Abstract
Pseudomonas aeruginosa biofilms employ a variety of strategies to hijack the host immune defense system to achieve chronic infection. However, the bacterial components that are involved in this process are not yet fully understood. PcrV, a needle tip protein of the P. aeruginosa type III secretion system (T3SS), was downregulated during P. aeruginosa biofilm infection. The impaired expression of the P. aeruginosa pcrV gene is associated with attenuated immune activation and an increased percentage of M2 macrophages following P. aeruginosa biofilm infection. Treatment with exogenous PcrV produced from Escherichia coli elevated tissue inflammation and the percentage of M1 macrophages, resulting in reduction in the biofilm burden. Further analyses demonstrated that the potential of PcrV to induce classically activated M1 macrophages as evidenced by the increased production of proinflammatory cytokines and anti-bacterial mediators, including inducible nitric oxide synthase (iNOS) and reactive oxygen species (ROS), as well as increased phagocytosis of bacteria. Mechanistically, PcrV-mediated promotion of macrophage M1 polarization and phagocytosis occurs through the activation of mitogen-activated protein kinases (MAPKs) and NF-κB signaling pathways. Collectively, these findings reveal a potential role of PcrV in skewing host immune defense to promote P. aeruginosa biofilm infection and provide new insights into the therapeutic strategies for P. aeruginosa biofilm infection.
Keywords: Pseudomonas aeruginosa, biofilm, PcrV, immune evasion, macrophage polarization, biofilm eradication
Introduction
Pseudomonas aeruginosa-mediated chronic infections are commonly associated with biofilm formation on host tissues and indwelling medical devices. Bacterial biofilm communities are encased in a self-produced extracellular matrix which precludes antibiotic penetration, which impedes eradication of biofilm bacteria by conventional antibiotic treatment. To combat biofilm infections, various strategies have been investigated, including utilization of antimicrobial peptides/lipids (Verderosa et al., 2019), quorum sensing inhibitors (Chang et al., 2019), and bacteriophages (Geredew Kifelew et al., 2019). In addition to the methods that directly target bacterial components, promising new strategies to control biofilm infection rely on improving host anti-bacterial immune responses, such as the induction of proinflammatory cytokines and antibacterial mediators, for instance, nitric oxide (NO) and reactive oxygen species (ROS), and the enhancement of phagocytosis activity (Campoccia and Mirzaei, 2019).
Macrophages act as the first line of defense against bacterial infection by secreting proinflammatory cytokines and bactericides, as well as by increasing phagocytotic activity against bacteria (Keewan and Naser, 2020). However, differences in the polarization of macrophage phenotypes determine the diverse efficacy in the eradication of bacteria. The classically activated M1 macrophages release large amounts of proinflammatory cytokines, such as tumor necrosis factor (TNFα), interleukin 6 (IL6), and IL12, and antibacterial mediators, such as NO, ROS, and reactive nitrogen species (RNS). As such, M1 macrophages mediate efficient phagocytosis and are actively involved in microbicidal action (Qian and Pollard, 2010; Shapouri-Moghaddam et al., 2018). Conversely, the alternatively activated M2 macrophages are characterized by enhanced production of anti-inflammatory cytokines, IL4 and IL10 and arginase. As such, M2 macrophages are involved in attenuating microbicidal activity (Panagi et al., 2019). Although M1 macrophages play critical roles in eliminating planktonic bacteria, studies have demonstrated that macrophages display an M2 phenotype following activation by Staphylococcus aureus biofilms (Thurlow et al., 2011), suggesting that biofilms play a role in inducing an anti-inflammatory activation of macrophages that benefits biofilm persistence. Conversely, treatment of biofilms with M1-activated macrophages indicates the potential importance for controlling biofilm infections. However, the issues regarding the bacterial elements that affect biofilm persistence following P. aeruginosa biofilm infection, as well as the efficacy of M1 macrophages against P. aeruginosa biofilm infections, have not yet been reported.
PcrV, which is a critical needle tip protein of the type III secretion system (T3SS) of P. aeruginosa, is an indispensable factor that allows the translocator protein PopB/D to form pores on the host cell membrane through which effector proteins translocate into host cells. In addition to this biological function, PcrV has been reported to possess a possible proinflammatory function (Wangdi et al., 2010). As a V-antigen, a PcrV-originated vaccine elicited a multifactorial immune response and conferred broad protection in an acute P. aeruginosa pneumonia model (Hamaoka et al., 2017; Wan et al., 2019), indicating the potential of PcrV for the treatment of bacterial biofilm infection through activation of the host immune response. Nevertheless, the role of PcrV in modulating macrophage polarization and improving the efficacy of P. aeruginosa biofilm eradication remains to be elucidated.
In this study, we demonstrated that pcrV gene expression is reduced during P. aeruginosa biofilm infection. Injection of PcrV into the milieu surrounding the biofilm-infected tissues induced a proinflammatory response with increased infiltration of M1-polarized macrophages, decreasing the biofilm burden. Further analyses demonstrated that PcrV promotes biofilm eradication, macrophage M1 polarization, and phagocytosis via the mitogen-activated protein kinases (MAPKs) and NF-κB signaling pathways. Taken together, these results suggest that decreased expression of PcrV during P. aeruginosa biofilm infection promotes biofilm persistence and provide novel clues into the therapeutic strategies against P. aeruginosa biofilm infection.
Materials and Methods
Mice and Ethics Statement
Male mice on a C57BL/6 background were purchased from biocytogen CO., Ltd. (Beijing, China). Animal experiments were conducted according to the experimental animal guidelines of the Army Medical University of China.
Expression and Purification of PcrV Protein
Pseudonomas aeruginosa PcrV gene was cloned into pQE31 (Qiagen, Germany), which introduces an N-terminal fusion of the protein to a His6 tag. Escherichia coli JM109 strain carrying expression plasmids was propagated in LB medium containing 100 μg/ml ampicillin until the OD600 nm reached 0.5 and was induced using 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG) at 37°C for 4 h. Cells were harvested and resuspended in phosphate buffered saline (PBS) and lysed by sonication. The fusion protein was purified by affinity chromatography using His-Trap HP (GE healthcare, Germany). Endotoxin was removed from PcrV by using Detoxi-Gel endotoxin removing gel (Thermo fisher, USA) following the manufacturer’s instructions.
Isolation and Induction of BMDMs
Tibias and femurs from the euthanized C57BL/6 mice were excised, and marrow cells were washed out with a 25-gauge needle attached to a 5 ml-syringe. Cells were cultured in 10% FBS/dulbecco’s modified eagle medium (DMEM), penicillin, streptomycin, and 50 ng/ml recombinant murine macrophage colony stimulating factor (M-CSF) at 37°C and 5% CO2 for 3 days. At day 4, the medium was replaced, and cells were cultured at same condition for an additional 3 days.
Mouse Model of P. aeruginosa Catheter-Associated Biofilm Infection
A mouse model of catheter-associated biofilm infection was established as described previously with some modifications (Thurlow et al., 2011; Hanke et al., 2013). Briefly, a sterile 1 cm intravenous catheter was implanted subcutaneously into the flank of mice under pentobarbital sodium anesthesia. A suspension (20 μl) of log-phase PAO1 (1 × 105 colony forming unit, CFU) was injected through the skin into the catheter lumen. Biofilm formation was monitored throughout the course of infection, and mice were sacrificed on days 2, 5, and 8 post-infection. For scanning electron microscopy (SEM) analysis, biofilms on catheters were fixed and dehydrated according to a standard SEM protocol and were observed under a Crossbeam 340 SEM (Carl Zeiss, Germany). Tissues surrounding infected catheters were homogenized and weighed after freezing in liquid nitrogen. The bacterial burdens of catheters and surrounding tissues were enumerated using P. aeruginosa isolation agar (PIA) plates.
