo Y.-S., Lin S.-Y., Wang S.-M., Wang C.-T., Chiu Y.-L., Huang T.-H., Hou M.-H. Oligomerization of the carboxyl terminal domain of the human coronavirus 229E nucleocapsid protein. FEBS Lett. 2013;587:120–127. doi: 10.1016/j.febslet.2012.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Z., Sun Y., Rao Z. Current progress in antiviral strategies. Trends Pharmacol. Sci. 2014;35:86–102. doi: 10.1016/j.tips.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Wang Q., Gao G.F. Bat-to-human: spike features determining ‘host jump’of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015;23:468–478. doi: 10.1016/j.tim.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H. Drug treatment options for the 2019-new coronavirus (2019-nCoV) Biosci. Trends. 2020 doi: 10.5582/bst.2020.01020. [DOI] [PubMed] [Google Scholar]
- Madelain V., Nguyen T.H.T., Olivo A., De Lamballerie X., Guedj J., Taburet A.-M., Mentré F. Ebola virus infection: review of the pharmacokinetic and pharmacodynamic properties of drugs considered for testing in human efficacy trials. Clin. Pharmacokinet. 2016;55:907–923. doi: 10.1007/s40262-015-0364-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin P., Jensen D. Ribavirin in the treatment of chronic hepatitis C. J. Gastroenterol. Hepatol. 2008;23:844–855. doi: 10.1111/j.1440-1746.2008.05398.x. [DOI] [PubMed] [Google Scholar]
- Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride R., Van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortola E., Roy P. Efficient assembly and release of SARS coronavirus‐like particles by a heterologous expression system. FEBS Lett. 2004;576:174–178. doi: 10.1016/j.febslet.2004.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mothay D., Ramesh K. Binding site analysis of potential protease inhibitors of COVID-19 using AutoDock. Virus Dis. 2020:1. doi: 10.1007/s13337-020-00585-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulangu S., Dodd L.E., Davey R.T., Jr., Tshiani Mbaya O., Proschan M., Mukadi D., Lusakibanza Manzo M., Nzolo D., Tshomba Oloma A., Ibanda A. A randomized, controlled trial of Ebola virus disease therapeutics. N. Engl. J. Med. 2019;381:2293–2303. doi: 10.1056/NEJMoa1910993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Maeda A., Maeda J., Makino S. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J. Virol. 2000;74:8127–8134. doi: 10.1128/jvi.74.17.8127-8134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narkhede R.R., Cheke R.S., Ambhore J.P., Shinde S.D. The molecular docking study of potential drug candidates showing anti-COVID-19 activity by exploring of therapeutic targets of SARS-CoV-2. Screening. 2020;5:8. [Google Scholar]
- Netland J., DeDiego M.L., Zhao J., Fett C., Álvarez E., Nieto-Torres J.L., Enjuanes L., Perlman S. Immunization with an attenuated severe acute respiratory syndrome coronavirus deleted in E protein protects against lethal respiratory disease. Virology. 2010;399:120–128. doi: 10.1016/j.virol.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Kiss G., Kunding A.H., Bhella D., Baksh M.F., Connelly S., Droese B., Klaus J.P., Makino S., Sawicki S.G. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 2011;174:11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Torres J.L., DeDiego M.L., Álvarez E., Jiménez-Guardeño J.M., Regla-Nava J.A., Llorente M., Kremer L., Shuo S., Enjuanes L. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology. 2011;415:69–82. doi: 10.1016/j.virol.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Torres J.L., DeDiego M.L., Verdia-Baguena C., Jimenez-Guardeno J.M., Regla-Nava J.A., Fernandez-Delgado R., Castano-Rodriguez C., Alcaraz A., Torres J., Aguilella V.M. Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog. 2014:10. doi: 10.1371/journal.ppat.1004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opstelten D.J., Raamsman M., Wolfs K., Horzinek M.C., Rottier P. Envelope glycoprotein interactions in coronavirus assembly. J. Cell Biol. 1995;131:339–349. doi: 10.1083/jcb.131.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortego J., Ceriani J.E., Patiño C., Plana J., Enjuanes L. Absence of E protein arrests transmissible gastroenteritis coronavirus maturation in the secretory pathway. Virology. 2007;368:296–308. doi: 10.1016/j.virol.2007.