Abstract
N6-methyladenosine (m6A) regulators are involved in the progression of various cancers via regulating m6A modification. However, the potential role and mechanism of the m6A modification in osteosarcoma remains obscure. In this study, WTAP was found to be highly expressed in osteosarcoma tissue and it was an independent prognostic factor for overall survival in osteosarcoma. Functionally, WTAP, as an oncogene, was involved in the proliferation and metastasis of osteosarcoma in vitro and vivo. Mechanistically, M6A dot blot, RNA-seq and MeRIP-seq, MeRIP-qRT-PCR and luciferase reporter assays showed that HMBOX1 was identified as the target gene of WTAP, which regulated HMBOX1 stability depending on m6A modification at the 3′UTR of HMBOX1 mRNA. In addition, HMBOX1 expression was downregulated in osteosarcoma and was an independent prognostic factor for overall survival in osteosarcoma patients. Silenced HMBOX1 evidently attenuated shWTAP-mediated suppression on osteosarcoma growth and metastasis in vivo and vitro. Finally, WTAP/HMBOX1 regulated osteosarcoma growth and metastasis via PI3K/AKT pathway. In conclusion, this study demonstrated the critical role of the WTAP-mediated m6A modification in the progression of osteosarcoma, which could provide novel insights into osteosarcoma treatment.
Subject terms: Outcomes research, Oncogenesis
Introduction
Osteosarcoma is a primary malignant bone tumor that is common among childhood and adolescents worldwide1. Despite the improvements including multi-agent chemotherapy with surgery in recent years, the 5-year survival rate is ~70% for localized osteosarcoma and is ~30% for recurrent and metastatic osteosarcoma2,3. Therefore, a better understanding of molecular mechanism is urgent for developing novel therapeutic strategies for osteosarcoma.
N6-methyladenosine (m6A) is the prominent dynamic mRNA modification, which is involved in various biological process by regulating mRNA translocation, translation, and stability4. It is catalyzed by m6A writer (methyltransferase), removed by erasers (RNA demethylases) and recognized by m6A readers, involving in various biological progression5–7. The formation of m6A is catalyzed by a methyltransferase, METTL3 and METTL14 form a core catalytic complex of methyltransferases that is stabilized by WTAP8. Recently, METTL169,10, METTL5, ZCCHC411, and Zc3h1312 were showed to play a critical role in compositing methyltransferase complex and facilitating m6A methylation13. The eraser ALKBH5 and FTO has m6A demethylation activity to remove the m6A modification. The reader proteins are from YTH family, heterogeneous nuclear ribonucleoprotein (HNRP) family and insulin-like growth factor 2 mRNA-binding protein family, they recognize the m6A modification to adjust RNA metabolisms14.
Dynamic and reversible m6A regulation was reported to be involved in various physiological and pathological processes including stem cell differentiation, cardiac homeostasis, adipogenesis, neuronal disorders, and spermatogenesis15–19. Recent years, compelling evidence has revealed that m6A modification plays an important role in the tumorigenesis of various cancers20–24. For example, METTL3 was reported to play key role in the progression of colorectal carcinoma25, bladder cancer26,27, gastric cancer28, and pancreatic cancer29. YTHDF2 was involved the progression of Acute myeloid leukemia (AML) via regulating hematopoietic stem cells (HSCs) activity4. FTO was also reported to play a crucial role in the progression of melanoma30, breast cancer31, gastric cancer32, and intrahepatic cholangiocarcinoma. Nevertheless, the role of m6A modification in osteosarcoma is still poorly characterized.
WTAP, a Wilms’ tumor 1 (WT1) associated protein33, has been reported to be critical in the biological processes including G2/M transition and pre-mRNA splicing34–36. In addition, accumulated studies identified the important role of WTAP in the progression of various cancers37. For example, WTAP function as oncogene in cholangiocarcinoma38, acute myeloid leukemia39, colon cancer40, ovarian cancer41, and diffuse large B-cell lymphoma42. Recent studies reported that WTAP is strictly connected to a functional m6A methylation complex43. Here, we revealed the increased expression of WTAP in osteosarcoma tissue, which was associated with clinicopathological features and poor prognosis in osteosarcoma patients. WTAP function as an oncogenic gene and it promotes the m6A modification and progression of osteosarcoma. Mechanistically, WTAP induced the growth and metastasis of osteosarcoma via downregulating HMBOX1 expression in m6A-dependent manner. Further investigations demonstrated that WTAP/HMBOX1 regulated osteosarcoma growth and metastasis via PI3K/AKT pathway. Overall, these results imply that WTAP/HMBOX1 may be an important mechanism of osteosarcoma progression and serve as a novel therapeutically target of osteosarcoma.
Materials and methods
Clinical samples and ethics
One-hundred-and-four paired fresh osteosarcoma (40 metastatic OS sample and 64 non-metastatic OS sample) and normal tissues were obtained from patients without radiotherapy and/or chemotherapy at The Third Xiangya Hospital of Central South University and then immediately frozen in −80 °C for RNA and protein extraction, or formalin-fixed and paraffin-embedded for further used. Informed consent was obtained from each patient or their guardians, and the study were approved by the Ethics Committee of The Third Xiangya Hospital of Central South University.
Cell culture, transfection, and infection
The human hFOB1.19 cells and osteosarcoma cell lines (SJSA-1, MG-63, HOS, U2OS, and 143B) were obtained from the cell bank of the Chinese Academy of Sciences (Shanghai, China). The osteosarcoma cells were cultured in DMEM with 10% FBS (Gibco, Grand Island, NY, USA). The hFOB1.19 cells were cultured in DMEM/F-12 (DF-12) with 10% FBS.
The sequences of shRNA cloned into PLKO.1 vector, and then shWTAP-PLKO.1 or shHMBOX1-PLKO.1 were co-transfected with packing and PAX2 plasmids into 293TF cells. Forty-eight hours after transfection, the lentivirus was collected and infected HOS and U2OS cells for 24 h. The 1 μg/ml puromycin was used for screening infected cells. The primers were listed in Table S1.
Western blot
The proteins were obtained from cells and tissues being lysed with RIPA buffer and then separated by 10% SDS-PAGE. The anti-WTAP (ab195380, Abcam), anti-HMBOX1 (16123-1-AP, Proteintech), anti-YTHDF2 (ab220163, Abcam), anti-PI3K (ab191606, Abcam), anti-pPI3K (ab182651, Abcam), Akt (9272S, Cell Signaling), p-Akt (Ser473) (4051S, Cell Signaling), and anti-GAPDH (ab181602, Abcam) were used for PVDF membrane incubating overnight at 4 °C. The protein expression was detected by incubating with anti-rabbit secondary antibodies.
RNA-seq analysis and qRT-PCR
Total RNAs were extracted from osteosarcoma tissue and cells using Trizol (ThermoFisher, USA). For RNA-seq analysis, the library construction and Illumina sequencing using the Illumina TruSeq RNA Sample Prep Kit (San Diego, CA, USA). For qPCR validation, the cDNA was obtained using First Strand cDNA Synthesis Kit (TOYOBO). Finally, the mRNA expression was detected using SYBR GREEN (Bio-Rad, California, USA). All primers were in Table S1.
RNA m6A dot blot assay and RNA m6A quantification
Total RNAs were extracted from osteosarcoma cells by TRIzol (ThermoFisher, USA), and then NanoDrop3000 was used for detecting RNA quality. The m6A content was detected by EpiQuik m6A RNA Methylation Quantification Kit (Colorimetric, Epigentek, USA).