Macrophage Administration Into Biofilm Infections in vivo
To determine the efficacy of biofilm clearance by differentially polarized macrophages, 106 non-activated or PcrV-activated bone marrow-derived macrophages (BMDMs) were subcutaneously injected at the sites surrounding infected catheters on days 5, 6, and 7 post-infection. The infected catheters and surrounding tissues were harvested on day 8 for subsequent analysis.
Generation of P. aeruginosa Static Biofilms in vitro
Static biofilms were generated as previously described (Thurlow et al., 2011) with minor modifications. Briefly, sterile 24-well cell culture plates were treated with 20% human plasma in sterile carbonate-bicarbonate buffer (pH 9.6) overnight at 4°C to facilitate bacterial attachment. The PAO1 strain was cultured overnight at 37°C with shaking in DMEM supplemented with 10% FBS. The bacterial culture was adjusted to an initial OD600 of 0.05 and then incubated in the plasma pre-coated plate at 37°C under static aerobic condition for 3 days. Medium was carefully replenished every 24 h to prevent disruption of the biofilm structure.
Phagocytosis Assay
The phagocytic efficacy of macrophages against planktonic bacteria or biofilms was evaluated according to a previously described method with modifications (Thurlow et al., 2011). Briefly, planktonic bacteria or biofilms were co-cultured for 30 min with RAW264.7 cells at a ratio of 1:10 at 37°C under 5% CO2. Cells were then treated with gentamicin (final concentration 400 μg/ml) for 2 h at 37°C under 5% CO2 to remove extracellular bacteria. For immunofluorescence staining, the cytoskeleton and bacteria were visualized by phalloidin (red) and anti-PAO1 antibody (green), respectively. For evaluation of the intracellular bacteria, cells were lyzed with 0.5% TritonX-100/PBS and enumerated on PIA plates.
Western Blot
Cell pellets were lysed using RIPA buffer (Beyotime, China) supplemented with protease inhibitor cocktail (Roche, USA). Equal amounts of proteins were separated on 10% SDS-PAGE and then transferred electrophoretically to PVDF membranes (Millipore, USA). The membrane was blocked using 5% skim milk in TBST at real time (RT) for 1 h. Then, the membrane was incubated with the appropriate first antibody at 4°C overnight and horseradish peroxidase-conjugated secondary antibody at RT for 1 h.
Real-Time Quantitative PCR
Total RNA extraction and reverse transcription to cDNA were performed according to the manufacturer’s instructions. Quantitative PCR was performed using an ABI 7500 RT PCR system (Applied Biosystems, Germany). For macrophages, the relative gene expression levels of cd11c, inos, ptgs2, cd206, and pparγ were normalized to GAPDH. For PAO1 strain, the relative gene expression level of ndk was normalized to gyrB, rpoD, or rplU gene. The primers used in this study were provided in Supplementary Table S1.
Enzyme-Linked Immunosorbent Assay
Supernatants of stimulated macrophages or biofilm infected tissues were assessed for their levels of TNFα, IL 12 p40/p70, and IL6 by using the sandwich enzyme-linked immunosorbent assay (ELISA) kits (BD biosciences, USA) according to the manufacturer’s instructions.
Measurement of Intracellular ROS
Macrophages were harvested and incubated with 2'7'-dichlorodihydrofluorescein diacetate (H2DCFDA; Santa Cruz, USA) dye in DMEM medium at a final concentration of 5 μM for 30 min at RT. Intracellular ROS were measured by flow cytometry.
Statistical Analysis
Data were expressed as means ± standard errors of the means (SEM). The statistical analysis was carried out with GraphPad (GraphPad Software Inc., San Diego, CA). Data were analyzed by unpaired t test when comparing two groups and one-way analysis of variance (ANOVA) with Tukey’s multiple-comparison test for multiple groups. A value of p < 0.05 was considered statistically significant.
Results
P. aeruginosa Biofilm Persistence Is Associated With Attenuated Activation of Host Proinflammtory Responses and Decreased Bacterial Eradication Ability
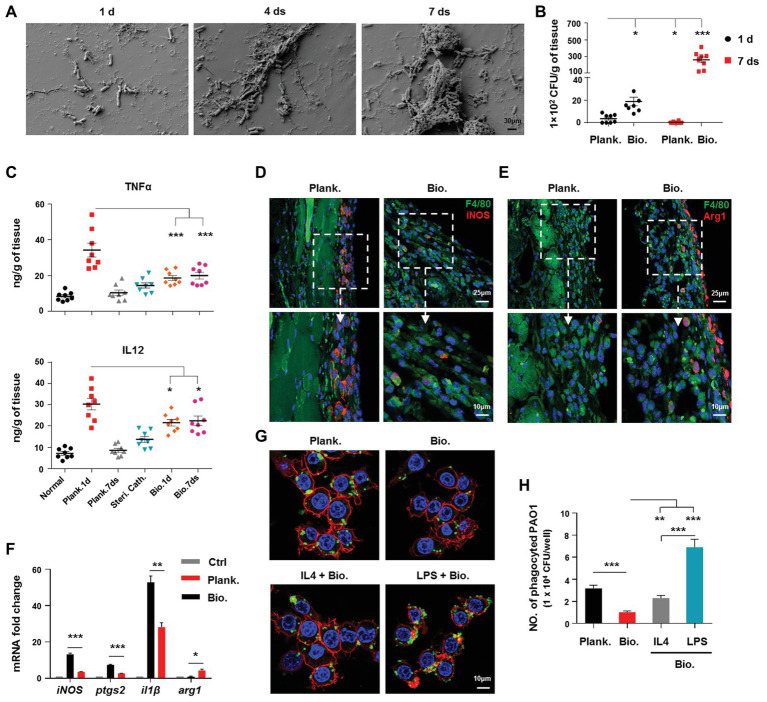
To evaluate the condition of the host immunity and bacterial killing efficacy in response to a P. aeruginosa biofilm, we used a mouse model of catheter-associated biofilms to mimic bacterial biofilm formation on medical devices in humans (Hanke et al., 2013). Bacterial biofilm formation involves a transformation from an immature to a mature structure involving bacterial surface attachment and bacterial colony formation in which bacteria are surrounded by matrix materials. In this study, we primarily observed the different stages of P. aeruginosa biofilm formation on catheters by SEM. Examination of the catheter lumen by SEM revealed that an immature biofilm was formed with few extracellular matrix-encompassed bacteria within 4 days of the initial infection (Figure 1A). With the extension of time post-infection (7 days), a mature biofilm was formed with a contiguous bacterial layer in which extracellular matrix and interior holes were found (Figure 1A), suggesting the successful establishment of a mature P. aeruginosa biofilm.
Figure 1.