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortego J., Escors D., Laude H., Enjuanes L. Generation of a replication-competent, propagation-deficient virus vector based on the transmissible gastroenteritis coronavirus genome. J. Virol. 2002;76:11518–11529. doi: 10.1128/JVI.76.22.11518-11529.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1–12. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecheur E., Borisevich V., Halfmann P., Morrey J., Smee D., Prichard M., Mire C., Kawaoka Y., Geisbert T., SJ P. The synthetic antiviral drug arbidol inhibits globally prevalent pathogenic viruses. J. Virol. 2016;90:3086–3092. doi: 10.1128/JVI.02077-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrella A., Carannante N., Berretta M., Rinaldi M., Maturo N., Rinaldi L. Editorial–Novel Coronavirus 2019 (Sars-CoV2): a global emergency that needs new approaches. Eur. Rev. Med. Pharmacol. 2020;24:2162–2164. doi: 10.26355/eurrev_202002_20396. [DOI] [PubMed] [Google Scholar]
- Plantone D., Koudriavtseva T. Current and future use of chloroquine and hydroxychloroquine in infectious, immune, neoplastic, and neurological diseases: a mini-review. Clin. Drug Investig. 2018;38:653–671. doi: 10.1007/s40261-018-0656-y. [DOI] [PubMed] [Google Scholar]
- Prabakaran P., Gan J., Feng Y., Zhu Z., Choudhry V., Xiao X., Ji X., Dimitrov D.S. Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody. J. Biol. Chem. 2006;281:15829–15836. doi: 10.1074/jbc.M600697200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradesh U., Upadhayay P.D.D., Vigyan P.C. Coronavirus infection in equines: a review. Asian J. An. Vet. Adv. 2014;9:164–176. [Google Scholar]
- Prajapat M., Sarma P., Shekhar N., Avti P., Sinha S., Kaur H., Kumar S., Bhattacharyya A., Kumar H., Bansal S. Drug targets for corona virus: a systematic review. Indian J. Pharmacol. 2020;52:56. doi: 10.4103/ijp.IJP_115_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Z., Dominguez S.R., Holmes K.V. Role of the spike glycoprotein of human Middle East respiratory syndrome coronavirus (MERS-CoV) in virus entry and syncytia formation. PLoS One. 2013:8. doi: 10.1371/journal.pone.0076469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Registry C.C.T. Favipiravir Tablets in Novel Coronavirus Pneumonia (COVID-19) Patients Who are Still Positive on Virus Detection Under the Current Antiviral Therapy. 2020. A randomized controlled trial for the efficacy and safety of Baloxavir Marboxil. [Google Scholar]
- Regla-Nava J.A., Nieto-Torres J.L., Jimenez-Guardeño J.M., Fernandez-Delgado R., Fett C., Castaño-Rodríguez C., Perlman S., Enjuanes L., DeDiego M.L. Severe acute respiratory syndrome coronaviruses with mutations in the E protein are attenuated and promising vaccine candidates. J. Virol. 2015;89:3870–3887. doi: 10.1128/JVI.03566-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rest J.S., Mindell D.P. SARS associated coronavirus has a recombinant polymerase and coronaviruses have a history of host-shifting. Infect. Genet. Evol. 2003;3:219–225. doi: 10.1016/j.meegid.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh C. A facile inhibitor screening of SARS coronavirus N protein using nanoparticle-based RNA oligonucleotide. Int. J. Nanomed. 2012;7:2173. doi: 10.2147/IJN.S31379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenke K., Feldmann H., Westover J.B., Hanley P.W., Martellaro C., Feldmann F., Saturday G., Lovaglio J., Scott D.P., Furuta Y. Use of favipiravir to treat Lassa virus infection in macaques. Emerg. Infect. Dis. 2018;24:1696. doi: 10.3201/eid2409.180233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruch T.R., Machamer C.E. The hydrophobic domain of infectious bronchitis virus E protein alters the host secretory pathway and is important for release of infectious virus. J. Virol. 2011;85:675–685. doi: 10.1128/JVI.01570-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruch T.R., Machamer C.E. The coronavirus E protein: assembly and beyond. Viruses. 2012;4:363–382. doi: 10.3390/v4030363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvatori G., Luberto L., Maffei M., Aurisicchio L., Roscilli G., Palombo F., Marra E. SARS-CoV-2 SPIKE PROTEIN: an optimal immunological target for vaccines. J. Transl. Med. 2020;18:1–3. doi: 10.1186/s12967-020-02392-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senanayake S.L. Drug repurposing strategies for COVID-19. Fut. Sci. 2020 [Google Scholar]