The poly(A) RNAs (600, 300, and 150 ng) were spotted onto a nylon membrane (GE Healthcare) for RNA m6A dot blot assay, and then incubated with m6A antibody (no. ABE572, Merck Millipore, USA) at 4 °C overnight. The m6A dots was analyzed using an imaging system (Bio-Rad, USA).
MeRIP-seq and MeRIP-qRT-PCR
Total RNAs were extracted from osteosarcoma cells by TRizol. The DNA-free fragmented RNAs were incubated with magnetic Dynabeads bounded anti-m6A antibody (Abcam, USA) to enrichment the mRNA with m6A. And then, beads were treated with Proteinase K and RNA was extracted for MeRIP-seq or validation by qRT-PCR. The primers are in Table S1.
Cell proliferation assays
The CCK-8 (Cell Counting Kit-8, C0038, Beyotime Biotechnology, China) was used for cell proliferation ability44.
Colony formation assays
The cells were seeded into 6-well plates with a density 2 × 103 cells per wells and cultured in DMEM medium for 2 weeks. And then, 4% paraformaldehyde (PFA) was used to fix the colonies and crystal violet was used to stain as previous described45.
Wound-healing assay
Osteosarcoma cells were cultured for 48 h to reach 80% confluency, and then a straight artificial wound was scraped with a 200 μl pipette tip. The cell migration ability was measured by photographing the distance at 0 and 24 h45.
Transwell invasion assays
The transwell chamber (BD Science, Bedford, MA, USA) was used for invasion assays as previously described1. In brief, 2 × 105 osteosarcoma cells were seeded into the upper chambers and incubated for 24 h. The invasive cells were counted and quantified for cell invasion as previous described45.
Luciferase reporter assays
The 3′UTR of HMBOX1 was cloned into pmiGLO vector (Promega, USA). The putative m6A sites bases (A) were mutated into bases (C) in 3′UTR using Site-Directed Mutagenesis Kit (Thermo, USA). The WT and Mut plasmids were transfected in osteosarcoma cells, and then the Dual-Luciferase Assay Kit (Promega) was used for detecting the luciferase activity44.
Animal experiments
Nude mice (4 weeks, male) were used for tumor model. All animal care and handling procedures were approved by the Institutional Animal Care and Use Committee of The Third Xiangya Hospital of Central South University, Changsha, China. For the subcutaneous tumor model, 1 × 106 shNC, shWTAP or shHMBOX1 or shWTAP/shHMBOX1 U2OS cells seeded into mice via subcutaneous injection. Tumor volume and tumor weight were measured to analyze tumor growth as previous described46. For orthotopic xenograft tumor model, shNC, shWTAP, shHMBOX1, or shWTAP/shHMBOX1 U2OS cells were labeled with a luminescent dye and GFP, and injected into the cavity of the tibia of mice. Thirty days after injection, tumor growth was detected. For the metastasis model, the cells were injected into the tail vein, and the lung metastasis were detected 30 days after injection using in vivo bioluminescence imaging system.
Immunohistochemistry
Immunohistochemistry (IHC) analysis was performed using anti-WTAP (ab195380, Abcam), anti-HMBOX1 (16123-1-AP, Proteintech) as pervious described26.
Bioinformatics analysis
The GEO dataset GSE87624 and GSE46705 were downloaded and analyzed by R (version 3.5.3, https://www.r-project.org/). The different expressed gene was calculated with edgeR package and identified by the threshold criteria of log2 Fold-change (FC) ≥ 1.75 and adj.p < 0.05. GO and KEGG analysis was performed to investigate the potential role of genes. Protein–protein interaction (PPI) network was analyzed by the STRING database.
Statistical analysis
The SPSS 22.0 (SPSS, Inc., Chicago, IL, USA) was used for the statistical analyses, using ANOVA or Student’s t-test in this study. Kaplan–Meier analysis was used for survival. The correlation between WTAP and HMBOX1 were analyzed using Pearson analysis. Statistical significance was defined p < 0.05.
Results
Elevated WTAP is associated with poor prognosis of osteosarcoma patients
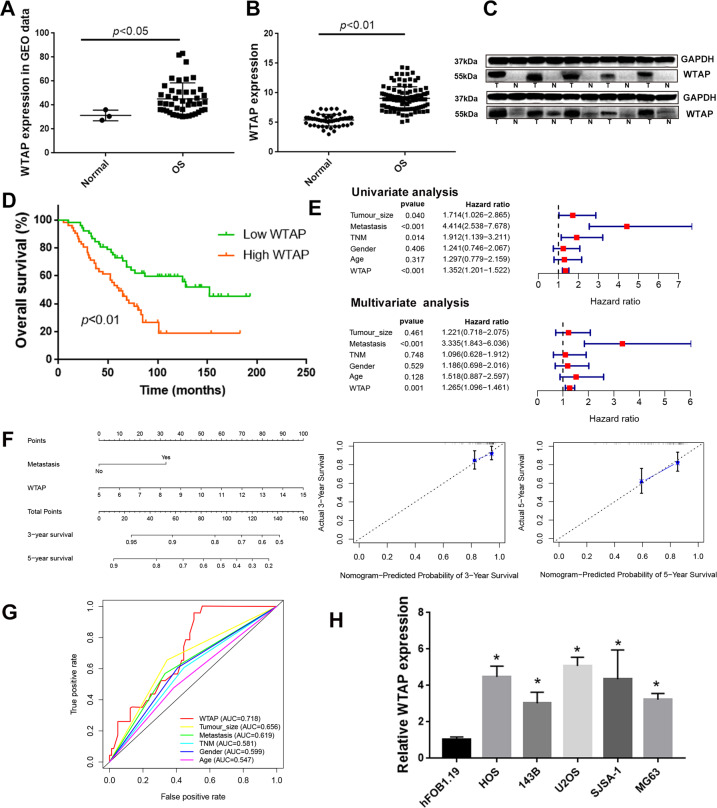
To reveal the important role of m6A modification in osteosarcoma, we explored the expression levels of 17 m6A-relative genes in osteosarcoma tissue and normal bone tissue (GSE87624: 3 normal and 44 osteosarcoma). As shown in Figs. 1a and S1A, we examined the expression of 17 m6A-related genes in GEO database, and found that WTAP, RBM15, YTHDF1, and YTHDF2 were differently expressed in tumor tissue compared with control tissue. The m6A writers were reported to play key role on the progression of various cancers, so WTAP was selected for further analysis. Next, we detected the expression of WTAP in 104 pairs of osteosarcoma tissue and the corresponding para-tumor tissues from The Third Xiangya Hospital of Central South University (TXHCSU). Consistent with the GEO results, the significant higher mRNA and protein levels of WTAP in osteosarcoma was detected using the qPCR and WB analysis (Fig. 1b, c). Moreover, the osteosarcoma patients with high WTAP showed poor overall survival in TXHCSU (Fig. 1d). And the univariate and multivariate cox analysis showed that WTAP and metastasis were the independent prognostic factor for overall survival in osteosarcoma patients (Fig. 1e). The prognostic nomogram was used to predict the probabilities of overall survival rates of osteosarcoma patients, and the calibration curves showed has a good consistency between the prediction and the actual observation (Fig. 1f). In addition, the 3 years ROC curve (AUC) of WTAP in osteosarcoma patients from TXHCSU was 0.718 (Fig. 1g). Besides, the overexpression of WTAP was associated with tumor size, metastasis, and TNM stage (Table 1 and Fig. S1B). Finally, the higher expression of WTAP was also detected in osteosarcoma cell lines (SJSA-1, MG-63, HOS, U2OS, and 143B) compared with that in the human osteoblast (hFOB1.19) cell (Fig. 1h). Collectively, these results demonstrated that WTAP is highly expressed in osteosarcoma and is correlated with its poor prognosis.