Pseudomonas aeruginosa biofilm persistence is associated with attenuated activation of host proinflammtory responses and decreased bacterial eradication ability, whereas increased programming of macrophages toward M2 phenotype. (A) P. aeruginosa biofilm formation on catheters was observed by scanning electron microscopy (SEM; 5000X magnification). Bacterial loads (B), production of tumor necrosis factor (TNFα) and IL12 p40/70 (C), and F4/80+/iNOS+ and F4/80+/Arg1+ macrophages (D,E) in planktonic P. aeruginosa (1 day)‐ or catheter biofilm (7 days)-infected tissues were determined by colony forming unit (CFU) enumeration, enzyme-linked immunosorbent assay (ELISA), and immunofluorescence staining, respectively. For immunofluorescence staining, M1 macrophages were counterstained with fluorescein isothiocyanate (FITC)-conjugated anti-F4/80 and AF647-conjugated anti-inducible nitric oxide synthase (iNOS) antibodies. M2 macrophages were counterstained with FITC-conjugated anti-F4/80 and AF647-conjugated anti-Arg1 antibodies; cellular nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). The static P. aeruginosa biofilms were co-cultured with Raw264.7 at 37°C, with 5% CO2 for 3 h (MOI = 10), and relative gene expression levels were analyzed by real-time quantitative PCR (RT-qPCR) using mouse GAPDH as a reference gene (F). To generate M1 and M2-polarized macrophages, Raw264.7 cells were pretreated with 100 ng/ml lipopolysaccharide (LPS) + 50 ng/ml IFNγ (named as LPS group) or 20 ng/ml IL4 for 6 h, respectively. The cells were then co-cultured with static P. aeruginosa biofilms at a ratio of 1:10 at 37°C, 5% CO2 for 30 min. Phagocytosis was detected by immunofluorescence staining (G) and CFU enumeration (H). Cytoskeleton was labeled with phalloidin (red); PAO1 was visualized by FITC anti-PAO1 antibody (green); cellular nuclei were stained with DAPI (blue). Error bars indicate the means ± standard errors of the means (SEM). An unpaired Student’s t test was used for statistical analysis (B,C,F,H). *p < 0.05; **p < 0.01; ***p < 0.001. Planktonic (Plank.), biofilm (Bio.), sterile catheter (steri cath).
To compare the host immune responses and bacterial eliminating efficacy between P. aeruginosa biofilms and planktonic bacteria-associated infections, we analyzed the production of inflammation-related cytokines, arginase, and inducible nitric oxide synthase (iNOS), as well as the bacterial burdens in the tissues surrounding the site of the infection. At 1 day post-infection, bacterial loads in planktonic bacteria-infected tissues were lower than biofilm bacteria-infected group (Figure 1B; p < 0.05). At 7 days post-infection, the planktonic bacteria were almost eradicated, while the bacterial loads in the biofilm remained high (Figure 1B). In contrast to the decreased bacterial loads, the levels of the proinflammatory cytokines, TNFα (Figure 1C; p < 0.001) and IL12 p40/70 (Figure 1C; p < 0.05), and the percentages of iNOS+ cells were higher in planktonic bacteria-infected tissues (1 day post-infection) than those in biofilm-infected tissues (Supplementary Figure S1). Arg1, which is involved in activating anti-inflammatory responses, was also higher in biofilm-infected tissues than in planktonic bacteria-infected tissues (Supplementary Figure S1), suggesting a lower ability of P. aeruginosa biofilms to activate proinflammatory responses to infection.
Phagocytosis by phagocytes, such as macrophages, accelerates the clearance of bacteria following infection; therefore, we compared the phagocytotic ability of macrophages against P. aeruginosa in the planktonic and biofilm forms in vitro. The results showed that the ability of macrophages to phagocytose biofilm bacteria was inferior to that observed for planktonic bacteria (Figures 1G,H; p < 0.001). Collectively, these results indicated that P. aeruginosa biofilms enhance bacterial chronic infection by circumventing the host immune response and anti-bacterial activities.
P. aeruginosa Biofilm-Mediated Programming of Macrophages Toward an M2 Phenotype Attenuates Immune Activation, and Promotes Biofilm Persistence
Based on the above results, we further investigated the association of the impaired activation of host immune responses to P. aeruginosa biofilm infection with alternatively activated M2 macrophages. Result revealed that a higher percentage of F4/80+/Arg1+ macrophages and a lower percentage of F4/80+/iNOS+ macrophages in biofilm-infected tissues (7 days post-infection) than those in planktonic bacteria-infected tissues (1 day post-infection; Figures 1D,E), reflecting a less M1-like phenotype of macrophages under a biofilm-associated infection. To further verify this phenomenon, we analyzed the gene expression profiles of Raw264.7 cells infected with P. aeruginosa planktonic bacteria or biofilms in vitro. In accordance with our in vivo findings, macrophages associated with the biofilm infection exhibited decreased expression of proinflammation-related genes, including inos, il1β, and ptgs2, compared with those associated with planktonic bacterial infection (Figure 1F), whereas the expression of arg1 was increased (Figure 1F; p < 0.05).
The decreased phagocytic ability of M2 macrophages skewed by bacterial biofilms is also responsible for the delayed bacterial clearance and biofilm persistence. To this end, we analyzed the phagocytic ability of differentially polarized macrophages against P. aeruginosa biofilms. Although, lipopolysaccharide (LPS)‐ or IL4-primed M1 or M2 macrophages both displayed higher phagocytic ability than non-activated macrophages (Figures 1G,H), the M1 macrophages exhibited a superior ability for phagocytosis of biofilm bacteria than M2 macrophages (Figures 1G,H; p < 0.001). Collectively, these results suggested that P. aeruginosa biofilm-mediated skewing of macrophages toward the M2 phenotype enhances biofilm persistence.
PcrV Production Is Attenuated in Chronic P. aeruginosa Biofilm Infection, and the Addition of Exogenous PcrV Accelerates Macrophage-Mediated P. aeruginosa Clearance in Biofilms
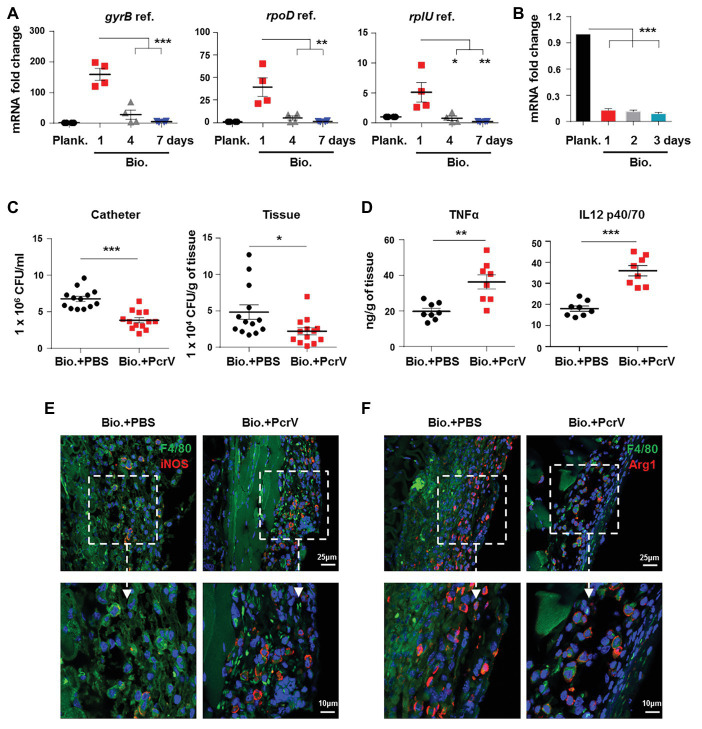
The reduced activation of host proinflammatory responses that allows P. aeruginosa biofilms to persist may be due to the inactivation of the T3SS, whereby components of the T3SS could be immunostimulatory. Previous research has shown that components of the P. aeruginosa needle tip complex, including PcrV, are involved in enhancing proinflammatory responses in mouse lung tissues following infection (Wangdi et al., 2010). Therefore, we speculated that pcrV gene expression is downregulated during P. aeruginosa biofilm infection, and the decrease in PcrV production in the milieu surrounding the infection accounts for the impaired activation of immune responses mediated by M2 macrophages following biofilm infection, which ultimately results in biofilm persistence. To verify this hypothesis, we first analyzed pcrV gene expression in P. aeruginosa biofilm-infected catheters in vivo by real-time quantitative PCR (RT-qPCR) analysis of three different P. aeruginosa reference genes. Compared to the immature biofilm bacteria (1 day post-infection), pcrV gene transcription was decreased both in the incompletely matured (4 days post-infection) and mature biofilm bacteria (7 days post-infection; Figure 2A). A similar trend was also observed in the in vitro static biofilm system (Figure 2B; p < 0.001). To further investigate the correlation between PcrV production and biofilm eradication, PcrV protein was subcutaneously injected into the tissues surrounding the biofilm-infected catheters. Bacterial burdens in PcrV-treated catheters (Figure 2C; p < 0.001) and surrounding tissues (Figure 2C; p < 0.05) were lower than those in the PBS-treated control group, whereas the levels of TNFα (Figure 2D; p < 0.01) and IL12 p40/70 (Figure 2D; p < 0.001) were elevated.