- Shah B., Modi P., Sagar S.R. In silico studies on therapeutic agents for COVID-19: Drug repurposing approach. Life Sci. 2020 doi: 10.1016/j.lfs.2020.117652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P., Sims A.C., Graham R.L., Menachery V.D., Gralinski L.E., Case J.B., Leist S.R., Pyrc K., Feng J.Y., Trantcheva I. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017:9. doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu B., Gong P. Structural basis of viral RNA-dependent RNA polymerase catalysis and translocation. Proc. Natl. Acad. Sci. 2016;113:E4005–E4014. doi: 10.1073/pnas.1602591113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J. Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu Y., Teoh K., Lo J., Chan C., Kien F., Escriou N., Tsao S., Nicholls J., Altmeyer R., Peiris J. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J. Virol. 2008;82:11318–11330. doi: 10.1128/JVI.01052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H.C., Seo M.-Y., Stadler K., Yoo B.J., Choo Q.-L., Coates S.R., Uematsu Y., Harada T., Greer C.E., Polo J.M. Synthesis and characterization of a native, oligomeric form of recombinant severe acute respiratory syndrome coronavirus spike glycoprotein. J. Virol. 2004;78:10328–10335. doi: 10.1128/JVI.78.19.10328-10335.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterling T., Irwin J.J. ZINC 15–ligand discovery for everyone. J. Chem. Inf. Model. 2015;55:2324–2337. doi: 10.1021/acs.jcim.5b00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stobart C.C., Sexton N.R., Munjal H., Lu X., Molland K.L., Tomar S., Mesecar A.D., Denison M.R. Chimeric exchange of coronavirus nsp5 proteases (3CLpro) identifies common and divergent regulatory determinants of protease activity. J. Virol. 2013;87:12611–12618. doi: 10.1128/JVI.02050-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Sanderson P.E., Zheng W. Drug combination therapy increases successful drug repositioning. Drug Discov. Today. 2016;21:1189–1195. doi: 10.1016/j.drudis.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundquist W.I., Kräusslich H.-G. HIV-1 assembly, budding, and maturation. Cold Spring Harb. Perspect. Med. 2012;2 doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surya W., Samsó M., Torres J. In: Respiratory Disease and Infection: A New Insight. Vats M., editor. 2013. Structural and functional aspects of viroporins in human respiratory viruses: respiratory syncytial virus and coronaviruses By Wahyu Surya, Montserrat Samsó and Jaume Torres. [Google Scholar]
- Tchesnokov E.P., Feng J.Y., Porter D.P., Götte M. Mechanism of inhibition of Ebola virus RNA-dependent RNA polymerase by remdesivir. Viruses. 2019;11:326. doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Te H., Randall G., Jensen D. Mechanism of action of ribavirin in the treatment of chronic hepatitis C. Gastroenterol. Hepatol. 2007;3:218–225. [PMC free article] [PubMed] [Google Scholar]
- Te Velthuis A.J., van den Worm S.H., Snijder E.J. The SARS-coronavirus nsp7+ nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res. 2012;40:1737–1747. doi: 10.1093/nar/gkr893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teissier E., Zandomeneghi G., Loquet A., Lavillette D., Lavergne J., Montserret R., Cosset F., Bockmann A., Meier B., F P., Pecheur E. Mechanism of inhibition of enveloped virus membrane fusion by the antiviral drug arbidol. PLoS ONE. 2011;6:e15874. doi: 10.1371/journal.pone.0015874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooze J., Tooze S., Warren G. Replication of coronavirus MHV-A59 in sac-cells: determination of the first site of budding of progeny virions. Eur. J. Cell Biol. 1984;33:281–293. [PubMed] [Google Scholar]
- Tremblay C.L. Combating HIV resistance–focus on darunavir. Ther. Clin. Risk Manage. 2008;4:759. doi: 10.2147/tcrm.s1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trials, N.C., 2020. Efficacy and Safety of Hydroxychloroquine for Treatment of COVID-19.
- Trials, N.C., 2020. A Prospective/Retrospective, Randomized Controlled Clinical Study of Antiviral Therapy in the 2019-nCoV Pneumonia.
- Trials, N.C., 2020. A Trial of Remdesivir in Adults With Mild and Moderate COVID-19.