Fig. 1. The expression levels of WTAP in osteosarcoma tissue and cell lines.
a WTAP expression in osteosarcoma tissue compared with normal bone tissue from GSE87624. b qPCR assay and c western blot assay revealed the WTAP expression in osteosarcoma tissue and the corresponding para-tumor tissues from TXHCSU. d The Kaplan–Meier survival analysis. e Univariate and multivariate survival analysis. f Establishment of the overall survival nomogram for osteosarcoma patients. g Time-dependent ROC analysis. h The mRNA expression levels of WTAP in osteosarcoma cell lines and hFOB1.19 cell line using qPCR.
Table 1.
The association of WTAP expression and clinicopathologic characteristics of osteosarcoma patients.
| Characteristics | Case number | WTAP expression | P value | |
|---|---|---|---|---|
| High (n = 52) | Low (n = 52) | |||
| Gender | p > 0.05 | |||
| Male | 48 | 21 | 27 | |
| Female | 56 | 31 | 25 | |
| Age | p > 0.05 | |||
| ≤20 | 60 | 29 | 31 | |
| >20 | 44 | 23 | 21 | |
| Tumor size | p < 0.01 | |||
| ≤8 cm | 60 | 17 | 42 | |
| >8 cm | 44 | 33 | 10 | |
| Metastasis | p < 0.01 | |||
| Yes | 40 | 30 | 10 | |
| No | 64 | 22 | 42 | |
| TNM | p = 0.015 | |||
| I/II | 55 | 21 | 33 | |
| III/IV | 49 | 31 | 19 | |
Silenced WTAP significantly represses osteosarcoma progression in vitro
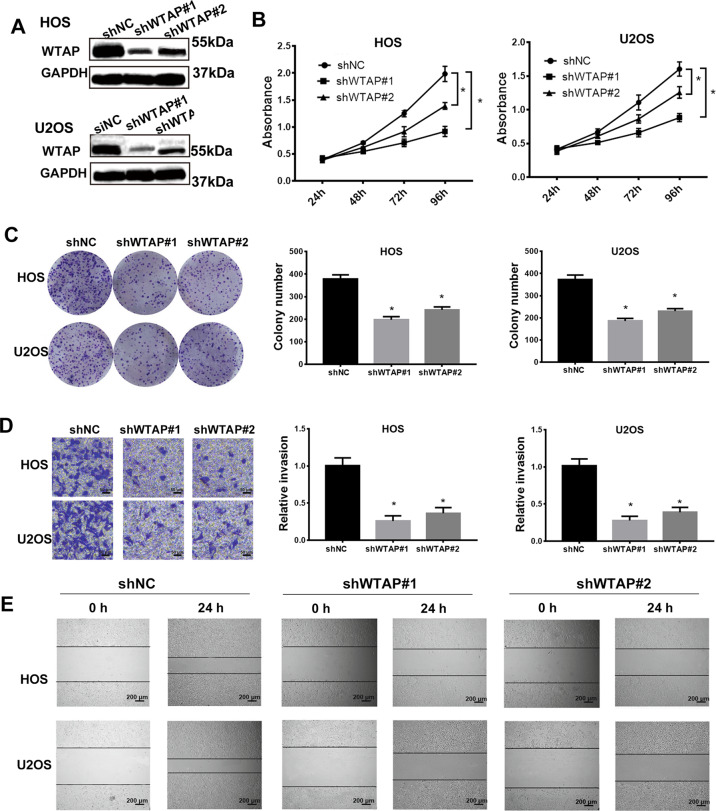
To further clarify the role of WTAP in osteosarcoma, we next used shRNA lentivirus (shWTAP#1 and shWTAP#2) to generate stable WTAP-knockdown osteosarcoma cells and analyzed the role of WTAP on cells migration, invasion and proliferation of osteosarcoma. The shWTAP#1 and shWTAP#2 significantly knockdown the expression levels of WTAP in osteosarcoma cells (Fig. 2a). The CCK-8 assay and colony formation assay showed that WTAP deficiency significantly reduced proliferative capacity of osteosarcoma cells (Fig. 2b, c). The transwell invasion and wound-healing assays showed that the invasion and migration ability of the osteosarcoma were significant reduced by silenced WTAP (Fig. 2d, e). These results determine that WTAP acts as an oncogene that promoted cell proliferation, migration, and invasion in osteosarcoma.
Fig. 2. Silenced WTAP significantly repressed osteosarcoma progression in vitro.
a The shWTAP#1 and shWTAP#2 downregulated WTAP expression. b CCK-8 assay revealed that silenced WTAP reduced the cell proliferation ability in osteosarcoma. c Colony formation assay showed that silenced WTAP decreased the cloning number of osteosarcoma cells. d Transwell invasion and e wound-healing assay revealed the inhibition of silenced WTAP on osteosarcoma cell invasion and migration.
HMBOX1 is potential target of WTAP in osteosarcoma
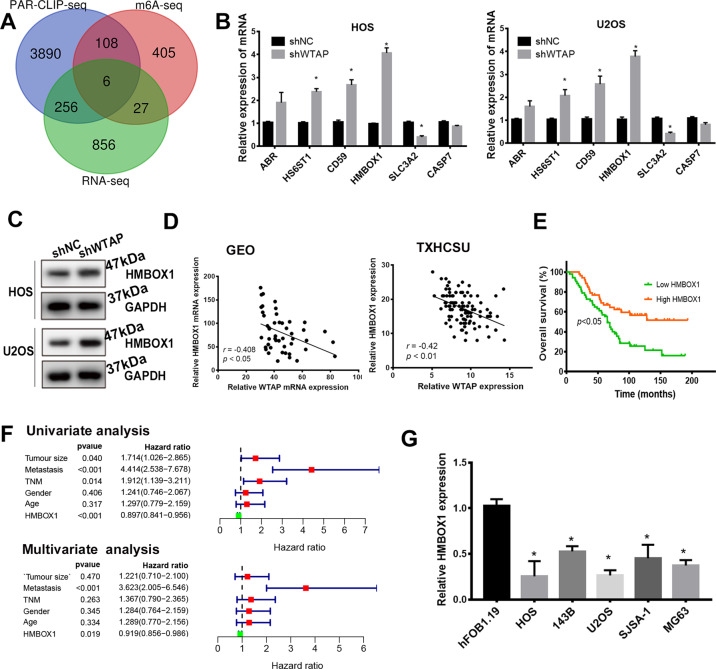
To reveal the potential mechanism by which WTAP regulates the progression of osteosarcoma, we performed RNA-seq, m6A-seq in shWTAP/sh-NC U2OS cell and CLIP-seq to reveal the potential genes regulated by WTAP-mediated m6A modification. The RNA-seq results revealed 521 downregulated DEGs and 624 upregulated DEGs with |logFC| > 1.75 and adj.p < 0.05 in shWTAP group compared with sh-NC group (Fig. S2A). Gene ontology (GO) showed that DEGs were enriched in neutrophil activation, cell cycle, and so on (Fig. S2B). KEGG analysis showed that DEGs were enriched in metabolism-related signal pathway (Fig. S2C). The m6A-seq analysis identified 546 differentially m6A modification genes in WTAP-silenced U2OS cell compared with normal U2OS cells (Table S2). The CLIP-seq (from GSE46705) revealed 4260 WTAP-targeted RNA in WTAP overexpressed cell using PAR-CLIP technology. And then, we obtained the overlapped genes in RNA-seq, m6A-seq, and CLIP-seq as shown in Fig. 3a, 6 genes were overlapped in three groups, including two upregulated gene (SLC3A2, CASP7) and four downregulated gene (ABR, HS6ST1, CD59, and HMBOX1). Consistent with these results, SLC3A2 and CASP7 were significantly upregulated and ABR, HS6ST1, CD59, and HMBOX1 were significantly downregulated in osteosarcoma tissues from GEO (GSE87624) database (Fig. S3A). These results were also confirmed in the osteosarcoma