Figure 2.
Decreased expression of P. aeruginosa PcrV gene following biofilm infection promotes biofilm persistence. (A) P. aeruginosa-infected catheters were subcutaneously implanted into C57BL/6 mice, and catheters were harvested after infection for 1, 4, and 7 days. The pcrV gene expression in biofilm catheter or planktonic bacteria harvested before infection was analyzed by RT-qPCR. P. aeruginosa gyrB, rpoD, and rplU genes were used as reference genes (abbreviated as ref.). (B) The pcrV gene expression in P. aeruginosa static biofilms or planktonic bacteria was analyzed by RT-qPCR using rplU genes as reference gene. After infection for 4 days, 10 μg of PcrV was daily injected into the tissues surrounding the biofilm-infected catheters for three times. Infected tissues and catheters were harvested at day 8. Bacterial loads in infected tissues or catheters were determined by counting CFU (C) and production of TNFα and IL12 p40/70 was detected by ELISA assay (D). F4/80+/iNOS+ and F4/80+/Arg1+ macrophages in tissues surrounding the biofilm-infected catheters were detected by immunofluorescence staining (E,F). For immunofluorescence staining, M1 macrophages were counterstained with FITC-conjugated anti-F4/80 and AF647-conjugated anti-iNOS antibodies. M2 macrophages were counterstained with FITC-conjugated anti-F4/80 and AF647-conjugated anti-Arg1 antibodies; cellular nuclei were stained with DAPI. One-way ANOVA (Tukey’s post hoc, A,B) or an unpaired Student’s t test (C,D) was used for statistical analysis. *p < 0.05; **p < 0.01; ***p < 0.001.
To explore the involvement of macrophages in PcrV-mediated elimination of biofilm bacteria, M1 and M2 polarization was analyzed in the macrophages associated with P. aeruginosa biofilm catheters-infected tissues following treatment with or without PcrV. The percentages of M1 macrophages (F4/80+/iNOS+) in the PcrV-treated groups were higher than those in the PBS-treated groups (Figure 2E), whereas the opposite trend was observed in M2 macrophages (F4/80+/Arg1+; Figure 2F). These results suggested that PcrV promotes the elimination of biofilm bacteria through polarization of macrophages toward an M1 phenotype.
PcrV Is Involved in Skewing Macrophages Toward an M1 Phenotype
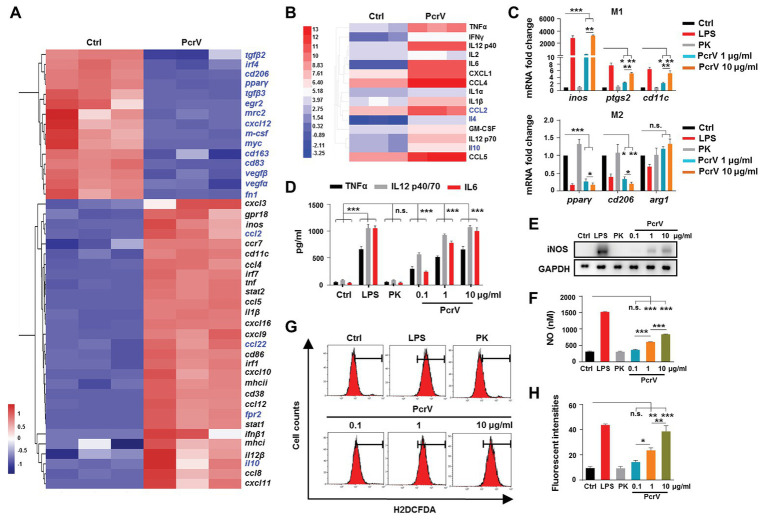
Given that subcutaneous injection of PcrV promoted biofilm clearance and increased the percentages of M1-polarized macrophages around the infected catheter, we investigated the ability of PcrV to directly drive macrophage differentiation toward an M1 phenotype. To this end, we first evaluated the extensive inflammatory modulation effects of PcrV on BMDMs by gene and protein chip analysis. Gene chip analysis showed that PcrV treatment extensively upregulated macrophage M1 activation-related genes including proinflammtory cytokines (e.g., tnf, il1β, il12, and il6), chemokines (e.g., cxcl3, cxcl9, and cxcl11), bacterial killing molecules (inos), antigen presentation (e.g., MHCI, MHCII, and CD86), and others (e.g., cd11c, ptgs2, egln3, and inhba), whereas M2 activation-related genes, such as cd206, pparγ, cd83, and egr2, were downregulated (Figure 3A). Protein chip assays revealed that PcrV-primed BMDMs displayed increased production of macrophage M1 polarization-related cytokines (e.g., GM-CSF, TNFα, IFNγ, IL12 p40/70, and IL1α/β), chemokines (e.g., CXCL1 and CCL4/6), and IL2, which is responsible for T cell activation (Figure 3B). Despite the extensive upregulation of M1 markers following PcrV treatment, and decreased expression of M2 markers in BMDMs, some of the M2 molecules, such as ccl2, ccl22, IL4, and IL10 were also elevated in PcrV-primed BMDMs (Figures 3A,B), suggesting a balance of immune responses following PcrV treatment.
Figure 3.
PcrV promotes macrophage M1 polarization. (A) Gene expression and (B) cytokine production in bone marrow-derived macrophages (BMDMs) treated with or without PcrV (10 μg/ml) for 24 h were analyzed by gene chip and protein chip, respectively. M1-related genes were marked in blank; M2-related genes were marked in blue. (C) Gene expression in BMDMs treated with LPS + IFNγ, hydrolyzed PcrV (PK), and PcrV (10 μg/ml) for 24 h was verified by RT-qPCR. (D) Levels of TNFα, IL12 p40/70, and interleukin 6 (IL6) in the culture supernatants of the treated BMDMs were assayed by ELISA assay. The production of iNOS (E) and reactive oxygen species (ROS; G,H) in Raw264.7 treated with LPS + IFNγ, PK, and PcrV for 6 h were detected by western blot and flow cytometry, respectively. (F) The concentration of nitric oxide (NO) in the culture supernatant of Raw264.7 treated with the indicated compound for 24 h was detected by NO detection kit. One-way ANOVA (Tukey’s post hoc, C,D,F,G) was used for statistical analysis. *p < 0.05; **p < 0.01; ***p < 0.001; n.s. indicates no significance.
To observe the dose-dependent effects of PcrV on macrophages, BMDMs and the RAW264.7 cells were treated with different concentrations of PcrV. Similar to LPS-primed M1 macrophages, PcrV-pulsed BMDMs and RAW264.7 cells showed upregulated M1-related gene expression (inos, ptgs2, and cd11c; Figure 3C; Supplementary Figure S2A), increased production of iNOS (Figure 3E) and NO (Figure 3F), TNFα, IL12 p40/70, IL6 (Figure 3D; Supplementary Figure S2A), and ROS (Figures 3G,H), whereas expression of M2-related genes, including cd206, pparγ (Figure 3C; Supplementary Figure S2B), and arg1 (Supplementary Figure S2B) was downregulated in a dose-dependent manner compared with the untreated‐ or hydrolyzed PcrV (PK)-treated groups. Taken together, these results suggested that PcrV plays a role in skewing macrophage differentiation toward an M1 phenotype.