- ul Qamar M.T., Alqahtani S.M., Alamri M.A., Chen L.-L. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J. Pharm. Anal. 2020 doi: 10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabret A., Dina J., Gouarin S., Petitjean J., Tripey V., Brouard J., Freymuth F. Human (non‐severe acute respiratory syndrome) coronavirus infections in hospitalised children in France. J. Paediatr. Child Health. 2008;44:176–181. doi: 10.1111/j.1440-1754.2007.01246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatagopalan P., Daskalova S.M., Lopez L.A., Dolezal K.A., Hogue B.G. Coronavirus envelope (E) protein remains at the site of assembly. Virology. 2015;478:75–85. doi: 10.1016/j.virol.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennema H., Godeke G.-J., Rossen J., Voorhout W., Horzinek M., Opstelten D., Rottier P. Nucleocapsid‐independent assembly of coronavirus‐like particles by co‐expression of viral envelope protein genes. EMBO J. 1996;15:2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdiá-Báguena C., Nieto-Torres J.L., Alcaraz A., DeDiego M.L., Enjuanes L., Aguilella V.M. Analysis of SARS-CoV E protein ion channel activity by tuning the protein and lipid charge. Biochim. Biophys. Acta. 2013;1828:2026–2031. doi: 10.1016/j.bbamem.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdiá-Báguena C., Nieto-Torres J.L., Alcaraz A., DeDiego M.L., Torres J., Aguilella V.M., Enjuanes L. Coronavirus E protein forms ion channels with functionally and structurally-involved membrane lipids. Virology. 2012;432:485–494. doi: 10.1016/j.virol.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent M.J., Bergeron E., Benjannet S., Erickson B.R., Rollin P.E., Ksiazek T.G., Seidah N.G., Nichol S.T. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2005;2:69. doi: 10.1186/1743-422X-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Graham R., Baric R., Li F. An analysis based on decade-long structural studies of SARS 3, JVI Accepted Manuscript Posted Online 29 January 2020. J. Virol. 2020 doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Zheng X., Gai W., Zhao Y., Wang H., Wang H., Feng N., Chi H., Qiu B., Li N. MERS-CoV virus-like particles produced in insect cells induce specific humoural and cellular imminity in rhesus macaques. Oncotarget. 2017;8:12686. doi: 10.18632/oncotarget.8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.-Y. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181 doi: 10.1016/j.cell.2020.03.045. 894-904.e899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V., Siegel D., Perron M., Bannister R., Hui H.C. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westover J., Mathis A., Taylor R., Wandersee L., Bailey K., Sefing E., Hickerson B., Jung K., Sheridan W., Gowen B. Galidesivir limits Rift Valley fever virus infection and disease in Syrian golden hamsters. Antiviral Res. 2018;156:38–45. doi: 10.1016/j.antiviral.2018.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Huang Y., Yuen K.-Y. Coronavirus diversity, phylogeny and interspecies jumping. Exp. Biol. Med. 2009;234:1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- World Health Organization, W. 2020. Coronavirus disease (COVID-19) Situation Report- 204.https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200519-covid-19-sitrep-120.pdf?sfvrsn=515cabfb_2 (accessed August 11, 2020) [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Wu T., Liu Q., Yang Z. The SARS-CoV-2 outbreak: what we know. Int. J. Infect. Dis.Int. J. Infect. Dis. 2020;94:44–48. doi: 10.1016/j.ijid.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., Yin H., Xiao Q., Tang Y., Qu X. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020;5:434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Yan L., Xu W., Agrawal A.S., Algaissi A., Tseng C.-T.K., Wang Q., Du L., Tan W., Wilson I.A. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci. Adv. 2019;5:eaav4580. doi: 10.1126/sciadv.aav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., Zhong W., Hao P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci.e China Life Sci. 2020;63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Xie W., Xue X., Yang K., Ma J., Liang W., Zhao Q., Zhou Z., Pei D., Ziebuhr J. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005:3. doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Xiong Z., Zhang S., Yan Y., Nguyen J., Ng B., Lu H., Brendese J., Yang F., Wang H. Bcl-xL inhibits T-cell apoptosis induced by expression of SARS coronavirus E protein in the absence of growth factors. Biochem. J. 2005;392:135–143. doi: 10.1042/BJ20050698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Wu Q., Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr. Biol. 2020;30 doi: 10.1016/j.cub.2020.03.063. 1346-1351.e1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Zhang, C., Zheng, W., 2020. Study shows pangolins may have passed new coronavirus from bats to humans. (accessed April 23, 2020).
- Zheng J. SARS-CoV-2: an emerging coronavirus that causes a global threat. Int. J. Biol. Sci. 2020;16:1678. doi: 10.7150/ijbs.45053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W., Sun W., Simeonov A. Drug repurposing screens and synergistic drug‐combinations for infectious diseases. Br. J. Pharmacol. 2018;175:181–191. doi: 10.1111/bph.13895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:1–18. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Wang L., Fang C., Peng S., Zhang L., Chang G., Xia S., Zhou W. Clinical analysis of 10 neonates born to mothers with 2019-nCoV pneumonia. Transl. Pediatr. 2020;9:51. doi: 10.21037/tp.2020.02.06. [DOI] [PMC free article] [PubMed] [Google Scholar]