tissues from our TXHCSU data (Fig. S3B). And then, we evaluated the regulation of WTAP on the six candidates in osteosarcoma cells using qPCR. Among which, HMBOX1 was the most significantly upregulated gene in shWTAP osteosarcoma cells (Fig. 3b) and was selected for further analysis. Consistent with the mRNA expression, the protein level of HMBOX1 was also remarkably increased by silenced WTAP (Fig. 3c). We next evaluated the relationship between WTAP and HMBOX1 expression in GSE87624 and TXHCSU. As our speculation, HMBOX1 expression was negatively correlated with WTAP expression (r = −0.408 in GSE87624 and r = −0.42 in TXHCSU) (Fig. 3d). Moreover, we analyzed the relationship between HMBOX1 expression clinicopathological features in 104 osteosarcoma patients from TXHCSU. HMBOX1 expression was significantly reduced with tumor size and metastasis (Table 2 and Fig. S3C). And the survival analysis results showed that the patients with low level HMBOX1 had poor overall survival in osteosarcoma (Fig. 3e). Moreover, the univariate and multivariate analysis demonstrated HMBOX1 as an independent prognostic factor for overall survival (p = 0.019) in osteosarcoma patients (Fig. 3f). We also revealed the lower expression of HMBOX1 in osteosarcoma cell lines (SJSA-1, MG-63, HOS, U2OS, and 143B) than that in the human osteoblast (hFOB1.19) cell line (Fig. 3g). In conclusion, these results indicated that HMBOX1 is a potential target of WTAP and is related to poor prognosis of osteosarcoma patients.
Fig. 3. HMBOX1 is a potential target of WTAP.
a The Venn diagram was generated from differentially expressed genes in RNA-seq, m6A-seq, and CLIP-seq in GSE46705. The expression of the overlapped genes in HOS and U2OS cells with silenced WTAP using (b) qPCR assay and (c) western blot assay. d Correlation analysis of WTAP and HMBOX1 in osteosarcoma from GSE87624 and TXHCSU. e The Kaplan–Meier survival analysis. f Univariate and multivariate survival analyses. g WTAP HMBOX1 expression in osteosarcoma cell lines and human osteoblast (hFOB1.19) cell line.
Table 2.
The association of HMBOX1 expression and clinicopathologic characteristics of osteosarcoma patients.
| Characteristics | Case number | HMBOX1 expression | P value | |
|---|---|---|---|---|
| High (n = 52) | Low (n = 52) | |||
| Gender | p > 0.05 | |||
| Male | 48 | 25 | 23 | |
| Female | 56 | 27 | 29 | |
| Age | p > 0.05 | |||
| ≤20 | 60 | 27 | 33 | |
| >20 | 44 | 25 | 19 | |
| Tumor size | p > 0.05 | |||
| ≤8 cm | 60 | 34 | 26 | |
| >8 cm | 44 | 18 | 26 | |
| Metastasis | p < 0.01 | |||
| Yes | 40 | 11 | 29 | |
| No | 64 | 41 | 23 | |
| TNM | p > 0.05 | |||
| I/II | 55 | 29 | 26 | |
| III/IV | 49 | 23 | 26 | |
WTAP regulates HMBOX1 expression via m6A modification in osteosarcoma
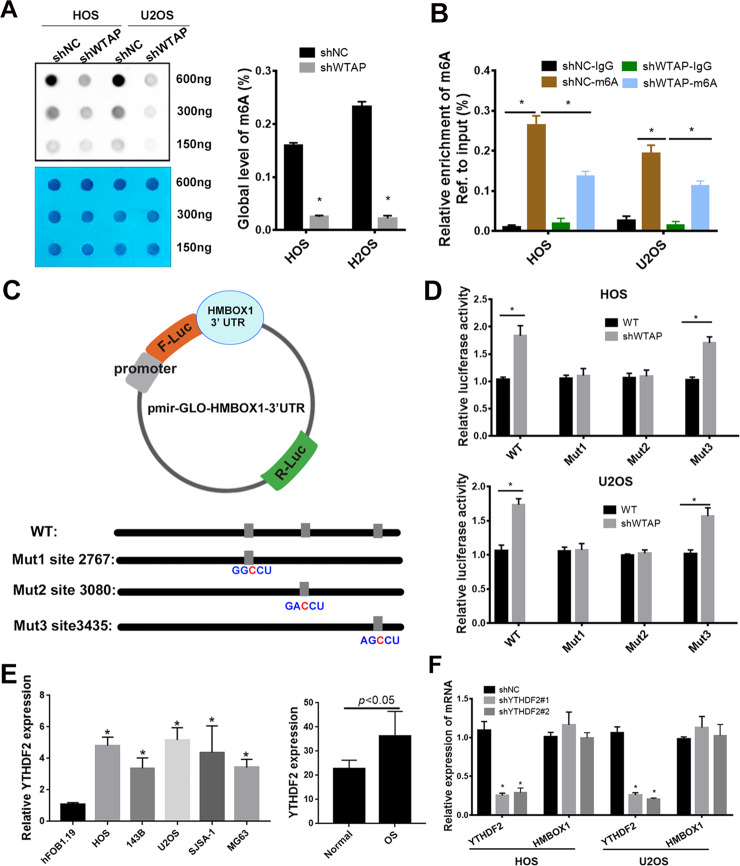
As MeRIP-seq data revealed different m6A modification of HMBOX1 in NC and WTAP-silenced cells, we next analyzed whether WTAP regulated the HMBOX1 expression in an m6A-dependent manner using m6A dot blot and RNA methylation quantification assay. As expected, m6A levels were substantially decreased in WTAP-knockdown osteosarcoma cells compared control osteosarcoma cells (Fig. 4a). Moreover, MeRIP-qPCR assay showed that HMBOX1 was effectively enriched by m6A-specific antibody, and enriched HMBOX1 was remarkably decreased by in WTAP-knockdown cells (Fig. 4b). Therefore, we supposed that WTAP could regulate m6A levels of HMBOX1. According to the m6A RNA-seq result, the m6A modification was at the 3′UTR of HMBOX1, and the SRAMP (http://www.cuilab.cn/sramp) predicted three very high confidence m6A sites at the 3′UTR of HMBOX1 (Fig. S4). To further prove the directed target role of WTAP on HMBOX1 with m6A modification, we cloned the HMBOX1 3′UTR containing these 3 m6A sites into pmirGLO vectors, and then we mutant the bases (A) into bases (C) in the predicted m6A sites (Fig. 3f). In Fig. 3g, the luciferase activity of HMBOX1 was significantly increased by silenced WTAP, however, the luciferase activity of Mut HMBOX1 did not affected by silenced WTAP in both HOS and U2OS cells (Fig. 4c).