PcrV Repolarizes Macrophages From M2 to M1 Phenotype
To evaluate the ability of PcrV to promote a macrophage switch from the M2 to M1 phenotype, BMDMs were pretreated with the M2 inducer IL4, before PcrV treatment. IL4 significantly increased the expression of M2-specific genes, including cd206, pparγ, and arg1 in BMDMs (Figure 4); however, the IL4-mediated upregulation of these genes was abolished by PcrV treatment (Figure 4). Despite the anti-inflammatory conditions of macrophages pretreated with IL4, PcrV treatment significantly elevated the expression of proinflammatory M1-related genes in BMDMs, such as inos, ptgs2, and cd11c (Figure 4), demonstrating that PcrV plays a role in inducing transformation from the M2 to M1 phenotype.
Figure 4.
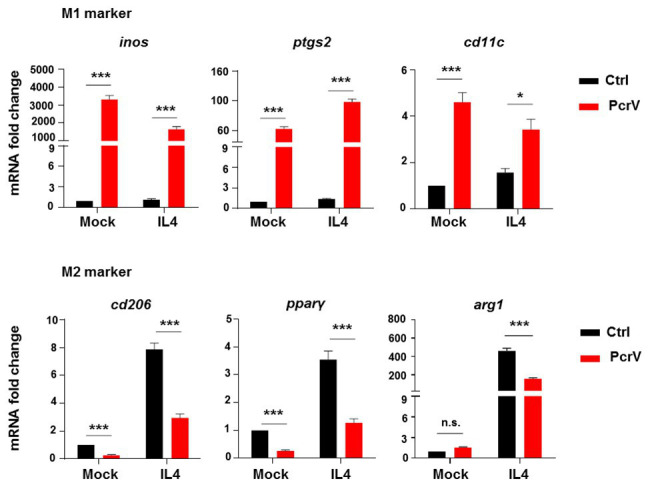
PcrV repolarizes macrophages from M2 to M1 phenotype. BMDMs pretreated with 20 ng/ml of IL4 for 12 h were primed by PcrV (10 μg/ml) for another 24 h. Macrophage M1 and M2-related genes were analyzed by RT-qPCR. An unpaired Student’s t test was used for statistical analysis. *p < 0.05; ***p < 0.001; n.s. indicates no significance.
The Proinflammtory M1 Macrophages Polarized by PcrV Are Involved in Biofilm Elimination
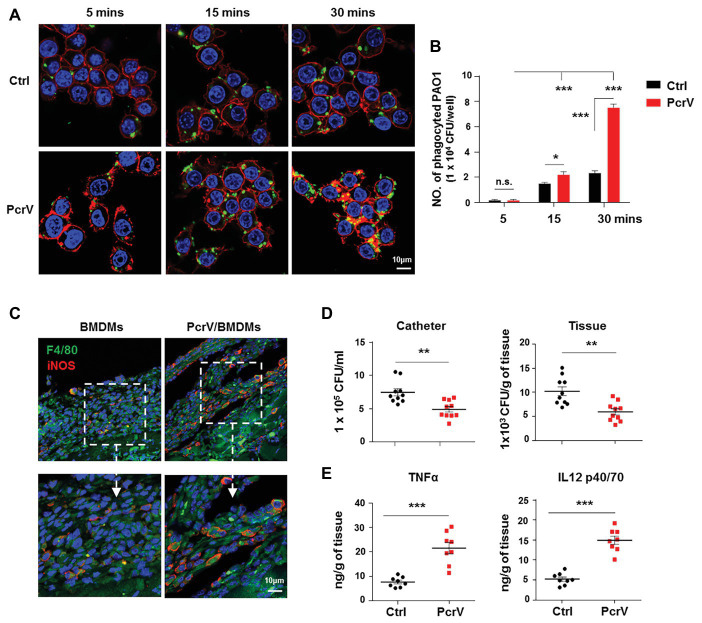
Considering that PcrV is involved in skewing macrophage toward the M1 phenotype, we further investigated the efficacy of PcrV-pulsed M1 macrophages in biofilm elimination both in vitro and in vivo. In vitro studies revealed that PcrV treatment significantly increased the phagocytosis ability of macrophages against biofilm bacteria in a time-dependent manner (Figures 5A,B; p < 0.001). Following injection of BMDMs pretreated with PcrV for 24 h into the tissues surrounding biofilm catheters in vivo, the percentage of F4/80+iNOS+ macrophages was higher in PcrV/BMDMs treated mice than that in mice treated with non-activated BMDMs (Figure 5C), suggesting that functional M1 macrophages were present at the site of infection. Analyses of bacterial burdens and tissue inflammation status revealed that PcrV-primed M1 macrophages reduced bacterial loads in catheters and infected tissues (Figure 5D; p < 0.01), whereas the production of TNFα and IL12 p40/70 was augmented (Figure 5E; p < 0.001). Collectively, these results suggested that PcrV-primed M1 macrophages are effective in accelerating biofilm clearance.
Figure 5.
PcrV-primed macrophages display increased phagocytosis, bacterial killing efficacy, and induction of proinflammtory cytokines. Raw264.7 cells pretreated with or without PcrV (10 μg/ml) for 6 h were co-cultured with P. aeruginosa biofilms (MOI = 10) for the indicated time point. The phagocyted bacteria were detected by immunofluorescence staining (A) and CFU enumeration (B). Cytoskeleton was labeled with phalloidin (red); PAO1 was visualized by FITC anti-PAO1 antibody (green); cellular nuclei were stained with DAPI (blue). BMDMs primed with or without PcrV (10 μg/ml) for 24 h were daily injected into the tissues surrounding biofilm-infected catheters after biofilm infection for 4 days. The infected tissues and catheters were harvested after the injection of PcrV for 3 days. F4/80+/iNOS+ macrophages in infected tissues (C), bacterial burdens (D), and production of TNFα and IL12 p40/70 (E) were analyzed by immunofluorescence staining, CFU enumeration, and ELISA, respectively. For immunofluorescence staining, M1 macrophages were counterstained with FITC-conjugated anti-F4/80 and AF647-conjugated anti-iNOS antibodies. M2 macrophages were counterstained with FITC-conjugated anti-F4/80 and AF647-conjugated anti-Arg1 antibodies; cellular nuclei were stained with DAPI. One-way ANOVA (Tukey’s post hoc, B) or unpaired Student’s t test (D,E) was used for statistical analysis. *p < 0.05; **p < 0.01; ***p < 0.001.
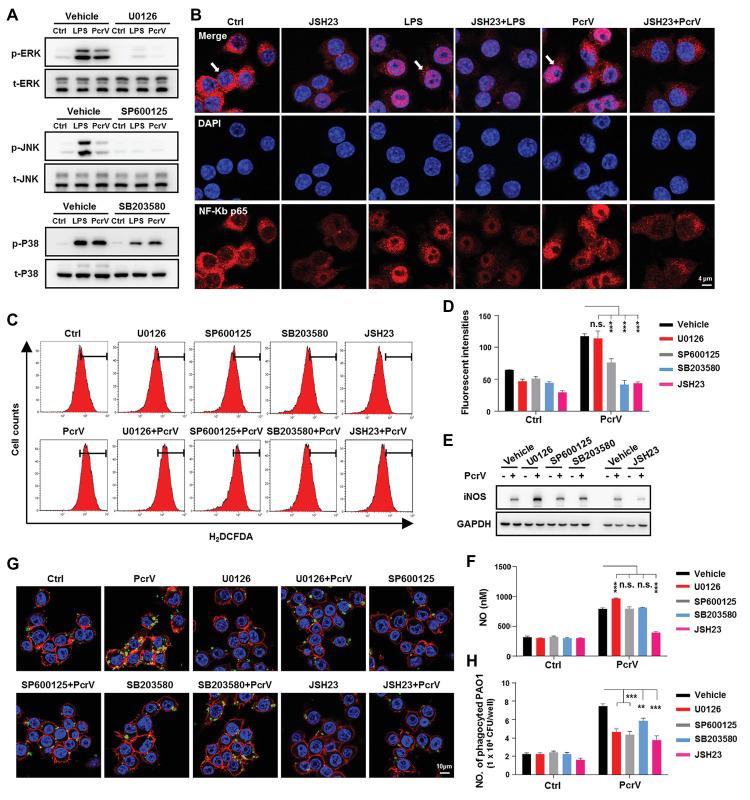
MAPKs and NF-κB Signaling Pathways Play Dominant Roles in Promoting PcrV-Mediated Macrophage M1 Activation and Phagocytosis
MAPK and NF-κB signaling pathways are among the most extensively reported pathways that are involved in activating anti-bacterial immune responses, involving macrophage M1 polarization (Akhtar et al., 2019; Justino et al., 2020). Our results demonstrated that PcrV promoted the phosphorylation of extracellular signal-regulated protein kinase (ERK), c-Jun N-terminal kinase (JNK), and p38 MAPKs and IKBα (an indicator of NF-κB pathway activation) in RAW264.7 cells in a dose‐ and time-dependent manner (Supplementary Figures S3A,B). Further utilization of the corresponding signal pathway inhibitors U0126, SP600125, SB203580 for ERK, JNK, and p38 MAPKs, respectively, revealed that PcrV-mediated activation of the three pathways was successfully inhibited (Figure 6A). Since the nuclear translocation of NF-κB p65 results in the activation of NF-κB signaling pathway, we used JSH23, an inhibitor of NF-κB p65 nuclear translocation and transcriptional activity, to treat the cells. Given NF-κB p65 might regulate the transcriptional level of itself, the JSH23 treatment also lead to a massive reduction of the expression of NF-κB p65 in the JSH23 treated groups (Figure 6B). In spite of that, LPS or PcrV-mediated promotion of nuclear translocation of NF-κB p65 was impaired by JSH23 (Figure 6B). Analyses of the downstream cytokines, gene expression, and phagocytosis showed that the levels of TNFα, IL6 (Supplementary Figure S3C; p < 0.001), and ROS (Figures 6C,D), expression of the M1-related genes, cd11c and ptgs2 (Supplementary Figure S3D), and phagocytosis (Figures 6G,H) were reduced in PcrV-treated RAW264.7 in the presence of both MAPK and NF-κB inhibitors; however, ERK inhibition elevated iNOS (Figure 6E; Supplementary Figure S3D) and NO (Figure 6F; p < 0.001), while JNK and p38 inhibition did not alter NO production (Figure 6F). Meanwhile, MAPK inhibition did not elevate pparγ expression in RAW264.7 cells primed with PcrV (Supplementary Figure S3D). In contrast, JSH23 treatment reduced the levels of iNOS (Figure 6E; Supplementary Figure S3D; p < 0.001) and NO (Figure 6F; p < 0.001) and reversed the decrease in pparγ expression in PcrV-stimulated RAW264.7 cells (Supplementary Figure S3D; p < 0.05). Collectively, these results indicated that the MAPK and NF-κB signaling pathways are involved in PcrV-mediated regulation of macrophage M1 polarization and phagocytosis.
Figure 6.
Mitogen-activated protein kinases (MAPKs) and NF-κB signaling pathways are involved in PcrV-mediated activation of M1 macrophages and increasing of phagocytosis. Raw264.7 cells pretreated with the corresponding inhibitors U0126 (5 μM), SP600125 (10 μM), SB203580 (5 μM), and JSH23 (15 μM) for extracellular signal-regulated protein kinase (ERK), c-Jun N-terminal kinase (JNK), p38 MAPKs, and NF-κB, respectively, were primed by LPS + IFNγ (named as LPS group) or PcrV (10 μg/ml) for 6 h. The total and phosphorylation levels of JNK, ERK, and p38 MAPKs were analyzed by western blot (A). The cellular translocation of NF-κB was visualized by immunofluorescence staining (B). NF-κB was labeled with AF647-conjugated anti-NF-κB p65 antibody (red); cellular nuclei were stained with DAPI (blue). ROS (C,D) and iNOS (E) production were analyzed by flow cytometry and western blot, respectively. (F) The concentration of NO in the culture supernatant of Raw264.7 treated with the indicated compound for 24 h was detected by NO detection kit. Raw264.7 pretreated with the corresponding inhibitors was primed by PcrV (10 μg/ml) for 6 h. The cells were then co-cultured with static PAO1 biofilms (MOI = 10) for 30 min. Phagocytosis was detected by immunofluorescence staining (G) and CFU enumeration (H). Cytoskeleton was labeled with phalloidin (red); PAO1 was visualized by FITC anti-PAO1 antibody (green); cellular nuclei were stained with DAPI (blue). Unpaired Student’s t test was used for statistical analysis (D,F,H). **p < 0.01; ***p < 0.001; n.s. indicates no significance.
Discussion
Bacterial biofilm formation on human tissues and implanted/indwelling devices provides a basis for persistent infections. The remodeling of host immune responses by biofilms following infection is one of the most important factors that benefit bacterial survival and chronicity of infection (Gonzalez et al., 2018; Campoccia and Mirzaei, 2019). Previous studies have revealed that S. aureus biofilms attenuated the production of proinflammatory cytokines/chemokines, including IL1β, TNFα, and CXCL2, as well as iNOS, and exacerbated bacterial biofilm infection (Thurlow et al., 2011). Chronic P. aeruginosa biofilm infections in cystic fibrosis patients are dominated by a Th2 response with increased and decreased levels of IL4 and IFNγ, respectively (Moser et al., 2000; Hartl et al., 2006), suggesting that P. aeruginosa biofilms persist by reducing host proinflammatory responses to infection. In vitro co-culture of P. aeruginosa biofilm matrix exopolysaccharides (EPS) and extracellular DNA (eDNA) with RAW264.7 cells induced a lower-grade inflammatory response than that induced by planktonic bacteria‐ or LPS-treated cells (Ramirez et al., 2019). In accordance with these reports, we have demonstrated that compared to planktonic bacteria, P. aeruginosa biofilms impaired the production of proinflammatory cytokines, as well as iNOS, while promoting the expression of the anti-inflammatory enzyme, Arg1, both in vitro and in vivo. Thus, our findings further confirm that P. aeruginosa biofilm-associated infection reduces proinflammatory responses to benefit biofilm bacterial survival.