Fig. 4. HMBOX1 is negative correlation WTAP expression and is associated to the poor prognosis of osteosarcoma.
a The m6A level in HOS and U2OS cells with silenced WTAP. b MeRIP-qPCR assay followed by qRT-PCR revealed the HMBOX1 m6A modification. c Wild-type or m6A site mutant HMBOX1 were cloned in pmirGLO. d Luciferase reporter assays revealed the target role of WTAP on the 3′UTR of HMBOX1. e YTHDF2 expression in osteosarcoma tissue and cells. f The effects of silenced YTHDF2 on HMBOX1 expression in osteosarcoma cells.
YTHDF2 is a well know m6A reader and plays an important role in the progression of several cancers via regulating mRNA degradation. Figure 1S showed that YTHDF2 was upregulated in osteosarcoma tissues in GSE87624. In Fig. 4e, YTHDF2 evidently upregulated in osteosarcoma tissue and cells. We next shed light on the expression of YTHDF2 in osteosarcoma and the role of YTHDF2 on HMBOX1 expression in osteosarcoma. Disappointedly, silenced YTHDF2 showed no effects on HMBOX1 expression in osteosarcoma cells (Fig. 4f), suggesting that WTAP regulated m6A-mediated HMBOX1 expression in YTHDF2-independent manner.
Together, these data suggested that WTAP-repressed HMBOX1 expression via regulating m6A modification of HMBOX1 at its 3′UTR.
HMBOX1 is involved in WTAP-mediated osteosarcoma proliferation and metastasis in vitro
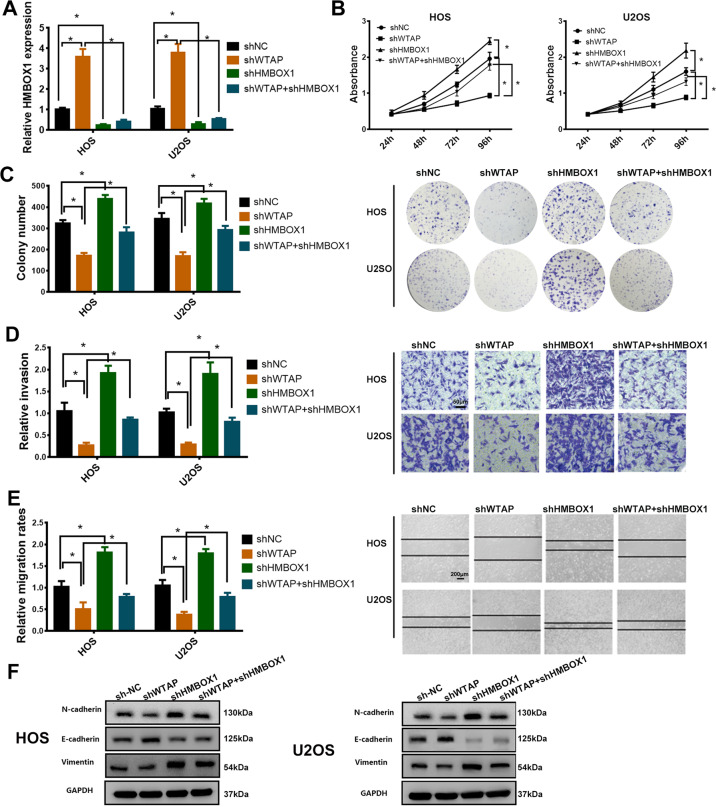
We next explored whether WTAP promoted osteosarcoma progression by regulating HMBOX1 expression. As shown in Fig. 5a, HMBOX1 expression was evidently increased by WTAP knockdown and was decreased by HMBOX1 knockdown in osteosarcoma cells. The CCK-8 results showed that the proliferation levels were increased by shHMBOX1 in WTAP-silenced HOS and U2OS cells (Fig. 5b). Consistent with the CCK-8 results, silenced HMBOX1 also alleviated shWTAP-mediated repression of cell proliferation in colony formation assay (Fig. 5c). The similar effects of HMBOX1 were also observed in WTAP-silenced osteosarcoma cell using wound-healing and transwell invasion assays (Fig. 5d and e). In addition, western blot results showed that silenced WTAP evidently repressed the expression of mesenchymal markers (cadherin and vimentin) and induced the expression of epithelial marker E-cadherin which was attenuated by shHMBOX1 in osteosarcoma cells (Fig. 5f). These data suggested WTAP promotes osteosarcoma cells proliferation and metastasis via repressing HMBOX1 expression.
Fig. 5. HMBOX1 as a tumor suppressor was involved in WTAP-mediated progression.
a HMBOX1 expression in osteosarcoma cell with shWTAP and shHMBOX1. Knockdown of HMBOX1 effectively alleviated WTAP-dependent b cell proliferation, c colony formation, d transwell invasion, and e wound-healing assay. f The protein expression level of EMT transition related protein. *p < 0.05.
HMBOX1 inhibits osteosarcoma growth and metastasis in vivo
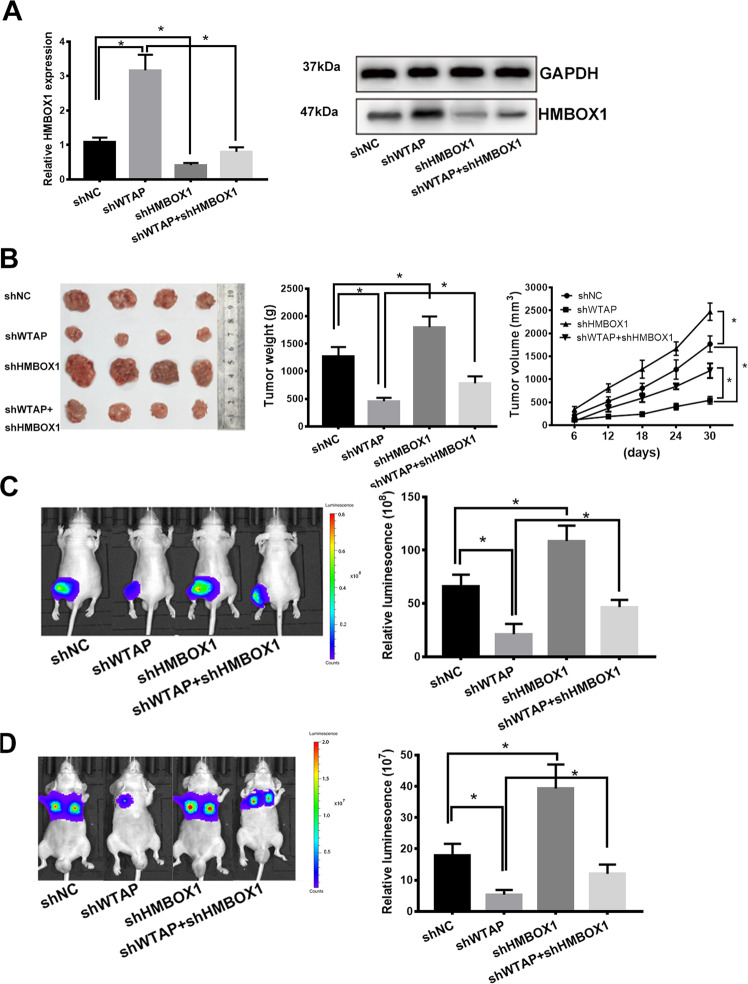
We next verified the role of HMBOX1 in vivo by injecting shNC, shWTAP, shHMBOX1, and shWTAP/shHMBOX1 U2OS cells to induce subcutaneous osteosarcoma mice model, orthotopic xenograft tumor model, and tail vein metastasis model. The expression levels of HMBOX1 was significantly upregulated by silenced WTAP in subcutaneous osteosarcoma tissue (Figs. 6a and S5 and S6). Moreover, silenced WTAP repressed the tumor size and tumor weight in subcutaneous osteosarcoma mice, which was rescued by silenced HMBOX1 (Fig. 6b). We next used a luminescent dye and GFP labeled U2OS cells to build an orthotopic xenograft tumor model. The bioluminescence imaging showed that the WTAP knockdown reduced the proliferation of U2OS cells in situ, while silenced HMBOX1 alleviated this repression (Fig. 6c). To further detect the role of WTAP and HMBOX1 on the metastatic ability of osteosarcoma in vivo, U2OS cells were injected into nude mice via the tail vein. The bioluminescence imaging showed that silenced HMBOX1 alleviated the repression of silenced WTAP on osteosarcoma metastasis (Fig. 6d). Taken together, these results suggest that HMBOX1 is involved in WTAP-mediated tumor growth and metastasis of osteosarcoma.