Accumulating evidence demonstrates that the biofilm-mediated hijacking of host immune defense relies on several processes, such as interference in the release of antimicrobial peptides (AMPs), enzymes, ROS, RNS, and NO from leukocytes; impaired phagocytosis; and the recruitment of immunosuppressive cells, such as MDSCs (Campoccia and Mirzaei, 2019). In addition, studies have demonstrated that the re-education of M1 macrophages to an anti-inflammatory M2 phenotype is also involved in biofilm-mediated immune suppression (Thurlow et al., 2011; Hanke et al., 2013). In accordance with this phenomenon observed in S. aureus, we demonstrated that P. aeruginosa biofilms obstructed the host immune response by activation of M2 macrophages. Mechanistically, it has been shown that the cyclic di-AMPs, alpha-toxin (Hla), and leukocidin AB (LukAB; Scherr et al., 2015), released from S. aureus biofilms promote biofilm persistence by enhancing macrophage anti-inflammatory polarization or inhibiting phagocytosis (Gries et al., 2016). In P. aeruginosa, the biofilm formation-related exopolysaccharide alginates (Leid et al., 2005) and rhamnolipids (Alhede et al., 2014) have been implicated in the protection of biofilm bacteria against macrophage-mediated phagocytosis or have been shown to exert direct cytotoxic effect against macrophages. However, the mechanisms by which P. aeruginosa biofilms skew macrophage phenotypes to favor their survival following infection still remain obscure. Although excessive activation of P. aeruginosa T3SS during acute infection might aggravate inflammation-mediated tissue damage and immune dysfunction, activation of the host immune response by T3SS might also accelerate bacterial recognition and eradication by host immune cells (Galle et al., 2012; Klockgether and Tummler, 2017). However, during a chronic infection, bacterial T3SS is inactivated (Jain et al., 2008), which enhances the ability of bacteria to evade host immune recognition and clearance, ultimately resulting in biofilm persistence. Due to the reverse correlation between T3SS and biofilms (Kuchma et al., 2005), it is highly possible that regulatory elements that control T3SS inhibition/activation might also regulate biofilm persistence. In this study, we found that the T3SS protein PcrV, which is involved in enhancing proinflammatory polarization and phagocytosis of macrophages, is downregulated during P. aeruginosa biofilm formation in vitro and in vivo. The addition of exogenous PcrV or PcrV-pulsed M1 macrophages reversed M2 macrophage-mediated immune inhibition and increased biofilm bacterial elimination, indicating that the decreased expression of PcrV during biofilm formation might impair the M1 macrophage-mediated proinflammatory response and bacterial clearance, ultimately promoting biofilm persistence. Studies have demonstrated that P. aeruginosa T3SS genes, including pcrV, are negatively regulated in bacteria by the intracellular second messenger cyclic di-GMPs (Romling and Galperin, 2017) and the three-component SadARS regulatory system (Kuchma et al., 2005). Given that cyclic di-GMPs and the SadARS system are also involved in positively regulating P. aeruginosa biofilm formation (Kuchma et al., 2005; Ha and O’Toole, 2015; Sharp and Rietsch, 2019), it is likely that these factors are involved in modulation of PcrV-mediated regulation of biofilm persistence during infection in vivo. Considering that PcrV, which is involved in inducing macrophage M1 polarization and enhancing phagocytosis, is downregulated during biofilm infection, the addition of exogenous PcrV shows promise as a therapeutic strategy in patients with biofilm infections or chronic immune suppression.
Macrophage polarization is influenced by a variety of factors, such as different types of inflammatory cytokines/chemokines and infiltrated immune cells, as well as cell membrane and intracellular molecule-related mechanisms (Zhou et al., 2014). In this study, we demonstrated that PcrV-mediated polarization of M1 macrophages is through the activation of MAPK and NF-κB signal pathways. Similar to our findings, the bacterial pathogenicity associated molecular patterns (PAMPs) derived from Gram-negative bacteria, such as Yersinia enterocolitica LcrV (Sing et al., 2002), Brucella abortus cell-surface protein 31 (BCSP31) protein (Li et al., 2014), and Vibrio cholerae porin OmpU (Khan et al., 2015), have also been demonstrated to induce macrophage M1 polarization via MAPK and NF-κB signaling pathways. Peroxisome proliferator activated receptor (PPARγ), which is mainly expressed in adipose tissue and immune cells, is a member of the nuclear receptor superfamily of ligand-inducible transcription factors that regulate a variety of biological activities, including adipogenesis, lipid metabolism, and insulin sensitization, as well as inflammation (Janani and Ranjitha Kumari, 2015). PPARγ-mediated regulation of inflammation often leads to an inhibitory effect on the activation of immune cells, as well as the production of inflammatory factors through the suppression of signaling pathways, such as NF-κB and JNK/p38 signal pathways (Wang et al., 2018). In this study, we found that PcrV treatment significantly decreased the expression of pparγ gene in macrophages, suggesting that the impaired pparγ expression might exacerbate NF-κB and MAPK pathway-mediated inflammation in PcrV-treated macrophages.
In summary, this study reveals a role for PcrV in altering biofilm persistence of P. aeruginosa, and subsequently the potential therapeutic effects of treating P. aeruginosa biofilm infections with PcrV to reverse biofilm-mediated immune suppression.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding authors.
Ethics Statement
The animal study was reviewed and approved by ethics committee of Army Medical University.
Author Contributions
HY and KZ conceived and designed the experiments. HY, JX, JQ, XH, HS, QD, DL, RX, LJ, QL, QC, JP, and MW performed the experiments. HY, XR, and KZ analyzed the data. HY, XR, and KZ wrote the paper. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Ming Li from the Department of Microbiology of Army Medical University, Chongqing, China, for providing the P. aeruginosa PAO1 strain (ATCC 15692).
Footnotes
Funding. This project was supported by grants from the National Natural Science Foundation of China (No. 31700129 and No. 31872634) and Special Financial Aid to Postdoctor Research Fellow in Chongqing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01971/full#supplementary-material.
References
- Akhtar M., Shaukat A., Zahoor A., Chen Y., Wang Y., Yang M., et al. (2019). Hederacoside-C inhibition of Staphylococcus aureus-induced mastitis via TLR2 & TLR4 and their downstream signaling NF-kappaB and MAPKs pathways in vivo and in vitro. Inflammation 43, 579–594. 10.1007/s10753-019-01139-2, PMID: [DOI] [PubMed] [Google Scholar]
- Alhede M., Bjarnsholt T., Givskov M., Alhede M. (2014). Pseudomonas aeruginosa biofilms: mechanisms of immune evasion. Adv. Appl. Microbiol. 86, 1–40. 10.1016/b978-0-12-800262-9.00001-9, PMID: [DOI] [PubMed] [Google Scholar]
- Campoccia D., Mirzaei R. (2019). Hijacking of immune defences by biofilms: a multifront strategy. Biofouling 35, 1055–1074. 10.1080/08927014.2019.1689964, PMID: [DOI] [PubMed] [Google Scholar]
- Chang A., Sun S., Li L., Dai X., Li H., He Q., et al. (2019). Tyrosol from marine Fungi, a novel quorum sensing inhibitor against chromobacterium violaceum and Pseudomonas aeruginosa. Bioorg. Chem. 91:103140. 10.1016/j.bioorg.2019.103140, PMID: [DOI] [PubMed] [Google Scholar]
- Galle M., Carpentier I., Beyaert R. (2012). Structure and function of the type III secretion system of Pseudomonas aeruginosa. Curr. Protein Pept. Sci. 13, 831–842. 10.2174/138920312804871210, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geredew Kifelew L., Mitchell J. G., Speck P. (2019). Mini-review: efficacy of lytic bacteriophages on multispecies biofilms. Biofouling 35, 472–481. 10.1080/08927014.2019.1613525, PMID: [DOI] [PubMed] [Google Scholar]
- Gonzalez J. F., Hahn M. M., Gunn J. S. (2018). Chronic biofilm-based infections: skewing of the immune response. Pathog. Dis. 76:fty023. 10.1093/femspd/fty023, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gries C. M., Bruger E. L., Moormeier D. E., Scherr T. D., Waters C. M., Kielian T. (2016). Cyclic di-AMP released from Staphylococcus aureus biofilm induces a macrophage type I interferon response. Infect. Immun. 84, 3564–3574. 10.1128/IAI.00447-16, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha D. G., O’Toole G. A. (2015). c-di-GMP and its effects on biofilm formation and dispersion: a Pseudomonas aeruginosa review. Microbiol. Spectr. 3:MB-0003–2014. 10.1128/microbiolspec.MB-0003-2014, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaoka S., Naito Y., Katoh H., Shimizu M., Kinoshita M., Akiyama K., et al. (2017). Efficacy comparison of adjuvants in PcrV vaccine against Pseudomonas aeruginosa pneumonia. Microbiol. Immunol. 61, 64–74. 10.1111/1348-0421.12467, PMID: [DOI] [PubMed] [Google Scholar]
- Hanke M. L., Heim C. E., Angle A., Sanderson S. D., Kielian T. (2013). Targeting macrophage activation for the prevention and treatment of Staphylococcus aureus biofilm infections. J. Immunol. 190, 2159–2168. 10.4049/jimmunol.1202348, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl D., Griese M., Kappler M., Zissel G., Reinhardt D., Rebhan C., et al. (2006). Pulmonary T(H)2 response in Pseudomonas aeruginosa-infected patients with cystic fibrosis. J. Allergy Clin. Immunol. 117, 204–211. 10.1016/j.jaci.2005.09.023, PMID: [DOI] [PubMed] [Google Scholar]
- Jain M., Bar-Meir M., McColley S., Cullina J., Potter E., Powers C., et al. (2008). Evolution of Pseudomonas aeruginosa type III secretion in cystic fibrosis: a paradigm of chronic infection. Transl. Res. 152, 257–264. 10.1016/j.trsl.2008.10.003, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janani C., Ranjitha Kumari B. D. (2015). PPAR gamma gene-a review. Diabetes Metab. Syndr. 9, 46–50. 10.1016/j.dsx.2014.09.015, PMID: [DOI] [PubMed] [Google Scholar]
- Justino P. F. C., Franco A. X., Pontier-Bres R., Monteiro C. E. S., Barbosa A. L. R., Souza M., et al. (2020). Modulation of 5-fluorouracil activation of toll-like/MyD88/NF-kappaB/MAPK pathway by Saccharomyces boulardii CNCM I-745 probiotic. Cytokine 125:154791. 10.1016/j.cyto.2019.154791, PMID: [DOI] [PubMed] [Google Scholar]
- Keewan E., Naser S. A. (2020). The role of notch signaling in macrophages during inflammation and infection: implication in rheumatoid arthritis? Cells 9:111. 10.3390/cells9010111, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan J., Sharma P. K., Mukhopadhaya A. (2015). Vibrio cholerae porin OmpU mediates M1-polarization of macrophages/monocytes via TLR1/TLR2 activation. Immunobiology 220, 1199–1209. 10.1016/j.imbio.2015.06.009, PMID: [DOI] [PubMed] [Google Scholar]
- Klockgether J., Tummler B. (2017). Recent advances in understanding Pseudomonas aeruginosa as a pathogen. F1000Res. 6:1261. 10.12688/f1000research.10506.1, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchma S. L., Connolly J. P., O’Toole G. A. (2005). A three-component regulatory system regulates biofilm maturation and type III secretion in Pseudomonas aeruginosa. J. Bacteriol. 187, 1441–1454. 10.1128/JB.187.4.1441-1454.2005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leid J. G., Willson C. J., Shirtliff M. E., Hassett D. J., Parsek M. R., Jeffers A. K. (2005). The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-gamma-mediated macrophage killing. J. Immunol. 175, 7512–7518. 10.4049/jimmunol.175.11.7512, PMID: [DOI] [PubMed] [Google Scholar]
- Li J. Y., Liu Y., Gao X. X., Gao X., Cai H. (2014). TLR2 and TLR4 signaling pathways are required for recombinant Brucella abortus BCSP31-induced cytokine production, functional upregulation of mouse macrophages, and the Th1 immune response in vivo and in vitro. Cell. Mol. Immunol. 11, 477–494. 10.1038/cmi.2014.28, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser C., Kjaergaard S., Pressler T., Kharazmi A., Koch C., Hoiby N. (2000). The immune response to chronic Pseudomonas aeruginosa lung infection in cystic fibrosis patients is predominantly of the Th2 type. APMIS 108, 329–335. 10.1034/j.1600-0463.2000.d01-64.x, PMID: [DOI] [PubMed] [Google Scholar]
- Panagi I., Jennings E., Zeng J., Günster R. A., Stones C. D., Mak H., et al. (2019). Salmonella effector SteE converts the mammalian serine/threonine kinase GSK3 into a tyrosine kinase to direct macrophage polarization. Cell Host Microbe 27, 41.e6–53.e6. 10.1016/j.chom.2019.11.002, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian B. Z., Pollard J. W. (2010). Macrophage diversity enhances tumor progression and metastasis. Cell 141, 39–51. 10.1016/j.cell.2010.03.014, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez T., Shrestha A., Kishen A. (2019). Inflammatory potential of monospecies biofilm matrix components. Int. Endod. J. 52, 1020–1027. 10.1111/iej.13093, PMID: [DOI] [PubMed] [Google Scholar]
- Romling U., Galperin M. Y. (2017). Discovery of the second messenger cyclic di-GMP. Methods Mol. Biol. 1657, 1–8. 10.1007/978-1-4939-7240-1_1, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherr T. D., Hanke M. L., Huang O., James D. B., Horswill A. R., Bayles K. W., et al. (2015). Staphylococcus aureus biofilms induce macrophage dysfunction through leukocidin AB and alpha-toxin. mBio 6, e01021–e01015. 10.1128/mBio.01021-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapouri-Moghaddam A., Mohammadian S., Vazini H., Taghadosi M., Esmaeili S. A., Mardani F., et al. (2018). Macrophage plasticity, polarization, and function in health and disease. J. Cell. Physiol. 233, 6425–6440. 10.1002/jcp.26429, PMID: [DOI] [PubMed] [Google Scholar]
- Sharp J. S., Rietsch A. (2019). RNase E promotes expression of type III secretion system genes in Pseudomonas aeruginosa. J. Bacteriol. 201, e00336–e00319. 10.1128/jb.00336-19, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing A., Rost D., Tvardovskaia N., Roggenkamp A., Wiedemann A., Kirschning C. J., et al. (2002). Yersinia V-antigen exploits toll-like receptor 2 and CD14 for interleukin 10-mediated immunosuppression. J. Exp. Med. 196, 1017–1024. 10.1084/jem.20020908, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurlow L. R., Hanke M. L., Fritz T., Angle A., Aldrich A., Williams S. H., et al. (2011). Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo. J. Immunol. 186, 6585–6596. 10.4049/jimmunol.1002794, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verderosa A. D., Totsika M., Fairfull-Smith K. E. (2019). Bacterial biofilm eradication agents: a current review. Front. Chem. 7:824. 10.3389/fchem.2019.00824, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan C., Zhang J., Zhao L., Cheng X., Gao C., Wang Y., et al. (2019). Rational design of a chimeric derivative of PcrV as a subunit vaccine against Pseudomonas aeruginosa. Front. Immunol. 10:781. 10.3389/fimmu.2019.00781, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., He S., Liu B., Liu C. (2018). MiR-27-3p regulates TLR2/4-dependent mouse alveolar macrophage activation by targetting PPARgamma. Clin. Sci. 132, 943–958. 10.1042/CS20180083, PMID: [DOI] [PubMed] [Google Scholar]
- Wangdi T., Mijares L. A., Kazmierczak B. I. (2010). In vivo discrimination of type 3 secretion system-positive and -negative Pseudomonas aeruginosa via a caspase-1-dependent pathway. Infect. Immun. 78, 4744–4753. 10.1128/IAI.00744-10, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Huang C., Lin Z., Zhan S., Kong L., Fang C., et al. (2014). Macrophage polarization and function with emphasis on the evolving roles of coordinated regulation of cellular signaling pathways. Cell. Signal. 26, 192–197. 10.1016/j.cellsig.2013.11.004, PMID: [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study are available on request to the corresponding authors.