Fig. 6. silenced HMBOX1 attenuated shWTAP-repressed osteosarcoma growth and metastasis in vivo.
a The expression levels of HMBOX1 in osteosarcoma mice models. b Knockdown of HMBOX1 effectively alleviated shWTAP-repressed osteosarcoma growth in mice. c The orthotopic xenograft tumor and d lung metastasis were detected using a vivo bioluminescence imaging system. Representative images and a histogram are shown (n = 8 each group).
WTAP/HMBOX1 regulates the proliferation and metastasis of osteosarcoma partly by PI3K/AKT pathway
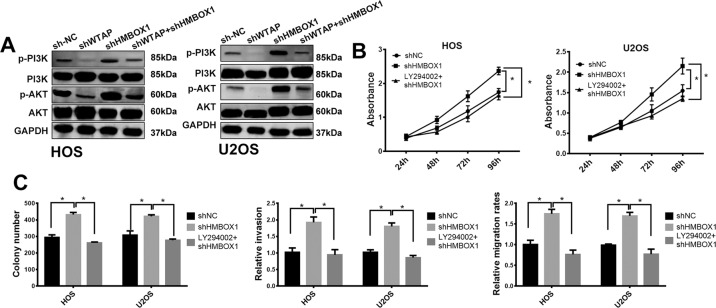
The KEGG results identified that PI3K/AKT pathways could be regulated by WTAP (Fig. S2B). PI3K/AKT signaling pathway promotes the growth, migration, and invasion of various cancers including osteosarcoma47,48. We hypothesized that WTAP/HMBOX1 was involved in the progression of osteosarcoma via regulating PI3K/AKT signaling pathway. As shown in Fig. 7a, phospho-PI3K and phospho-AKT were evidently repressed by shWTAP and induced by shHMBOX1, and shHMBOX1 significantly attenuated shWTAP-repressed phospho-PI3K and phospho-AKT in both HOS and U2OS cells. LY294002, a PI3K/AKT pathways inhibitor, remarkably reduced shHMBOX1-induced cell proliferation, migration, and invasion in both HOS and U2OS cells (Fig. 7b, c). Therefore, these results imply that WTAP/HMBOX1 regulates the proliferation and metastasis of osteosarcoma partly via PI3K/AKT pathway (Fig. 8).
Fig. 7. WTAP/ HMBOX1 was involved the progression of osteosarcoma via PI3K/AKT pathway.
a Western blot analysis for the expression of pPI3K, PI3K, pAKT, and AKT in osteosarcoma cells. b Cell proliferation, c colony formation, transwell invasion, and wound-healing assay of osteosarcoma cell treated with 5 μM Akt inhibitor LY294002 (Abcam, ab120243) for 12 h.
Fig. 8.
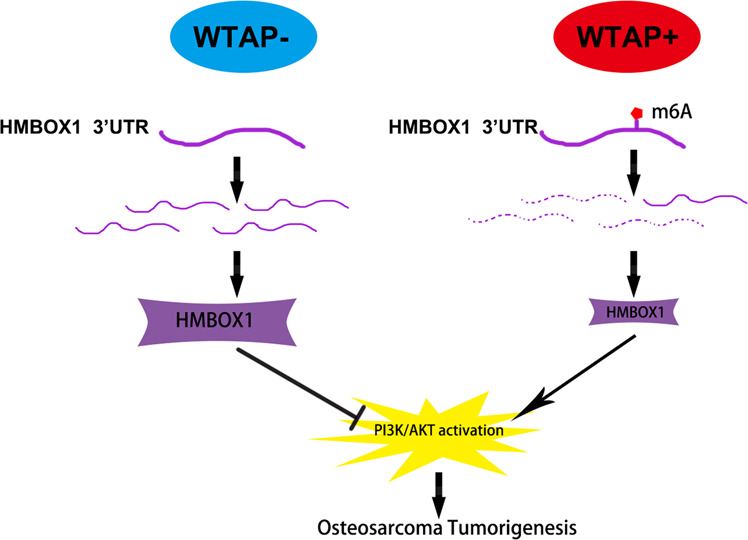
A schematic model illustrating WTAP-mediated m6A regulation in osteosarcoma.
Discussion
In the past several years, m6A modification is considered as a pervasive internal modification of mRNA and plays critical roles in the progression of a variety of human diseases including cancers20. However, the underlying involvement of m6A regulators in osteosarcoma progression is still unclear. In the present study, we focused on the role and underlying mechanism of WTAP and it-mediated m6A modification in the progression and metastasis of osteosarcoma. In this study, WTAP was firstly identified to upregulated which was associated with poor prognosis of osteosarcoma. Functionally, WTAP promoted the growth and metastasis of osteosarcoma in vitro and vivo. Mechanistically, HMBOX1 was identified as the target gene of WTAP, and it was regulated by WTAP with m6A modification at the 3′UTR. Finally, WTAP/HMBOX1 regulated osteosarcoma growth and metastasis in a PI3K/AKT-dependent pattern.
Actually, WTAP was reported as an oncogene in various cancers37–39,42. Recent studies reported that WTAP is strictly connected to a functional m6A methylation complex43. However, few study demonstrated the important role of WTAP as a m6A regulator. Here, we concluded that WTAP is not only upregulated but also plays key role on the m6A modification in osteosarcoma. Notably, the aberrant of m6A modification is related to various biological processes via regulating mRNA stability, splicing and translation. Next, we shed light on the downstream mRNA that modified by WTAP-mediated m6A modification by combining the data from RNA-seq, m6A-seq, and CLIP-seq. The results showed that HMBOX1 is a potential target gene of WTAP. Subsequently, MeRIP-qPCR, western blot and luciferase reporter assay results revealed that WTAP repressed HMBOX1 expressed with WTAP-dependent m6A modification at the 3′UTR of HMBOX1. Thus, WTAP involved in tumorigenesis depending on its role of m6A modification. Although YTHDF2 is a well-known m6A reader in several cancers. We found that YTHDF2 showed no effects on HMBOX1 expression, suggesting that WTAP regulated m6A-mediated HMBOX1 expression in YTHDF2-independent manner. And the m6A reader which was involved in the m6A modification of HMBOX1 need be further investigated.
HMBOX1, a homeobox containing protein49, was reported to be a transcriptional repressor, involving in the biological processes in bone marrow-derived stroma cells50, NK cells51,52, and vascular endothelial cells53. Recent studies demonstrated the dysregulated HMBOX1 in various cancers. For example, high expression of HMBOX1 was observed in gastric cancer, and it was associated with the poor prognosis of gastric cancer54. Moreover, HMBOX1 also observed as tumor suppressor in ovarian cancer55 and cervical cancer56. HMBOX1 repressed the progression of ovarian cancer via regulating cell proliferation and apoptosis55. HMBOX1 repressed liver cancer progression via regulating autophagy as well as and immune escape57. Consistently, HMBOX1 is downregulated and closely related to the poor prognosis of osteosarcoma in the present study. In addition, silenced HMBOX1 evidently alleviated WTAP-knockdown-mediated repression of osteosarcoma progression, which implied the import roles of HMBOX1 in WTAP-driven osteosarcoma development. Finally, we analyzed the downstream pathway of WTAP/HMBOX1 in osteosarcoma. PI3K/AKT pathway was reported to an important role in the progression of various cancers including osteosarcoma58. We found that PI3K/AKT pathway were evidently repressed by shWTAP, which was abolished by HMBOX1 knockdown. Inhibiting PI3K/AKT pathways by LY294002 remarkably reduced shHMBOX1-promoted cell proliferation, migration, and invasion. Therefore, these results imply that WTAP/HMBOX1 regulates the proliferation and metastasis of osteosarcoma partly via regulating PI3K/AKT pathway. However, it remains unclear how HMBOX1, as a transcriptional repressor, regulates PI3K/AKT pathway. Nonetheless, silenced HMBOX1 only partly alleviated WTAP-mediated OS progression. It means that there are other potential molecular mechanisms regulated by WTAP-mediated epigenetic modulation in OS.
In summary, we identified the elevated WTAP in osteosarcoma and which is associated with poor clinical outcome and serves as an independent prognostic factor for osteosarcoma patients. WTAP dramatically promoted osteosarcoma proliferation and metastasis via regulating HMBOX1 mRNA stability in a m6A-dependent manner. WTAP/HMBOX1 regulated osteosarcoma growth and metastasis via PI3K/AKT pathway. Altogether, our results determined WTAP/ HMBOX1 as a potential therapeutic target for osteosarcoma.
Supplementary information
Acknowledgements
This study was supported by National Natural Science Foundation of China (Grant Nos. 81772866); National Natural Science Foundation of China (Grant Nos. 81502331); Natural Science Foundation of Hunan Province (Grant Nos. 2018JJ2617); Natural Science Foundation of Hunan Province (Grant Nos. 2016JJ3176); National Key Research and Development Program of China (No. 2016YFC1201800); Natural Science Foundation of Hunan Province (No. 2018SK2090).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by A. Stephanou
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jinglei Miao, Email: miaojinglei@126.com.
Jinsong Li, Email: jinsongli_csu@163.com.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41419-020-02847-6).
References
- 1.El-Naggar AM, et al. HACE1 is a potential tumor suppressor in osteosarcoma. Cell Death Dis. 2019;10:21. doi: 10.1038/s41419-018-1276-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim H, et al. Oncogenic role of SFRP2 in p53-mutant osteosarcoma development via autocrine and paracrine mechanism. Proc. Natl Acad. Sci. USA. 2018;115:E11128–e11137. doi: 10.1073/pnas.1814044115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie L, et al. Deep RNA sequencing reveals the dynamic regulation of miRNA, lncRNAs, and mRNAs in osteosarcoma tumorigenesis and pulmonary metastasis. Cell Death Dis. 2018;9:772. doi: 10.1038/s41419-018-0813-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paris J, et al. Targeting the RNA m(6)A reader YTHDF2 selectively compromises cancer stem cells in acute myeloid leukemia. Cell Stem Cell. 2019;25:137–148.e136. doi: 10.1016/j.stem.2019.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fustin JM, et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155:793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 6.Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell. 2016;62:335–345. doi: 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016;534:575–578. doi: 10.1038/nature18298. [DOI] [PubMed] [Google Scholar]
- 9.Warda AS, et al. Human METTL16 is a N(6)-methyladenosine (m(6)A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017;18:2004–2014. doi: 10.15252/embr.201744940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mendel M, et al. Methylation of structured RNA by the m(6)A writer METTL16 is essential for mouse embryonic development. Mol. Cell. 2018;71:986–1000.e1011. doi: 10.1016/j.molcel.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Tran N, et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 2019;47:7719–7733. doi: 10.1093/nar/gkz619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knuckles P, et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes Dev. 2018;32:415–429. doi: 10.1101/gad.309146.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wen J, et al. Zc3h13 regulates nuclear RNA m(6)A methylation and mouse embryonic stem cell self-renewal. Mol. Cell. 2018;69:1028–1038.e1026. doi: 10.1016/j.molcel.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ries RJ, et al. m(6)A enhances the phase separation potential of mRNA. Nature. 2019;571:424–428. doi: 10.1038/s41586-019-1374-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang CX, et al. METTL3-mediated m6A modification is required for cerebellar development. PLoS Biol. 2018;16:e2004880. doi: 10.1371/journal.pbio.2004880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dorn LE, et al. The N(6)-methyladenosine mRNA methylase METTL3 controls cardiac homeostasis and hypertrophy. Circulation. 2019;139:533–545. doi: 10.1161/CIRCULATIONAHA.118.036146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engel M, et al. The role of m(6)A/m-RNA methylation in stress response regulation. Neuron. 2018;99:389–403.e389. doi: 10.1016/j.neuron.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Z, et al. Suppression of m(6)A reader Ythdf2 promotes hematopoietic stem cell expansion. Cell Res. 2018;28:904–917. doi: 10.1038/s41422-018-0072-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin Z, Tong MH. m(6)A mRNA modification regulates mammalian spermatogenesis. Biochimica et biophysica acta. Gene Regul. Mech. 2019;1862:403–411. doi: 10.1016/j.bbagrm.2018.10.016. [DOI] [PubMed] [Google Scholar]
- 20.Chen XY, Zhang J, Zhu JS. The role of m(6)A RNA methylation in human cancer. Mol. cancer. 2019;18:103. doi: 10.1186/s12943-019-1033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vu LP, Cheng Y, Kharas MG. The Biology of m(6)A RNA Methylation in Normal and Malignant Hematopoiesis. Cancer Disco. 2019;9:25–33. doi: 10.1158/2159-8290.CD-18-0959. [DOI] [PubMed] [Google Scholar]
- 22.Li Y, et al. Molecular characterization and clinical relevance of m(6)A regulators across 33 cancer types. Mol. Cancer. 2019;18:137. doi: 10.1186/s12943-019-1066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lang F, et al. EBV epitranscriptome reprogramming by METTL14 is critical for viral-associated tumorigenesis. PLoS Pathog. 2019;15:e1007796. doi: 10.1371/journal.ppat.1007796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panneerdoss S, et al. Cross-talk among writers, readers, and erasers of m(6)A regulates cancer growth and progression. Sci. Adv. 2018;4:eaar8263. doi: 10.1126/sciadv.aar8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li T, et al. METTL3 facilitates tumor progression via an m(6)A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol. Cancer. 2019;18:112. doi: 10.1186/s12943-019-1038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han J, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol. Cancer. 2019;18:110. doi: 10.1186/s12943-019-1036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng M, et al. The m(6)A methyltransferase METTL3 promotes bladder cancer progression via AFF4/NF-kappaB/MYC signaling network. Oncogene. 2019;38:3667–3680. doi: 10.1038/s41388-019-0683-z. [DOI] [PubMed] [Google Scholar]
- 28.Wang, Q. et al. METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut10.1136/gutjnl-2019-319639 (2019). [DOI] [PubMed]
- 29.Zhang J, et al. Excessive miR-25-3p maturation via N(6)-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat. Commun. 2019;10:1858. doi: 10.1038/s41467-019-09712-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang S, et al. m(6)A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat. Commun. 2019;10:2782. doi: 10.1038/s41467-019-10669-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niu Y, et al. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol. Cancer. 2019;18:46. doi: 10.1186/s12943-019-1004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su Y, Huang J, Hu J. m(6)A RNA methylation regulators contribute to malignant progression and have clinical prognostic impact in gastric cancer. Front. Oncol. 2019;9:1038. doi: 10.3389/fonc.2019.01038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Little NA, Hastie ND, Davies RC. Identification of WTAP, a novel Wilms’ tumour 1-associating protein. Hum. Mol. Genet. 2000;9:2231–2239. doi: 10.1093/oxfordjournals.hmg.a018914. [DOI] [PubMed] [Google Scholar]
- 34.Horiuchi K, et al. Wilms’ tumor 1-associating protein regulates G2/M transition through stabilization of cyclin A2 mRNA. Proc. Natl Acad. Sci. USA. 2006;103:17278–17283. doi: 10.1073/pnas.0608357103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horiuchi K, et al. Identification of Wilms’ tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J. Biol. Chem. 2013;288:33292–33302. doi: 10.1074/jbc.M113.500397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ortega A, et al. Biochemical function of female-lethal (2)D/Wilms’ tumor suppressor-1-associated proteins in alternative pre-mRNA splicing. J. Biol. Chem. 2003;278:3040–3047. doi: 10.1074/jbc.M210737200. [DOI] [PubMed] [Google Scholar]
- 37.Xie, W. et al. Physiological functions of Wilms’ tumor 1-associating protein and its role in tumourigenesis. J. Cell. Biochem. 10.1002/jcb.28402 (2019). [DOI] [PubMed]
- 38.Jo HJ, et al. WTAP regulates migration and invasion of cholangiocarcinoma cells. J. Gastroenterol. 2013;48:1271–1282. doi: 10.1007/s00535-013-0748-7. [DOI] [PubMed] [Google Scholar]
- 39.Bansal H, et al. WTAP is a novel oncogenic protein in acute myeloid leukemia. Leukemia. 2014;28:1171–1174. doi: 10.1038/leu.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang J, et al. Carbonic anhydrase IV inhibits colon cancer development by inhibiting the Wnt signalling pathway through targeting the WTAP-WT1-TBL1 axis. Gut. 2016;65:1482–1493. doi: 10.1136/gutjnl-2014-308614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu HL, et al. WTAP is a prognostic marker of high-grade serous ovarian cancer and regulates the progression of ovarian cancer cells. OncoTargets Ther. 2019;12:6191–6201. doi: 10.2147/OTT.S205730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuai Y, et al. Wilms’ tumor 1-associating protein plays an aggressive role in diffuse large B-cell lymphoma and forms a complex with BCL6 via Hsp90. Cell Commun. Signal. 2018;16:50. doi: 10.1186/s12964-018-0258-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sorci M, et al. METTL3 regulates WTAP protein homeostasis. Cell Death Dis. 2018;9:796. doi: 10.1038/s41419-018-0843-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Y, et al. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol. Cancer. 2019;18:127. doi: 10.1186/s12943-019-1053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ren T, et al. Osteosarcoma cell intrinsic PD-L2 signals promote invasion and metastasis via the RhoA-ROCK-LIMK2 and autophagy pathways. Cell Death Dis. 2019;10:261. doi: 10.1038/s41419-019-1497-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang S, et al. Up-regulation of PCOLCE by TWIST1 promotes metastasis in Osteosarcoma. Theranostics. 2019;9:4342–4353. doi: 10.7150/thno.34090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu Q, et al. Acylglycerol kinase promotes tumour growth and metastasis via activating the PI3K/AKT/GSK3beta signalling pathway in renal cell carcinoma. J. Hematol. Oncol. 2020;13:2. doi: 10.1186/s13045-019-0840-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zou Q, et al. miR-19a-mediated downregulation of RhoB inhibits the dephosphorylation of AKT1 and induces osteosarcoma cell metastasis. Cancer Lett. 2018;428:147–159. doi: 10.1016/j.canlet.2018.04.027. [DOI] [PubMed] [Google Scholar]
- 49.Chen S, et al. Isolation and functional analysis of human HMBOX1, a homeobox containing protein with transcriptional repressor activity. Cytogenetic Genome Res. 2006;114:131–136. doi: 10.1159/000093328. [DOI] [PubMed] [Google Scholar]
- 50.Su L, et al. Role of Hmbox1 in endothelial differentiation of bone-marrow stromal cells by a small molecule. ACS Chem. Biol. 2010;5:1035–1043. doi: 10.1021/cb100153r. [DOI] [PubMed] [Google Scholar]
- 51.Wu L, Zhang C, Zhang J. HMBOX1 negatively regulates NK cell functions by suppressing the NKG2D/DAP10 signaling pathway. Cell. Mol. Immunol. 2011;8:433–440. doi: 10.1038/cmi.2011.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu L, Zhang C, Zheng X, Tian Z, Zhang J. HMBOX1, homeobox transcription factor, negatively regulates interferon-gamma production in natural killer cells. Int. Immunopharmacol. 2011;11:1895–1900. doi: 10.1016/j.intimp.2011.07.021. [DOI] [PubMed] [Google Scholar]
- 53.Ma H, et al. HMBOX1 interacts with MT2A to regulate autophagy and apoptosis in vascular endothelial cells. Sci. Rep. 2015;5:15121. doi: 10.1038/srep15121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Diao N, et al. High expression of HMBOX1 contributes to poor prognosis of gastric cancer by promoting cell proliferation and migration. Biomed. Pharmacother. 2019;115:108867. doi: 10.1016/j.biopha.2019.108867. [DOI] [PubMed] [Google Scholar]
- 55.Yu YL, et al. Low expression level of HMBOX1 in high-grade serous ovarian cancer accelerates cell proliferation by inhibiting cell apoptosis. Biochem. Biophys. Res. Commun. 2018;501:380–386. doi: 10.1016/j.bbrc.2018.04.203. [DOI] [PubMed] [Google Scholar]
- 56.Zhou S, et al. Knockdown of homeobox containing 1 increases the radiosensitivity of cervical cancer cells through telomere shortening. Oncol. Rep. 2017;38:515–521. doi: 10.3892/or.2017.5707. [DOI] [PubMed] [Google Scholar]
- 57.Zhao H, Jia H, Han Q, Zhang J. Homeobox containing 1 inhibits liver cancer progression by promoting autophagy as well as inhibiting stemness and immune escape. Oncol. Rep. 2018;40:1657–1665. doi: 10.3892/or.2018.6551. [DOI] [PubMed] [Google Scholar]
- 58.Pei, T. et al. MUC13 promotes intrahepatic cholangiocarcinoma progression via EGFR/PI3K/AKT pathways. J. Hepatol. 10.1016/j.jhep.2019.11.021 (2019).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.